Abstract
The development of drugs to inhibit glioblastoma (GBM) growth requires reliable preclinical models. To date, proteomic level validation of widely used patient-derived glioblastoma xenografts (PDGX) has not been performed. In the present study, we characterized 20 PDGX models according to subtype classification based on The Cancer Genome Atlas (TCGA) criteria, TP53, PTEN, IDH 1/2 and TERT promoter genetic analysis, EGFR amplification status, and examined their proteomic profiles against those of their parent tumors. The 20 PDGXs belonged to three of four TCGA subtypes: 8 classical, 8 mesenchymal, and 4 proneural; none neural. Amplification of EGFR gene was observed in 9 out of 20 xenografts, and of these, 3 harbored the EGFRvIII mutation. We then performed proteomic profiling of PDGX, analyzing expression/activity of several proteins including EGFR. Levels of EGFR phosphorylated at Y1068 vary considerably between PDGX samples, and this pattern was also seen in primary GBM. Partitioning of 20 PDGX into high (n=5) and low (n=15) groups identified a panel of proteins associated with high EGFR activity. Thus, PDGX with high EGFR activity represent an excellent preclinical model to develop therapies for a subset of GBM patients whose tumors are characterized by high EGFR activity. Further, the proteins found to be associated with high EGFR activity can be monitored to assess the effectiveness of targeting EGFR.
Keywords: Glioblastoma, Xenografts, RPPA, EGFR, signaling
Introduction
Glioblastoma (GBM), WHO grade IV astrocytoma, is an aggressive malignant brain tumor for which an effective treatment does not yet exist. GBM carries a grim prognosis, having a median survival of 15 months after diagnosis (Ostrom et al. 2013, Dunn et al. 2012, Lima et al. 2012, Louis et al. 2007). A major limiting factor in the efforts to develop effective therapies for GBMs is the lack of experimental models that faithfully recapitulate the molecular features of the primary tumors. As discussed by Carlson et al., patient-derived cell lines, including those used in GBM research, are known to gradually lose the genetic and morphologic characteristics of their parent tumors as a result of prolonged culturing and serial passaging, thus becoming unreliable experimental models overtime (Carlson et al. 2011). With the advent of tumor xenografts generated via direct subcutaneous transfer of primary tumors into the flank of immunodeficient mice, known as patient-derived xenografts, it seems that we may be able to establish models that more reliably mimic the molecular features of the original tumor (Tentler et al. 2012, Williams et al. 2013, Siolas & Hannon 2013, Mattie et al. 2013, Higgins et al. 2013). Furthermore, the establishment of subcutaneous patient-derived xenografts allows for straightforward isolation of cells for in vitro studies and provides the researcher with a constant source of viable tissue in cases where the parent tumor tissue is scarce. In addition, in the case of central nervous system tumors, they can be used to generate intracranial xenografts for further in vivo studies that require careful attention to the effects of the blood-brain barrier on drug delivery (Carlson et al. 2011). In the case of GBM, patient-derived glioblastoma xenografts (PDGX) lines have been found to retain the key genetic features of the corresponding parent tumors (Carlson et al. 2011, Joo et al. 2013). However, PDGX have not been characterized for their proteomic profiling. This information is important because the cellular proteome and phosphoproteome of a tumor, both of which are critical drivers of cell proliferation, cell death and other important cellular processes, play a critical role in determining the response or failure of cancer therapy. The present study was undertaken to characterize PDGX at the proteomic level and to assess whether activity of key drivers of GBM growth, such as EGFR (Frederick et al. 2000), in PDGX is similar to that seen in the parent tumor. Toward this goal, we established a panel of 20 PDGXs, classified them into subtypes using the criteria established by The Cancer Genome Atlas (TCGA) network (Verhaak et al. 2010, Parsons et al. 2008), characterized them for key molecular abnormalities common in GBMs, namely EGFR gene amplification, and PTEN, TP53, IDH 1/2 and TERT promoter mutations, and obtained their proteomic profiles. We found that 5 out of 20 PDGX express elevated EGFR activity and this pattern is seen in corresponding parent GBM. We discuss the utility of the PDGX with elevated EGFR activity for drug development against a subset of GBMs that are characterized by elevated EGFR activity.
Methods
Establishment of patient-derived GBM xenografts
To establish xenograft lines, fresh tumors were obtained, under an IRB-approved protocol, from GBM patients undergoing surgery at Duke University Medical Center. Under sterile conditions in a laminar flow containment hood, the tumor was minced, passed through a modified tissue press, sieved through two layers of mesh, and the homogenate passed through a 19-gauge needle. The coat of an athymic mouse (nu/nu genotype, BALB/c background) was disinfected with betadine, and the tumor homogenate (< 500 uL) was injected subcutaneously into the right flank (Friedman et al. 1988). Both male and female animals, which were obtained from the DLAR (Division of Laboratory Animal Resources) breeding facility at Duke University Medical Center, were used for this study. Tumor tissue is considered viable up to 48 hours after resection (Carlson et al. 2011). Therefore, all homogenate injections required to establish all xenografts described in this study were performed within this timeframe. After implantation, animals were monitored daily for tumor growth, and after attaining a size of approximately 500 mm3 (~1 cm in diameter), tumors were removed and analyzed. A diameter of 1 cm was chosen in accordance to commonly accepted practice in order to prevent any central necrosis, which could interfere with proteomic studies (Carlson et al. 2011).
Mutation analysis
DNA was extracted from xenografts using the Puregene extraction kit (Qiagen Inc.). Mutations in the TP53, PTEN, IDH 1/2, and TERT promoter genes were detected by Sanger sequencing using primers, as described by Parsons et al. (Parsons et al. 2008).
EGFR RNA amplification assay
Total RNA was extracted from xenografts using the Qiagen RNeasy kit (Qiagen, Inc., Valencia, CA) according to the manufacturer’s instructions, and RNA concentration was measured with the Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE). Synthesis of cDNA was accomplished using random hexamers (Invitrogen Superscript III kit). EGFR quantitative PCR (qPCR) was carried out, using a cDNA equivalent to 10 ng RNA input, with the TaqMan assay system (Applied Biosystems, Life Technologies) in an ABI 7900 real-time PCR system (Applied Biosystems, Life Technologies). Three different primer pairs and a corresponding FAM-MGB probe for each were designed to differentiate between PCR signals produced following amplification of EGFRvIII (exon 1 joined to exon 8), non vIII-type EGFR (intact exons 2–7), and carboxyl end amplifications.
To detect EGFRvIII, primers 5’TCCTGGCGCTGCTGGCTG 3’ (exon 1) and 5’ CCTCCATCTCATAGCTGTCG 3’ (exon 8), and the corresponding probe FAM-5’ AGGAAAAGAAAGGTAATTATGTG 3’-MGB (exon 1–8 junction) were used. To detect non-vIII-type EGFR (which has intact exons 2–7), amplification primers, 5’ GCGGGACATAGTCAGCAGTG 3’ (exon 4) and 5’ TGGTCAGTTTCTGGCAGTTCTC 3’ (exon 6), and the corresponding probe FAM-5’ CACCTGGGCAGCTGCCAAAAGT 3’-MGB (exon 4–5 junction) were used. Amplification of carboxyl end was assayed using primers 5’ TGACTGAGGACAGCATAGACG 3’ (exon 27) and 5’ AGAGGCTGATTGTGATAGACAGGAT 3’ (exon 28), and detected with probe FAM-5’ CAGTGCCTGAATACAT 3’-MGB (exon 27–28 junction).
Real-time PCR was done in triplicate using individual wells for each primer pair. Quantitative PCR results are expressed as relative quantity (RQ), fold difference in expression measured against a calibrator (normal brain RNA), and normalized for input cDNA quantity by measuring a reference gene. Human GAPDH (Applied Biosystems Pre-developed TaqMan reagent kit) in triplicate was used to obtain the endogenous reference CT (threshold cycle number) value. Reverse transcribed cDNA from normal brain RNA (Clontech, MountainView, CA) was used as the calibrator sample. The PCR conditions were an initial incubation at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 sec and combined annealing and amplification at 60°C for 1 min. The real-time PCR data was collected and analyzed with ABI SDS (v2.3, Applied Biosystems, Life Technologies). The RQ Manager (v1.2) feature of the software automatically calculates RQ using the formula RQ = 2−ÄÄCT. The ÄCT value is obtained by subtracting each sample’s average reference (GAPDH) CT value from average target (EGFR) CT value. The ÄÄCT is derived by subtracting ÄCT of the calibrator (normal brain) from the ÄCT of the sample. For normal brain, which has zero EGFRvIII, a CT value of 40, the highest cycle number, was used in EGFRvIII ÄCT calculations. This approach yielded undetectable amounts of EGFRvIII in PDGX expressing non-vIII variants. EGFR was considered amplified if we observed a RQ value > 3 during evaluation of the carboxyl terminus or exons 2–7 (non-vIII).
Classification of PDGX according to TCGA-defined subtypes
Total RNA was extracted using Qiagen’s RNeasy Mini Kit (Valencia, CA) according to the manufacturer’s directions. Total RNA was used to obtain global gene expression data using the GeneChip Human Genome U133A 2.0 array from Affymetrix (Santa Clara, CA). Microarray data was normalized and probes were summarized as gene expression levels using RMA (Irizarry et al. 2003). Data was log2 transformed, and genes were median centered. Xenografts were classified into one of the four subtypes (Verhaak et al. 2010) using Classification to the Nearest Centroid (ClaNC) (Dabney 2006) with the TCGA GBM data as the training dataset.
Reverse Phase Protein Array
Reverse Phase Protein Array (RPPA) is a high throughput, antibody-based screening tool that can quantify the levels of several hundred proteins and their post-translational modifications within a sample (Charboneau et al. 2002, Espina et al. 2009, Pin et al. 2014). All RPPA analyses in this study were performed at the Functional Proteomics RPPA Core Facility at MD Anderson, and all preliminary sample preparations were conducted according to the Core Facility’s ‘Preparation of tumor lysates from xenograft’s protocol, located on their website (http://www.mdanderson.org/education-and-research/resources-for-professionals/scientific-resources/core-facilities-and-services/functional-proteomics-rppa-core/index.html).
Tissue lysates from PDGX and primary GBM were examined separately in two analyses. Twenty PDGX samples were flash frozen in liquid nitrogen at the time of removal from mice and were subsequently homogenized in RPPA lysis buffer [1% Triton X-100, 50mM HEPES, pH 7.4, 150mM NaCl, 1.5mM MgCl2, 1mM EGTA, 100mM NaF, 10mM Na pyrophosphate, 1mM Na3VO4, 10% glycerol, protease and phosphatase inhibitors] (Roche Applied Science, #05056489001 and #04906837001)]. Sample protein concentrations were adjusted to 1 mg/mL after being measured with the Bio-Rad protein assay (Bio-Rad, #500-0006). Afterwards, samples were treated with a 4X SDS buffer [40% Glycerol, 8% SDS, 0.25M Tris.HCl, pH 6.8, 10% beta-mercaptoethanol] and boiled for 5 minutes. These lysates were stored at −80°C until analyzed. Of the 20 PDGXs, primary tumor samples were available for only 15, and these were processed in the same manner as the xenografts and analyzed separately in the RPPA system.
All PDGX and primary tumor lysates were serially diluted and printed onto nitrocellulose-coated slides, using an Aushon 2470 Arrayer (Aushon BioSystems, Billerica, MA, USA) and probed with a panel of 133 and 206 primary antibodies, respectively, followed by biotin-conjugated secondary antibodies. Signal intensities for each sample were amplified using the Autostainer Plus system (Dako, Carpinteria, CA, USA), analyzed and quantitated with the Microvigene software (VigeneTech, Carlisle, MA, USA). Each dilution curve was fitted with a logistic model (“Supercurve Fitting” developed by the Department of Bioinformatics and Computational Biology, MD Anderson Cancer Center, Houston, Texas (“http://bioinformatics.mdanderson.org/OOMPA”). All samples were normalized for protein loading and the results transformed into linear values, and these values used as the quantitative expression of the individual proteins in the tumors or xenografts.
Because PDGX and primary GBMs were analyzed by RPPA at different times without internal controls, the normalized linear values of protein expression between the two data sets cannot be compared directly. Therefore, we compared the patterns, instead of absolute levels, of protein expression in PDGX and primary GBMs separately.
Western Blotting
10 µg of xenograft lysate was loaded per well into a 4–12% Novex Bis-Tris gel. The gel was transferred to a PVDF membrane, blocked with 5% milk and probed with a rabbit polyclonal EGFR antibody directed against the c-terminus (Cell signaling catalog #4267P) at 1 µg/mL in 5% BSA, followed by goat anti-rabbit and anti-biotin (Cell Signalling #7074 and 7075, respectively) secondary antibodies at 1 µg/mL. The membrane was developed on Biomax MR film. Following the development, the membrane was stripped with beta-mercaptoethanol and the probing procedure was repeated using phospho-EGFR antibody specific for tyrosine phosphorylation at residue 1068 (Cell Signalling catalog # 2234) at 2 µg/mL in 5% BSA and 2 µg/mL for goat anti-rabbit and anti-biotin antibodies, respectively. The blot was then developed as described above.
Statistical Analysis
To identify PDGX and primary tumor samples with activated EGFR (assessed by its phosphorylation at Y1068) we performed hierarchical cluster analysis based on the RPPA expression data. Among the 20 PDGX samples we identified an EGFR non-activated group of 15 with lower expression levels, and an EGFR activated group of 4 with higher expression levels. One PDGX (08-0430) did not cluster with any other samples; upon further investigation we found it contained an EGFR mutation and excluded it from subsequent analysis. Among the 15 primary tumor samples we identified an EGFR non-activated group of 10 that had lower expression levels, and an EGFR activated group of 4 combining 2 small clusters that had elevated expression levels. We identified one primary tumor (10-0171) that did not cluster with any other samples, and treated it as an outlier due to its extremely high EGFR activity (5.5-fold higher activity than the sample with the second highest activity). Clustering was carried out using JMP software (SAS Institute, Cary, NC).
To assess whether EGFR activation is associated with changes in expression of other proteins, we performed Wilcoxon Rank Sum tests comparing the protein expression levels in the two identified EGFR activation groups (low and high EGFR activation) for the PDGX samples. The Benjamini-Hochberg procedure was used to control the false discovery rate (FDR). We identified proteins that had FDR values less than 0.05 as highly significant and proteins with FDR less than 0.30 as significant to an exploratory analysis in a small sample. Statistical testing was performed in R version 3.1.1.
Results
PDGX Characterization using the genomic approach
We established and characterized a panel of twenty PDGX. The mean age for the patients whose glioblastoma parent tumor was used to derive the xenografts was 62.25 ± 9.67 years. As Table 1 shows, our PDGX fall into three of the four TCGA subtypes: classical (8 samples), mesenchymal (8 samples), and proneural (4 samples). Table 1 also lists their demographic data, TP53, PTEN, TERT promoter and EGFR genes status. The genetic abnormalities seen in our PDGX are as follows: 1) Mutations in TP53 and PTEN were observed in 5 and 14 PDGX, respectively 2) None of our PDGX had mutations in either IDH 1 or IDH 2; 3) Mutations in the TERT promoter were seen in all 20 PDGX, 7 having a C250T substitution, and the rest having a C228T substitution 4) EGFR amplification observed in 9 PDGX; and 5) EGFRvIII, the constitutively active deletion mutant of EGFR (Banaszczyk et al. 2013, Gan et al. 2013), observed in 3 PDGX.
Table 1.
PDGX Characteristics
| PDGX # |
ID # | Mouse Passage # |
Dx | Sex/Age | TCGA Subtype |
TP53 | PTEN | TERT | EGFR* |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| vIII | Wt | Amplified | |||||||||
| 1 | 08-0308 | 10 | GBM | M/76 | C | Wt | Wt | C228T | − | + | Y |
| 2 | 08-0500 | 14 | GBM | M/75 | C | Mut | Mut | C250T | − | + | Y |
| 3 | 08-0549 | 14 | GBM | M/62 | C | Wt | Wt | C228T | + | + | Y |
| 4 | 09-0155 | 20 | GBM | F/52 | C | Wt | Wt | C228T | + | + | Y |
| 5 | 09-0211 | 26 | GBM | F/53 | C | Wt | Wt | C228T | − | + | N |
| 6 | 09-0394 | 33 | GBM | M/73 | C | Wt | Mut | C228T | − | + | N |
| 7 | 09-0584 | 7 | GBM | M/64 | C | Wt | Mut | C228T | − | + | Y |
| 8 | 10-0171 | 12 | GBM | M/54 | C | Wt | Wt | C228T | + | + | Y |
| 9 | 08-0430 | 11 | GBM | M/59 | M | Wt | Wt | C250T | − | + | Y‡ |
| 10 | 08-0679 | 21 | GBM | F/47 | M | Wt | Mut | C250T | − | + | Y |
| 11 | 09-0337 | 19 | GBM | M/52 | M | Wt | Mut | C228T | − | + | N |
| 12 | 09-0362 | 39 | GBM | M/50 | M | Mut | Mut | C228T | − | + | N |
| 13 | 09-0477 | 27 | GBM | M/73 | M | Wt | Mut | C228T | − | + | N |
| 14 | 09-0500 | 21 | GBM | M/58 | M | Wt | Mut | C228T | − | + | N |
| 15 | 09-0627 | 9 | GBM | M/61 | M | Wt | Mut | C228T | − | + | Y |
| 16 | 11-0040 | 11 | GBM | M/65 | M | Mut | Mut | C250T | − | + | N |
| 17 | 08-0478 | 13 | GBM | M/62 | P | Mut | Mut | C250T | − | + | N |
| 18 | 08-0499 | 20 | GBM | M/66 | P | Mut | Mut | C250T | − | + | N |
| 19 | 08-0624 | 11 | GBM | M/63 | P | Wt | Mut | C228T | − | + | N |
| 20 | 10-0021 | 12 | GBM | F/82 | P | Wt | Wt | C250T | − | + | N |
Mut – Mutation; Wt – Wild-type; TCGA – The Cancer Genome Atlas; C – Classical; M – Mesenchymal; P – Proneural; TP53 – Tumor Protein p53; PTEN - Phosphatase and Tensin homolog; TERT – Telomerase Reverse Transcriptase; EGFR – Epidermal Growth Factor Receptor.
EGFR qPCR data expressed as relative quantity (RQ; not shown), fold difference in expression measured against normal brain RNA as described in the methods section. Those having an RQ > 3 were considered to be amplified. None of the PDGX showed mutations in Isocitrate Dehydrogenase1 or 2.
Presence of transcript and amplification was observed only with primers specific for the carboxyl-end, consistent with a truncated variant detected by western blotting (see Figure 1).
PDGX Characterization using the proteomic approach
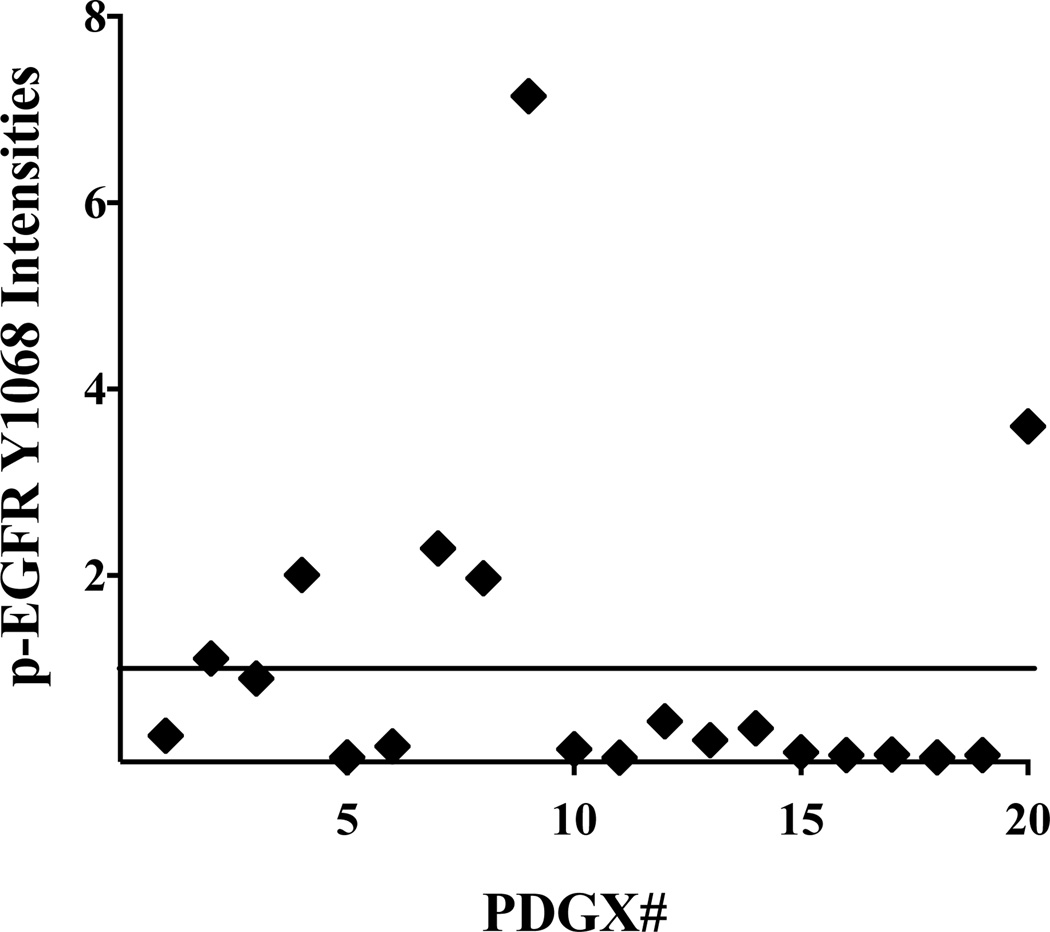
We used RPPA to examine 133 unique proteins/phosphoproteins, including EGFR in its unphosphorylated and phosphorylated forms, in our panel of 20 PDGXs. We found that PDGXs differ considerably from each other in the expression of total EGFR and autophosphorylated EGFR at tyrosine 1068 (p-EGFR Y1068) and 1173 (p-EGFR Y1173) (Table 2). Plotting of EGFR pY1068 expression levels (Figure 1), which is a measure of EGFR activity (Wheeler et al. 2012), demonstrates that phosphor-EGFR values are near 1 or higher in only seven PDGX (marked with “**”). We labeled these PDGX as PDGX with activated EGFR. Out of seven PDGX with higher EGFR activity, five are of the classical subtype (Table 2). This is not surprising given that abnormalities in the EGFR gene are the hallmark of the classical subtype (Verhaak et al. 2010). What is surprising, however, is that higher EGFR activity can also be seen in mesenchymal and proneural subtypes (Table 2).
Table 2.
Expression of Total EGFR, p-EGFR Y1068, and p-EGFR Y1173 in 20 PDGX
| PDGX # | ID # | TCGA Subtype |
EGFR Amplification |
RPPA* |
||
|---|---|---|---|---|---|---|
| EGFR Total | p-EGFR Y1068 | p-EGFR Y1173 | ||||
| 1 | 08-0308 | C | Y | 0.43 | 0.28 | 0.75 |
| 2** | 08-0500 | C | Y | 1.17 | 1.11 | 0.77 |
| 3** | 08-0549 | C | Y | 1.66 | 0.89 | 0.88 |
| 4** | 09-0155 | C | Y | 5.82 | 2 | 1.26 |
| 5 | 09-0211 | C | N | 0.5 | 0.05 | 0.82 |
| 6 | 09-0394 | C | N | 0.35 | 0.17 | 0.72 |
| 7** | 09-0584 | C | Y | 3.24 | 2.29 | 1.19 |
| 8** | 10-0171 | C | Y | 2.15 | 1.97 | 1.23 |
| 9** | 08-0430 | M | Y | 2.27 | 7.14 | 2.1 |
| 10 | 08-0679 | M | Y | 0.56 | 0.14 | 0.73 |
| 11 | 09-0337 | M | N | 0.5 | 0.05 | 0.75 |
| 12 | 09-0362 | M | N | 0.37 | 0.44 | 0.7 |
| 13 | 09-0477 | M | N | 0.67 | 0.24 | 0.79 |
| 14 | 09-0500 | M | N | 0.44 | 0.36 | 0.76 |
| 15 | 09-0627 | M | Y | 0.57 | 0.1 | 0.81 |
| 16 | 11-0040 | M | N | 0.3 | 0.07 | 0.87 |
| 17 | 08-0478 | P | N | 0.3 | 0.08 | 0.81 |
| 18 | 08-0499 | P | N | 0.24 | 0.05 | 0.79 |
| 19 | 08-0624 | P | N | 0.28 | 0.07 | 0.88 |
| 20** | 10-0021 | P | N | 4.32 | 3.6 | 1.4 |
Intensity of Expression;
Activity (phosphorylation) of EGFR near 1 or higher; C – Classical; M – Mesenchymal; P – Proneural.
Figure 1.
Expression of EGFR phosphorylated at tyrosine 1068 (pEGFRY1068) in PDGX. The expression levels of p-EGFRY1068 were obtained by RPPA analysis of 20 PDGX. PDGX #9, corresponding to sample ID 08-0430, has the highest EGFR activity (auto-phosphorylation). This PDGX expresses a mutant EGFR with N-terminus deletion as indicated in the legend of Figure 2.
Western blotting of selected PDGX matches EGFR phosphorylation data obtained using reverse phase protein array
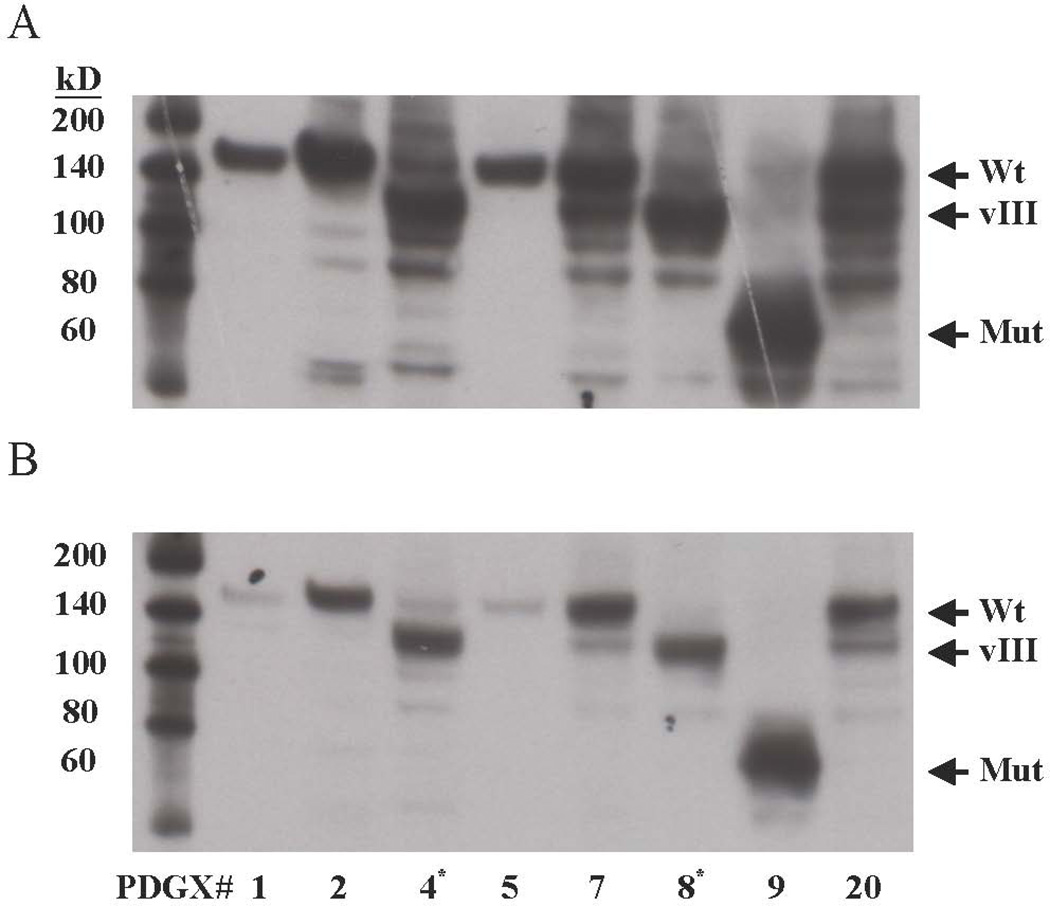
For validation of the results obtained using RPPA, we examined the expression of EGFR and EGFR-pY1068 in a subset of our PDGX samples by western blotting. The results, shown in Figure 2, indicate PDGX #1 (08-0308) and #5 (09-0211) to have lower levels of EGFR and p-EGFR Y-1068, and these data are consistent with the RPPA data (Table 2 and Figure 1). PDGX #4 (09-0155) and #8 (10-0171), which harbor EGFRvIII (Table 1), both show high levels of expression of EGFR Y1068, which again is in accord with the RPPA data (Table 2). As expected, the EGFR band in these EGFRvIII expressing PDGX is lower than that seen in PDGX with the wild-type EGFR (Figure 2). The wild-type EGFR expressing PDGXs #7 (09-0584) and #20 (10-0021) also display a phosphorylated band corresponding to the molecular weight of EGFRvIII (Figure 2, panel B), but this is probably a proteolytic product of EGFR as EGFRvIII was not detected in these xenografts. Finally, the EGFR in PDGX #08-0430 has a very high phosphorylation at Y1068, which is again consistent with the RPPA data (Table 2), but its size is much lower. This truncation of EGFR is consistent with the observation that in this xenograft, high EGFR amplification was seen only with PCR primers directed against the last exon (Table 1).
Figure 2.
EGFR and EGFR Y1068 expression in selected PDGX by western blotting. Of the PDGX examined, 6 are Classical (#1, 2, 4, 5, 7, and 8); 1 is Mesenchymal (#9); and 1 is Proneural (#20). (A) Total EGFR: the anti-EGFR antibody used was against the intracellular domain of EGFR. (B) p-EGFRY1068: the anti-pEGFR antibody used was selective for EGFR phosphorylated at tyrosine 1068 near the c-terminus. PDGX #9 expresses a lower molecular weight mutant EGFR with high auto-phosphorylation. This mutant EGFR is probably deleted at the N-terminus because its presence and amplification was detected only with primers directed against the carboxyl terminal part of the receptor. Exposure times were 2 and 5 minutes for panel A and panel B, respectively. *PDGX expressing EGFRvIII.
EGFR activity in PDGX and primary GBM have a similar pattern
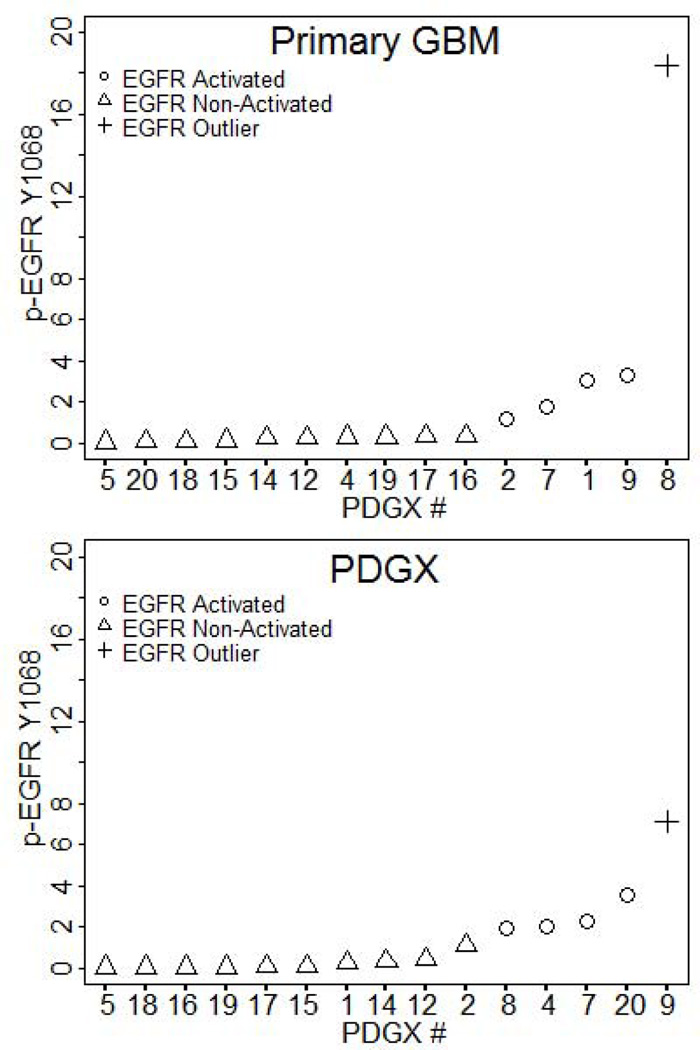
We next compared whether the pattern of EGFR activity seen in PDGX is similar to that seen in corresponding primary GBM. We compared the EGFR activation group assignments of the 15 pairs of PDGX/primary tumor as described under the statistical analysis section. Figure 3 shows the EGFR activity patterns in PDGX and primary GBM. We found that of the 10 PDGXs with low EGFR activity all but 1 primary tumor also have low EGFR activity, indicating that EGFR activity in PDGX accurately reflects EGFR activity in primary GBM. The agreement between PDGX and primary GBM expressing high EGFR activity shows a lower concordance and this may be due to the small sample size of PDGX with high EGFR activity.
Figure 3.
EGFR activity in 15 PDGX and primary GBM pairs exhibits similar patterns. Primary GBMs of nine of the 10 PDGX with lower EGFR activity also have lower EGFR activity.
High activity of EGFR is correlated with high activity of several other proteins
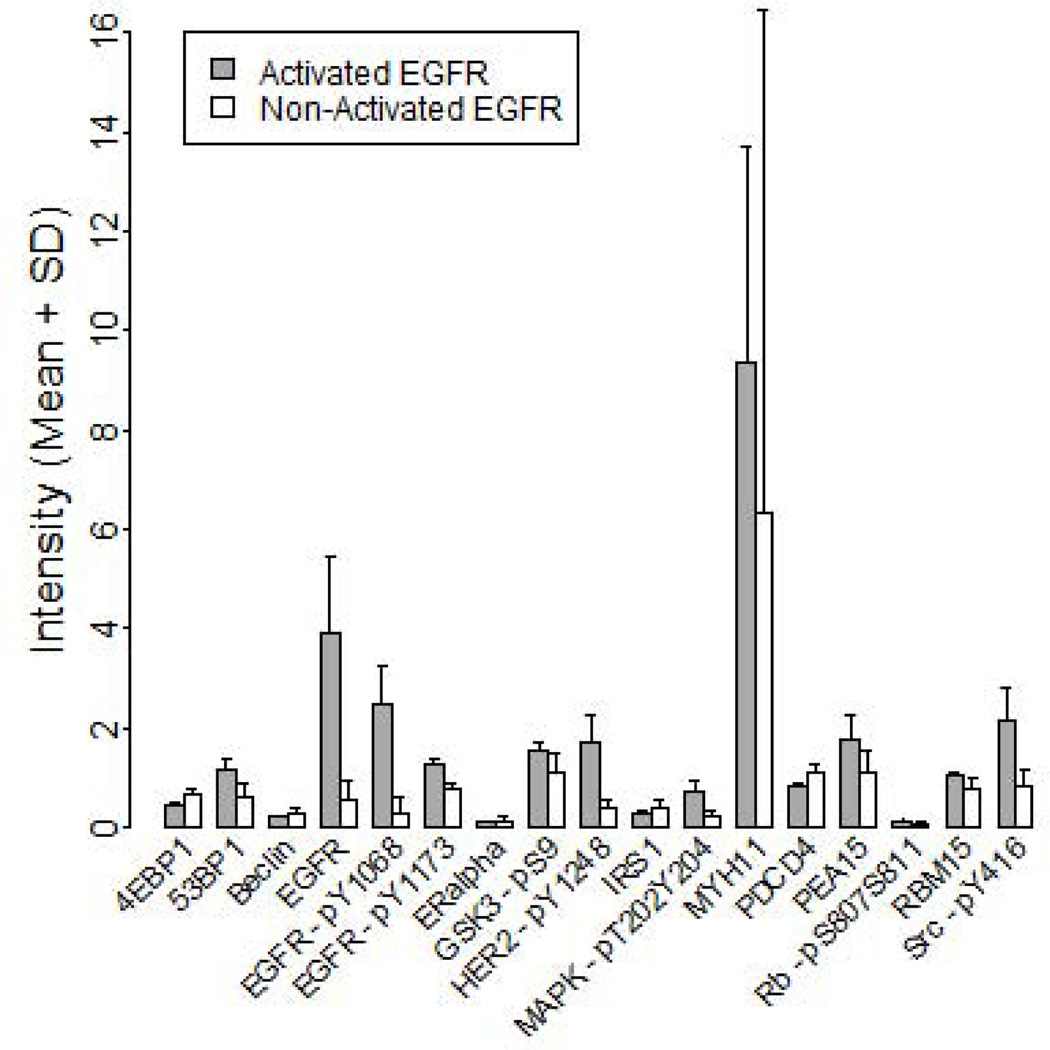
To identify whether PDGXs with EGFR activation, as identified by hierarchical clustering, have significant changes in the expression of other proteins, we compared the expression levels of the 133 unique proteins obtained through RPPA between the two EGFR activation groups, using Wilcoxon Rank Sum tests. The results, Figure 4, revealed that at FDR < 0.05, PDGXs with high EGFR phosphorylation display significantly higher expression of the following 6 proteins: EGFR, pEGFR-Y1068, pEGFR-Y1163, pHER2-Y1248, PDCD4, and pSrc-Y416. When the analysis was extended to FDR < 0.3, association with an additional 11 proteins (4E-BP1, 53BP1, Beclin, ER-alpha, pGSK3-pS9, IRS1, pMAPK-T202/Y204, MYH11, PEA15, pRb-S807/S811, and RBM15) was detected. If confirmed in future studies, this would suggest that the status of these proteins could be used as a marker of the efficacy of EGFR kinase inhibitors employed to silence EGFR signaling as a brain tumor therapeutic.
Figure 4.
Proteins whose expression was found to be significantly different (FDR < 0.3) in PDGX with activated EGFR (n=4) when compared to those having low activation of EGFR (n=15). The expression levels of these proteins were obtained by the RPPA analysis.
In this issue, we have characterized a panel of patient derived glioblastoma xenografts (PDGXs) at genetic and proteomic level. Since proteomic profiling provides information on activated growth pathways, PDGXs could be used to identify an arsenal of signal transduction modulators that can be targeted to effectively arrest glioblastoma growth.
Discussion
Our characterization of a panel of 20 PDGX according to TCGA criteria represents the first such study performed by a laboratory outside of the TCGA group. Although TCGA-defined four subgroups of primary GBMs consisting of classical, mesenchymal, proneural, and neural, we detected only three subtypes in our 20 PDGX. Apparently, neural subtype is difficult to detect in PDGX as this subtype was also not detected by Verhaak et al. (Verhaak et al. 2010), a TCGA-associated group, in their characterization of PDGX. In this connection, it is interesting to point out that Huse et al. (Huse et al. 2013), in their recent review of GBM subtypes, state that most studies have been able to classify GBM into three major subtypes: classical, mesenchymal, and proneural.
The genetic abnormalities seen in key genes in our 20 PDGX are qualitatively similar to those reported by Carlson et al. for their collection of 20 PDGX lines (Carlson et al. 2011), in which they found 10 of 20 with EGFR amplification, 3 of which were expressing EGFRvIII, 7 of 20 with mutations in TP53, and 8 of 20 with mutations or deletions in PTEN (Carlson et al. 2011). Similarly, our findings regarding the TERT promoter mutations were on par with those of Killela et al. who found mutations in a remarkably high number of tumors (83% of 78 tumors) (Killela et al. 2013). Additionally, they reported that TERT promoter mutations were associated with EGFR amplification but since 100% of our PDGX are positive for TERT promoter mutations, we were unable to test this association.
In this study, we focused on EGFR and made some interesting observations. First, high EGFR activity is not only seen in classical subtypes, but also in other subtypes. Second, while 6 out of 7 PDGX with higher EGFR activity have EGFR gene amplification, higher EGFR activity is also seen in the absence of gene amplification as shown by the data with PDGX #20 (Tables 1, 2 and Figure 1). Conversely, PDGX with EGFR gene amplification may not have elevated EGFR activity as indicated by data on PDGX #10 (08-0679) and 15 (09-0627; Tables 1, 2 and Figure 1). Third, we have identified three types of EGFR in our xenografts; 1) wild-type, 2) EGFRvIII and 3) a highly active truncated EGFR that remains to be characterized. Whether or not this truncated mutant of EGFR has already been described in GBM is not clear (Frederick et al. 2000), but it may resemble the EGFRvI mutant described by Nicholas et al in their review (Nicholas et al. 2006). Further studies are needed to fully characterize this truncated EGFR mutant present in our PDGX #08-0430 (Tables 1 and 2). Interestingly, a recent report documented that the diversity of EGFR seen in bulk GBM is also seen at the single cell level (Francis et al. 2014, Gini & Mischel 2014).
Since EGFR is dysregulated in a large number of GBM and has been a target for drug development, we wanted to determine whether EGFR activity seen in PDGX is also seen in primary GBMs. To this end, we also obtained proteomic profiles of 15 primary GBMs that were available corresponding to our PDGX. To our knowledge, this is the first time that such a comparison has been made (Siolas & Hannon 2013). A high concordance of EGFR activity between PDGX and their parent tumors implies that PDGX could be used for personalized therapy and/or targeted drug development. We also examined whether other proteins important in GBM biology, such as Akt, exhibit similar patterns of expression/activity in PDGX and primary GBMs. However, in this case, we found that there was a poor correlation between PDGX and primary GBMs. While there could be several reasons for this discrepancy, the main one appears to be that primary GBM used for PDGX generation were fresh whereas primary GBMs used for RPPA analysis have been in storage for different lengths of time and may have also experienced different freeze-thaw cycles. Future studies should store a part of the primary GBM used for xenograft generation and obtain its proteomic profile at the same time as PDGX. Given that proteomic analysis, despite limitations in the currently available software-based analytic tools, has become an important tool guiding the development of anticancer therapy (Dawson & Carragher 2014), future studies should also compare whether stem cells isolated from primary GBMs retain the proteomic profile of primary GBMs.
The findings of the present study underscore the importance of using proteomic data as a tool for segregating GBM patients for drug development and clinical trials. Heterogeneity among GBMs is well known and contributes to failure in clinical trials performed without segregating GBM patients. To address this problem, the TCGA network identified four distinct subtypes based on gene expression data and attempts are being made to group GBM patients according to the TCGA-defined subtype of their tumor (Verhaak et al. 2010, Olar & Aldape 2014, Cloughesy et al. 2014). However, it is our belief that this approach will be limited as a strategy for testing targeted therapies, because, as shown through the EGFR example, the same target may be active in more than one subtype. We suggest that a better approach is to segregate GBM patients based on the presence of the protein modulator that is being targeted.
The use of PDGX models is an improvement over previous models that generated xenografts using cell lines with limited relevance to the primary GBMs (Kola & Landis 2004). However, one drawback of our study is that our PDGX models were not generated by intracranial transplantation. The development of patient-specific orthotopic models of GBM are clearly superior (Joo et al. 2013), and future studies should compare proteomic profiling of orthotopic PDGX and primary GBMs.
In summary, we have characterized a panel of 20 PDGX and shown that the proteomic, and not the genomic, profile provides an accurate picture of EGFR activation and that PDGXs can be a helpful tool for designing targeted therapies in the future.
Supplementary Material
Acknowledgements
We would like to thank Ms. Lisa Ehinger for her help with brain tumor samples, and Mr. Cory Nanni and Ms. Drew Masters for their assistance with the preparation of figures.
Financial support: This research is supported by a grant from the NIH (R21NS078642) and a grant from the Musella Foundation to MMK.
Abbreviations used
- GBM
glioblastoma
- PDGX
patient-derived glioblastoma xenografts
- TCGA
The Cancer Genome Atlas
- TP53
tumor protein p53
- PTEN
phosphatase and tensin homolog
- IDH 1/2
isocitrate dehydrogenase 1/2
- TERT
telomerase reverse transcriptase
- EGFR
epidermal growth factor receptor
- WHO
World Health Organization
- IRB
Institutional Review Board
- DLAR
Division of Laboratory Animal Resources
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- CT
threshold cycle number
- RPPA
Reverse Phase Protein Array
- FDR
false discovery rate
Footnotes
Disclosures: The authors disclose no potential conflicts of interest.
References
- Banaszczyk M, Stoczynska-Fidelus E, Winiecka-Klimek M, Bienkowski M, Och W, Rieske P, Piaskowski S. EGFRvIII - A Stable Target for Anti-EGFRvIII Therapy. Anticancer Res. 2013;33:5343–5348. [PubMed] [Google Scholar]
- Carlson BL, Pokorny JL, Schroeder MA, Sarkaria JN. Establishment, maintenance and in vitro and in vivo applications of primary human glioblastoma multiforme (GBM) xenograft models for translational biology studies and drug discovery. Chapter 14, Unit 14 16. Curr Protoc Pharmacol. 2011 doi: 10.1002/0471141755.ph1416s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charboneau L, Tory H, Chen T, Winters M, Petricoin EF, 3rd, Liotta LA, Paweletz CP. Utility of reverse phase protein arrays: applications to signalling pathways and human body arrays. Brief Funct Genomic Proteomic. 2002;1:305–315. doi: 10.1093/bfgp/1.3.305. [DOI] [PubMed] [Google Scholar]
- Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: from molecular pathology to targeted treatment. Annu Rev Pathol. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- Dabney AR. ClaNC: point-and-click software for classifying microarrays to nearest centroids. Bioinformatics. 2006;22:122–123. doi: 10.1093/bioinformatics/bti756. [DOI] [PubMed] [Google Scholar]
- Dawson JC, Carragher NO. Quantitative phenotypic and pathway profiling guides rational drug combination strategies. Frontiers in pharmacology. 2014;5:118. doi: 10.3389/fphar.2014.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Rinne ML, Wykosky J, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espina V, Liotta LA, Petricoin EF., 3rd Reverse-phase protein microarrays for theranostics and patient tailored therapy. Methods Mol Biol. 2009;520:89–105. doi: 10.1007/978-1-60327-811-9_7. [DOI] [PubMed] [Google Scholar]
- Francis JM, Zhang CZ, Maire CL, et al. EGFR Variant Heterogeneity in Glioblastoma Resolved through Single-Nucleus Sequencing. Cancer discovery. 2014;4:956–971. doi: 10.1158/2159-8290.CD-13-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- Friedman HS, Colvin OM, Skapek SX, Ludeman SM, Elion GB, Schold SC, Jr, Jacobsen PF, Muhlbaier LH, Bigner DD. Experimental chemotherapy of human medulloblastoma cell lines and transplantable xenografts with bifunctional alkylating agents. Cancer Res. 1988;48:4189–4195. [PubMed] [Google Scholar]
- Gan HK, Cvrljevic AN, Johns TG. The epidermal growth factor receptor variant III (EGFRvIII): where wild things are altered. The FEBS journal. 2013;280:5350–5370. doi: 10.1111/febs.12393. [DOI] [PubMed] [Google Scholar]
- Gini B, Mischel PS. Greater Than the Sum of Its Parts: Single-Nucleus Sequencing Identifies Convergent Evolution of Independent EGFR Mutants in GBM. Cancer discovery. 2014;4:876–878. doi: 10.1158/2159-8290.CD-14-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DM, Wang R, Milligan B, et al. Brain tumor stem cell multipotency correlates with nanog expression and extent of passaging in human glioblastoma xenografts. Oncotarget. 2013;4:792–801. doi: 10.18632/oncotarget.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse JT, Holland E, DeAngelis LM. Glioblastoma: molecular analysis and clinical implications. Annu Rev Med. 2013;64:59–70. doi: 10.1146/annurev-med-100711-143028. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo KM, Kim J, Jin J, et al. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell reports. 2013;3:260–273. doi: 10.1016/j.celrep.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nature reviews. Drug discovery. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- Lima FR, Kahn SA, Soletti RC, et al. Glioblastoma: therapeutic challenges, what lies ahead. Biochim Biophys Acta. 2012;1826:338–349. doi: 10.1016/j.bbcan.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattie M, Christensen A, Chang MS, et al. Molecular characterization of patient-derived human pancreatic tumor xenograft models for preclinical and translational development of cancer therapeutics. Neoplasia (New York, N.Y.) 2013;15:1138–1150. doi: 10.1593/neo.13922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas MK, Lukas RV, Jafri NF, Faoro L, Salgia R. Epidermal growth factor receptor - mediated signal transduction in the development and therapy of gliomas. Clin Cancer Res. 2006;12:7261–7270. doi: 10.1158/1078-0432.CCR-06-0874. [DOI] [PubMed] [Google Scholar]
- Olar A, Aldape KD. Using the molecular classification of glioblastoma to inform personalized treatment. J Pathol. 2014;232:165–177. doi: 10.1002/path.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro-oncology. 2013;15(Suppl 2):ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin E, Federici G, Petricoin EF., 3rd Preparation and use of reverse protein microarrays. Curr Protoc Protein Sci. 2014;75:27, 27, 21–27, 27, 29. doi: 10.1002/0471140864.ps2707s75. [DOI] [PubMed] [Google Scholar]
- Siolas D, Hannon GJ. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res. 2013;73:5315–5319. doi: 10.1158/0008-5472.CAN-13-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, Arcaroli JJ, Messersmith WA, Eckhardt SG. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9:338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler S, Siwak DR, Chai R, et al. Tumor epidermal growth factor receptor and EGFR PY1068 are independent prognostic indicators for head and neck squamous cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:2278–2289. doi: 10.1158/1078-0432.CCR-11-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Anderson WC, Santaguida MT, Dylla SJ. Patient-derived xenografts, the cancer stem cell paradigm, and cancer pathobiology in the 21st century. Lab Invest. 2013;93:970–982. doi: 10.1038/labinvest.2013.92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.