Abstract
Since their first description in mammalian cells, more than 2,500 microRNA molecules have been predicted or verified within human cells. Recently, extracellular microRNAs have been described, protected from degradation by specialized packaging in extracellular vesicles or RNA-binding proteins. Such microRNAs, circulating in the bloodstream and extracellular space, have been proposed as attractive candidates as both diagnostic and prognostic biomarkers in various diseases, including a spectrum of cardiovascular conditions. Moreover, consistent with our evolving appreciation of the role of exosomes and microvesicles in intercellular communication, it has been proposed that delivery of active microRNAs to recipient tissues may serve as a primary mode of intercellular communication. Indeed, the transfer of functional microRNAs has been demonstrated in in vitro models and has been reported in a few in vivo contexts. In this review, we will discuss the recent data of circulating microRNAs in cardiovascular disease with an emphasis on their potential roles as diagnostic and prognostic biomarkers as well as the challenges of proving their potential clinical utility. In addition, we will discuss the evidence regarding the role of circulating microRNAs in intercellular communication as well as known molecular factors affecting their packaging, transfer, and uptake in recipient cardiovascular cell types.
Keywords: circulating microRNAs, exosome, Argonaute 2, disease biomarker, intercellular communication
Introduction
MicroRNAs (miRNAs) are small non-coding RNA molecules that mediate post-transcriptional gene regulation by binding the 3’ untranslated regions of messenger RNA [1]. Since the first description of their biologic activity in mammalian cells [2-4], more than 2,500 miRNAs have been identified in the human genome, with >1,500 miRNAs with more defined gene regulatory functions [5]. MiRNAs participate in a wide range of biological processes, including numerous aspects of normal cardiovascular development and physiology as well as cardiovascular disease [6, 7]. Regarding the latter, dysregulated or altered miRNA expression has been implicated in cardiovascular disease such as myocardial infarction, hypertrophy, atherosclerosis, and hypertension, to name a few [6-8].
More recently, Mitchell and colleagues [9] were first to describe the presence of miRNAs in the extracellular space and circulating in mammalian plasma, packaged in remarkably stable forms either in microvesicles or in RNA-protein complexes. Since then, a great variety of studies have been reported regarding the dynamic alterations of circulating miRNA expression in a variety of cardiovascular conditions, leading to speculation of their utility in clinical diagnosis and prognosis [10-13]. Furthermore, the functions of circulating miRNAs as endocrine messengers to allow for intercellular communication have been increasingly interrogated. In this review, we will discuss recent data regarding circulating miRNAs, their potential role as novel biomarkers and intercellular communicators, as well as future challenges of studying and applying such novel biology, particularly in the cardiovascular system.
Stability and packaging of extracellular miRNAs
Although circulating messenger RNAs have previously been detected in human plasma [14], it was not until 2008 that the presence of extracellular miRNAs was described in the human bloodstream by Mitchell et al. [9]. In the same year, Chim et al. [15] described placental miRNAs in the plasma of pregnant women, and Lawrie et al. [16] identified an elevation of tumor-associated miRNAs in the sera of lymphoma patients. The presence of circulating miRNAs in other bodily fluids (i.e., urine, saliva, etc.) was subsequently identified, marked by distinct compositions according to the different fluid types [17, 18].
These initial reports emphasized the notable stability of extracellular miRNAs, even under harsh conditions such as boiling, low/high pH, extended storage at room temperature, and multiple freeze-thaw cycles [9, 17]. These studies also found that naked, synthetic miRNAs directly added to plasma are rapidly degraded by existing RNase activity [9], suggesting endogenous protection against these enzymes. In contrast, detergents or proteinase K facilitate extracellular miRNAs degradation [19, 20], thus indicating protection by lipid membrane encapsulation and/or RNA-binding proteins [21]. Subsequently, several mechanisms for packaging of extracellular miRNAs for transport were defined (Fig. 1), including the encapsulation of miRNAs in membrane-derived vesicles (exosomes [22], microvesicles [23], apoptotic bodies [24]), RNA-binding proteins (Argonaute 2 protein; AGO2 [20] or nucleophosmin I [25]), or lipoprotein complexes such as high-density lipoprotein (HDL) [26].
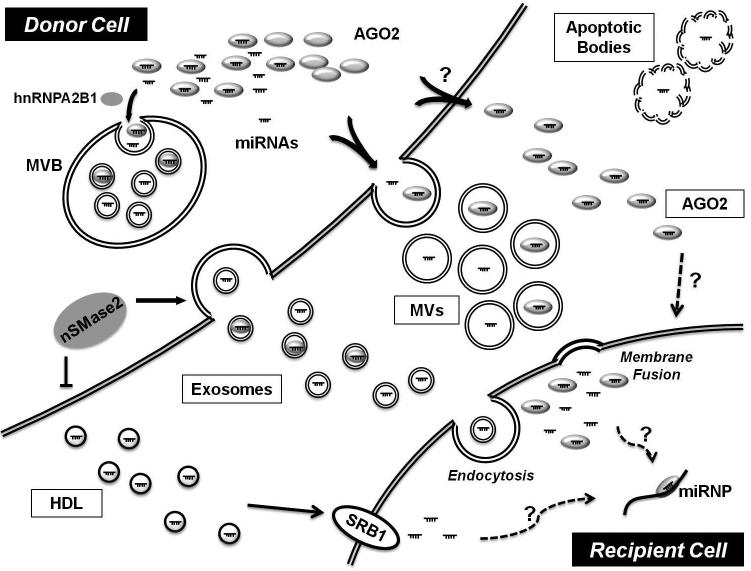
Figure 1. Schematic representation of intercellular communication of miRNAs.
Circulating miRNAs are protected from degradation by several extracellular miRNA carriers, which include membrane-derived vesicles such as exosomes, microvesicles (MVs), or apoptotic bodies, and RNA-binding proteins such as Argonaute 2 protein (AGO2), or high-density lipoprotein (HDL). Exosomes are secreted by the fusion of multivesicular bodies (MVB) and the plasma membrane. MVs are released through outward budding of the plasma membrane. Sumoylated heterogenous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) controls the loading of miRNAs into exosomes through the recognition of specific motif present in miRNAs. Moreover, neutral sphingomyelinase 2 (nSMase2) regulates the ceramide-dependent release of exosomes carrying these miRNAs. Membrane vesicles are delivered by endocytosis or membrane fusion. HDL-associated miRNAs are selectively transferred via scavenger receptor class B type 1 (SR-B1). miRNP = microRNA ribonucleoprotein complex.
Exosomes are small (40 ~ 100 nm in diameter) secreted membrane vesicles that originate from intracellular endosomes and are released upon fusion of multivesicular bodies with the plasma membrane [27, 28]. Exosomes serve as carriers for various miRNAs [22, 29-31]. Notably, expression levels of miRNAs in exosomes can differ substantially as compared with expression in donor cells [22, 32], thus suggesting the existence of an active cellular mechanism that packages specific miRNAs and other cellular contents into these secreted bodies. Similarly, microvesicles are larger (50 ~ 1,000 nm in diameter) membrane vesicles that are produced by budding directly from the plasma membrane [28]. Plasma microvesicles also package miRNAs, and their biology is thought to be integral in cell-to-cell transfer of their miRNA cargo [23, 33, 34]. Apoptotic bodies are even larger particles (1 ~ 5 μm in diameter) released from apoptotic cells and can transport active miRNAs involved in gene repression in recipient cells. For instance, in atherosclerotic vascular disease, endothelial cell-derived apoptotic bodies, enriched with microRNA-126 (miR-126), are produced and trigger the production of the CXC chemokine CXCL12 in the recipient vascular cells, thus mediating, at least in part, the atheroprotective effect of apoptotic bodies [24].
In addition to these membrane-encapsulated vesicles, a significant portion of circulating miRNAs is associated with RNA-binding proteins. Nucleophosmin 1 is a nucleophosphoprotein and involved in the nuclear export of the ribosome [35]. Nucleophosmin 1 protects miRNA degradation by RNase and was the first protein identified as crucial for miRNA transport [25]. Since then, it has been reported that a majority of circulating miRNAs are bound to proteins of the Argonaute family (mainly AGO2), important catalytic components of the RNA induced silencing complex of proteins (RISC) which mediate canonical miRNA engagement with intracellular target transcripts [20, 36]. Secreted miRNAs have also been found in association with lipoproteins [26], including the high- and low-density lipoproteins (HDL and LDL) that enable the transport of lipids and fat-soluble vitamins through the bloodstream [37, 38]. An extracellular miRNA frequently associated with HDL particles is miR-223, a miRNA enriched in hematologic cells such as monocytes and macrophages [23]. This suggests that lipoproteins may be able to be loaded with miRNAs derived from cells other than those responsible for the biogenesis of the lipoprotein itself [39]. Alternatively, it has been suggested that the association of miRNAs with HDL could be, at least in part, nonspecific [40].
The exact molecular processes regulating packaging and release of extracellular miRNAs either via vesicle-mediated or non-vesicle-mediated mechanisms are largely unknown. Some extracellular miRNAs, such as let-7a, have been identified primarily in vesicles, while others, such as miR-16 and miR-92a, have been detected mainly in complexes carrying non-vesicular AGO2 ribonucleoproteins [20]. In the bloodstream, one high-throughput analysis indicated that a majority of miRNAs was found in non-vesicle-associated complexes bound to AGO2 [20]. In contrast, other investigators have reported that extracellular miRNAs detectable in both serum and saliva are mainly concentrated in exosomes [29]. This discrepancy may arise from differences in technique for microvesicle isolation and RNA extraction. Indeed, miRNA plasma content can be heavily contaminated by platelets and blood cells remaining after plasma processing -- factors which must be addressed prior to accurately assessment of expression [41, 42]. Nonetheless, in both vesicles and RNA-protein complexes alike, the association of AGO2 with many miRNAs has been reported to play a critical role in stabilizing these molecules in extracellular space [43, 44]. Alternatively, AGO2 may be crucial for directing transport and/or activity of miRNAs delivered to recipient tissues. For instance, Laffont et al. [45] demonstrated that platelet microparticles specifically containing AGO2/miR-223 complexes could regulate reporter gene expression in recipient endothelial cells. Interestingly, Lv et al. [44] have shown that AGO2 overexpression in HeLa cells facilitates the packaging of miR-16 into cell-secreted microvesicles, and only after such packaging can miR-16 be delivered and significantly reduce Bcl2 protein levels in recipient HEK293 cells. Therefore, it was proposed that non-AGO2-associated miRNAs in the microvesicles are degraded in the recipient cells, while functional AGO2-bound miRNAs are preserved and can be transferred to recipient cells [44].
Beyond complexing with AGO2, several mechanisms have been suggested to control the packaging and sorting of miRNAs for release (Fig. 1). Specific miRNAs may be selectively sorted into the vesicles for extracellular release, based on cellular needs or environmental stimulation [19]. Level of miRNA expression may be a key determinant of such sorting. Recent RNA profiling of macrophages and their released exosomes demonstrated that artificially increasing the cellular levels of a miR-511-3p or its target sequences favors sorting of miR-511-3p into multivesicular bodies for secretion as opposed to P bodies for canonical intracellular activity. Thus, alterations of miRNA expression appear to activate transport mechanisms to relocate miRNAs from one cellular compartment to another and change in the balance between endogenous miRNA activity and secretion through the exosomal pathway [46]. Cellular energetics also contribute to regulation, given findings by Wang et al. [25] that intracellular ATP levels control the exportation of most extracellular miRNAs. Protein components of the RISC beyond AGO2, such as GW182, have also been detected in multivesicular bodies [47] and may participate in the process of miRNAs sorting into exosomes [48]. Alternatively, the sumoylated heterogenous nuclear ribonucleoprotein A2B1 has been reported to control the loading of miRNAs into exosomes through the recognition of specific sequence motifs present in miRNAs [49]. Such recognition may interface with ceramide-dependent secretory machinery, which has also been implicated in packaging of miRNAs into exosomes [50, 51]. Specifically, the sphingolipid ceramide is required for the sorting of exosome-associated domains into multivesicular endosomes, independent of the endosomal sorting complex required for transport system [52]. Inhibition of neutral sphingomyelinase 2 (nSMase2), which regulates the biosynthesis of ceramide, reduces miRNAs secretion, and overexpression of nSMase2 increases extracellular miRNA levels [50, 51]. Interestingly, contrary to the effect on the export of exosomal miRNAs, inhibition of nSMase2 significantly increases cellular export of miR-223 with HDL [26]. Therefore, current data indicate a specific and complex set of pathways must exist that regulates the packaging and release of miRNAs. We now await the more comprehensive identification of such putative actions for many more molecular factors thought to be involved in these processes.
Circulating miRNAs as potential biomarkers in cardiovascular disease
Due to their robust stability and reasonable ease of detectability in the bloodstream, circulating miRNAs are emerging as attractive diagnostic biomarker candidates in a wide range of cardiovascular diseases (Table 1). Of these circulating miRNAs, cardiac-enriched miRNAs, such as miR-1, miR-133, miR-208, and miR-499, have become the most extensively investigated miRNAs, particularly for the diagnosis of acute coronary syndrome (ACS) and acute myocardial infarction (AMI), as compared with conventional markers of myocardial damage such as creatine kinase (CK) or cardiac troponin [10, 53]. For example, cardiac-specific miR-208a, which is exclusively expressed in the heart, was reported as an attractive candidate miRNA consistently observed in a rat myocardial injury model [54] and a study of human AMI cases [55]. However, characterization of a single miRNA as a reliable cardiac biomarker can be challenging; for instance, in other studies, plasma miR-208a concentration was too low to be detected either at baseline or after myocardial injury [53, 56, 57]. Furthermore, in the recent large-scale studies performed in suspected ACS patients, the diagnostic accuracy of using single miRNAs for detecting MI was lower than that of cardiac troponin T [58, 59]. In contrast, the use of a panel of multiple miRNAs or a combination of miRNAs with cardiac troponin has been reported to improve the discriminatory power in ACS diagnosis [60, 61]. The kinetics of release of these miRNAs may also allow for early diagnosis of AMI as compared with traditional biomarkers. Recently, Libetrau et al. [62] demonstrated that serum miR-1 and miR-133a are released within 15 minutes after induction of AMI in patients with hypertrophic cardiomyopathy who underwent transcoronary ablation for septal hypertrophy. These findings are consistent with other studies reporting the release of cardiac-specific miRNAs prior to CK or troponin after prolonged aerobic exercise (i.e., marathon run) [63]. Together, these data suggest the usefulness of circulating cardiac miRNAs in the early diagnosis of AMI and myocardial injury. Finally, to confirm the source of release of miRNAs from cardiac tissue, De Rosa et al. [64] demonstrated the presence of a transcoronary concentration gradient of cardiac-enriched miRNAs proportional to the degree of myocardial injury in ACS, thereby indicating that damaged myocardium was the likely source of released miRNAs. Gidlöf et al. [58] also supported this idea by demonstrating the presence of miR-208b and miR-499-5p in the coronary sinus immediately after, but not before, cardioplegia in patients undergoing coronary artery bypass graft surgery. These findings suggested that those miRNAs are released from injured myocardium. However, it is still unclear whether these released miRNAs are just by-product from damaged myocardium or has additional roles as intercellular messengers.
Table 1.
Circulating miRNAs as diagnostic biomarkers for cardiovascular disease
| Circulating miRNAs |
Expression in cardiovascular disease | References |
|---|---|---|
| miR-1 | Up-regulation in AMI | [91, 125, 126] |
| Up-regulation in ACS | [60] | |
| miR-16 | Down-regulation in CAD | [127] |
| miR-17 | Down-regulation in CAD | [128] |
| miR-19a | Up-regulation in AMI | [129] |
| miR-21 | Up-regulation in NSTEMI in elderly | [130] |
| Up-regulation in ACS | [60] | |
| miR-22 | Up-regulation in HF | [131] |
| miR-23b | Up-regulation in PH | [74] |
| miR-26a | Down-regulation in PAH | [77] |
| miR-30a | Up-regulation in AMI | [132] |
| miR-30b | Down-regulation in HF vs. non-HF dyspnea or control |
[66] |
| miR-31 | Down-regulation in CAD | [127] |
| miR-92a | Down-regulation in CAD | [128] |
| miR-92b | Up-regulation in HF | [131] |
| miR-103 | Down-regulation in HF vs. non-HF dyspnea or control |
[66] |
| miR-122 | Down-regulation in AMI | [53] |
| miR-125b | Down-regulation in AMI | [133] |
| miR-126 | Down-regulation in AMI | [126] |
| Down-regulation in CAD | [128] | |
| miR-130a | Up-regulation in PH | [74] |
| miR-132 | Down-regulation in UA | [61] |
| miR-133a | Up-regulation in AMI | [53, 134] |
| Up-regulation in ACS | [57, 64] | |
| Up-regulation in CAD | [134] | |
| miR-133b | Up-regulation in AMI | [53] |
| miR-134 | Up-regulation in AMI | [135] |
| Up-regulation in APE vs. non-APE or control |
[78] | |
| miR-142-3p | Down-regulation in HF vs. non-HF dyspnea or control |
[66] |
| miR-145 | Down-regulation in CAD | [127, 128] |
| miR-150 | Down-regulation in UA | [61] |
| Down-regulation in PAH | [73] | |
| Down-regulation in atrial fibrillation | [80] | |
| miR-155 | Down-regulation in CAD | [128] |
| miR-181a | Down-regulation in CAD | [127] |
| miR-186 | Up-regulation in UA | [61] |
| miR-191 | Up-regulation in PH | [74] |
| miR-195 | Up-regulation in AMI | [132] |
| miR-208a | Up-regulation in AMI | [55] |
| Up-regulation in ACS | [64] | |
| miR-208b | Up-regulation in AMI | [10, 58-60, 136] |
| miR-320a | Up-regulation in AMI | [59] |
| Up-regulation in HF | [131] | |
| miR-320b | Down-regulation in AMI | [133] |
| miR-323-3p | Up-regulation in ACS | [87] |
| miR-328 | Up-regulation in AMI | [135] |
| miR-342-3p | Down-regulation in HF vs. non-HF dyspnea or control |
[66] |
| miR-375 | Down-regulation in AMI | [53] |
| miR-423-5p | Up-regulation in HF vs. non-HF dyspnea or control |
[65] |
| Up-regulation in HF | [131] | |
| miR-433 | Up-regulation in CAD | [137] |
| miR-451 | Down-regulation in PH | [74] |
| miR-485-3p | Up-regulation in CAD | [137] |
| miR-499 | Up-regulation in AMI | [10, 53, 58, 59, 136] |
| Up-regulation in NSTEMI in elderly | [130] | |
| Up-regulation in ACS | [60, 64] | |
| Up-regulation in HF | [136] | |
| miR-1246 | Down-regulation in PH | [74] |
| let-7b | Down-regulation in AMI | [132] |
| Down-regulation in CTEPH | [79] |
ACS, Acute coronary syndrome; AMI, acute myocardial infarction; APE, acute pulmonary embolism; CAD, coronary artery disease; CTEPH, chronic thromboembolic pulmonary hypertension; HF, heart failure; NSTEMI, non-ST-segment elevation myocardial infarction; PAH, pulmonary arterial hypertension; PH, pulmonary hypertension; UA, unstable angina.
In addition to the diagnosis of coronary artery disease, alterations in the expression of several circulating miRNAs including miR-423-5p have been reported to discriminate heart failure (HF) from dyspnea of different etiologies [65, 66]. Notably, circulating miR-423-5p was not helpful in identifying patients with right HF [67], suggesting that perhaps any miRNA-dependent regulatory mechanism for right HF likely differs from those of left HF. Circulating miRNAs have begun to be assessed in patients with HF with preserved ejection fraction (EF) [68, 69]. Although no single miRNA was better than B-natriuretic peptide (BNP) in discriminating HF with preserved EF from HF with reduced EF, the assessment of multiple plasma miRNAs or combination of miRNAs with BNP significantly improved the discriminative power compared with BNP alone. Recently, circulating miRNAs have been evaluated in the response to treatment in patients with end-stage HF. Akat et al. [70] demonstrated that increased levels of certain myomiRs such as miR-208a, miR-208b, miR-499, and miR-1, were nearly completely reversed after the initiation of a left ventricular assist device (LVAD) in advanced HF, suggesting the usefulness of these myomiRs as biomarkers monitoring cardiac injury. Morley-Smith et al. [71] also found that miR-1202 was useful to discriminate between good and poor response to LVAD placement. In addition, a differential expression of certain miRNAs was observed in allograft rejection, suggesting their potential utility to monitor progress after heart transplantation [72].
Other vascular diseases such as pulmonary hypertension have also been studied in this regard. Rhodes et al. reported that circulating levels of miR-150 were reduced in patients with pulmonary arterial hypertension (PAH), and reduced plasma miR-150 levels were associated with poor survival in these patients [73]. Wei et al. [74] demonstrated that a certain set of circulating miRNAs were dysregulated in patients with pulmonary hypertension (PH) and were proportional to the degree of PH determined by mean pulmonary arterial pressure. Other miRNAs such as the miR-130/301 family [75] and miR-210 [76], which have known causative actions in the pathogenesis of PH have been reported to be elevated in the pulmonary circulation of PH patients. Circulating miR-26a has also been identified to be reduced in PAH and directly related with functional severity of this disease [77]. Differential expression of plasma miRNAs has been reported in acute pulmonary embolism [78] and chronic thromboembolic PH [79].
Presence of arrhythmias such as atrial fibrillation (AF) has been associated with alterations of circulating miRNAs. Liu et al. [80] reported that plasma miR-150 levels were significantly lower in patients with AF compared with healthy controls. These findings were independently validated in a larger population by McManus et al. [81]. Interestingly, in this study, miR-21 and miR-150 levels were lower in persistent AF than in paroxysmal AF, and increased after catheter ablation of AF [81]. Moreover, several attempts have been made to create miRNAs signature using circulating miRNAs for various cardiovascular disease including peripheral arterial disease [82] or congenital heart disease such as ventricular septal defect [83].
In addition to their roles as putative diagnostic biomarkers, the prognostic value of circulating miRNAs in cardiovascular disease has also been investigated with mixed utility (Table 2). It has been demonstrated that the circulating levels of miR-133a and miR-208b were related with all-cause mortality at 6 months in ACS patients [84]. However, both miRNAs added little incremental prognostic value to high-sensitive troponin. In accordance with this result, Eitel et al. [85] also reported that circulating concentration of miR-133a could not independently predict cardiovascular events in ST segment elevation MI patients after adjustment for traditional markers. The association of increased levels of miR-208b and miR-499-5p with increased risk of mortality or HF in MI patients was lost after adjustment for troponin T [58]. On the other hand, low concentration of circulating miR-150 was reported to predict LV remodeling after AMI and outperformed N-terminal proBNP in this regard [86]. Pilbrow et al. [87] reported that lower levels of miR-652 can independently predict HF after AMI. Moreover, the use of a panel of multiple miRNAs or a combination of miRNAs with existing prognostic markers such as BNP or cardiac troponin appeared to improve risk stratification in this context [87-89].
Table 2.
Circulating miRNAs as prognostic biomarkers for cardiovascular disease
| Circulating miRNAs |
Outcome parameter | Expression associated with poor outcome |
References |
|---|---|---|---|
| miR-10 | Allograft rejection after heart transplantation |
Down- regulation |
[72] |
| miR-16 | LV contractility at 6 months post-MI |
Up-regulation | [88] |
| miR-27a | LV contractility at 6 months post-MI |
Up-regulation | [88] |
| miR-29a | LVEDV at 90 days post-MI | Up-regulation | [138] |
| miR-31 | Allograft rejection after heart transplantation |
Up-regulation | [72] |
| miR-34a | HF within 1 year post-MI | Up-regulation | [139] |
| Mortality or HF within 6 months post-MI |
Up-regulation | [89] | |
| LVEDD at 1 year post-MI | Up-regulation | [139] | |
| LVEDV at 6 months post-MI | Up-regulation | [89] | |
| miR-92a | Allograft rejection after heart transplantation |
Up-regulation | [72] |
| miR-101 | LV contractility at 6 months post-MI |
Down-regulation | [88] |
| miR-126 | Incident MI within 10 years | Up-regulation | [140] |
| miR-133a | All-cause mortality within 6 months after ACS |
Up-regulation | [84] |
| MACE within 6 months post-MI | Up-regulation | [85] | |
| miR-133b | Early myocardial injury and recovery after heart transplantation |
Up-regulation | [141] |
| miR-134 | Cardiac death or HF within 6 months post-MI |
Up-regulation | [135] |
| miR-150 | LVEDV post-MI | Down- regulation |
[86] |
| LV contractility at 6 months post-MI |
Down- regulation |
[88] | |
| Survival in PAH | Down- regulation |
[73] | |
| miR-155 | Cardiac death within 1 year post-MI |
Up-regulation | [142] |
| Allograft rejection after heart transplantation |
Up-regulation | [72] | |
| miR-192 | HF within 1 year post-MI | Up-regulation | [139] |
| miR-194 | HF within 1 year post-MI | Up-regulation | [139] |
| LVEDD at 1 year post-MI | Up-regulation | [139] | |
| miR-197 | Incident MI within 10 years | Down- regulation |
[140] |
| miR-208b | Mortality or HF within 30 days post-MI |
Up-regulation | [58] |
| Mortality or HF within 6 months post-MI |
Up-regulation | [89] | |
| LVEDV at 6 months post-MI | Up-regulation | [89] | |
| All-cause mortality within 6 months after ACS |
Up-regulation | [84] | |
| miR-223 | Incident MI within 10 years | Down- regulation |
[140] |
| miR-328 | Cardiac death or HF within 6 months post-MI |
Up-regulation | [135] |
| miR-380* | Cardiac death within 1 year post-MI |
Up-regulation | [142] |
| miR-499-5p | Mortality or HF within 30 days post-MI |
Up-regulation | [58] |
| miR-652 | Readmission for HF post-ACS | Down- regulation |
[87] |
| miR-1202 | Response to LVAD therapy | Up-regulation | [71] |
ACS, Acute coronary syndrome; HF, heart failure; LV, left ventricle; LVAD, left ventricular assist device; LVEDD, left ventricular end-diastolic dimension; LVEDV, left ventricular end-diastolic volume; MACE, major cardiovascular event; MI, myocardial infarction; PAH, pulmonary arterial hypertension.
Despite the attraction of measuring circulating miRNAs, many challenges exist in proving their utility as ideal diagnostic or prognostic biomarkers in cardiovascular disease (Fig. 2). First, many of these miRNAs are ubiquitous, so the definite source of these miRNAs cannot be identified in most cases, with the exception of some cardiac or muscle-specific miRNAs (so-called “myomiRs”) [6]. In spite of recent efforts to identify more definitively the tissue source of released miRNAs [58, 64], many questions remain such as whether alteration of miRNAs levels in cardiovascular disease can be attributed to increased expression or simple release from damaged tissue, and whether released miRNAs are merely by-products of cell injury or play an actual biologic purpose in disease progression or manifestation. Second, there are numerous examples of the same circulating miRNA or myomiR altered in a variety of clinical situations (Table 1). Thus, a consideration of context specificity will be necessary in the future determination of the utility of these miRNAs as true biomarkers of disease. Third, due to their low expression, some cardiac- and muscle-specific circulating miRNAs can be difficult to detect and quantify accurately with currently available methods [90]. Coupled with the possibility of contamination from blood cells, the measurement of these miRNAs can be fraught with error [42]. As such, a number of the alterations in circulating miRNAs that have previously been reported in a variety of cardiovascular disease conditions (Tables 1 and 2) require independent validation in larger cohorts of patients for real application to clinical practice. Fourth, there are no standardized endogenous controls for normalization. Although the spike-in of exogenous miRNAs (e.g., miRNAs derived from the worm Caenorhabditis elegans) is widely used in such analyses [9], alternative methods for normalization using endogenous miRNA controls [53] or plasma volume [91] have been utilized, often with varying effects on final quantitations. Fifth, inter-individual and intra-individual variations in circulating miRNA expression certainly exist, notably dependent upon time of day, diet or nutrition, or gender [92, 93], as well as activity level or physical fitness [63, 94]. Therefore, there exist no reference values for “normal expression” for facile clinical interpretation. Finally, it is still unclear which type of assay system for miRNA measurements would be ideal for clinical use, with possibilities including quantitative polymerase chain reaction vs. next generation sequencing. Recent advances in both platforms have allowed for the simultaneous evaluation of multiple miRNAs with improved sensitivity. However, the decision on the diagnosis of cardiovascular disease is usually required urgently, especially in ACS. Therefore, more rapid and sensitive quantification techniques may need to be developed in the future if circulating miRNAs are to be useful as clinically applicable biomarkers [95].
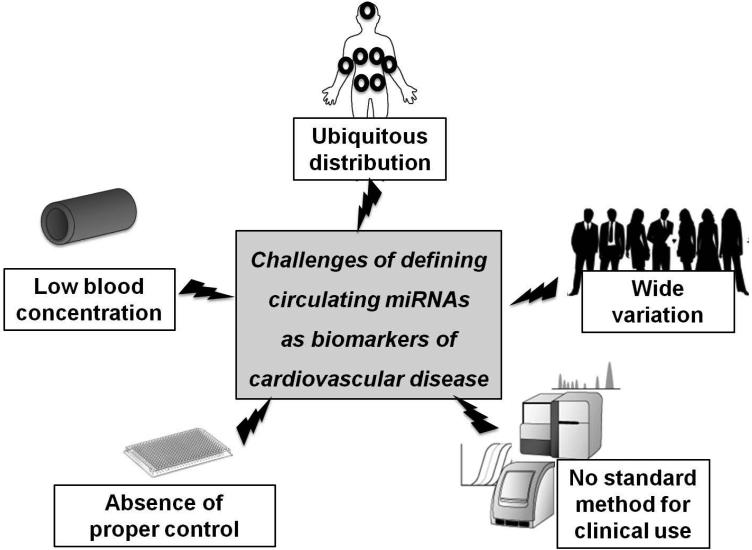
Figure 2. Challenges in defining circulating miRNAs as useful biomarkers of cardiovascular disease.
Although miRNAs have many attractive features for study in the circulating bloodstream, there are still many challenges for establishing circulating miRNAs as clinically useful biomarkers. These include non-specific tissue- or organ-distribution, low serum/plasma concentration, wide inter- or intra-individual variations, absence of controls for normalization, and absence of standardized quantification methods.
Circulating miRNAs as endocrine or paracrine messengers in cardiovascular disease
Beyond the diagnostic and prognostic potential for these molecules, circulating miRNAs may carry substantial functions as endocrine or paracrine messengers in cardiovascular health and disease. Namely, the stability of circulating miRNAs offered by packaging in membrane vesicles or in RNA-protein complexes coupled by an increasing appreciation of the role of microvesicles in facilitating cell-to-cell communication between the cells [28] has raised the possibility of the primary role of miRNAs as intercellular communicators. Several pathways have been suggested as possible mechanisms for cellular uptake of miRNAs into recipient tissue. First, consistent with the known biology of membrane-derived vesicles, Yuan et al. [96] presented early in vitro evidence of intercellular transfer of miRNAs by demonstrating that a subset of miRNAs within microvesicles of mouse embryonic stem cells can be delivered to mouse embryonic fibroblasts. Such microvesicles have been proposed to deliver miRNAs by endocytosis [97], membrane fusion [98], or phagocytosis [99]. Delivery of non-vesicle associated miRNAs has been studied in less depth, but examples exist for effective delivery to recipient tissues. For instance, HDL-associated miRNAs have been demonstrated to be selectively transferred by engaging the scavenger receptor class B type 1 on recipient cells [26]. Alternatively, the hypoxia-induced miR-210 can be delivered to recipient endothelial cells via AGO2-associated RNA-protein complexes outside of vesicles [100]. It remains plausible that extracellular miRNA packaging may modulate the efficiency of recipient tissue delivery, but any differences in delivery efficiency of miRNAs within or outside of microvesicles have yet to be described. Alternatively, it remains unclear whether such packaging may also affect clearance of circulating miRNAs out of the bloodstream into urine, feces, or other excretory substances.
In the context of cardiovascular disease, several in vitro studies have proposed that delivered miRNAs can regulate gene expression through the same canonical action on target messenger RNAs as endogenous miRNAs [22, 26]. For instance, Zernecke et al. [24] reported endogenous transfer of functional miRNAs via apoptotic bodies enriched with the endothelial-specific miRNA, miR-126. Delivery of miR-126 from endothelial cells was reported to convey paracrine signals to recipient vascular cells, thus triggering the production of CXCL12, a cytokine that counteracts apoptosis and recruits progenitor cells. In a separate study, extracellular AGO2-associated miR-126 was also found to transfer from endothelial cells to smooth muscle cells and regulate vascular smooth muscle cell turnover [101]. Halkein et al. [102] also demonstrated that miR-146a-loaded exosomes from endothelial cells can be absorbed by cardiomyocytes, after which miR-146a levels are increased while target gene expression and metabolic activity are decreased. Alternatively, we found that functional hypoxia-induced miR-210, complexed to AGO2 but packaged independently from microvesicles, can be delivered to recipient endothelial cells [100]. Interestingly, we found that prolyl-hydroxylation of AGO2 acts as a molecular switch that not only controls miR-210 release and activity in source cells but also regulates intracellular activity in recipient cells. Native HDL incorporated with exogenous miR-375 or miR-223 can also be delivered to recipient cells, leading to reduction of transcripts encoding RHOB and EFNA1, both of which are direct target of miR-223 [26]. More recently, Tabet et al. [103] demonstrated translational repression of intercellular adhesion molecule 1 in human coronary artery endothelial cells by HDL-transferred miR-223, suggesting a novel mechanism for the known anti-inflammatory properties of HDL.
Although many investigators have reported the effect of transferred miRNAs in target cells, the exact molecular events that facilitate biological activity of mature delivered miRNAs are still unknown. Specifically, it is unclear if RISC proteins already complexed with delivered miRNAs participate in the post-transcriptional regulation of target gene in recipient cells [104]. Chen et al. [17] found that mesenchymal stem cells secrete precursor form of miRNAs (pre-miRNAs) enriched in microvesicles. However, Dicer and AGO2 were not found in the secreted particles and, therefore, RISC proteins derived from recipient cells were assumed to be crucial for functionality of these delivered miRNAs [105]. More recently, the presence of pre-miRNAs with Dicer, AGO2, and TRBP in exosomes has been reported, and cell-independent maturation of miRNAs in the exosomes has been suggested as an effective way of functional miRNA delivery [78]. Alternatively, delivery of AGO2-complexed circulating miRNAs such as miR-210 has been described, suggesting the importance of AGO2 from source cells to allow for canonical function of delivered miRNAs in recipient cells [100]. Finally, while most studies have interrogated the principle of canonical actions of delivered miRNAs, other mechanisms may exist for such miRNA functionality. Fabbri et al. [106] recently found that miR-21 and miR-29a secreted from lung tumor cells via exosomes can be transferred to recipient macrophages. In macrophages, such miRNAs can be transported to endosomes, where they act as ligands of Toll-like receptors and induce secretion of prometastatic inflammatory cytokines. Therefore, delivered miRNAs may carry specific biologic functions, facilitated through both canonical and non-canonical methods that have yet to be elucidated completely.
The biologic importance of miRNA delivery in the cardiovascular system has been demonstrated by several in vivo studies. For instance, consistent with their in vitro findings, Zernecke et al. [24] demonstrated that intravenous injection of endothelial apoptotic bodies promotes recruitment of endothelial progenitor cells in aortic root plaques of apolipoprotein E (ApoE)-deficient mice on a high fat diet. In that context, administration of apoptotic bodies or miR-126 reduced the size and macrophage content in atherosclerotic plaques, while apoptotic bodies from miR-126-deficient mice induced negligible atheroprotective effects. These data suggested that the atheroprotective effects of apoptotic bodies are mediated by delivered miR-126. Hergenreider et al. [31] also demonstrated that extracellular vesicles secreted by Krüppel-like factor 2 (KLF2)-transduced or shear-stress-stimulated endothelial cells are enriched with miR-143/145. These extracellular vesicles controlled target gene expression in co-cultured smooth muscle cells. When vesicles from KLF2-transduced mouse endothelial cells were injected into ApoE-deficient mice on an atherogenic diet, fatty lesions in the aorta were significantly decreased compared with control mice. However, when miR-143/145 expression was depleted from these vesicles, the atheroprotective effects of these vesicles were lost. In another study, Jansen et al. [107] revealed that microvesicles from apoptotic endothelial cells promoted endothelial cell migration and proliferation in recipient endothelial cells. These authors found that miR-126 was predominantly expressed in these microvesicles, was transferred to recipient cells, and downregulated expression of direct target genes. In correlation, administration of miR-126-enriched microvesicles promoted re-endothelialization after electrical denudation of the common carotid artery in wildtype mice, while treatment with miR-126-depleted microparticles abrogated such re-endothelialization.
Such in vitro and in vivo studies support the notion of functional delivery of circulating miRNAs to a variety of cardiovascular tissues, but the real contribution of such biology to cardiovascular pathogenesis remains to be determined. In part, this question persists due to the challenges of interpreting contemporary studies (Fig. 3). Notably, many current reports rely upon quantifying levels of “delivered” miRNAs in recipient tissue that otherwise are ubiquitously expressed. Thus, even though increased miRNA expression is often noted in recipient cells after exposure to extracellular miRNA molecules, these studies often do not rule out the possibility of up-regulation of endogenous miRNA expression rather than actual delivery, per se. Rarely have contemporary studies studied recipient tissues that are genetically deficient in specific endogenous miRNAs [100], and to our knowledge, such data have been limited to in vitro analyses. Second, in cases where recipient cells can already express a delivered miRNA endogenously, often the amount that is already present far exceeds the calculated level of delivery possible during most physiologic conditions. Thus, unless exogenous and endogenous miRNAs carry differing biologic actions once inside a cell, delivery of an already expressed miRNA would not be expected to contribute significantly to overall target gene regulation. On the other hand, delivery of miRNAs may be more effective in the context of non-endogenous miRNAs or poorly expressed miRNAs at baseline. Thus, stemming from these points, rigorous studies to demonstrate transfer and activity of miRNAs in vivo are challenging and have not been common in current studies.
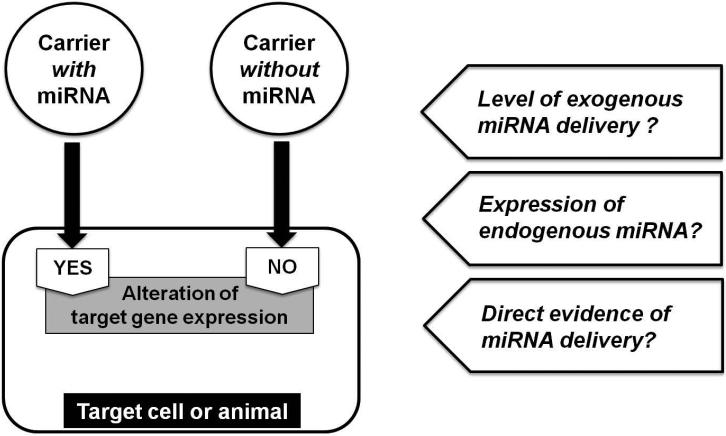
Figure 3. Model of circulating microRNAs (miRNAs) as intercellular messengers and the challenges of interrogating this model.
Recently, the functional delivery of circulating miRNAs has been reported in many in vitro and in vivo studies. Many of these studies have proposed the function of delivered specific miRNAs by relying upon the subtractive logic that the carrier without those miRNAs could not alter the expression of target gene in recipient cells or tissues. Caveats in interpreting these data should be considered, which include the possibility of endogenous ubiquitous miRNAs expression and the efficiency of delivered exogenous miRNAs, among others. Furthermore, robust delivery of a labeled and active miRNA in vivo, particularly in rodents genetically deficient in that given miRNA, has not been demonstrated to date in cardiovascular tissue.
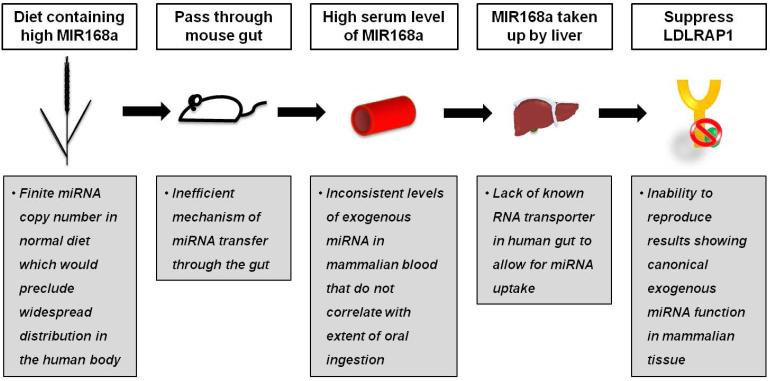
A recent example of such challenges in data interpretation have stemmed from a surprising report by Zhang et al. [108], claiming that ingested exogenous plant (rice)-derived MIR168a can survive and pass through the mammalian gut, enter the circulation, and is delivered to the liver, where it suppresses LDL receptor adapter protein 1 expression and consequently decreases hepatic LDL uptake. These findings suggested a new possibility of a “cross-kingdom” platform by which plant miRNAs can regulate mammalian gene expression via oral nutrition. However, although the detectability of fungal, bacterial, and plant-derived miRNAs in mammalian blood has been reported by multiple groups [109-111], substantial delivery of diet-derived miRNAs into mammalian circulation or tissues and any subsequent effects on putative target gene expression have been unable to be replicated by independent groups [112-114]. It may be possible to increase gut uptake of miRNAs via artificially increasing ingested levels or in settings of substantial gut damage [115]. Yet, given the exceptionally high sensitivity of miRNA detection techniques, the confounding issue of miRNA contamination during tissue processing has been proposed as a more likely explanation of detectable levels of foreign miRNAs in human tissue [116]. These inconsistencies in data imply the presence of various obstacles preventing significant and reliable delivery of orally ingested diet-derived miRNAs (Fig. 4). Consequently, any further studies in this regard will require an exceptional level of scientific rigor in order to prove definitively the uptake of exogenous and active miRNAs via the digestive tract in a biologically relevant context [117].
Figure 4. Controversy regarding the model of functional delivery of diet-derived microRNAs (miRNAs).
Recently, functional cross-kingdom transfer of food-derived miRNAs has been proposed (white boxes) [108]. However, current evidence demonstrates limitations to the original data and the poor efficacy of delivery of diet-derived miRNAs after typical dietary ingestion, in general (gray boxes) [110, 112-114]. LDLRAP1 = Low-density lipoprotein receptor adaptor protein 1.
Future therapeutic perspectives
As the diverse roles of circulating miRNAs in cardiovascular disease have been identified, therapeutic application of extracellular miRNAs has also been explored. For example, Hu et al. [118] demonstrated that administration of miR-210 using nonviral vectors improved cardiac function after MI in mice. They suggested Efna3 and Ptp1b, which are involved in angiogenesis and apoptosis, respectively, as potential targets of miR-210. It has also been reported that exogenous administration of hsa-miR-590 and hsa-miR-199a using lipid transfection reagent or viral vector significantly increases cardiomyocyte proliferation and, therefore, reduces infarct size and improves cardiac function in a murine model of MI [119].
Although these studies suggest the therapeutic potential of extracellular miRNAs in the cardiovascular system, it is not determined yet which methods will be most suitable for their pharmacologic delivery. Engineered exosomes have been considered as ideal carriers for miRNAs due to their small size, stability, and ability to cross biological membranes [120, 121]. Furthermore, modifications of exosomal surfaces by engineering of donor cells have made target cell delivery of exosomes more efficient. For example, Alvarez-Erviti et al. [122] demonstrated neuron-specific targeting of exosomal vesicles in vivo by generating dendritic cells that express the membrane protein Lamp2b fused to the neuron-specific rabies viral glycoprotein peptide. More recently, Ohno et al. [123] reported that let-7a miRNA could be delivered specifically to epidermal growth factor receptor-expressing breast cancer cells using modified exosomes. Yet, in spite of these promising results, challenges still remain, including the development of a more effective purification method for the study and handling of exosomes, the determination of the half-life and clearance of engineered exosomes in the bloodstream in vivo, and continued improvements in the specificity of delivery of exosomal cargo [124].
Conclusions
Over recent years, increasing attention has been paid to the role of circulating miRNAs in cardiovascular disease. As biomarkers and intercellular communicators, circulating miRNAs could play important roles in the prediction, diagnosis, and tailored treatment of cardiovascular diseases in the near future. For successful clinical application, however, it is hoped that a better understanding of circulating miRNAs from packaging and release to uptake will be forthcoming in the next phase of scientific investigation.
Acknowledgments
This work was supported by the NIH [grants HL096834, HL124021]; McArthur-Radovsky, Lerner, Harris, and Watkins Funds; Pulmonary Hypertension Association; and the American Heart Association (14GRNT19600012).
Footnotes
Disclosures
None
References
- 1.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 2.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 3.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 5.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–42. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdellatif M. Differential expression of microRNAs in different disease states. Circ Res. 2012;110:638–50. doi: 10.1161/CIRCRESAHA.111.247437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–8. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devaux Y, Vausort M, Goretti E, Nazarov PV, Azuaje F, Gilson G, et al. Use of circulating microRNAs to diagnose acute myocardial infarction. Clin Chem. 2012;58:559–67. doi: 10.1373/clinchem.2011.173823. [DOI] [PubMed] [Google Scholar]
- 11.Pleister A, Selemon H, Elton SM, Elton TS. Circulating miRNAs: novel biomarkers of acute coronary syndrome? Biomark Med. 2013;7:287–305. doi: 10.2217/bmm.13.8. [DOI] [PubMed] [Google Scholar]
- 12.Rotkrua P, Shimada S, Mogushi K, Akiyama Y, Tanaka H, Yuasa Y. Circulating microRNAs as biomarkers for early detection of diffuse-type gastric cancer using a mouse model. Br J Cancer. 2013;108:932–40. doi: 10.1038/bjc.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–87. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopreski MS, Benko FA, Kwak LW, Gocke CD. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clin Cancer Res. 1999;5:1961–5. [PubMed] [Google Scholar]
- 15.Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–90. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 16.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–5. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 18.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–41. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133–44. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu L, Yang BF, Ai J. MicroRNA transport: a new way in cell communication. J Cell Physiol. 2013;228:1713–9. doi: 10.1002/jcp.24344. [DOI] [PubMed] [Google Scholar]
- 22.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 23.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 25.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–59. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thery C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–83. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang M, Chen J, Su F, Yu B, Su F, Lin L, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, Zeiher AM, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–56. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 32.Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5:e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collino F, Deregibus MC, Bruno S, Sterpone L, Aghemo G, Viltono L, et al. Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PLoS One. 2010;5:e11803. doi: 10.1371/journal.pone.0011803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maggi LB, Jr., Kuchenruether M, Dadey DY, Schwope RM, Grisendi S, Townsend RR, et al. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the Mammalian ribosome. Mol Cell Biol. 2008;28:7050–65. doi: 10.1128/MCB.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown WV. High-density lipoprotein and transport of cholesterol and triglyceride in blood. J Clin Lipidol. 2007;1:7–19. doi: 10.1016/j.jacl.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Babin PJ, Gibbons GF. The evolution of plasma cholesterol: direct utility or a "spandrel" of hepatic lipid metabolism? Prog Lipid Res. 2009;48:73–91. doi: 10.1016/j.plipres.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Rayner KJ, Hennessy EJ. Extracellular communication via microRNA: lipid particles have a new message. J Lipid Res. 2013;54:1174–81. doi: 10.1194/jlr.R034991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner J, Riwanto M, Besler C, Knau A, Fichtlscherer S, Roxe T, et al. Characterization of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arterioscler Thromb Vasc Biol. 2013;33:1392–400. doi: 10.1161/ATVBAHA.112.300741. [DOI] [PubMed] [Google Scholar]
- 41.Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5:492–7. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng HH, Yi HS, Kim Y, Kroh EM, Chien JW, Eaton KD, et al. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS One. 2013;8:e64795. doi: 10.1371/journal.pone.0064795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, Zhu D, Huang L, Zhang J, Bian Z, Chen X, et al. Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS One. 2012;7:e46957. doi: 10.1371/journal.pone.0046957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lv Z, Wei Y, Wang D, Zhang CY, Zen K, Li L. Argonaute 2 in cell-secreted microvesicles guides the function of secreted miRNAs in recipient cells. PLoS One. 2014;9:e103599. doi: 10.1371/journal.pone.0103599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laffont B, Corduan A, Ple H, Duchez AC, Cloutier N, Boilard E, et al. Activated platelets can deliver mRNA regulatory Ago2*microRNA complexes to endothelial cells via microparticles. Blood. 2013;122:253–61. doi: 10.1182/blood-2013-03-492801. [DOI] [PubMed] [Google Scholar]
- 46.Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–46. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 47.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–9. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 48.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–32. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 49.Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–52. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–7. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 53.D'Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765–73. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009;55:1944–9. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- 55.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–66. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 56.Adachi T, Nakanishi M, Otsuka Y, Nishimura K, Hirokawa G, Goto Y, et al. Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin Chem. 2010;56:1183–5. doi: 10.1373/clinchem.2010.144121. [DOI] [PubMed] [Google Scholar]
- 57.Kuwabara Y, Ono K, Horie T, Nishi H, Nagao K, Kinoshita M, et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet. 2011;4:446–54. doi: 10.1161/CIRCGENETICS.110.958975. [DOI] [PubMed] [Google Scholar]
- 58.Gidlof O, Smith JG, Miyazu K, Gilje P, Spencer A, Blomquist S, et al. Circulating cardio-enriched microRNAs are associated with long-term prognosis following myocardial infarction. BMC Cardiovasc Disord. 2013;13:12. doi: 10.1186/1471-2261-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Devaux Y, Mueller M, Haaf P, Goretti E, Twerenbold R, Zangrando J, et al. Diagnostic and prognostic value of circulating microRNAs in patients with acute chest pain. J Intern Med. 2015;277:260–71. doi: 10.1111/joim.12183. [DOI] [PubMed] [Google Scholar]
- 60.Oerlemans MI, Mosterd A, Dekker MS, de Vrey EA, van Mil A, Pasterkamp G, et al. Early assessment of acute coronary syndromes in the emergency department: the potential diagnostic value of circulating microRNAs. EMBO Mol Med. 2012;4:1176–85. doi: 10.1002/emmm.201201749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeller T, Keller T, Ojeda F, Reichlin T, Twerenbold R, Tzikas S, et al. Assessment of microRNAs in patients with unstable angina pectoris. Eur Heart J. 2014;35:2106–14. doi: 10.1093/eurheartj/ehu151. [DOI] [PubMed] [Google Scholar]
- 62.Liebetrau C, Mollmann H, Dorr O, Szardien S, Troidl C, Willmer M, et al. Release kinetics of circulating muscle-enriched microRNAs in patients undergoing transcoronary ablation of septal hypertrophy. J Am Coll Cardiol. 2013;62:992–8. doi: 10.1016/j.jacc.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 63.Baggish AL, Park J, Min PK, Isaacs S, Parker BA, Thompson PD, et al. Rapid upregulation and clearance of distinct circulating microRNAs after prolonged aerobic exercise. J Appl Physiol. 2014;116:522–31. doi: 10.1152/japplphysiol.01141.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Rosa S, Fichtlscherer S, Lehmann R, Assmus B, Dimmeler S, Zeiher AM. Transcoronary concentration gradients of circulating microRNAs. Circulation. 2011;124:1936–44. doi: 10.1161/CIRCULATIONAHA.111.037572. [DOI] [PubMed] [Google Scholar]
- 65.Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, et al. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106:1035–9. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 66.Ellis KL, Cameron VA, Troughton RW, Frampton CM, Ellmers LJ, Richards AM. Circulating microRNAs as candidate markers to distinguish heart failure in breathless patients. Eur J Heart Fail. 2013;15:1138–47. doi: 10.1093/eurjhf/hft078. [DOI] [PubMed] [Google Scholar]
- 67.Tutarel O, Dangwal S, Bretthauer J, Westhoff-Bleck M, Roentgen P, Anker SD, et al. Circulating miR-423_5p fails as a biomarker for systemic ventricular function in adults after atrial repair for transposition of the great arteries. Int J Cardiol. 2013;167:63–6. doi: 10.1016/j.ijcard.2011.11.082. [DOI] [PubMed] [Google Scholar]
- 68.Watson CJ, Gupta SK, O'Connell E, Thum S, Glezeva N, Fendrich J, et al. MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Eur J Heart Fail. 2015 Mar 4; doi: 10.1002/ejhf.244. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wong LL, Armugam A, Sepramaniam S, Karolina DS, Lim KY, Lim JY, et al. Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur J Heart Fail. 2015 Jan 23; doi: 10.1002/ejhf.223. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 70.Akat KM, Moore-McGriff D, Morozov P, Brown M, Gogakos T, Correa Da Rosa J, et al. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc Natl Acad Sci U S A. 2014;111:11151–6. doi: 10.1073/pnas.1401724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morley-Smith AC, Mills A, Jacobs S, Meyns B, Rega F, Simon AR, et al. Circulating microRNAs for predicting and monitoring response to mechanical circulatory support from a left ventricular assist device. Eur J Heart Fail. 2014;16:871–9. doi: 10.1002/ejhf.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duong Van Huyen JP, Tible M, Gay A, Guillemain R, Aubert O, Varnous S, et al. MicroRNAs as non-invasive biomarkers of heart transplant rejection. Eur Heart J. 2014;35:3194–202. doi: 10.1093/eurheartj/ehu346. [DOI] [PubMed] [Google Scholar]
- 73.Rhodes CJ, Wharton J, Boon RA, Roexe T, Tsang H, Wojciak-Stothard B, et al. Reduced microRNA-150 is associated with poor survival in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2013;187:294–302. doi: 10.1164/rccm.201205-0839OC. [DOI] [PubMed] [Google Scholar]
- 74.Wei C, Henderson H, Spradley C, Li L, Kim IK, Kumar S, et al. Circulating miRNAs as potential marker for pulmonary hypertension. PLoS One. 2013;8:e64396. doi: 10.1371/journal.pone.0064396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bertero T, Lu Y, Annis S, Hale A, Bhat B, Saggar R, et al. Systems-level regulation of microRNA networks by miR-130/301 promotes pulmonary hypertension. J Clin Invest. 2014;124:3514–28. doi: 10.1172/JCI74773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.White K, Lu Y, Annis S, Hale AE, Chau BN, Dahlman JE, et al. Genetic and hypoxic alterations of the microRNA-210-ISCU1/2 axis promote iron–sulfur deficiency and pulmonary hypertension. EMBO Mol Med. 2015 doi: 10.15252/emmm.201404511. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schlosser K, White RJ, Stewart DJ. miR-26a linked to pulmonary hypertension by global assessment of circulating extracellular microRNAs. Am J Respir Crit Care Med. 2013;188:1472–5. doi: 10.1164/rccm.201308-1403LE. [DOI] [PubMed] [Google Scholar]
- 78.Xiao J, Jing ZC, Ellinor PT, Liang D, Zhang H, Liu Y, et al. MicroRNA-134 as a potential plasma biomarker for the diagnosis of acute pulmonary embolism. J Transl Med. 2011;9:159. doi: 10.1186/1479-5876-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo L, Yang Y, Liu J, Wang L, Li J, Wang Y, et al. Differentially expressed plasma microRNAs and the potential regulatory function of Let-7b in chronic thromboembolic pulmonary hypertension. PLoS One. 2014;9:e101055. doi: 10.1371/journal.pone.0101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Z, Zhou C, Liu Y, Wang S, Ye P, Miao X, et al. The expression levels of plasma micoRNAs in atrial fibrillation patients. PLoS One. 2012;7:e44906. doi: 10.1371/journal.pone.0044906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McManus DD, Tanriverdi K, Lin H, Esa N, Kinno M, Mandapati D, et al. Plasma microRNAs are associated with atrial fibrillation and change after catheter ablation (the miRhythm study) Heart Rhythm. 2015;12:3–10. doi: 10.1016/j.hrthm.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stather PW, Sylvius N, Wild JB, Choke E, Sayers RD, Bown MJ. Differential microRNA expression profiles in peripheral arterial disease. Circ Cardiovasc Genet. 2013;6:490–7. doi: 10.1161/circgenetics.111.000053. [DOI] [PubMed] [Google Scholar]
- 83.Li D, Ji L, Liu L, Liu Y, Hou H, Yu K, et al. Characterization of circulating microRNA expression in patients with a ventricular septal defect. PLoS One. 2014;9:e106318. doi: 10.1371/journal.pone.0106318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Widera C, Gupta SK, Lorenzen JM, Bang C, Bauersachs J, Bethmann K, et al. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. J Mol Cell Cardiol. 2011;51:872–5. doi: 10.1016/j.yjmcc.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 85.Eitel I, Adams V, Dieterich P, Fuernau G, de Waha S, Desch S, et al. Relation of circulating MicroRNA-133a concentrations with myocardial damage and clinical prognosis in ST-elevation myocardial infarction. Am Heart J. 2012;164:706–14. doi: 10.1016/j.ahj.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 86.Devaux Y, Vausort M, McCann GP, Zangrando J, Kelly D, Razvi N, et al. MicroRNA-150: a novel marker of left ventricular remodeling after acute myocardial infarction. Circ Cardiovasc Genet. 2013;6:290–8. doi: 10.1161/CIRCGENETICS.113.000077. [DOI] [PubMed] [Google Scholar]
- 87.Pilbrow AP, Cordeddu L, Cameron VA, Frampton CM, Troughton RW, Doughty RN, et al. Circulating miR-323-3p and miR-652: Candidate markers for the presence and progression of acute coronary syndromes. Int J Cardiol. 2014;176:375–85. doi: 10.1016/j.ijcard.2014.07.068. [DOI] [PubMed] [Google Scholar]
- 88.Devaux Y, Vausort M, McCann GP, Kelly D, Collignon O, Ng LL, et al. A panel of 4 microRNAs facilitates the prediction of left ventricular contractility after acute myocardial infarction. PLoS One. 2013;8:e70644. doi: 10.1371/journal.pone.0070644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lv P, Zhou M, He J, Meng W, Ma X, Dong S, et al. Circulating miR-208b and miR-34a are associated with left ventricular remodeling after acute myocardial infarction. Int J Mol Sci. 2014;15:5774–88. doi: 10.3390/ijms15045774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.D'Alessandra Y, Pompilio G, Capogrossi MC. Letter by D'Alessandra et al regarding article, "Circulating microRNA-208b and microRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. 2011;4:e7. doi: 10.1161/CIRCGENETICS.110.958769. author reply e8. [DOI] [PubMed] [Google Scholar]
- 91.Cheng Y, Tan N, Yang J, Liu X, Cao X, He P, et al. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond) 2010;119:87–95. doi: 10.1042/CS20090645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sredni ST, Gadd S, Jafari N, Huang CC. A parallel study of mRNA and microRNA profiling of peripheral blood in young adult women. Front Genet. 2011;2:49. doi: 10.3389/fgene.2011.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One. 2011;6:e20769. doi: 10.1371/journal.pone.0020769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baggish AL, Hale A, Weiner RB, Lewis GD, Systrom D, Wang F, et al. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J Physiol. 2011;589:3983–94. doi: 10.1113/jphysiol.2011.213363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnson BN, Mutharasan R. Biosensor-based microRNA detection: techniques, design, performance, and challenges. Analyst. 2014;139:1576–88. doi: 10.1039/c3an01677c. [DOI] [PubMed] [Google Scholar]
- 96.Yuan A, Farber EL, Rapoport AL, Tejada D, Deniskin R, Akhmedov NB, et al. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS One. 2009;4:e4722. doi: 10.1371/journal.pone.0004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Morelli AE, Larregina AT, Shufesky WJ, Sullivan ML, Stolz DB, Papworth GD, et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104:3257–66. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- 98.Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–66. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF, et al. Cellular internalization of exosomes occurs through phagocytosis. Traffic. 2010;11:675–87. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- 100.Hale A, Lee C, Annis S, Min PK, Pande R, Creager MA, et al. An Argonaute 2 switch regulates circulating miR-210 to coordinate hypoxic adaptation across cells. Biochim Biophys Acta. 2014;1843:2528–42. doi: 10.1016/j.bbamcr.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou J, Li YS, Nguyen P, Wang KC, Weiss A, Kuo YC, et al. Regulation of vascular smooth muscle cell turnover by endothelial cell-secreted microRNA-126: role of shear stress. Circ Res. 2013;113:40–51. doi: 10.1161/CIRCRESAHA.113.280883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Halkein J, Tabruyn SP, Ricke-Hoch M, Haghikia A, Nguyen NQ, Scherr M, et al. MicroRNA-146a is a therapeutic target and biomarker for peripartum cardiomyopathy. J Clin Invest. 2013;123:2143–54. doi: 10.1172/JCI64365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tabet F, Vickers KC, Cuesta Torres LF, Wiese CB, Shoucri BM, Lambert G, et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat Commun. 2014;5:3292. doi: 10.1038/ncomms4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem Sci. 2005;30:106–14. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 105.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483–95. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 106.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–6. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jansen F, Yang X, Hoelscher M, Cattelan A, Schmitz T, Proebsting S, et al. Endothelial microparticle-mediated transfer of MicroRNA-126 promotes vascular endothelial cell repair via SPRED1 and is abrogated in glucose-damaged endothelial microparticles. Circulation. 2013;128:2026–38. doi: 10.1161/CIRCULATIONAHA.113.001720. [DOI] [PubMed] [Google Scholar]
- 108.Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–26. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Munch EM, Harris RA, Mohammad M, Benham AL, Pejerrey SM, Showalter L, et al. Transcriptome profiling of microRNA by Next-Gen deep sequencing reveals known and novel miRNA species in the lipid fraction of human breast milk. PLoS One. 2013;8:e50564. doi: 10.1371/journal.pone.0050564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang Y, Wiggins BE, Lawrence C, Petrick J, Ivashuta S, Heck G. Analysis of plant-derived miRNAs in animal small RNA datasets. BMC Genomics. 2012;13:381. doi: 10.1186/1471-2164-13-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang K, Li H, Yuan Y, Etheridge A, Zhou Y, Huang D, et al. The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PLoS One. 2012;7:e51009. doi: 10.1371/journal.pone.0051009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Snow JW, Hale AE, Isaacs SK, Baggish AL, Chan SY. Ineffective delivery of diet-derived microRNAs to recipient animal organisms. RNA Biol. 2013;10:1107–16. doi: 10.4161/rna.24909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Witwer KW, McAlexander MA, Queen SE, Adams RJ. Real-time quantitative PCR and droplet digital PCR for plant miRNAs in mammalian blood provide little evidence for general uptake of dietary miRNAs: limited evidence for general uptake of dietary plant xenomiRs. RNA Biol. 2013;10:1080–6. doi: 10.4161/rna.25246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dickinson B, Zhang Y, Petrick JS, Heck G, Ivashuta S, Marshall WS. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol. 2013;31:965–7. doi: 10.1038/nbt.2737. [DOI] [PubMed] [Google Scholar]
- 115.Yang J, Farmer LM, Agyekum AA, Hirschi KD. Detection of dietary plant-based small RNAs in animals. Cell Res. 2015 Feb 27; doi: 10.1038/cr.2015.26. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tosar JP, Rovira C, Naya H, Cayota A. Mining of public sequencing databases supports a non-dietary origin for putative foreign miRNAs: underestimated effects of contamination in NGS. RNA. 2014;20:754–7. doi: 10.1261/rna.044263.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Witwer KW, Hirschi KD. Transfer and functional consequences of dietary microRNAs in vertebrates: concepts in search of corroboration: negative results challenge the hypothesis that dietary xenomiRs cross the gut and regulate genes in ingesting vertebrates, but important questions persist. Bioessays. 2014;36:394–406. doi: 10.1002/bies.201300150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hu S, Huang M, Li Z, Jia F, Ghosh Z, Lijkwan MA, et al. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122:S124–31. doi: 10.1161/CIRCULATIONAHA.109.928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–81. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 120.Vrijsen KR, Sluijter JP, Schuchardt MW, van Balkom BW, Noort WA, Chamuleau SA, et al. Cardiomyocyte progenitor cell-derived exosomes stimulate migration of endothelial cells. J Cell Mol Med. 2010;14:1064–70. doi: 10.1111/j.1582-4934.2010.01081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen L, Wang Y, Pan Y, Zhang L, Shen C, Qin G, et al. Cardiac progenitor-derived exosomes protect ischemic myocardium from acute ischemia/reperfusion injury. Biochem Biophys Res Commun. 2013;431:566–71. doi: 10.1016/j.bbrc.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–5. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 123.Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21:185–91. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hagiwara K, Ochiya T, Kosaka N. A paradigm shift for extracellular vesicles as small RNA carriers: from cellular waste elimination to therapeutic applications. Drug Deliv Transl Res. 2014;4:31–7. doi: 10.1007/s13346-013-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ai J, Zhang R, Li Y, Pu J, Lu Y, Jiao J, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. 2010;391:73–7. doi: 10.1016/j.bbrc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 126.Long G, Wang F, Duan Q, Chen F, Yang S, Gong W, et al. Human circulating microRNA-1 and microRNA-126 as potential novel indicators for acute myocardial infarction. Int J Biol Sci. 2012;8:811–8. doi: 10.7150/ijbs.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Satoh M, Takahashi Y, Tabuchi T, Tamada M, Takahashi K, Itoh T, et al. Circulating Toll-like receptor 4-responsive microRNA panel in patients with coronary artery disease: results from prospective and randomized study of treatment with renin-angiotensin system blockade. Clin Sci (Lond) 2015;128:483–91. doi: 10.1042/CS20140417. [DOI] [PubMed] [Google Scholar]
- 128.Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–84. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 129.Zhong J, He Y, Chen W, Shui X, Chen C, Lei W. Circulating microRNA-19a as a potential novel biomarker for diagnosis of acute myocardial infarction. Int J Mol Sci. 2014;15:20355–64. doi: 10.3390/ijms151120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Olivieri F, Antonicelli R, Lorenzi M, D'Alessandra Y, Lazzarini R, Santini G, et al. Diagnostic potential of circulating miR-499-5p in elderly patients with acute non ST-elevation myocardial infarction. Int J Cardiol. 2013;167:531–6. doi: 10.1016/j.ijcard.2012.01.075. [DOI] [PubMed] [Google Scholar]
- 131.Goren Y, Kushnir M, Zafrir B, Tabak S, Lewis BS, Amir O. Serum levels of microRNAs in patients with heart failure. Eur J Heart Fail. 2012;14:147–54. doi: 10.1093/eurjhf/hfr155. [DOI] [PubMed] [Google Scholar]
- 132.Long G, Wang F, Duan Q, Yang S, Chen F, Gong W, et al. Circulating miR-30a, miR-195 and let-7b associated with acute myocardial infarction. PLoS One. 2012;7:e50926. doi: 10.1371/journal.pone.0050926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang S, Chen M, Li L, He M, Hu D, Zhang X, et al. Circulating MicroRNAs and the occurrence of acute myocardial infarction in Chinese populations. Circ Cardiovasc Genet. 2014;7:189–98. doi: 10.1161/CIRCGENETICS.113.000294. [DOI] [PubMed] [Google Scholar]
- 134.Wang F, Long G, Zhao C, Li H, Chaugai S, Wang Y, et al. Plasma microRNA-133a is a new marker for both acute myocardial infarction and underlying coronary artery stenosis. J Transl Med. 2013;11:222. doi: 10.1186/1479-5876-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.He F, Lv P, Zhao X, Wang X, Ma X, Meng W, et al. Predictive value of circulating miR-328 and miR-134 for acute myocardial infarction. Mol Cell Biochem. 2014;394:137–44. doi: 10.1007/s11010-014-2089-0. [DOI] [PubMed] [Google Scholar]
- 136.Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, et al. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. 2010;3:499–506. doi: 10.1161/CIRCGENETICS.110.957415. [DOI] [PubMed] [Google Scholar]
- 137.D'Alessandra Y, Carena MC, Spazzafumo L, Martinelli F, Bassetti B, Devanna P, et al. Diagnostic potential of plasmatic MicroRNA signatures in stable and unstable angina. PLoS One. 2013;8:e80345. doi: 10.1371/journal.pone.0080345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zile MR, Mehurg SM, Arroyo JE, Stroud RE, DeSantis SM, Spinale FG. Relationship between the temporal profile of plasma microRNA and left ventricular remodeling in patients after myocardial infarction. Circ Cardiovasc Genet. 2011;4:614–9. doi: 10.1161/CIRCGENETICS.111.959841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Matsumoto S, Sakata Y, Suna S, Nakatani D, Usami M, Hara M, et al. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ Res. 2013;113:322–6. doi: 10.1161/CIRCRESAHA.113.301209. [DOI] [PubMed] [Google Scholar]
- 140.Zampetaki A, Willeit P, Tilling L, Drozdov I, Prokopi M, Renard JM, et al. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol. 2012;60:290–9. doi: 10.1016/j.jacc.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 141.Wang E, Nie Y, Zhao Q, Wang W, Huang J, Liao Z, et al. Circulating miRNAs reflect early myocardial injury and recovery after heart transplantation. J Cardiothorac Surg. 2013;8:165. doi: 10.1186/1749-8090-8-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Matsumoto S, Sakata Y, Nakatani D, Suna S, Mizuno H, Shimizu M, et al. A subset of circulating microRNAs are predictive for cardiac death after discharge for acute myocardial infarction. Biochem Biophys Res Commun. 2012;427:280–4. doi: 10.1016/j.bbrc.2012.09.039. [DOI] [PubMed] [Google Scholar]