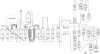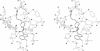Abstract
The Thermus aquaticus DNA methyltransferase M.Taq I (EC 2.1.1.72) methylates N6 of adenine in the specific double-helical DNA sequence TCGA by transfer of --CH3 from the cofactor S-adenosyl-L-methionine. The x-ray crystal structure at 2.4-A resolution of this enzyme in complex with S-adenosylmethionine shows alpha/beta folding of the polypeptide into two domains of about equal size. They are arranged in the form of a C with a wide cleft suitable to accommodate the DNA substrate. The N-terminal domain is dominated by a nine-stranded beta-sheet; it contains the two conserved segments typical for N-methyltransferases which form a pocket for cofactor binding. The C-terminal domain is formed by four small beta-sheets and alpha-helices. The three-dimensional folding of M.Taq I is similar to that of the cytosine-specific Hha I methyltransferase, where the large beta-sheet in the N-terminal domain contains all conserved segments and the enzymatically functional parts, and the smaller C-terminal domain is less structured.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker D., Bystroff C., Fletterick R. J., Agard D. A. PRISM: topologically constrained phased refinement for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 1993 Sep 1;49(Pt 5):429–439. doi: 10.1107/S0907444993004032. [DOI] [PubMed] [Google Scholar]
- Barany F., Slatko B., Danzitz M., Cowburn D., Schildkraut I., Wilson G. G. The corrected nucleotide sequences of the TaqI restriction and modification enzymes reveal a thirteen-codon overlap. Gene. 1992 Mar 1;112(1):91–95. doi: 10.1016/0378-1119(92)90307-b. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991 Nov 19;30(46):11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- Cheng X., Kumar S., Posfai J., Pflugrath J. W., Roberts R. J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell. 1993 Jul 30;74(2):299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- Ho D. K., Wu J. C., Santi D. V., Floss H. G. Stereochemical studies of the C-methylation of deoxycytidine catalyzed by HhaI methylase and the N-methylation of deoxyadenosine catalyzed by EcoRI methylase. Arch Biochem Biophys. 1991 Feb 1;284(2):264–269. doi: 10.1016/0003-9861(91)90294-s. [DOI] [PubMed] [Google Scholar]
- Jack W. E., Greenough L., Dorner L. F., Xu S. Y., Strzelecka T., Aggarwal A. K., Schildkraut I. Overexpression, purification and crystallization of BamHI endonuclease. Nucleic Acids Res. 1991 Apr 25;19(8):1825–1829. doi: 10.1093/nar/19.8.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janulaitis A., Vaisvila R., Timinskas A., Klimasauskas S., Butkus V. Cloning and sequence analysis of the genes coding for Eco57I type IV restriction-modification enzymes. Nucleic Acids Res. 1992 Nov 25;20(22):6051–6056. doi: 10.1093/nar/20.22.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Kumar S., Roberts R. J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994 Jan 28;76(2):357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Nelson J. L., Roberts R. J. The sequence specificity domain of cytosine-C5 methylases. Nucleic Acids Res. 1991 Nov 25;19(22):6183–6190. doi: 10.1093/nar/19.22.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S., Timinskas A., Menkevicius S., Butkienè D., Butkus V., Janulaitis A. Sequence motifs characteristic of DNA[cytosine-N4]methyltransferases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res. 1989 Dec 11;17(23):9823–9832. doi: 10.1093/nar/17.23.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., Roberts R. J. How M.MspI and M.HpaII decide which base to methylate. Nucleic Acids Res. 1992 Sep 25;20(18):4811–4816. doi: 10.1093/nar/20.18.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogolotti A. L., Jr, Ono A., Subramaniam R., Santi D. V. On the mechanism of DNA-adenine methylase. J Biol Chem. 1988 Jun 5;263(16):7461–7464. [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatko B. E., Benner J. S., Jager-Quinton T., Moran L. S., Simcox T. G., Van Cott E. M., Wilson G. G. Cloning, sequencing and expression of the Taq I restriction-modification system. Nucleic Acids Res. 1987 Dec 10;15(23):9781–9796. doi: 10.1093/nar/15.23.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman S. M. DNA methylation: a phoenix rises. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):8761–8762. doi: 10.1073/pnas.90.19.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P., Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- Wilson G. G. Amino acid sequence arrangements of DNA-methyltransferases. Methods Enzymol. 1992;216:259–279. doi: 10.1016/0076-6879(92)16026-g. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4778–4786. [PubMed] [Google Scholar]