Abstract
North Carolina is the second leading state in pork production in the United States, with over 10 million swine. Swine manure in NC is typically collected and stored in open-pit lagoons before the liquid waste is sprayed onto agricultural fields for disposal. Components of this waste may be able to impact surface water quality with the potential for human exposure. This study examined viruses of public health concern in creeks adjacent to swine concentrated animal feeding operation (CAFO) spray fields. Surface water samples (n = 154) were collected from public access waters in proximity to swine CAFO spray fields for six months and were tested for hepatitis E virus (HEV) and coliphages. HEV was detected in one sample. Somatic coliphages were detected in 98% of samples (geometric mean 24 ± 4.1 PFU per 100 ml), and F+ coliphages were detected in 85% of samples (geometric mean 6.8 ± 5.0 PFU per 100 ml). Only 3% (21) of the F+ coliphage isolates were RNA phage, and all of the F+ RNA coliphages belonged to genogroup I. Although the pervasiveness of swine CAFOs in this area prevented a comparison with samples from un-impacted sites, the near ubiquity of coliphages, as well as the presence of HEV, suggests that current waste management practices may be associated with the dissemination of viruses of public health concern in waters proximal to CAFO spray fields.
Keywords: Hepatitis E virus, Concentrated animal feeding operation, Coliphage
GRAPHICAL ABSTRACT
1. Introduction
North Carolina is the second leading state in national pork production with over 10 million swine (Edwards and Ladd, 2000). Five adjacent counties in eastern NC (Bladen, Duplin, Greene, Sampson, and Wayne) were estimated to have a population of over 7.5 million swine in 2002 (USGAO, 2008). This number of swine can produce up to 15.5 million tons of manure annually (USGAO, 2008). Swine manure in NC is typically collected and stored in open-pit lagoons before the liquid waste is sprayed onto agricultural fields for disposal. As a result of runoff and percolation events, components of manure, including zoonotic and human pathogens, may impact surface water quality proximal to swine concentrated animal feeding operations (CAFOs) (Anderson and Sobsey, 2006; Campagnolo et al., 2002; Sayah et al., 2005; Thurston-Enriquez et al., 2005). Pathogens potentially present include Salmonella, Campylobacter, Listeria, enteropathogenic Escherichia coli, Cryptosporidium, Giardia, and viruses such as enteric calicivirus, rotavirus, and hepatitis E virus (HEV).
In industrialized countries, little is known about possible sources and transmission routes for endemic human HEV infections. Research is often impeded by the rare detection of outbreaks, occurrence of asymptomatic infections, and a long and variable incubation period for the pathogen (Lewis et al., 2010). However, previous research has suggested the possibility of zoonotic transmission routes for HEV (Meng, 2009; Pavio et al., 2008). A systematic review found that in industrialized countries, specifically Europe, zoonotic transmission seemed likely (Lewis et al., 2010), and a meta-analysis found a significant association between occupational exposure to swine and human HEV IgG seropositivity (Wilhelm et al., 2011).
The potential dissemination of HEV in association with swine waste has not been well characterized. Studies in NC have documented HEV in swine wastes including swine feces and liquid wastes from lagoons (Kase et al., 2009), however nearby surface waters were not tested. A study from the Midwestern US evaluated surface waters for HEV but all samples were negative (Kasorndorkbua et al., 2005). Separately, a report from Slovenia has documented waterborne HEV in the vicinity of a pig farm (Steyer et al., 2011).
Similarly, only a few studies have documented the presence and subtypes of F+ RNA coliphage in surface waters surrounding agricultural areas (Brion et al., 2002; Cole et al., 2003; Rahman et al., 2009; Thurston-Enriquez et al., 2005). F+ RNA coliphages are promising indicators for enteric viruses, due to their similarity in size, shape, structure, and genetic makeup to many human enteric viruses. A meta-analysis found that F+ coliphages, as well as coliphages as a whole, are positively correlated to multiple pathogens [odds ratio for total coliphage and F+ coliphage of 1.29 (95% CI = 0.82–2.05) and 1.27 (95% CI = 0.48–3.35), respectively; Wu et al., 2011]. Moreover, genotyping of F+ RNA coli-phage may be used to predict the source of fecal pollution, as animal and human feces have been shown to contain different genogroups. Genogroups I and IV are generally associated with nonhuman animals, while genogroup II isolates are generally associated with human and pig feces, and genogroup III isolates are generally associated with human feces (Griffin et al., 2001).
The goal of this study was to systematically evaluate surface waters adjacent to swine CAFO spray fields for the presence of HEV and coli-phages, potential indicators of enteric viruses.
2. Materials and methods
2.1. Study sites and sample collection
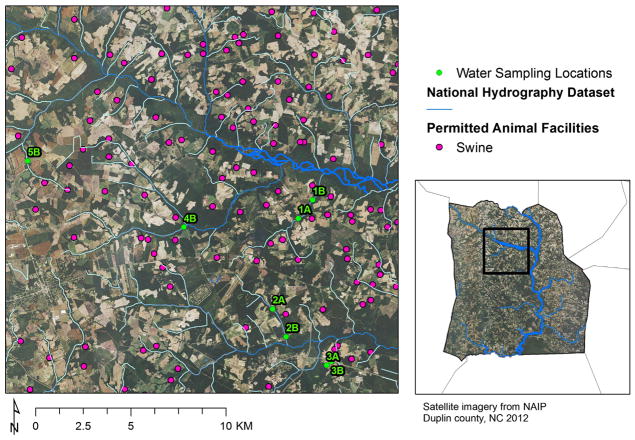
The study area is in Duplin County, North Carolina, USA, where there is a high density of swine CAFOs (Wing et al., 2000). To determine the impact of swine lagoon spray fields on adjacent water quality, sampling locations were situated in public access waters upstream (A) and downstream (B) of three spray fields (Sites 1–3, Fig. 1). Distances between the A and B sites were 1.3 km (Site 1), 1.7 km (Site 2), and 0.4 km (Site 3). Sites 4 and 5 were sampled as B locations only. Spray fields 1, 2, 3, 4, and 5 were approximately 0.3, 0.2, 0.03, 0.3, and 0.2 km2 in size and, at their closest points, were within 42, 66, 15, 76, and 26 m of the receiving water bodies, respectively. Spray fields 1, 2, and 3 were within 9 km of each other, and all sites were within 20 km of each other (Fig. 1). Surface water samples were collected from sampling sites weekly from mid-February to mid-August 2010. Sterile 4-l polycarbonate bottles were used for sample collection, and the bottles were coded so sample processors were blinded to sample locations during analysis. Water samples were taken in the late morning or early afternoon and transported on ice to the laboratory.
Fig. 1.
Study area showing sampling points within public access waters upstream (A) and downstream (B) of 5 swine CAFO spray fields in Duplin County, NC (USA). Sampling points are overlaid onto the National Agricultural Imagery Program overlay, NAIP 2012 GE. Creeks are indicated by blue lines, and permitted swine animal facilities by pink dots. Distances between the A and B sites were 1.3 km (Site 1), 1.7 km (Site 2), and 0.4 km (Site 3). Sites 4 and 5 were sampled as B locations only. Spray fields 1, 2, and 3 were within 9 km of each other, and all sites were within 20 km of each other. The receiving water bodies were within 10–76 m of the edge of the land application fields. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Rainfall data was obtained through the State Climate Office of North Carolina from the Williamsdale Field Lab station (North Carolina Environment and Climate Observing Network) located in Wallace, North Carolina (Lat: 34.7658; Long: −78.10117). This station was between 25 and 36.5 km south of the sampling sites. Hourly increments of rainfall were combined to compare the amount of precipitation 24 and 48 h before sampling to microbial concentrations.
2.2. Detection of hepatitis E virus
Viruses were concentrated using an adsorption–elution method described previously (Gentry et al., 2009), and viral concentrates were stored at −80 °C. RNA was extracted from 200 μl of viral concentrate using the QIAamp One-For-All Nucleic Acid kit (Qiagen, Valencia, CA), following the protocol for liquid transport media, to extract nucleic acids into 100-μl buffer AVE using a BioRobot Universal System (Qiagen). Nucleic acid samples were stored at −80 °C overnight. RNA was reverse-transcribed using the Applied Biosystems High Capacity cDNA Reverse Transcription Kit (Life Technologies, Carlsbad, CA). The cDNA synthesis mixture contained 5 μl nucleic acid, 1 mM of the specific reverse primer JVHEVR (Jothikumar et al., 2006), 10 mM dNTP mix, 2 μl 10× RT buffer, 1 μl MultiScribe reverse transcriptase, and nuclease-free water for a total reaction mixture of 20 μl. The reaction mixture was subjected to reverse transcription on an Applied Biosystems 7900 (Life Technologies) using the following conditions: 10 min at 25 °C, 120 min at 37 °C, and 5 min at 85 °C. The cDNA product was stored at −80 °C.
TaqMan primers and probes, described in Jothikumar et al. (2006), were used to assay for HEV cDNA. The qPCR reaction mixture contained 2 μl cDNA, 400 nM of each primer, 100 nM probe, 10 μl 2× Probe PCR Mix, 400 ng/μl BSA, 150 ng/μl T4 gene 32 protein, and nuclease-free water for a total reaction mixture of 20 μl (Qiagen Quantitect Probe PCR kit). The reaction mixture was subjected to qPCR on an Applied Biosystems 7900 using the following conditions: 1 min at 60 °C, 15 min at 95 °C, 45 cycles of 15 s at 94 °C and 1 min at 60 °C. All amplification reactions were carried out in duplicate. Samples that gave a positive result in either or both of the duplicate reactions were amplified by qPCR again. Only after a sample gave a second positive result was it counted as a presumptive positive.
To confirm the presence of HEV in samples, the original concentrated water sample was shipped, on dry ice, to a second laboratory at Johns Hopkins University (JHU) where it was analyzed for HEV using the RT-qPCR method described above. In both laboratories, conventional and nested RT-PCR protocols were utilized to amplify larger genome fragments for sequencing. At the University of North Carolina, nested RT-PCR protocols described in Kase et al. (2009) and Inoue et al. (2006) were utilized to amplify segments of ORF2 and ORF2/ORF3, respectively. At JHU, RT-PCR protocols described in Inoue et al. (2006) and Dong et al. (2011) were utilized to amplify segments of ORF2/ORF3 and ORF1, respectively.
2.3. Coliphage detection and isolation
Water samples were analyzed for somatic and F+ coliphage using the Single Agar Layer (SAL) Method (USEPA, 2001a). Up to 8 F+ coliphage plaques from each sample were isolated for characterization in 2 ml tryptic soy broth (TSB) and stored at −80 °C. Samples in which no coliphages were detected were assigned a concentration of <1 plaque forming unit (PFU) per 100 ml. For regression analyses, the qualified values of <1 and ‘too numerous to count (TNTC)’ were converted to 0.5 PFU and 1000 PFU per 100 ml (because the highest reported count was 766 PFU per 100 ml) in a manner similar to that of Levy et al. (2012).
For samples in which no F+ coliphages were detected, an enrichment technique was used to detect F+ coliphages following USEPA Method 1601 (USEPA, 2001b). One plaque from each of three dilutions (300 ml, 30 ml, and 3 ml) was isolated for further characterization in 2 ml TSB and stored at −80 °C.
To distinguish F+ RNA and F+ DNA coliphages, 5 μl of all F+ isolates was spotted on two plates, one containing E. coli Famp and one containing E. coli Famp plus 10 mg/ml RNase A. Plaque formation on the E. coli Famp and the E. coli Famp plus RNase plates indicated an F+ DNA coliphage. Plaque formation on the E. coli Famp plate but not the E. coli Famp plus RNase plate indicated an F+ RNA coliphage.
2.4. Typing of F+ RNA coliphage isolates
RNA was extracted from 200 μl of viral isolation or enrichment using the RNeasy Mini kit (Qiagen, Valencia, CA) to elute the RNA sample in 50-μl nuclease-free sterile water. The RNA was immediately subjected to RT-PCR according to Friedman et al. (2009) using a MasterCycler gradient (Eppendorf, Hauppauge, NY). Isolates were first classified into their respective genera, Levivirus or Allolevivirus, using the MJV82 forward and either the Levivirus JV41 reverse or the Allolevivirus JV81 reverse primer, respectively (Vinjé et al., 2004). Each isolate was then assayed using genogroup specific primers (Friedman et al., 2009). The prototype strains MS2 (GI), GA (GII), QB (GIII), and SP (GIV) were used as positive controls. Amplicons were separated by gel electrophoresis in 1.5% agarose, stained with ethidium bromide, and visualized under UV light.
2.5. Statistical analyses
All statistical analyses were conducted in SAS 9.1 (SAS Inc., Cary, NC) statistical software. To test the significance of difference between coli-phage concentrations in the A and B sampling locations at sites 1, 2, and 3, the Mann–Whitney U test was performed. To test the significance of differences in coliphage concentrations between seasons, Kruskal–Wallis one-way ANOVAs were used. Relationships were determined to be significant at p < 0.05. The association between coliphage concentrations and rainfall amount was determined using fixed effects linear models. The results of the models determine whether a 1-unit increase in the dependent variable (coliphage concentration) is associated with a coefficient change in the independent variable (rainfall). The relationship of F+ RNA coliphages to antecedent rainfall was determined using generalized logistic regressions.
3. Results
A total of 154 samples were collected for this study. Samples were collected from site 1 thirteen times at both A and B locations, and from sites 2 and 3 twenty-six times at both A and B locations. Site 1 became too dry to obtain samples after early June 2010 so sites 4 and 5 were added on June 8, 2010 as B locations only and were sampled 12 times each.
3.1. Hepatitis E virus
HEV was detected once during the study period at site 3A in March, 2010. The presence of HEV in this sample was confirmed at JHU. Unfortunately, larger fragments of the HEV genome could not be amplified in either laboratory, a problem that has been reported in previous studies examining HEV in environmental samples (Steyer et al., 2011), so the strain could not be genotyped. For the one sample positive for HEV, no precipitation was recorded during the 48 h preceding sample collection. The somatic coliphage concentration within the HEV-positive sample was ‘too numerous to count.’ This sample and a water sample from site 3B on the same date were the only 2 samples that had a somatic coliphage concentration so high that they could not be confidently enumerated. The F+ coliphage concentration within the HEV-positive sample was 5 PFU/ml.
3.2. Coliphages
98% of samples were positive for somatic coliphages, with a range of 1 to 1000 PFU per 100 ml, and 85% of samples were positive for F+ coliphages, with a range of 1 to 99 PFU per 100 ml (Table 1). Of the 660 F+ coliphages isolated and subjected to RNase testing, 21 isolates (3%) were RNA phage. Genotyping revealed that all of the F+ RNA coliphages belonged to genogroup I.
Table 1.
Percentage of surface water samples positive for somatic and F+ coliphages and geometric mean concentrations (PFU per 100 ml) by site.
| Site | Somatic coliphages
|
F+ coliphages
|
||||
|---|---|---|---|---|---|---|
| % positive (na) | Geo. mean (PFU per 100 ml) | Min, max | % positive (na) | Geo. mean (PFU per 100 ml) | Min, max | |
| All A samples | 97 (63) | 20 | <1, 1000 | 83 (54) | 6.3 | <1, 82 |
| All B samples | 99 (88) | 28 | <1, 1000 | 87 (77) | 7.2 | <1, 99 |
| 1A | 85 (11) | 5.5 | <1, 187 | 85 (11) | 2.5 | <1, 18 |
| 1B | 100 (13) | 18 | 3.0, 104 | 77 (13) | 3.0 | <1, 69 |
| 2A | 100 (26) | 18 | 2.0, 67 | 77 (20) | 9.2 | <1, 82 |
| 2B | 100 (26) | 22 | 1.0, 172 | 88 (23) | 8.6 | <1, 83 |
| 3A | 100 (26) | 45 | 4.0, 1000 | 88 (23) | 6.7 | <1, 63 |
| 3B | 96 (25) | 35 | <1, 1000 | 77 (20) | 4.5 | <1, 99 |
| 4B | 100 (12) | 12 | 2.0, 30 | 100 (12) | 20 | 3.0, 84 |
| 5B | 100 (12) | 96 | 8.0, 766 | 100 (12) | 13 | 4.0, 74 |
n = number of samples positive for coliphage.
3.2.1. Comparison by sample location
There was no significant difference in the somatic or F-specific coliphage concentrations between the A and B locations at site 1, 2, or 3. F+ RNA coliphage isolates were detected at least once at all sites examined except site 4B. The largest percentage of F+ RNA coliphages was isolated in samples from site 3A (24%).
3.2.2. Temporal and seasonal distribution
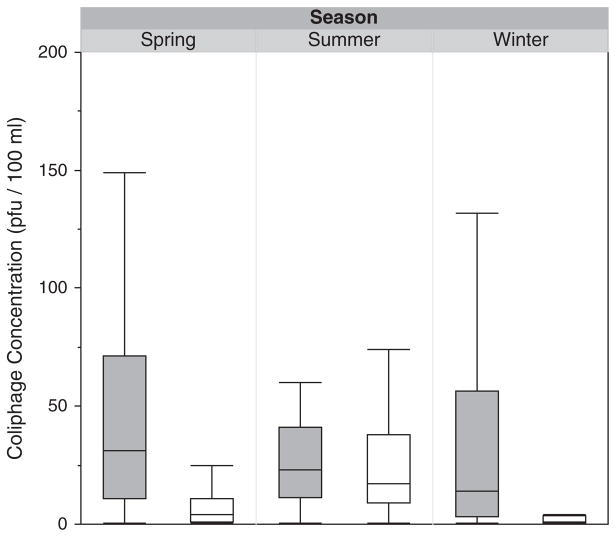
Somatic coliphage concentrations were similar for the 3 seasons examined (winter, spring, and summer; Fig. 2). F+ coliphage concentrations were higher in the summer (July–September) than in the winter or spring (p < 0.0001; Fig. 2). All F+ RNA coliphages were isolated from sampling time-points in February, April, and August 2010, and 65% were isolated from coliphage enrichments in February 2010.
Fig. 2.
Box and whisker plot depicting the concentration (in PFU per 100 ml) of somatic (gray) and F+ coliphages (white) sorted by season. The lower boundary of the box indicates the 25th percentile, the line within the box represents the median, and the boundary of the box farthest from zero indicates the 75th percentile. Whiskers below and above the box indicate the 10th and 90th percentiles, respectively.
3.2.3. Correlation to rainfall
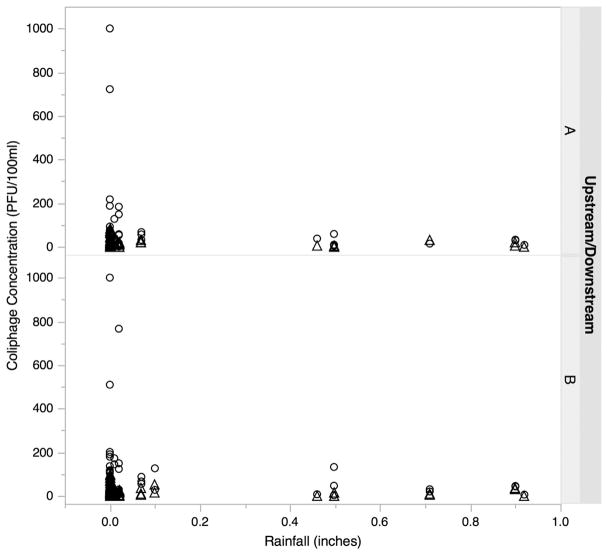
Very little rainfall occurred during the study period, on average less than 0.1 in. a day (Fig. 3). Somatic and F+ coliphage concentrations were not associated with rainfall across all sites combined or at individual sites. Additionally, generalized logistic regressions revealed that the presence of F+ RNA coliphages was not correlated to antecedent rainfall.
Fig. 3.
Concentration (in PFU per 100 ml) of somatic (circle) and F+ coliphages (triangle) at upstream (A) and downstream (B) sites in relation to levels of rainfall occurring in the 24 h before sample collection.
4. Discussion
In this study we investigated surface waters proximal to swine CAFO lagoon waste spray fields for the presence of hepatitis E virus (HEV), and for coliphages, potential indicators of enteric viruses. HEV was detected only once during the study period. Previous studies have detected HEV in swine (Choi et al., 2003; Dell’Amico et al., 2011; Huang et al., 2002; Kase et al., 2008; Takahashi et al., 2003) and swine lagoons (Kasorndorkbua et al., 2005; McCreary et al., 2008; Pina et al., 2000), but few studies have examined CAFO-impacted surface waters for HEV (Karetnyi et al., 1999; Kasorndorkbua et al., 2005; Steyer et al., 2011). One conference abstract (Karetnyi et al., 1999) reported detecting HEV in a tile outlet draining a field to which manure had been applied in the Midwestern United States and a report from Slovenia detected HEV in the vicinity of a pig farm (Steyer et al., 2011). Because HEV in developed countries is thought to be predominantly of zoonotic origin (when travel to developing countries can be ruled out) (Nelson et al., 2011), HEV in our study may have originated from swine. However, without knowing the genotype of the HEV strain, we cannot infer whether the HEV isolate is more closely related to swine or human HEV. Thus, the presence of HEV in surface waters adjacent to a swine CAFO spray field is intriguing, but additional studies are required to determine if HEV present in swine waste is transmitted to CAFO spray fields and adjacent surface waters.
The low prevalence of HEV in our study is similar to results from previous studies (Kasorndorkbua et al., 2005; Steyer et al., 2011), and may be related to challenges in detecting HEV (and viruses in general) in environmental samples, including low virus concentrations, low virus recovery, matrix effects, and the presence of PCR inhibitors (Julian and Schwab, 2012). With these challenges in mind, this study also examined water samples for the presence of F+ RNA coliphages, potential indicators for human enteric viruses. Of 660 F+ isolates, only 21 (3%) were F+ RNA coliphage. This low prevalence was not surprising, given that a previous study examining surface waters impacted by swine feces found that F+ RNA coliphages represented only 18% of F+ coliphage isolates (Cole et al., 2003). The low percentage of F+ RNA coliphages may be due to higher inactivation rates of F+ RNA than F+ DNA coliphages; previous studies have documented higher inactivation rates of F+ RNA coliphages than F+ DNA at warmer temperatures (Cole et al., 2003; Rahman et al., 2009). The majority of our samples were collected during the spring and summer months, and 65% of F+ RNA coliphages were isolated in February 2010.
All of the F+ RNA isolates belonged to genogroup I (GI), indicating an animal source of fecal pollution (Osawa et al., 1981). In contrast to this, previous studies have found GI F+ RNA coliphages to represent only 19–60% of F+ RNA isolates in swine wastewaters (Cole et al., 2003; Lee et al., 2009) and 0% (0 of 3) of F+ RNA isolates in surface waters impacted by swine feces (Cole et al., 2003). In addition to the impact of swine waste in the creeks, the higher percentage of GI in our water samples than previous studies could be a product of the enhanced persistence of GI over other genogroups at higher temperatures (Brion et al., 2002; Long and Sobsey, 2004; Schaper et al., 2002) or a factor of selective enrichment. Genogroup I has a larger burst size and produces more progeny during enrichment (Furuse, 1987), which can result in an overestimation of the frequency of that group when isolating and typing individual plaques from low dilutions (Sobsey et al., 2006). 65% of the F+ RNA coliphages were detected in coliphage enrichments.
F+ RNA coliphages were not detected in the HEV-positive water sample nor were F+ coliphage concentrations elevated in this water sample. Conversely, the HEV-positive sample did contain the highest concentration of somatic coliphage in this study. Previous studies have found correlations between somatic coliphages and adenoviruses (Aw and Gin, 2010) and enteroviruses (Mocé-Llivina et al., 2005). However, as only one sample was positive for HEV in this study, additional studies are required to determine if a potential association between increased somatic coliphage concentrations and HEV exists.
To determine the impact of individual swine lagoon spray fields on water quality in adjacent streams, coliphage concentrations were compared at the A and B locations. Our results indicated that, on average, somatic and F+ coliphage concentrations were not different at A and B sites. We suspect that these results are due to diffuse contamination of surface waters with swine waste due to the high density of CAFOs in this area. Sampling sites in this study are located in Duplin County, NC, with an estimated swine population of over 2 million according to the most recent available data (USDA, 2007). In fact, given the ubiquity of swine CAFOs in this area, we could not categorize our upstream, A, sites as un-impacted and thus we could not assess whether the microbial concentrations at these sites are different from those in un-impacted surface waters. However, it is also possible that other fecal sources could be responsible for the diffuse concentrations of coliphages. There are numerous poultry CAFOs and cattle grazing in open fields in this area, in addition to the ubiquitous swine CAFOs. Moreover, some rural homes in the area use septic systems for sewage disposal. Detailed information on the number of poultry CAFOs and cattle fields, the density of homes, the number of septic systems, and land use data in the sampling area was not collected at the time of the study. This limitation of our study restricts our ability to confidently state that coliphages primarily resulted from swine CAFO spray field runoff. Nevertheless, swine and poultry CAFOs are estimated to be the largest contributors to fecal waste in this area (Steve Wing, personal communication), and all of the F+ RNA coliphage isolates belong to genogroup I (GI), indicating an animal-source (e.g., pigs, cattle, sheep) of fecal pollution in the surface waters (Osawa et al., 1981). Similar concentrations of coliphages at A and B sites may also be due to low levels of rainfall during the study period. Precipitation levels exceeded 1 in. only three times during the study. The detection of HEV and coliphages in surface waters proximal to swine CAFOs warrants further investigation to address sources of fecal pollution in areas of high swine CAFO density.
In sum, the presence of HEV, as well as the near ubiquity of coli-phages, suggests that current CAFO waste management practices may be associated with the dissemination of viruses of public health concern in waters proximal to CAFO spray fields. Nevertheless, the ubiquity of swine CAFOs prevented us from being able to compare samples with those from un-impacted sites in this same area, and additional studies incorporating land use data are necessary to better understand the impact of spray fields on the presence of these viruses in adjacent surface waters.
HIGHLIGHTS.
Hepatitis E virus was detected in one sample.
Somatic coliphages were detected in 98% of samples.
F+ coliphages were detected in 85% of samples.
21 F+ coliphage isolates (3%) were RNA phage, which all belonged to genogroup I.
Acknowledgments
This work was supported in part by a Gillings Innovation Laboratory award from the UNC Gillings School of Global Public Health, the W.K. Kellogg Health Scholars Program — Community Track, and Debevoise & Plimpton, LLP. The funders had no role in data collection and analysis, decision to publish, or preparation of the manuscript.
We would like to thank REACH for their assistance in this study. We would also like to thank Prof. Steve Wing for his guidance in statistical analyses; Hunter Story and Neil Blathela for their help in processing samples for coliphages; and the UNC Microbiome Core, especially Dr. M. Andrea Azcarate-Peril, for their help with qPCR analysis. Jeff Essic kindly provided assistance on the sampling sites figure.
Footnotes
Conflict of interest
All authors attest that they have no actual or potential conflict of interest including any financial, personal or other relationships with other people or organizations that could inappropriately influence, or be perceived to influence, their work.
Contributor Information
Jennifer Gentry-Shields, Email: jen_shields@ncsu.edu.
Kevin Myers, Email: kevwmyers@gmail.com.
Nora Pisanic, Email: npisanic@jhsph.edu.
Christopher Heaney, Email: cheaney@jhsph.edu.
Jill Stewart, Email: jill.stewart@unc.edu.
References
- Anderson ME, Sobsey MD. Detection and occurrence of antimicrobially resistant E. coli in groundwater on or near swine farms in eastern North Carolina. Water Sci Technol. 2006;54:211–8. doi: 10.2166/wst.2006.471. [DOI] [PubMed] [Google Scholar]
- Aw TG, Gin KY. Environmental surveillance and molecular characterization of human enteric viruses in tropical urban wastewaters. J Appl Microbiol. 2010;109:716–30. doi: 10.1111/j.1365-2672.2010.04701.x. [DOI] [PubMed] [Google Scholar]
- Brion GM, Meschke JS, Sobsey MD. F-specific RNA coliphages: occurrence, types, and survival in natural waters. Water Res. 2002;36:2419–25. doi: 10.1016/s0043-1354(01)00547-4. [DOI] [PubMed] [Google Scholar]
- Campagnolo ER, Johnson KR, Karpati A, Rubin CS, Kolpin DW, Meyer MT, et al. Antimicrobial residues in animal waste and water resources proximal to large-scale swine and poultry operations. Sci Total Environ. 2002;299:89–95. doi: 10.1016/s0048-9697(02)00233-4. [DOI] [PubMed] [Google Scholar]
- Choi I-S, Kwon H-J, Shin N-R, Yoo HS. Identification of swine hepatitis E virus (HEV) and prevalence of anti-HEV antibodies in swine and human populations in Korea. J Clin Microbiol. 2003;41:3602–8. doi: 10.1128/JCM.41.8.3602-3608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D, Long SC, Sobsey MD. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl Environ Microbiol. 2003;69:6507. doi: 10.1128/AEM.69.11.6507-6514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Amico MC, Cavallo A, Gonzales JL, Bonelli SI, Valda Y, Pieri A, et al. Hepatitis E virus genotype 3 in humans and swine, Bolivia. Emerg Infect Dis. 2011;17:1488–90. doi: 10.3201/eid1708.100769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Meng J, Dai X, Liang J-H, Feagins A, Meng XJ, et al. Restricted enzooticity of hepatitis E virus genotypes 1 to 4 in the United States. J Clin Microbiol. 2011;49:4164–72. doi: 10.1128/JCM.05481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards B, Ladd AE. Environmental justice, swine production and farm loss in North Carolina. Sociol Spectr. 2000;20:286. [Google Scholar]
- Friedman SD, Cooper EM, Casanova L, Sobsey MD, Genthner FJ. A reverse transcription-PCR assay to distinguish the four genogroups of male-specific (F+) RNA coliphages. J Virol Methods. 2009;159:47–52. doi: 10.1016/j.jviromet.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Furuse K. Distribution of coliphages in the environment. In: Goyal SM, Gerba CP, Bitton G, editors. Phage Ecology. New York: Wiley-Interscience; 1987. pp. 87–124. [Google Scholar]
- Gentry JB, Vinje J, Guadagnoli D, Lipp EK. Norovirus distribution within an estuarine environment. Appl Environ Microbiol. 2009;75:5474–80. doi: 10.1128/AEM.00111-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DW, Lipp EK, McLaughlin MR, Rose JB. Marine recreation and public health microbiology: quest for the ideal indicator. Bioscience. 2001;51:817–25. [Google Scholar]
- Huang FF, Haqshenas G, Guenette DK, Halbur PG, Schommer SK, Pierson FW, et al. Detection by reverse transcription-PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J Clin Microbiol. 2002;40:1326–32. doi: 10.1128/JCM.40.4.1326-1332.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue J, Masaharu T, Yazaki Y, Tsuda F, Okamoto H. Development and validation of an improved RT-PCR assay with nested universal primers for detection of hepatitis E virus strains with significant sequence divergence. J Virol Methods. 2006;137:325–33. doi: 10.1016/j.jviromet.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Jothikumar N, Cromeans TL, Robertson BH, Meng XJ, Hill VR. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J Virol Methods. 2006;131:65–71. doi: 10.1016/j.jviromet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Julian TR, Schwab KJ. Challenges in environmental detection of human viral pathogens. Curr Opin Virol. 2012;2:78–83. doi: 10.1016/j.coviro.2011.10.027. [DOI] [PubMed] [Google Scholar]
- Karetnyi YV, Moyer N, Gilchrist MJR, Naides SJ. Swine hepatitis E virus contamination in hog operation waste streams — an emerging infection?. Workshop on the effects of animal feeding operations on hydrologic resources and the environment; Fort Collins, CO: USGS; 1999. [Google Scholar]
- Kase J, Correa M, Luna C, Sobsey MD. Isolation, detection and characterization of swine hepatitis E virus from herds in Costa Rica. Int J Environ Health Res. 2008;18:165–76. doi: 10.1080/09603120701498311. [DOI] [PubMed] [Google Scholar]
- Kase J, Correa MT, Sobsey MD. Detection and molecular characterization of swine hepatitis E virus in North Carolina swine herds and their faecal wastes. J Water Health. 2009;7:344–57. doi: 10.2166/wh.2009.137. [DOI] [PubMed] [Google Scholar]
- Kasorndorkbua C, Opriessnig T, Huang FF, Guenette DK, Thomas PJ, Meng XJ, et al. Infectious swine hepatitis E virus is present in pig manure storage facilities on United States farms, but evidence of water contamination is lacking. Appl Environ Microbiol. 2005;71:7831–7. doi: 10.1128/AEM.71.12.7831-7837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Lim MY, Kim SY, Lee S, Lee H, Oh H-M, et al. Molecular characterization of bacteriophages for microbial source tracking in Korea. Appl Environ Microbiol. 2009;75:7107–14. doi: 10.1128/AEM.00464-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy K, Nelson KL, Hubbard A, Eisenberg JNS. Rethinking indicators of microbial drinking water quality for health studies in tropical developing countries: case study in northern coastal Ecuador. Am J Trop Med Hyg. 2012;86:499–507. doi: 10.4269/ajtmh.2012.11-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HC, Wichmann O, Duizer E. Transmission routes and risk factors for autochthonous hepatitis E virus infection in Europe: a systematic review. Epidemiol Infect. 2010;138:145–66. doi: 10.1017/S0950268809990847. [DOI] [PubMed] [Google Scholar]
- Long S, Sobsey M. A comparison on the survival of F+ RNA and F+ DNA coliphages in lake water microcosms. J Water Health. 2004;2:15–22. [PubMed] [Google Scholar]
- McCreary C, Martelli F, Grierson S, Ostanello F, Nevel A, Banks M. Excretion of hepatitis E virus by pigs of different ages and its presence in slurry stores in the United Kingdom. Vet Rec. 2008;163:261–5. doi: 10.1136/vr.163.9.261. [DOI] [PubMed] [Google Scholar]
- Meng XJ. Hepatitis E, virus: animal reservoirs and zoonotic risk. Vet Microbiol. 2009;140:256–65. doi: 10.1016/j.vetmic.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocé-Llivina L, Lucena F, Jofre J. Enteroviruses and bacteriophages in bathing waters. Appl Environ Microbiol. 2005;71:6838–44. doi: 10.1128/AEM.71.11.6838-6844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Kmush B, Labrique AB. The epidemiology of hepatitis E virus infections in developed countries and among immunocompromised patients. Expert Rev Anti Infect Ther. 2011;9:1133–48. doi: 10.1586/eri.11.138. [DOI] [PubMed] [Google Scholar]
- Osawa S, Furuse K, Watanabe I. Distribution of ribonucleic-acid coliphages in animals. Appl Environ Microbiol. 1981;41:164–8. doi: 10.1128/aem.41.1.164-168.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavio N, Renou C, Di Liberto G, Boutrouille A, Eloit M. Hepatitis E: a curious zoonosis. Front Biosci. 2008;1:7172–83. doi: 10.2741/3219. [DOI] [PubMed] [Google Scholar]
- Pina S, Buti M, Cotrina M, Piella J, Girones R. HEV identified in serum from humans with acute hepatitis and in sewage of animal origin in Spain. J Hepatol. 2000;33:826–33. doi: 10.1016/s0168-8278(00)80316-5. [DOI] [PubMed] [Google Scholar]
- Rahman R, Alum A, Ryu H, Abbaszadegan M. Identification of microbial faecal sources in the New River in the United States–Mexican border region. J Water Health. 2009;7:267–75. doi: 10.2166/wh.2009.025. [DOI] [PubMed] [Google Scholar]
- Sayah RS, Kaneene JB, Johnson Y, Miller R. Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl Environ Microbiol. 2005;71:1394–404. doi: 10.1128/AEM.71.3.1394-1404.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaper M, Duran AE, Jofre J. Comparative resistance of phage isolates of four genotypes of F-specific RNA bacteriophages to various inactivation processes. Appl Environ Microbiol. 2002;68:3702–7. doi: 10.1128/AEM.68.8.3702-3707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobsey MD, Love DC, Lovelace GL. F+ RNA coliphages as source tracking viral indicators of fecal contamination. Chapel Hill, NC: NOAA; 2006. [Google Scholar]
- Steyer A, Naglič T, Močilnik T, Poljšak-Prijatelj M, Poljak M. Hepatitis E virus in domestic pigs and surface waters in Slovenia: prevalence and molecular characterization of a novel genotype 3 lineage. Infect Genet Evol. 2011;11:1732–7. doi: 10.1016/j.meegid.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Nishizawa T, Miyajima H, Gotanda Y, Iita T, Tsuda F, et al. Swine hepatitis E virus strains in Japan form four phylogenetic clusters comparable with those of Japanese isolates of human hepatitis E virus. J Gen Microbiol. 2003;84:851–62. doi: 10.1099/vir.0.18918-0. [DOI] [PubMed] [Google Scholar]
- Thurston-Enriquez JA, Gilley JE, Eghball B. Microbial quality of runoff following land application of cattle manure and swine slurry. J Water Health. 2005;3:157–71. [PubMed] [Google Scholar]
- USDA; Service NAS, editor. Census of agriculture: 2007 census; 2007. Washington, D.C: 2011. [Google Scholar]
- USEPA. Method 1602: male-specific (F+) and somatic coliphage in water — April 2001 version. Washington, DC: US Environmental Protection Agency; 2001a. pp. 1–38. [Google Scholar]
- USEPA. Method 1601: male-specific (F+) and somatic coliphage in water by two-step enrichment procedure. Washington, DC: US Environmental Protection Agency, Office of Water; 2001b. [Google Scholar]
- USGAO. Report to congressional requestors — concentrated animal feeding operations —EPA needs more information and a clearly defined strategy to protect air and water quality from pollutants of concern. United States Government Accountability Office; 2008. [Google Scholar]
- Vinjé J, Oudejans SJG, Stewart JR, Sobsey MD, Long SC. Molecular detection and genotyping of male-specific coliphages by reverse transcription-PCR and reverse line blot hybridization. Appl Environ Microbiol. 2004;70:5996. doi: 10.1128/AEM.70.10.5996-6004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BJ, Rajic A, Greig J, Waddell L, Trottier G, Houde A, et al. A systematic review/meta-analysis of primary research investigating swine, pork or pork products as a source of zoonotic hepatitis E virus. Epidemiol Infect. 2011;139:1127–44. doi: 10.1017/S0950268811000677. [DOI] [PubMed] [Google Scholar]
- Wing S, Cole D, Grant G. Environmental injustice in North Carolina’s hog industry. Environ Health Perspect. 2000;108:225–31. doi: 10.1289/ehp.00108225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Long SC, Das D, Dorner SM. Are microbial indicators and pathogens correlated? A statistical analysis of 40 years of research. J Water Health. 2011;9:265–78. doi: 10.2166/wh.2011.117. [DOI] [PubMed] [Google Scholar]