SUMMARY
Small Ubiquitin-like Modifiers play critical roles in the DNA Damage Response (DDR). To increase our understanding of SUMOylation in the mammalian DDR, we employed a quantitative proteomics approach to identify dynamically regulated SUMO-2 conjugates and modification sites upon treatment with the DNA damaging agent MMS. We have uncovered a dynamic set of 20 upregulated and 33 downregulated SUMO-2 conjugates, and 755 SUMO-2 sites, of which 362 were dynamic in response to MMS. In contrast to yeast, where a response is centered on homologous recombination, we identified dynamically SUMOylated interaction networks of chromatin modifiers, transcription factors, DNA repair factors and nuclear body components. SUMOylated chromatin modifiers include JARID1B/KDM5B, JARID1C/KDM5C, p300, CBP, PARP1, SetDB1 and MBD1. Whereas SUMOylated JARID1B was ubiquitylated by the SUMO-targeted ubiquitin ligase RNF4 and degraded by the proteasome in response to DNA damage, JARID1C was SUMOylated and recruited to the chromatin to demethylate histone H3K4.
INTRODUCTION
Posttranslational modifications (PTMs) significantly increase the functional repertoire of proteomes. PTMs range from small chemical modifications such as phosphorylation, acetylation and methylation, to small protein modifiers such as ubiquitin and ubiquitin-like family members (Ubls). Ubls are covalently attached to lysines in target proteins through an isopeptide bond, and thereby regulate the functions of these proteins. Moreover, modification by Ubls is a reversible process and therefore provides cells with a mechanism to facilitate a rapid response to dynamic conditions. Conjugation and removal of Ubls is coordinated by a subset of specialized and often context-specific enzymes.
Small ubiquitin-like modifiers (SUMOs) are members of the Ubl family, and have been implicated in orchestration of biological processes ranging from control of cell cycle progression and transcriptional regulation to chromatin remodeling and DNA repair (Flotho and Melchior, 2013; Hickey et al., 2012; Ulrich and Walden, 2010; Vertegaal, 2011). Compared to ubiquitin, the machinery responsible for SUMO conjugation and removal consists of a relatively small subset of enzymes, even though SUMOs modify in the range of 1,500 proteins in mammalian cells (Hendriks et al., 2014). Furthermore, whereas ubiquitin functions in all cellular compartments, SUMOs are predominantly located in the nucleus and enriched in Promyelocytic Leukemia (PML) nuclear bodies, playing a pivotal role in the regulation of this critical subcellular domain. SUMOs display some specificity in conjugation, with the predominant consensus motif being [VIL]KxE, although SUMOylation on alternate or non-consensus motifs also occurs (Matic et al., 2010).
Our knowledge on the coordination of the DNA damage response by SUMOylation has significantly improved over the last decade (Jackson and Durocher, 2013). As SUMOs are naturally abundant in the nucleus, they provide an effective cellular mechanism for regulating the function of proteins involved in the response to DNA damage. A significant number of studies have been published on SUMOylation with regard to regulating singular DNA damage response proteins (Galanty et al., 2009; Hoege et al., 2002; Jackson and Durocher, 2013; Morris et al., 2009; Stelter and Ulrich, 2003; Ulrich and Walden, 2010). Whereas insight into the direct mechanistic effect of SUMO conjugation on a protein is highly interesting, there are cases where the effect of deregulation of SUMOylation on a singular target only causes a modest defect and does not always lead to a significant phenotype (Silver et al., 2011). Instead, SUMOs are likely to regulate the function of many proteins simultaneously, which altogether is required for efficient functioning of the cell. Indeed, disruption of the SUMO machinery at the level of conjugating and deconjugating enzymes leads to embryonic lethality, associated with genome instability (Geiss-Friedlander and Melchior, 2007).
Advances in the field of mass spectrometry and bio-informatics have increasingly facilitated the system-wide study of PTMs. Large studies on modifications such as phosphorylation, acetylation and ubiquitylation have been published, with many thousands of target proteins being quantitatively investigated, and tens of thousands specific conjugation sites being mapped (Choudhary and Mann, 2010; Silva et al., 2013). Due to the relatively low abundance of SUMOylation and purification challenges of SUMO-conjugated proteins, SUMOs still elude in-depth investigation. Progress has been made in large-scale analysis of SUMOylated proteins, especially through application of stable isotope labeling of amino acids in culture (SILAC), allowing SUMOylation to be quantitatively studied at the protein level (Schimmel et al., 2014; Vertegaal, 2011; Yang et al., 2012). More recently, there have been large advances in the identification of SUMO-2 conjugation sites, allowing hundreds of these sites to be mapped (Hendriks et al., 2014; Tammsalu et al., 2014).
We utilized a SUMO purification method combining harsh lysis with efficient and high-yield purification of SILAC-labeled FLAG-SUMO-conjugated proteins (Schimmel et al., 2014), and additionally applied a label-free site-specific method for identification of SUMO sites utilizing His10-tagged SUMO (Hendriks et al., 2014). This combined methodology was employed to study the role of SUMOylation in the cellular response to methyl methanesulfonate (MMS), a commonly utilized alkylating agent. MMS provokes the formation of DNA adducts which impair replication fork progression (Alabert et al., 2009; Vazquez et al., 2008). Recruitment of DNA repair proteins to these damaged replication forks is necessary to bypass the lesions (Gonzalez-Prieto et al., 2013).
RESULTS
A Strategy to Purify FLAG-SUMO-2 Conjugates by Immunoprecipitation
SUMO-specific proteases (SENPs) are responsible for deconjugation of SUMOs from their target proteins. SUMO conjugates are notoriously difficult to purify from cells due to the robustness and high activity of SENPs upon cell lysis. To date, there are no effective and cell-permeable SENP-specific inhibitors. Thus, lysis of cells in standard buffers makes isolation of SUMO-conjugates difficult as SENPs are stable under a wide range of conditions, and act very swiftly and promiscuously to remove SUMOs from all target proteins.
In order to study SUMOylated proteins, a common approach is usage of an epitope-tagged exogenous SUMO in order to facilitate purification after lysis under highly denaturing conditions. We utilized FLAG-tagged SUMO-2 stably expressed at near-endogenous levels in HeLa cells (Figure 1A). Investigation of the FLAG-SUMO-2 cell line by confocal fluorescent microscopy revealed a proper nuclear localization of FLAG-SUMO-2, as well as characteristic nuclear bodies scattered around the nucleus (Figure 1B). SUMO-2, which is virtually identical to SUMO-3 (Wang and Dasso, 2009), was chosen in the context of our experiment because it is the most dynamic and abundant SUMO family member (Saitoh and Hinchey, 2000).
Figure 1. Generation of cell lines stably expressing FLAG-tagged SUMO-2, and purification of FLAG-SUMO-2 conjugates following MMS treatment.
A) HeLa cells were infected with a lentivirus encoding FLAG-SUMO-2. Cells stably expressing low levels of FLAG-SUMO-2 were selected by flow cytometry. Total lysates were analyzed by immunoblotting to confirm the expression of FLAG-SUMO-2. Ponceau-S staining is shown as a loading control.
B) Stable cell lines were investigated by Z-stacked confocal fluorescent microscopy to confirm nuclear localization of FLAG-SUMO-2. Characteristic SUMO nuclear bodies are indicated with arrows. Upon MMS treatment (0.02% for 90 minutes), SUMO nuclear bodies dispersed. Scale bars represent 5 μm.
C) Schematic representation of the SILAC proteomics workflow. One set of parental HeLa cells and two sets of HeLa cells expressing FLAG-SUMO-2 were differentially SILAC labeled (K0R0/K4R6/K8R10). One labeled FLAG-SUMO-2 set was treated with 0.02% MMS for 90 minutes. The experiment was repeated with reversal of SILAC labels.
D) Coomassie analysis of total lysate sample versus FLAG-purified SUMOylated proteins, prior to in-gel digestion and analysis by mass spectrometry.
E) Schematic representation of the label-free proteomics workflow for identification of SUMO sites. Four sets of His10-SUMO-2-K0-Q87R cells were treated with 0.02% MMS for 90 minutes, and four sets were control treated, prior to SUMO site enrichment, identification and quantification.
SUMOylation Dynamics in Response to the DNA Damaging Agent MMS
SUMOylation plays a key role in the DNA Damage Response (DDR) (Jackson and Durocher, 2013), e.g., it was shown that the SUMOylation system is required for cellular responses to MMS (Hoege et al., 2002; Maeda et al., 2004). Despite the identification of a significant number of DDR components as SUMO target proteins, global insight in SUMOylation dynamics in response to DNA damage is limited.
In order to quantitatively detect changes in SUMOylation of proteins during the DDR, we utilized a SILAC approach to apply differential metabolic labeling to FLAG-SUMO-2 cell lines as well as the parental HeLa cell line (Figure 1C). Subsequently, we treated the FLAG-SUMO-2 cells with MMS, and left another pool of FLAG-SUMO-2 cells and the parental HeLa cells untreated, prior to lysis and subsequent immunoprecipitation of FLAG-SUMO-2 (Figure 1D). Upon treatment with MMS, SUMO nuclear bodies rapidly dispersed, indicating a role for the dissociation of these SUMO-enriched nuclear bodies in response to DNA damage (Figure 1B).
In addition to studying the changes in the SUMOylated proteome at the protein-level, we also investigated changes at the site-specific level in response to MMS (Figure 1E). To this end, we employed a label-free methodology utilizing a HeLa cell line stably expressing lysine-deficient His10-tagged Q87R SUMO-2 (Hendriks et al., 2014). In order to increase accurate quantitative coverage of SUMOylation dynamics at the site-specific level, we cross-matched peptides detected by MS2 in any replicate to all others, through alignment of retention time and m/z characteristics at the MS1 level (Figure 1E).
LC-MS Identification of Dynamically Regulated SUMOylated Proteins in Response to DNA Damage
We identified 1,431 putative SUMO targets using the SILAC approach, and 844 putative SUMO targets using the label-free approach. By overlapping proteins detected in both approaches, we identified 317 SUMOylated proteins with very high confidence (Figure 2A and Table S1). Furthermore, we mapped SUMOylation sites in 194 of these proteins. In total, we mapped 755 SUMO sites (Table S2) in 352 proteins (Table S3).
Figure 2. Analysis of the mass spectrometry data reveals highly enriched interaction clusters.
A) Overview of proteins identified by the SILAC protein-level workflow, by the label-free sites-level workflow, and proteins in which sites were identified. Only proteins which were found by both approaches, shaded in blue, were used for further analysis.
B) Scatter plot analysis of protein SUMOylation ratio (log2) in response to MMS, versus protein SUMOylation SILAC ratio (log2) enrichment over the parental control. The parental control refers to cells not expressing FLAG-SUMO-2. “Dynamic” proteins were found consistently and significantly changed across all proteomic analyses and replicates. Proteins of interest which were confirmed by immunoblotting analysis are indicated.
C) Volcano plot displaying SUMOylation site ratio (log2) in response to MMS, versus the negative P value (log10) resulting from permutation-based FDR. Red dots indicate downregulated sites, blue dots indicate upregulated sites. Sites in proteins of interest are indicated.
D) Term enrichment analysis of identified up- and down-regulated SUMOylated proteins in response to MMS. Relative score of regulated SUMOylated proteins versus the human proteome is indicated for the Gene Ontology (GO) Molecular Functions, Cellular Compartments and Biological Processes categories. A full term enrichment analysis is available for upregulated SUMO targets (Table S8) and downregulated SUMO targets (Table S9).
E) STRING network analysis of all regulated SUMOylated proteins in response to MMS. Enrichment ratio refers to the amount of interactions observed versus the amount of expected interactions (p>0.4).
We identified 20 proteins with upregulated SUMOylation and 33 proteins with downregulated SUMOylation in response to MMS treatment (Figure 2B and Table S4). Proteins were only considered if they were enriched after purification from the FLAG-SUMO-2 cell line as compared to the parental control cell line. In order for proteins to be considered dynamic, we filtered for a significant and consistent change across all proteomic analyses and replicates. We observed a notable correlation between the SILAC and label-free approaches (Figure S1A), regardless of different experimental setup.
Interestingly, while a modest decrease in the total SUMOylated pool of proteins was observed, various SUMOylated proteins were upregulated, including p300, CBP, PARP1 and JARID1C (Figure 2B and S1B). One of the most striking changes we observed was the downregulation of SUMOylated demethylase JARID1B, whereas SUMOylation of the closely related family member JARID1C was upregulated in response to MMS.
At the sites level, 71 sites were observed to be upregulated in response to MMS, whereas 291 sites were found to be downregulated (Figure 2C and Table S5). We identified sites in 36 out of 53 of the dynamic SUMO targets, and in general found site dynamics to closely match protein dynamics. Although more downregulated sites were observed than upregulated sites, several key DNA damage proteins such as BRCA1, Rap80, MDC1, PARP1 and XPA were found to be increased for SUMOylated.
We performed a comparison of all SUMOylated proteins and sites we identified to other MS/MS studies (Becker et al., 2013; Bruderer et al., 2011; Golebiowski et al., 2009; Hendriks et al., 2014; Matic et al., 2010; Schimmel et al., 2014; Tammsalu et al., 2014). Overall, 93% of the proteins we identified were previously reported in the literature, and 94% of the MMS-dynamic subset (Figure S1C and Table S6). In addition, 92% of SUMOylated proteins identified by site (Figure S1D and Table S6), and 86% of all SUMOylation sites, were previously reported (Figure S1D and Table S7). As such, the high accuracy of our combined approach is demonstrated by re-identification of many known SUMO target proteins.
Characterization of a SUMO-regulated System-wide Response to DNA Damage
Term enrichment analysis of SUMO-regulated proteins provided more insight into the dynamic regulation of the DNA damage response by SUMO (Figure 2D). Proteins with increased SUMOylation showed enrichment in categories such as signal transduction in response to DNA damage, double strand break repair, and methyltransferase activity. Conversely, proteins with decreased SUMOylation were found to be involved in regulation of transcription, transcription cofactors, and a wide range of chromatin modifications and organization. Overall, SUMO targets were enriched for localization in nucleus and in the chromatin.
STRING network analysis of all SUMO-regulated proteins in response to MMS identified 57% of these proteins to be known or predicted interactors to each other (Figure 2E), giving strong backing to the theory of dynamic regulation by SUMO of groups of functionally related proteins (Johnson, 2004; Psakhye and Jentsch, 2012). Addition of SUMO-2 itself to the cluster connects 70% of all proteins together (Figure S1E). Of special note is the PML/CBP/p300/PARP1 main cluster, which functionally connects 38% of all identified SUMO-regulated proteins to each other. This cluster is composed of 12 up- and 8 down-regulated SUMOylated proteins, displaying SUMO’s ability to modulate proteins in a balanced manner.
STRING network analysis of all proteins containing dynamic SUMO sites linked 75% of these proteins together in one functional cluster (Figure S1F). Many proteins which contained multiple dynamic sites generally displayed the same dynamics for all sites. However, certain proteins such as PML, TRIM24, TRIM28 and TRIM33 contained sites that were inversely regulated in response to MMS, indicative of the dynamic nature of SUMO-2 modification.
Differential SUMOylation of Chromatin Modifiers in Response to MMS
On further investigation of selected target proteins by immunoblotting, we confirmed the differential SUMO-regulation of these proteins, as initially identified by mass spectrometry (Figure 3A). P300 and CBP, two important transcriptional co-activators, were found to be more highly SUMOylated in response to MMS. A higher SUMOylation of these proteins is indicative of reduced function, which would lead to overall transcriptional repression (Girdwood et al., 2003). We verified the increase in Poly ADP-Ribose Polymerase 1 (PARP-1) SUMOylation (Figure 3A), and also observed a slight decrease in total SUMO (Figure 3B).
Figure 3. Confirmation of mass spectrometry results.
A) HeLa cells and HeLa cells stably expressing FLAG-SUMO-2 were either mock treated or treated with MMS. Cells were lysed, and FLAG-IP was performed to enrich SUMOylated proteins. Total lysates and SUMO-enriched fractions were analyzed by immunoblotting to confirm the SUMO regulation of the indicated proteins.
B) Immunoblotting verification of overall level of SUMO-conjugated proteins in the total lysates and SUMO-enriched fractions after MMS treatment and FLAG-IP.
C) Cell lines stably expressing FLAG-SUMO-2 were investigated by confocal fluorescent microscopy for the presence of PML nuclear bodies. Upon MMS treatment, PML bodies rapidly dispersed. Scale bars represent 5 μm.
Strikingly, we identified the histone 3 lysine 4 di- and tri-methylation specific demethylases JARID1B and JARID1C as being differentially regulated by SUMO in response to MMS (Figure 3A). Whereas JARID1C SUMOylation was increased after MMS treatment, JARID1B SUMOylation was no longer detectable. Furthermore, upon confirmation that the upper band on the immunoblot corresponded to full-length endogenous JARID1B by shRNA-mediated knockdown of the protein in HeLa and U2OS cells (Figure S2), we discovered that the JARID1B protein was swiftly degraded in its entirety during MMS treatment.
The SUMOylated form of Promyelocytic Leukemia (PML) protein, a tumor suppressor protein, has a key function in structuring nuclear bodies (Ishov et al., 1999; Muller and Dejean, 1999; Shen et al., 2006). We confirmed the rapid loss of SUMOylation after MMS treatment by immunoblotting, and the dissociation of PML nuclear bodies by confocal fluorescent microscopy (Figure 3C). We also verified the decreased SUMOylation of Methyl-CpG-binding domain protein 1 (MBD1) after MMS treatment by immunoblotting (Figure 3A).
SUMOylated JARID1B and PML are Degraded by the Ubiquitin-Proteasome in Response to MMS
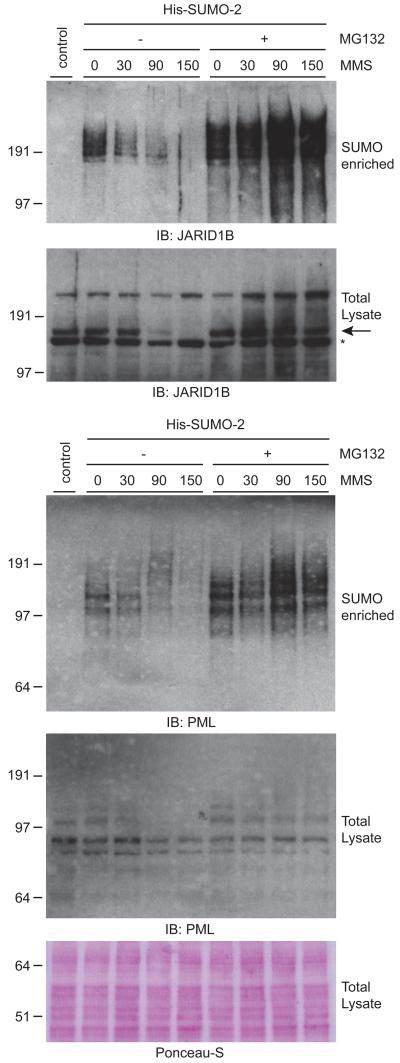
We investigated the involvement of the ubiquitin-proteasome by combining MMS treatment of cells with the proteasome inhibitor MG132. For this purpose, we used a U2OS cell line stably expressing His10-SUMO-2 to investigate SUMOylated proteins. In response to MMS, SUMOylated JARID1B and PML were rapidly degraded, as observed earlier in HeLa cells (Figure 4). When pre-treating cells with MG132 before addition of MMS, the degradation of JARID1B protein was blocked entirely. Furthermore, the degradation of SUMOylated JARID1B and PML was prevented.
Figure 4. SUMOylated JARID1B and PML are rapidly degraded by the proteasome upon MMS treatment.
U2OS cells stably expressing His10-tagged SUMO-2 were treated with 0.02% MMS for the indicated amount of time. One set of cells was additionally treated with the proteasome inhibitor MG132 at 10 μM. Cells were lysed, and His10-pulldown was performed to enrich SUMOylated proteins. Total lysates and SUMO-enriched fractions were analyzed by immunoblotting using antibodies against JARID1B and PML. Ponceau-S staining was performed on total lysate fractions as a loading control.
Degradation of SUMOylated JARID1B and PML is Controlled by the SUMO-specific Ubiquitin E3 Ligase RNF4
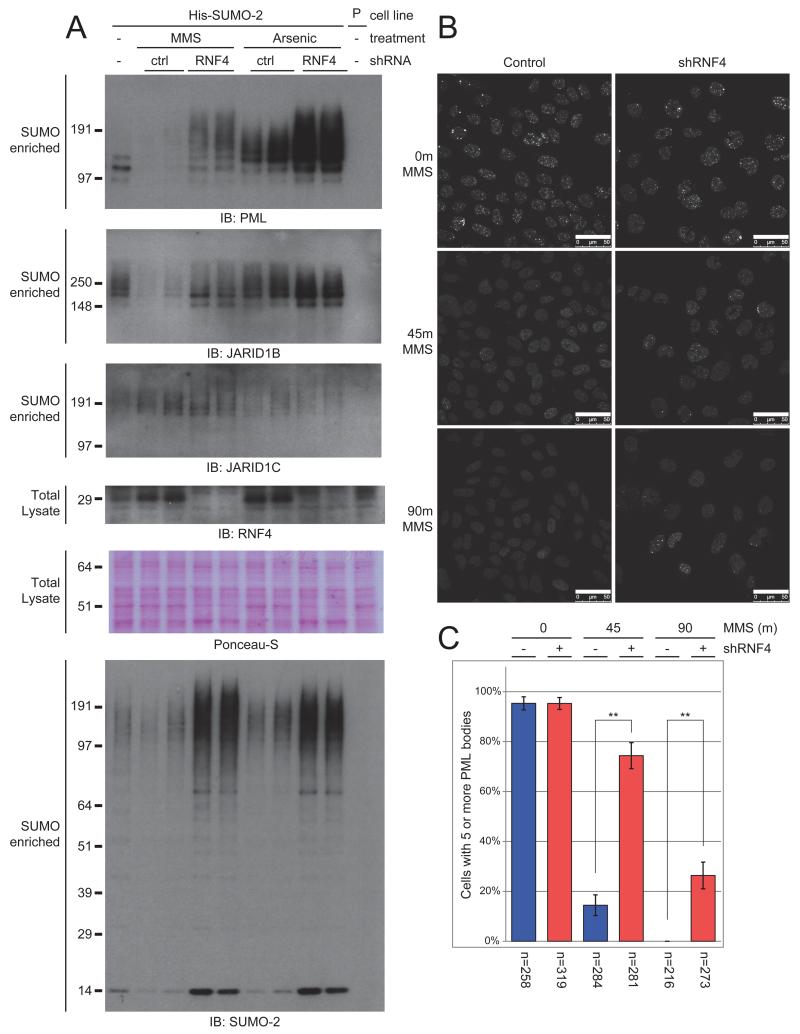
RNF4 is a SUMO-specific ubiquitin E3 ligase that recognizes poly-SUMOylated target proteins through its SUMO Interaction Motifs (SIMs), and subsequently facilitates the ubiquitylation and degradation of these proteins (Sun et al., 2007). We investigated the involvement of RNF4 by using shRNA-mediated depletion of endogenous RNF4 prior to MMS treatment (Figure 5A). We observed an accumulation of highly SUMOylated PML when depleting endogenous RNF4 prior to treatment with MMS. When treating cells with arsenic trioxide as a control (Tatham et al., 2008), PML was found to be more heavily poly-SUMOylated, and additional depletion of endogenous RNF4 resulted in a further increase of highly SUMOylated forms of PML. Similarly, SUMOylated JARID1B was retained after MMS treatment when endogenous RNF4 was depleted. In contrast to PML, we observed no notable change in JARID1B’s SUMOylation state in response to arsenic treatment. Depletion of RNF4 did not further increase JARID1C SUMOylation in response to MMS. We noted a considerable increase of these proteins when endogenous RNF4 was depleted. RNF4 regulates many poly-SUMOylated proteins, and its absence would result in an accumulation of its SUMOylated targets.
Figure 5. Degradation of SUMOylated JARID1B and PML is mediated by the SUMO-targeted ubiquitin ligase RNF4.
A) U2OS cells stably expressing His10-tagged SUMO-2 were treated with 0.02% MMS or 1 μM arsenic trioxode for 90 minutes or 4 hours, respectively. One set of cells was additionally depleted for RNF4 by shRNA-mediated knockdown. After treatment, cells were lysed, and SUMOylated proteins were enriched. SUMO-enriched fractions were analyzed by immunoblotting using the indicated antibodies. Total lysate fractions were analyzed by immunoblotting for depletion of RNF4.
B) U2OS cells were treated with MMS for the indicated amount of time. One set of cells was additionally depleted for RNF4 by shRNA-mediated knockdown. Groups of cells were investigated by Z-stacked confocal fluorescence microscopy for the presence of PML bodies. Scale bars represent 50 μm.
C) Multiple fields of cells were recorded as in Figure 5B. Cells were counted, PML bodies were quantified in every cell, and scored for the presence of at least five PML bodies. The number of quantified cells is indicated below each experiment. Error bars represent 2× SEM. ** p < 1*10−15
Dissociation of PML Nuclear Bodies in Response to MMS Treatment is Modulated by SUMO-specific Ubiquitin E3 Ligase RNF4
Dissociation of PML bodies in response to DNA damage has been reported before (Brouwer et al., 2009), and is thought to mediate redistribution of proteins from these nuclear depots to sites of DNA damage (Bernardi and Pandolfi, 2007). We observed that depletion of RNF4 prevents poly-SUMOylated PML from being ubiquitylated and degraded (Figure 5A). Similarly, we investigated the dissociation of PML bodies in response to MMS using Z-stacked confocal microscopy, in a control setting and with depletion of endogenous RNF4 by shRNA-mediated knockdown (Figure 5B). We additionally selected multiple fields in an unbiased fashion and quantified cells and their respective PML bodies (Figure 5C). When RNF4 was depleted in cells prior to MMS treatment, as opposed to the control, the large majority of cells still retained their PML bodies after 45 minutes of MMS treatment, and after 90 minutes over a quarter of all cells retained a moderate amount of PML bodies. These findings show that PML bodies dissociate through RNF4-mediated ubiquitylation and subsequent degradation of SUMOylated PML in response to MMS.
Cells enter a transcriptionally-repressed state in response to MMS treatment
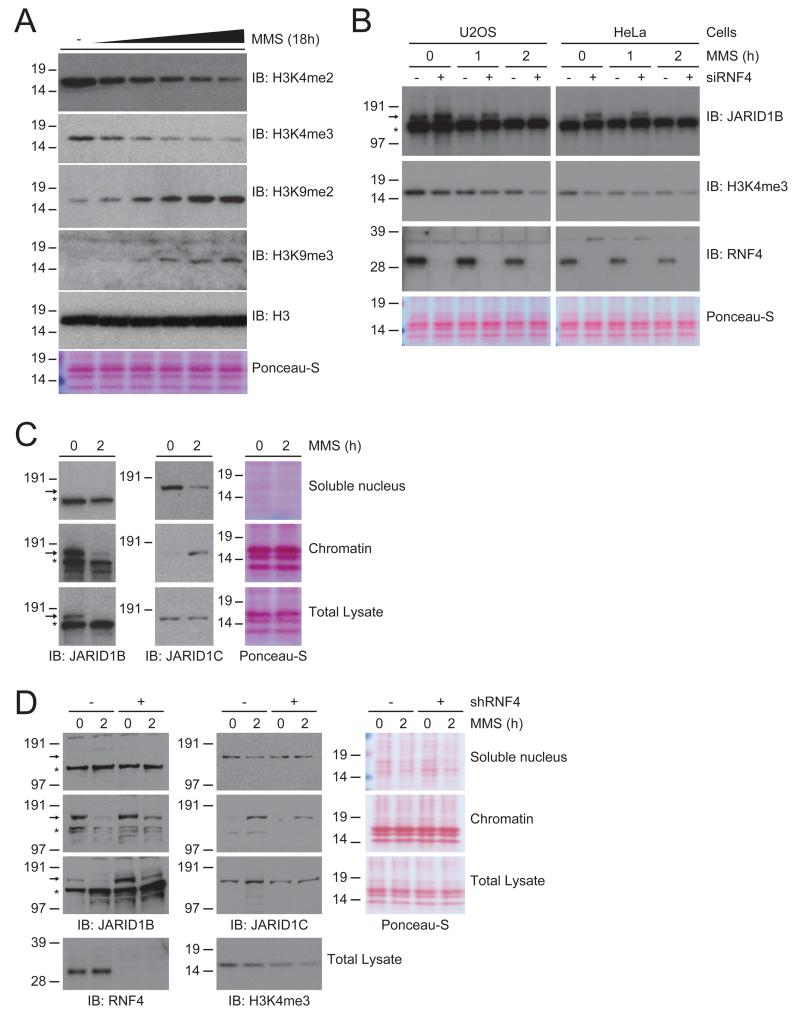
As JARID1B and JARID1C are H3K4me2/3-specific demethylases, we investigated the effect of MMS on the global level of the transcriptional activity markers H3K4me2 and -me3 in U2OS cells (Figure 6A). We observed that with increasing amounts of DNA damage induced by treating cells with increasing dosage of MMS, the global H3K4me2 and H3K4me3 levels drop in a dynamic fashion in concurrence with the amount of DNA damage, indicating that the differential regulation of JARID1B and JARID1C resulted in increased demethylase activity.
Figure 6. H3K4me2/3-specific demethylase JARID1C is recruited to the chromatin in response to DNA damage.
A) U2OS cells were treated with an increasing dose of MMS for 18 hours. Cells were lysed and analyzed by immunoblotting with the indicated antibodies.
B) U2OS and HeLa cells were treated with 0.02% MMS for the indicated amount of time, and one set of cells was additionally depleted for RNF4 using siRNA-mediated knockdown. Cells were lysed and analyzed by immunoblotting with the indicated antibodies.
C) U2OS cells were mock treated or treated with 0.02% MMS for 90 minutes, and subsequently cellular fractionation was performed. Cellular fractions and total lysates were analyzed by immunoblotting for JARID1B and JARID1C. Ponceau-S staining of the histones is shown as a loading and fractionation control.
D) As Figure 6C, with one set of cells additionally depleted for RNF4 using shRNA-mediated knockdown. Additionally, total lysates were analyzed by immunoblotting for H3K4me3 and RNF4.
In these experiments, we also verified the levels of the transcriptional repressive marks H3K9me2 and -me3, since our screen also indicated the dynamic regulation by SUMOylation of the H3K9 methylases SETDB1 and MBD1 (Figure 3A, Tables S2 and S3). Interestingly, global levels of H3K9me2 and H3K9me3 were elevated as more DNA damage is incurred. Overall, our results show that SUMOylation orchestrates multiple histone demethylases and methylases in response to DNA damage, resulting in a state of transcriptional inhibition, as cells are likely to decrease transcriptional activity to prioritize DNA repair processes (Svejstrup, 2010).
JARID1B levels are regulated by RNF4 under standard growth conditions
We demonstrated that SUMOylated forms of JARID1B can be stabilized in response to MMS by depletion of RNF4 (Figure 5A). The effect of RNF4 depletion on the total levels of JARID1B was further investigated through siRNA-mediated knockdown (Figure 6B). We observed a large increase in the base level of JARID1B in both HeLa and U2OS cells in the absence of RNF4. After 1 hour of MMS treatment, there is a stabilization of JARID1B in response to RNF4 knockdown, although eventually non-modified JARID1B is degraded after 2 hours of MMS treatment. H3K4me3 was found to be decreased after depletion of RNF4, which could be a direct result of stabilization of JARID1B (Figure 6B). In response to MMS, however, a more significant drop in H3K4me3 levels was observed, and even though aggravated by absence of RNF4, the decline in H3K4me3 was likely orchestrated by additional demethylases other than JARID1B.
H3K4me2/3-specific demethylase JARID1C is recruited to the chromatin in response to DNA damage
Investigation of JARID1C total protein levels after MMS treatment revealed no substantial decrease, as opposed to JARID1B. In order to explain the decreased levels of H3K4me2/3, we studied the subcellular localization of these JARID1 family members (Figure 6C). As anticipated, we found JARID1B to be located exclusively in the chromatin-associated fraction prior to DNA damage. Furthermore, MMS treatment resulted in a swift chromatin-associated loss of JARID1B as expected from the global drop in JARID1B. Similar results were acquired in HeLa cells (Figure S3B). Consistently, we found the known JARID1B targets JUN and MCL1 to be upregulated in response to MMS treatment by quantitative PCR (Figure S3A).
Remarkably, we found JARID1C to be localized mainly in the soluble nucleoplasmic fraction prior to DNA damage (Figure 6B). It is therefore plausible to assume that JARID1C exists in a mostly inactive form in cells under normal conditions, whereas JARID1B continuously modulates the H3K4me2/3 state of its targets. Strikingly, upon MMS treatment of cells we found that JARID1C is relocated from the nucleoplasmic fraction to the chromatin-associated fraction, indicating a novel activation of this H3K4me2/3 demethylase in response to DNA damage (Figure 6B). Furthermore, the total level of JARID1C remained mostly unchanged, indicative of a relocation of a considerable fraction of JARID1C. Similar results were obtained in HeLa cells (Figure S3B).
We investigated the SUMOylation state of JARID1C in the chromatin-associated fraction, and found that there is a small amount of SUMOylated JARID1C present at the chromatin before MMS treatment (Figure S3C). Upon MMS treatment, the amount of SUMOylated JARID1C in the chromatin-associated fraction increased considerably, in line with the increase in JARID1C SUMOylation in the total protein pool. Similar results were found in HeLa cells stably expressing His10-SUMO-2 (Figure S3C). Our findings imply that the SUMOylation of JARID1C may be a result of its localization to the chromatin, or conversely may play a role in its localization to the chromatin.
In order to investigate the effect of RNF4 on the regulation and localization of JARID1B and JARID1C in response to DNA damage, we performed a cellular fractionation assay in combination with shRNA-mediated RNF4 depletion and in response to MMS treatment (Figure 6D). As before, we observed a stabilization of total JARID1B by depletion of RNF4. Localization of JARID1C from the soluble nucleoplasmic fraction to the chromatin did not depend on the presence of RNF4. We further confirmed that RNF4 had no effect on the localization of JARID1B and JARID1C by siRNA-mediated depletion of RNF4 and cellular fractionation (Figure S3D). As such, the stronger decline in H3K4me3 by a combination of MMS treatment and RNF4 depletion may be a result of both JARID1B and JARID1C actively demethylating their targets (Figure 6D).
JARID1C demethylates global H3K4me2/3 in response to DNA damage
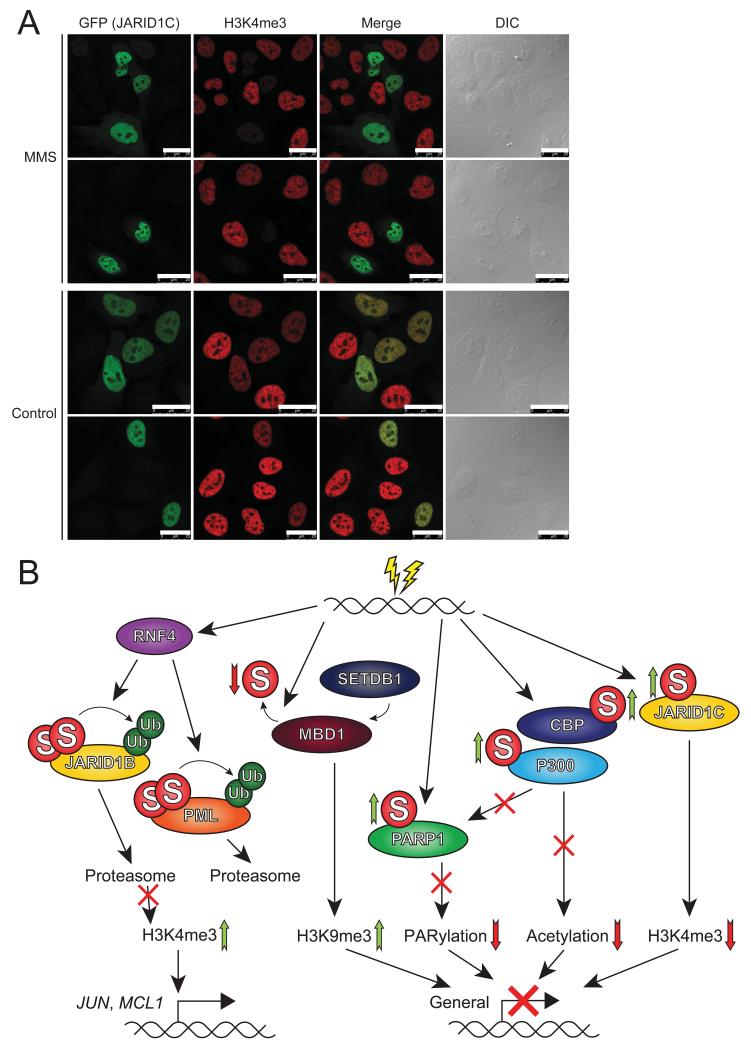
In U2OS cells overexpressing GFP-JARID1C, we observed that the protein localized exclusively to the nucleus, as anticipated (Figure 7A). Overexpression of GFP-JARID1C by itself resulted in a slight decrease of global H3K4me3. However, overexpression of GFP-JARID1B at similar or lower levels resulted in a near complete removal of H3K4me3 (Figure S4), indicative of efficient localization of JARID1B to the chromatin under regular cell culture conditions. Furthermore, MMS treatment of U2OS cells resulted in a similar slight decrease of global H3K4me3. Strikingly, when cells overexpressing GFP-JARID1C were treated with MMS, we observed a dramatic drop of H3K4me3 (Figure 7A). We propose that upon treatment with MMS, GFP-JARID1C is recruited to the chromatin analogous to endogenous JARID1C, where it then actively removes H3K4me3.
Figure 7. JARID1C demethylates global H3K4me3, and SUMOylation mediates chromatin remodeling and transcriptional regulation, in the cellular response to alkylating DNA damage.
A) U2OS cells were transfected with GFP-JARID1C, and either mock treated or treated with 0.02% MMS for 90 minutes. Subsequently, cells were fixed and investigated by confocal fluorescent microscopy to check for the presence of JARID1C (GFP) and the level of H3K4me3. Scale bars represent 25 μm.
B) Schematic overview of the effect of SUMOylation on various key proteins involved in the cellular response to DNA damage, and the downstream effect on chromatin remodeling and transcriptional regulation.
Overall, our study indicates that SUMOylation orchestrates a complex network of chromatin modifiers to regulate transcriptional responses to DNA damage (Figure 7B).
DISCUSSION
SUMO Orchestrates the Cellular Response to DNA damage
Small Ubiquitin-like Modifiers play critical roles in the DNA Damage Response (Jackson and Durocher, 2013; Ulrich and Walden, 2010). To increase our understanding of the roles of SUMOylation in the DDR, we have performed a combined SILAC and label-free quantitative proteomics approach to identify dynamically regulated SUMO-2 conjugates and sites upon treatment with MMS. We have uncovered a set of 20 SUMO-2 conjugates and 71 sites that were upregulated in response to MMS, and 33 conjugates and 291 sites that were downregulated Identified dynamic SUMO target proteins included interaction networks of chromatin modifiers, transcription factors, DNA repair factors and nuclear body components.
SUMO Coordinates Global Transcriptional Repression in Response to DNA Damage
Cells respond to DNA damaging agents by globally altering their transcriptional programs (Fry et al., 2005). Transcriptome-wide studies have indicated that around 30% of mRNAs are altered in response to MMS treatment (Begley and Samson, 2004). This response includes the downregulation of genes involved in protein synthesis to enable cells to focus on nucleic acid metabolism. Moreover, genes involved in the DDR are upregulated in response to MMS (Jelinsky and Samson, 1999).
Our results indicate that SUMO plays an important role in the orchestration of global transcriptional reprogramming in response to MMS at the chromatin level. Firstly, trimethylation of histone H3K4 is a key chromatin mark of active genes (Chi et al., 2010) associated with SUMOylation (Neyret-Kahn et al., 2013) and was found to be decreased upon MMS treatment. Mechanistically, this could be explained by recruitment of JARID1C to the chromatin upon MMS treatment to reduce global H3K4me3. Our findings indicate a novel role for JARID1C in the DNA damage response.
Secondly, global levels of the transcriptional repressive marks H3K9me2/3 (Chi et al., 2010), were found to be increased upon MMS treatment. Mechanistically, this could be explained by reduced SUMOylation of Methyl-CpG-Binding domain 1 (MBD1) in response to MMS. MBD1 forms a repressive complex with the methylase SETDB1 to facilitate methylation of H3K9, and SUMOylation of MBD1 has been reported to counteract its interaction with SETDB1 and thus prevent its repressive function (Lyst et al., 2006). The decrease in MBD1 SUMOylation that we observed in our study is therefore expected to increase the interaction with SETDB1, concomitantly increasing the methylation of H3K9. We also discovered an increased SUMOylation of SETDB1 itself.
Thirdly, we observed increased SUMOylation of the acetyl transferases CBP and p300, which was previously shown to decrease the activity of these proteins (Girdwood et al., 2003). CBP and p300 acetylate all four core histones, which is expected to cause nucleosome instability (Sterner and Berger, 2000). Interestingly, in addition to histones, another target proteins is (Poly [ADP-ribose] Polymerase 1) PARP-1, which requires its acetylated state in order to PARylate its targets (Messner et al., 2009), including histone H3 (Messner et al., 2010), thus functioning as a transcriptional co-activator by organizing a permissive chromatin environment. Correspondingly, we identified PARP1 itself to be increasingly SUMOylated after MMS treatment, which has been implied to further inhibit its acetylation by P300 (Messner et al., 2009), in turn leading to additional diminishment of PARP1’s activity. The decrease of PARP1 activity would then lead to loss of histone H3 PARylation, in turn paving the way for the JARID1 family members to demethylate H3K4me3 (Krishnakumar and Kraus, 2010), leading to inhibition of transcription.
Combined, orchestration of these chromatin modifiers by SUMOylation contributes to a transcriptional repressive environment by decreasing H3K4me2/3, increasing H3K9me2/3 and decreasing histone acetylation. Although transcriptional inhibition was the dominant effect orchestrated by SUMOylation, it was not an exclusive phenomenon as JUN and MCL1 were upregulated (Figure S3A). Mechanistically, this could be explained by the degradation of JARID1B, since it actively controls a set of cell cycle and DNA damage response proteins (Bueno and Richard, 2013). Whether the regulation of JARID1B and 1C in response to MMS is a more general feature of the DNA damage response remains to be addressed. JARID1B and 1C deregulation has been associated with various types of cancer (Kandoth et al., 2013; Mitra et al., 2011; Roesch et al., 2008; Xiang et al., 2007).
SUMO Group Modification in Response to DNA Damage
STRING analysis revealed that over half of the identified SUMO-2 target proteins that were responsive to DNA damage are known to functionally interact. Over one third of all identified proteins resided in one single interaction cluster. This most striking cluster we identified contains many functionally relevant proteins such as PML, SP100, TDG, CBP, P300, PARP1, TOPORS, MDC1, HSF1, MITF, SETDB1 and MBD1, among others, only highlighting a subsection from this cluster. Coordination of these protein networks by SUMOylation is an efficient manner for rapid regulation of functional protein groups via post-translational modification. Group modification by SUMO in response to MMS in yeast was described previously (Johnson, 2004; Psakhye and Jentsch, 2012), but differs from our findings as in yeast the DNA damage response was centered around homologous recombination whereas in human cells we observed a response tailored towards DNA repair, transcriptional regulation and chromatin remodeling.
SUMO-Targeted Ubiquitin Ligases in the DNA Damage Response
We have shown here that proteasomal degradation of SUMOylated JARID1B and SUMOylated PML in response to MMS is mediated by the SUMO-targeted ubiquitin ligase RNF4. Previously, the Mediator of DNA-damage checkpoint 1 (MDC1) protein was identified as a SUMOylated target protein that is subsequently ubiquitylated by RNF4 in response to ionizing radiation (Galanty et al., 2012; Luo et al., 2012; Vyas et al., 2013; Yin et al., 2012). RNF4 ubiquitylated SUMOylated MDC1 leading to its subsequent degradation by the proteasome, an event that is necessary to facilitate proper progression of homologous recombination repair. In our study, we also observed an increase of MDC1 SUMOylation in response to MMS (Table S4).
Summary and future perspective
In summary, we have studied the role of SUMOylation in response to DNA damage, uncovering a co-regulated group of SUMO target proteins, including a significant number of chromatin modifiers that cooperate to decrease global transcription upon DNA damage. Our study uncovers a tight link between the role of SUMOylation in the DNA damage response and the role of SUMOylation in transcriptional regulation, the two key areas of SUMOylation research. Detailed functional analysis of proteins identified in this project, and further studies on the role of SUMO in response to different types of DNA damage, will undoubtedly further increase our insight in the role of SUMOylation in the DNA damage response and genome stability.
EXPERIMENTAL PROCEDURES
Plasmids
Plasmids are described in Supplemental Experimental Procedures.
Cell culture & cell line generation
HeLa and U2OS cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% FBS and 100 U/mL penicillin and streptomycin (Invitrogen). HeLa cells stably expressing FLAG-SUMO-2 were generated through lentiviral infection with a virus encoding FLAG-SUMO-2-IRES-GFP. U2OS cells stably expressing His10-SUMO-2 were generated in a similar fashion using a lentiviral construct carrying His10-SUMO-2-IRES-GFP. Two weeks after infection, cells were fluorescence-sorted for a low expression level of GFP using a FACSAria II (BD Biosciences). His10-SUMO-2-K0-Q87R cells were described previously (Hendriks et al., 2014).
SILAC labeling & MMS treatment
HeLa cells were cultured in DMEM supplemented with 10% dialyzed FBS and containing either light ([12C6 14N2]lysine/[12C6,14N4]arginine), medium ([2H4,12C6,14N2]lysine/[13C6,14N4]arginine) or heavy ([13C6,15N2]lysine/[13C6,15N4]arginine) lysines and arginines. Light label is referred to as K0/R0, medium label as K4/R6, and heavy label as K8/R10. Cells were cultured in SILAC DMEM for eight days, during which they were passaged twice, prior to treatment and lysis. The SILAC MMS experiment was carried out in biological duplicate with a label-swap, with the MMS-treated FLAG-SUMO-2 cell line being heavy-labeled and light-labeled in separate biological replicates, and the mock treated FLAG-SUMO-2 cell line being light-labeled and medium-labeled, respectively. The HeLa parental control was included in the second replicate using heavy-label. The label-free MMS experiment was carried out in biological quadruplicate. For proteomics, all MMS-treated cells were incubated with 0.02% MMS for a duration of 90 minutes.
Primary antibodies
Mouse α SUMO-2 (8A2, Abcam), Mouse α FLAG (M2, Sigma), Mouse α PML (5E10, kind gift from Prof. R van Driel, University of Amsterdam (Stuurman et al., 1992)), Rabbit α JARID1B (A301-813A, Bethyl), Rabbit α JARID1C (A301-035A, Bethyl), Rabbit α p300 (a kind gift from Dr. A. Zantema, Leiden University Medical Center (Ramos et al., 2010)), Rabbit α CBP (A-22, Santa Cruz), Rabbit α H3K4me2 (C64G9, Cell Signaling Technology), Rabbit α PARP1 (9542L, Cell Signaling Technology), Mouse α MBD1 (IMG-306, IMGENEX), Rabbit α H3K4me3 (C42D8, Cell Signaling Technology), Rabbit α H3K9me2 (4658P, Cell Signaling Technology), Rabbit α H3K9me3 (07-442, Millipore), Rabbit α Histone H3 (06-755, Millipore), Rabbit α RNF4 (raised against GST-RNF4 (Vyas et al., 2013)) and Mouse α GFP (11 814 460 001, Roche).
Mass spectrometric and bioinformatics analysis
The mass spectrometry analysis and subsequent bioinformatics analyses are described in Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Netherlands Organization for Scientific Research (NWO) project 70058425 (NWO) (A.C.O.V.), ZonMW project 93511037 (A.C.O.V.), the European Research Council project 310913 (A.C.O.V) and the research career program FSS Sapere Aude (J.V.O.) from the Danish Research Council. The NNF Center for Protein Research is supported by a generous donation from the Novo Nordisk Foundation. We want to thank René Overmeer, Sander Tuit and Jer-gung Chang for technical assistance. We are grateful to Drs. H.T. Timmers, P. de Graaf, R. van Driel and A. Zantema for providing critical reagents.
Footnotes
Supplemental information includes four figures, nine tables, supplemental experimental procedures and supplemental references.
REFERENCES
- Alabert C, Bianco JN, Pasero P. Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. EMBO J. 2009;28:1131–1141. doi: 10.1038/emboj.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Barysch SV, Karaca S, Dittner C, Hsiao HH, Berriel DM, Herzig S, Urlaub H, Melchior F. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat. Struct. Mol. Biol. 2013;20:525–531. doi: 10.1038/nsmb.2526. [DOI] [PubMed] [Google Scholar]
- Begley TJ, Samson LD. Network responses to DNA damaging agents. DNA Repair (Amst) 2004;3:1123–1132. doi: 10.1016/j.dnarep.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Brouwer AK, Schimmel J, Wiegant JC, Vertegaal AC, Tanke HJ, Dirks RW. Telomeric DNA mediates de novo PML body formation. Mol. Biol. Cell. 2009;20:4804–4815. doi: 10.1091/mbc.E09-04-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT. Purification and identification of endogenous polySUMO conjugates. EMBO Rep. 2011;12:142–148. doi: 10.1038/embor.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno MT, Richard S. SUMOylation negatively modulates target gene occupancy of the KDM5B, a histone lysine demethylase. Epigenetics. 2013;8:1162–1175. doi: 10.4161/epi.26112. [DOI] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- Fry RC, Begley TJ, Samson LD. Genome-wide responses to DNA-damaging agents. Annu. Rev. Microbiol. 2005;59:357–377. doi: 10.1146/annurev.micro.59.031805.133658. [DOI] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 2012;26:1179–1195. doi: 10.1101/gad.188284.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT. P300 transcriptional repression is mediated by SUMO modification. Mol. Cell. 2003;11:1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Prieto R, Munoz-Cabello AM, Cabello-Lobato MJ, Prado F. Rad51 replication fork recruitment is required for DNA damage tolerance. EMBO J. 2013;32:1307–1321. doi: 10.1038/emboj.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks IA, D’Souza RC, Yang B, Verlaan-de VM, Mann M, Vertegaal AC. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 2014;21:927–936. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 2012;13:755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, III, Maul GG. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MD, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell. 2010;39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Zhang H, Wang L, Yuan J, Lou Z. Sumoylation of MDC1 is important for proper DNA damage response. EMBO J. 2012;31:3008–3019. doi: 10.1038/emboj.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyst MJ, Nan X, Stancheva I. Regulation of MBD1-mediated transcriptional repression by SUMO and PIAS proteins. EMBO J. 2006;25:5317–5328. doi: 10.1038/sj.emboj.7601404. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Maeda D, Seki M, Onoda F, Branzei D, Kawabe Y, Enomoto T. Ubc9 is required for damage-tolerance and damage-induced interchromosomal homologous recombination in S. cerevisiae. DNA Repair (Amst) 2004;3:335–341. doi: 10.1016/j.dnarep.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Matic I, Schimmel J, Hendriks IA, van Santen MA, van de Rijke F, van DH, Gnad F, Mann M, Vertegaal AC. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell. 2010;39:641–652. doi: 10.1016/j.molcel.2010.07.026. [DOI] [PubMed] [Google Scholar]
- Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38:6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner S, Schuermann D, Altmeyer M, Kassner I, Schmidt D, Schar P, Muller S, Hottiger MO. Sumoylation of poly(ADP-ribose) polymerase 1 inhibits its acetylation and restrains transcriptional coactivator function. FASEB J. 2009;23:3978–3989. doi: 10.1096/fj.09-137695. [DOI] [PubMed] [Google Scholar]
- Mitra D, Das PM, Huynh FC, Jones FE. Jumonji/ARID1 B (JARID1B) protein promotes breast tumor cell cycle progression through epigenetic repression of microRNA let-7e. J. Biol. Chem. 2011;286:40531–40535. doi: 10.1074/jbc.M111.304865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, Ng T, Solomon E. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- Muller S, Dejean A. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 1999;73:5137–5143. doi: 10.1128/jvi.73.6.5137-5143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyret-Kahn H, Benhamed M, Ye T, Le GS, Cossec JC, Lapaquette P, Bischof O, Ouspenskaia M, Dasso M, Seeler J, Davidson I, Dejean A. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 2013 doi: 10.1101/gr.154872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151:807–820. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- Ramos YF, Hestand MS, Verlaan M, Krabbendam E, Ariyurek Y, van GM, van DH, van Ommen GJ, den Dunnen JT, Zantema A, ’t Hoen PA. Genome-wide assessment of differential roles for p300 and CBP in transcription regulation. Nucleic Acids Res. 2010;38:5396–5408. doi: 10.1093/nar/gkq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Mueller AM, Stempfl T, Moehle C, Landthaler M, Vogt T. RBP2-H1/JARID1B is a transcriptional regulator with a tumor suppressive potential in melanoma cells. Int. J. Cancer. 2008;122:1047–1057. doi: 10.1002/ijc.23211. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- Schimmel J, Eifler K, Sigurethsson JO, Cuijpers SA, Hendriks IA, Verlaan-de VM, Kelstrup CD, Francavilla C, Medema RH, Olsen JV, Vertegaal AC. Uncovering SUMOylation Dynamics during Cell-Cycle Progression Reveals FoxM1 as a Key Mitotic SUMO Target Protein. Mol. Cell. 2014;53:1053–1066. doi: 10.1016/j.molcel.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol. Cell. 2006;24:331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A, Vitorino R, Domingues MR, Spickett CM, Domingues P. Post-translational Modifications and Mass Spectrometry Detection. Free Radic. Biol. Med. 2013;65:925–941. doi: 10.1016/j.freeradbiomed.2013.08.184. [DOI] [PubMed] [Google Scholar]
- Silver HR, Nissley JA, Reed SH, Hou YM, Johnson ES. A role for SUMO in nucleotide excision repair. DNA Repair (Amst) 2011;10:1243–1251. doi: 10.1016/j.dnarep.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N, de GA, Floore A, Josso A, Humbel B, de JL, van DR. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 1992;101(Pt 4):773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup JQ. The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem. Sci. 2010;35:333–338. doi: 10.1016/j.tibs.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Tammsalu T, Matic I, Jaffray EG, Ibrahim AF, Tatham MH, Hay RT. Proteome-Wide Identification of SUMO2 Modification Sites. Sci. Signal. 2014;7:rs2. doi: 10.1126/scisignal.2005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat. Rev. Mol. Cell Biol. 2010;11:479–489. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- Vazquez MV, Rojas V, Tercero JA. Multiple pathways cooperate to facilitate DNA replication fork progression through alkylated DNA. DNA Repair (Amst) 2008;7:1693–1704. doi: 10.1016/j.dnarep.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Vertegaal AC. Uncovering ubiquitin and ubiquitin-like signaling networks. Chem. Rev. 2011;111:7923–7940. doi: 10.1021/cr200187e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas R, Kumar R, Clermont F, Helfricht A, Kalev P, Sotiropoulou P, Hendriks IA, Radaelli E, Hochepied T, Blanpain C, Sablina A, van AH, Olsen JV, Jochemsen AG, Vertegaal AC, Marine JC. RNF4 is required for DNA double-strand break repair in vivo. Cell Death Differ. 2013;20:490–502. doi: 10.1038/cdd.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dasso M. SUMOylation and deSUMOylation at a glance. J. Cell Sci. 2009;122:4249–4252. doi: 10.1242/jcs.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP, Chen CD. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Thompson JW, Wang Z, Wang L, Sheng H, Foster MW, Moseley MA, Paschen W. Analysis of oxygen/glucose-deprivation-induced changes in SUMO3 conjugation using SILAC-based quantitative proteomics. J. Proteome. Res. 2012;11:1108–1117. doi: 10.1021/pr200834f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 2012;26:1196–1208. doi: 10.1101/gad.189274.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.