Abstract
Approximately 10–15% of the human prion disease is inherited and one of the important genetic mutations occurs in the octapeptide repeat region of prion protein gene. One of the variants, one octapeptide repeat deletion (1-OPRD), existed in several gastric cancer cell lines and its mutation frequency was higher in gastric cancer cases. However, the biological functions of it remain unknown. Wild-type and mutation forms of PrPC were cloned and transfected into gastric cancer cells. Cell apoptosis, adhesion, invasion, multidrug resistance (MDR) and proliferation were, respectively, investigated. Different expressed genes were screened by gene array and proved by PT-PCR. Further, luciferase report assay was used to explore the transcriptional activation of target genes. Forced overexpression PrPC (1-OPRD) could promote the gastric cancer cells SGC7901 growth through facilitating G1- to S-phase transition in the cell cycle. PrPC (1-OPRD) could also inhibit apoptosis, and promote adhesion, invasion and MDR in SGC7901. However, it exhibited no significant difference between wild-type PrPC (1-OPRD) and PrPC on apoptosis, invasion or MDR effects. Further experiments indicated that PrPC (1-OPRD) could trigger the transactivation of cyclinD3 besides cyclinD1 to promote cell transition and proliferation. Overexpression of PrPC (1-OPRD) might promote the proliferation of gastric cancer cells at least partially through transcriptional activation of cyclinD3 to accelerate the G1-/S-phase transition. The promoting proliferation effect of PrPC (1-OPRD) was more than that of wild-type PrPC. However, they showed no difference on apoptosis, adhesion, invasion or MDR effects of gastric cancer cells.
Keywords: PrPC (1-OPRD), proliferation, apoptosis, invasion, MDR, gastric cancer
Introduction
The prion protein gene PRNP encoded PrPC and PrPSc is the infectious pathogen causing disorders including Creutzfeldt–Jakob disease in human beings and bovine spongiform encephalopathy. Approximately 10–15% of the human prion disease is inherited and one of the important genetic mutations occurs in the octapeptide repeat region of PRNP. It has reported one to nine extra insertions of octapeptide repeats of the fundamental sequence PHGGGWGQ in this region [1], which is associated with younger age at onset [2]. One octapeptide repeat deletion (1-OPRD) is another variant in this region and has been described to be less common [3]. However, our previous work found homozygous or heterozygous for 1-OPRD existed in several gastric cell lines, whose mutation frequency was higher in gastric cancer than in normal ones [4].
Despite the abundant studies on the function of misfolding prion protein PrPSc[5], relatively little is known about the characteristics of PrPC[6]. One of the well-studied functions of PrPC is its ability to selectively bind to copper ions through the octapeptide repeat region [7], which also plays a lead function in PrPC physiology and neuroprotection against oxidative stress in vivo[8]. Other emerging functions of PrPC include its protective role in cell survival [9]. Our previous work demonstrated that PrPC was overexpressed in gastric cancer [10], and that forced expression of PrPC could inhibit apoptosis [11], and promote proliferation [12], metastasis [13] and multidrug resistance (MDR) [14] in gastric cancer cells. So it is interesting to investigate if the mutation form of PrPC (1-OPRD) is also involved in apoptosis, proliferation, invasion or MDR in gastric cancer.
In present study, we found that ectopic expression of the mutation form PrPC (1-OPRD) showed the similar effects on apoptosis, adhesion, invasion and MDR as PrPC in gastric cancer cells. However, this mutation form promoted cell proliferation more significantly than wild-type PrPC itself. Differentially expressed genes between PrPC and PrPC (1-OPRD) were selected by gene array and then proved by RT-PCR and Western blot. CyclinD3 was found to be specifically up-regulated by PrPC (1-OPRD) compared with PrPC. Collectively, this study demonstrated that overexpression of PrPC (1-OPRD) would affect the cell proliferation, apoptosis, adhesion, invasion and MDR in gastric cancer cells. However, its effects on apoptosis, adhesion, invasion or MDR showed no significant difference with PrPC. Further study into the mechanisms of these relationships might enrich our knowledge of PrP and better our understanding of the nature of PrP in gastric carcinoma.
Materials and methods
Cell culture
Human gastric cancer cell line SGC7901, MKN28 and human normal gastric epithelial cell line GES (SV40-transformed gastric epithelial cell line) were obtained from Beijing Institute of Oncology and kept in our institute. Cells were cultured in RPMI 1640 medium (Roswell Park Memorial Institute basal medium 1640) (Life Technologies, Inc., Gaithersburg, MD, USA) supplemented with 10% foetal calf serum in a 37°C humidified CO2 incubator.
Plasmid construction and cell transfection
Target sequences were aligned to the human genome database in a BLAST search to ensure that the chosen sequences were not highly homologous with other genes. The primers were designed with Primer.5 software as in Table 1[10–14]. Full length PRNP gene was cloned from human normal gastric epithelial cell line GES and deletion of one octapeptide repeat region PRNP gene was cloned from gastric cancer cell line MKN28. Using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) 2 g of pcDNA3.1-PrPC or pcDNA3.1-PrPC (1-OPRD) plasmids were transfected into SGC7901 cells according to the manufacturer’s instructions. The cells transfected with pcDNA3.1 vector alone was served as respective negative control. The G418-resistant multiple combined clones were selected and expression of PrPC were evaluated by Western blot analysis. Gastric cancer cell line SGC7901 transfected with PrPC, PrPC (1-OPRD) and pcDNA3.1 were designated as SGC7901/PrPC, SGC7901/PrPC (1-OPRD) and SGC7901/pcDNA3.1, respectively.
Table 1.
Primers for plasmids construction
| Primers for PrPc | |
|---|---|
| F1 | 5′-CCCAAGCTTGGGATGGCGAACCTTGGCTGCT-3′ |
| R1 | 5′- CGGGATCCTCCCACATCAGGAAGATGAGGA-3′ |
| Primers for CCND Promoter | |
| CCND2 Promoter F2 | 5′- GGGGTACCGAAGTTATCAGGAACACAGA-3′ |
| R2 | 5′- CCCAAGCTTTTCCCCTGACCTCCTTC-3′ |
| CCND3 Promoter F3 | 5′- GGGGTACCTATTGGAGGTCTTTTTCGGC-3′ |
| R3 | 5′- CCCAAGCTTCAGCGAACAGGCAGGG-3′ |
FACS analysis of apoptosis
FACS analysis of apoptosis was performed as previously described [10]. The cancer cells were induced by serum deprivation for 24 hrs and then trypsinized. Annexin V/FITC apoptosis detection kit I (BD PharMingen) was used to identify the apoptotic and viable cells following the manufacturer’s instructions. The percentage of early apoptotic (FITC : positive and proliferative indexes [PI] : negative) cells was calculated from the data originated from flow cytometry.
Analysis of apoptosis by immunofluorescence microscope
The cancer cells were induced by serum deprivation for 24 hrs and then trypsinized. After staining for 10 min. at 37°C with 10 ug/ml Hoechest 33258, the cells were counterstained for 1 min. with 10 ug/ml PI. Apoptotic cell numbers was counted with immunofluorescence microscope (Olympus, Fukushima, Japan). Relatively apoptotic rate of cells was calculated according to the following formula: R=B 1/B 2 × 100% in which R is the relative apoptotic rate of cell growth; B 1 is the amount of apoptotic cells per 200 cells and B 2 is two hundred cells counted randomly. Each experiment was independently repeated for at least three times.
Adhesion assay
The ability of gastric cancer cells to adhere to matrigel was determined in 24-well plates as previously reported [13]. The plate surface was covered by 0.2 ml of 50 μg/ml matrigel, incubated for 2 hrs, and the supernatant was removed. A total of 0.5 ml suspension of tumour cells (1 × 105/ml) was transferred into the covered wells. After 0.5, 1, 2 and 4 hrs of incubation at 37°C, the adhesive cells were washed with PBS twice and then counted under a microscope at ×200 magnification on 10 random fields in each well. Each experiment was performed in triplicate.
Invasion assay
Cell invasion assays were performed with Transwell (8-μm pore size, Corning Costar Corp., New York, NY, USA) as previously reported [13]. Freshly trypsinized and washed cells were suspended at 2 × 105/ml in RPMI 1640 containing 1% bovine serum. The cell suspension (150 μl) was placed into upper compartments, and the cells were allowed to invade for 24 hrs at 37°C in a 5% CO2 humidified incubator. After incubation, the cells were removed from the upper surface of the filter with the cotton swat; the cells that had invaded into the bottom surface of the filter were fixed with methanol and stained with haematoxylin. The invasiveness was determined by counting of the penetrating cells under a microscope at ×200 magnification on 10 random fields in each well. Each experiment was performed in triplicate.
In vitro drug sensitivity assay
In vitro drug sensitivity assay was performed as previously reported [14, 15]. Adriamycin (ADR), vincristine (VCR), cisplatin (CDDP), 5-fluorouracil (5-FU), Etoposide (VP-16), cyclophosphamide (CTX), arabinosylcytosine (Ara) and methotrexate (MTX) were all freshly prepared before each experiment. Drug sensitivity was evaluated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay. The absorbance at 490 nm (A490) of each well was read on a spectrophotometer. Cell survival rates were calculated according to the following formula: survival rate (mean A490 of treated wells/mean A490 of untreated wells) ×100%. Finally, dose-effect curves of the anticancer drugs were drawn on semilogarithm coordinate paper and IC50 values were determined. Each experiment was performed in triplicate.
Fluorescence intensity assay of intracellular ADR
The fluorescence intensity of intracellular ADR was determined by FCM. Briefly, cells in log phase were planted into six-well plates (1 × 106 cells/well) and cultured overnight. After addition of ADR to a final concentration of 5 μg/ml, cells continued to be cultured for 1 hr. Cells were then trypsinized and harvested (for detection of ADR accumulation) or, alternatively, cultured in drug-free RPMI 1640 for another 30 min. followed by trypsinization and harvesting (for detection of ADR retention). The fluorescence intensity of intracellular ADR was determined using FCM with an exciting wavelength of 488 nm and emission wavelength of 575 nm. Finally, the ADR releasing index of gastric cancer cells was calculated according to the following formula: releasing index = (accumulation value-retention value)/accumulation value.
MTT assay
MTT assay was performed to evaluate cell proliferation as previously described [12]. The absorbance at 490 nm (A490) of each well was read on a microplate reader BP800 (BIOHIT, Neptune, NJ, USA). Each experiment was performed in quadruplicates and repeated three times.
Plate colony formation assay
Log-phase cells were trypsinized into single cell suspension and passaged into 90-mm2 plates at a density of 1 × 103 cells/well. The colonies were stained with Giemsa and total numbers of colonies were counted. Each assay was performed in triplicate.
Soft agar colony formation assay
Gum agar (2%) was melted in a microwave and cooled to 50–60°C in a water bath. The top agar was prepared with 2% agar, RPMI 1640 and FCS to give 0.3% agar and 10% FCS. Cells were harvested, washed and mixed with the top-agarose suspension at a final concentration of 0.3%, which was then layered onto the bottom agar. The dish was overlaid with 1 ml of RPMI 1640 containing supplements. Cells were incubated for 2 weeks at 37°C in 5% CO2 before colony counting. Each assay was performed in triplicate.
Cell cycle analysis
Subconfluent cells were washed with ice-cold PBS, suspended in 0.5 ml ethanol and then kept at 4°C for 30 min. The suspension was filtered through a 50-μm nylon mesh, and the DNA content of stained nuclei was analysed by a flow cytometer (EPICS XL, Coulter, Miami, FL, USA). The cell cycle was analysed using Multicycle-DNA Cell Cycle Analyzed Software. The PI was calculated as follows: PI = (S + G2)/(S + G2 + G1).
Cell cycle synchronization
Transfected cells were synchronized by double thymidine block as previously described [16]. Briefly, cells with approximately 50% confluent were treated with 2 mM thymidine for 16 hrs, then washed two times and incubated for an additional 8 hrs in the absence of thymidine. Cells were again incubated a second time with 2 mM thymidine for 16 hrs to arrest cells at the G1/S boundary of the cell cycle, and then harvested at 4 hrs intervals for 16 hrs. Then cells at each time point were washed twice with PBS and fixed with 70% ethanol. A total of 1 × 104 cells were then analysed for fluorescence intensity by a flow cytometer (EPICS XL) using Multicycle Software.
Gene array
The total RNA was extracted from all transfected cells using Trizol (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol. DNase was used to decrease the contamination of genomic DNA. Compared samples SGC7901/PrPC and SGC7901/PrPC (1-OPRD) were labelled with cy3 and cy5, and then hybridized into Aligent Ltd. (Foster City, CA, USA) Human 1A Microarray (V2) G4110B (22,575 human cDNA chip). They were scanned with the Agilent Scanner (Scan resolution 10 μm, PMT 100%) and normalized with Genespring. The different expressed genes were standardized with cy3/cy5 ration ≤ 2 as up-regulated genes and cy3/cy5 ≤ 0.5 as down-regulated genes.
RNA extraction and semiquantitative RT-PCR
Total RNA was extracted and DNase was used to decrease the contamination of genomic DNA. The PCR primers and reaction parameters that were used for cyclin and CDK family genes amplification are listed in Table 2. The reaction condition of PCR (e.g. cyclinD1) was as follows: Initial initial denaturation at 94°C for 10 min, Thirty-five cycles of denaturation at 94°C for 45 sec., annealing at 59°C for 30 sec. and extension at 72°C for 45 sec. on a Touchgene Gradient thermal cycler (Techne, Cambridge, UK). Appropriate cycles were chosen to assure the termination of PCR amplification before reaching stable stage in each reaction. Gene expression was presented by the relative yield of the PCR product from target sequences to that from the β-actin gene. PCR products were loaded onto a 1.5% agarose gel and electrophoretically separated. The gel was then visualised under ultraviolet light following ethidium bromide staining.
Table 2.
PCR primers and reaction parameters that were used for cyclin and CDK family genes
| Gene | Primers | Denaturation | Annealing | Extension | Cycles |
|---|---|---|---|---|---|
| CyclinD1 | F:5′- GGAGCTGCTCCTGGTGAACA -3′ | 94°C; 45′ | 59°C; 30′ | 72°C; 45′ | 35 |
| R:5′- TGTTGGGGCTCCTCAGGTTCA -3′ | |||||
| CyclinD2 | F:5′- CCAGCAGGATGAGGAAGTGA -3′ | 94°C; 45′ | 59°C; 30′ | 72°C; 45′ | 35 |
| R:5′- GACGGTACTGCTGCAGGCTATT -3′ | |||||
| CyclinD3 | F:5′- CATCCATGATCGCCACG -3′ | 94°C; 45′ | 59°C; 30′ | 72°C; 45′ | 35 |
| R:5′- GGAGCTGGTCTGAGAGGCT -3′ | |||||
| CDK4 | F:5′- GCATCCCAATGTTGTCCG-3′ | 94°C; 45′ | 50°C; 30′ | 72°C; 45′ | 26 |
| R:5′- GGCAGCCCAATCAGGTCA-3′ | |||||
| b-actin | F:5′-ATG ATA TCG CCG CGC TCG TC-3′ | 94°C; 45′ | 50°C; 30′ | 72°C; 45′ | 23 |
| R:5′-CGC TCG GTG AGG ATC TTC A-3′ |
Western blot
The Western blot was done as previously described [14]. In brief, total cellular proteins were prepared and then quantified by Bradford method. A measure of 50 g of lysates was electrophoresed in 12% SDS-PAGE and blotted on a nitrocellulose membrane (Immoblin-P, Millipore, Bedford, MA, USA). The membranes were blocked with 10% fat-free milk powder at room temperature for 2 hrs and incubated overnight with monoclonal antibody specific for PrP (clone 3F4 Monoclonal antibodies that bind epitopes comprising residues 96–104, 1:1000 Sigma, St. Louis, MO, USA), cyclinD1 (1:500, Upstate, Billerica, MA, USA), cyclinD3 (1:500, Upstate), anti-CDK4 (1:500, Upstate) and anti-β-actin antibody (1:2000, Santa Cruz Biotech, Santa Cruz, CA, USA) at 4°C. After three washes for 15 min. in PBS-T, the membrane was incubated with the HRP-conjugated goat antimouse IgG antibody (Boshide, Hubei, China) for 1 hr at room temperature. The membrane was washed again in PBST; enhanced chemiluminescence (Amersham Life Science, Piscataway, NJ, USA) was added and monitored for the development of colour.
Reporter assay
Sequences of CCND2 and CCND3 promoters were amplified by PCR from genomic DNA of peripheral blood mononuclear cells [17]. CCND1 promoter sequence was given by Dr. Richard G. Pestell and Chenguang Wang (Georgetown, USA). These promoter sequences were then cloned into pGL3 enhancer vector (Promega, Madison, WI, USA) to give the reporter vectors (designated pGL-CCND1, pGL-CCND2 and pGL-CCND3, respectively). PrPC (1-OPRD), PrPC or empty pcDNA3.1 plasmids were co-transfected into SGC7901 cells with cells with pGL-CCND1 or pGL-CCND2, pGL-CCND3. pRL-TK was used as a control for transfection efficiency. Luciferase reporter assays were performed with the dual-luciferase reporter assay system (Promega) following the vendor’s manual. Each experiment was performed in triplicate and repeated twice.
Statistical analysis
All the values of the in vitro assays were expressed as means ± S.D. anova analysis was performed with statistics package SPSS (version 10.0; SPSS, Chicago, IL, USA). Differences were considered significant when P < 0.05.
Results
PrPC (1-OPRD) had the similar effect on gastric cancer cells apoptosis as PrPC
Our previous work has shown that PrPC could be detected in several human gastric cancer cell lines [10], among which SGC7901 took heterozygous for 1-OPRD [4]. In order to show the differential effects of PrPC (1-OPRD) and PrPC expression on cell apoptosis, metastasis, MDR and proliferation, PRNP genes with or without one octapeptide repeat region deletion mutation were cloned by PCR from MKN28 and GES cells, respectively, and then transfected into human gastric cancer cell SGC7901. Gastric cancer cells MKN28 exhibited homozygotes for 1-OPRD, and GES without mutation. After cell transfection and antibiotic selection for more than 2 months, multiple drug-resistant clones were selected and the expression of PrPC in the cells were confirmed by Western blot with the antibody 3F4 (Sigma), which could recognize the epitope aa 109–112 of PRNP not containing the octapeptide repeat region (Fig. 1A).
Figure 1.
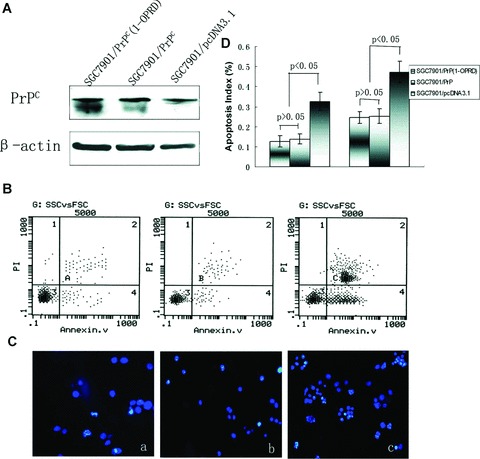
Effects of PrPC (1-ORPD) on apoptosis of gastric cancer cells. (A) Western blot analysis of the vector transfectants and PrPC (1-OPRD), PrPC transfectants. β-actin was used as a loading control. (B) Detection of cells apoptosis by flow cytometry with annexin V/PI staining. (a) SGC7901/PrPC (1-OPRD); (b) SGC7901/PrPC and (c) SGC7901/pcDNA3.1. (C) Detection of cells apoptosis by immunofluorescence microscopy with Hoechest 3325/PI staining. (a) SGC7901/PrPC (1-OPRD); (b) SGC7901/PrPC and (c) SGC7901/pcDNA3.1. (D) Apoptosis index of transfected cells.
In vitro activity of apoptosis in SGC7901/PrPC (1-OPRD), SGC7901/PrPC and SGC7901/pcDNA3.1 were determined by flow cytometry with annexin V/PI staining (Fig. 1B) and immunofluorescence microscopy with Hoechest 3325/PI staining (Fig. 1C) after induction of serum deprivation for 24 hrs. In vitro transfectant of SGC7901/PrPC (1-OPRD) and SGC7901/PrPC showed the similar responses to serum deprivation induced apoptosis, which were both higher than that in SGC7901/pcDNA3.1 cells (Fig. 1D). PrPC (1-OPRD) had the similar effect on apoptosis as PrPC, which showed no significant difference (P > 0.05).
PrPC (1-OPRD) had the similar effect on gastric cancer cells adhesion and invasion as PrPC
Our previous work had shown that gastric cancer cell line SGC7901 had invasive and metastatic abilities by in vitro invasion assay and in vivo nude mice assay [13]. To evaluate the effect of PrPC (1-OPRD) and PrPC on cell adhesion, the abilities of SGC7901/PrPC (1-OPRD) and SGC7901/PrPC to adhere to matrigel were investigated by adhesive assay. All the gastric cancer cells bound to matrigel with a time-dependent manner. Both SGC7901/PrPC (1-OPRD) and SGC7901/PrPC increased their adhesive abilities to matrigel than SGC7901/pcDNA3.1. However, the adhesive abilities between those in SGC7901/PrPC (1-OPRD) and SGC7901/PrPC exhibited no significant difference (P > 0.05) (Fig. 2A).
Figure 2.
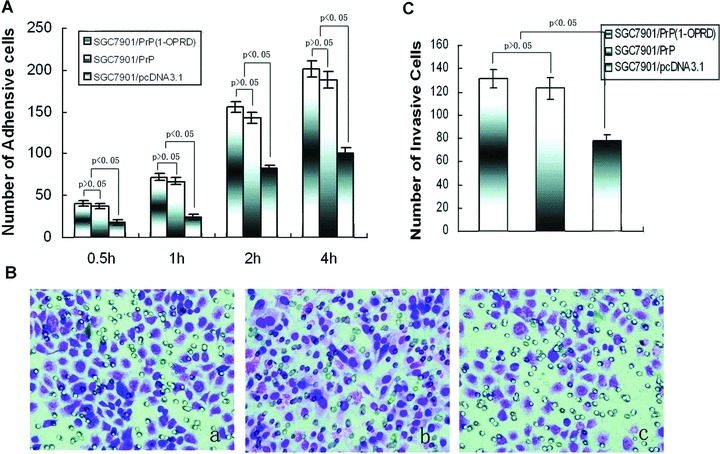
Effects of PrPC (1-ORPD) on adhesion and invasion of gastric cancer cells. (A) After 0.5, 1, 2 and 4 hrs of incubation, the cells attachè to matrigel were counted under a microscope. (B) Invasive ability was evaluated by counting cells invading through matrigel and membrane with 8-μm-pore Transwell. (a) SGC7901/PrPC (1-OPRD); (b) SGC7901/PrPC and (c) SGC7901/pcDNA3.1. (C) Number of invasive cells.
Because invasive potential is a common feature in the process of tumour metastasis, we then studied the influence of PrPC (1-OPRD) and PrPC on the invasive abilities of gastric cancer cells in vitro invasion assay. As shown in Fig. 2B, PrPC (1-OPRD) and PrPC transfection produced a marked increase of invasion of SGC7901/pcDNA3.1 cells through matrigel on Boyden chamber assay, with an average increasing rate of 48.6% and 45.7%, respectively. However, between SGC7901/PrPC (1-OPRD) and SGC7901/PrPC, it took no significant difference (P > 0.05) (Fig. 2C).
PrPC (1-OPRD) had the similar effect on gastric cancer cells MDR as PrPC
To determine the effect of PrPC (1-OPRD) and PrPC on MDR of gastric cancer, in vitro effects of chemotherapeutics on the growth of SGC7901 transfected cells were evaluated by MTT assay. Ectopic expression of PrPC (1-OPRD) and PrPC conferred the cells more resistance to ADR, VCR, VP-16, 5-FU and CDDP than controlling vector transfected cells (Table 3). IC50 values in SGC7901/PrPC (1-OPRD) and SGC7901/PrPC to these drugs were increased significantly compared with controlling SGC7901/pcDNA3.1 (P < 0.05). However, IC50 values between SGC7901/PrPC (1-OPRD) and SGC7901/PrPC showed no significant difference (P > 0.05).
Table 3.
IC50 values of the transfected cells to chemotherapeutic drugs
| Cell line | IC50 (μg/ml) | |||||||
|---|---|---|---|---|---|---|---|---|
| ADR | VCR | VP-16 | 5-FU | CDDP | CTX | Ara | MTX | |
| SGC7901/PrPC (1-OPRD) | 7.36 ± 0.82a,b | 6.59 ± 0.71a,b | 8.02 ± 0.79a,b | 2.58 ± 0.44a,b | 3.05 ± 0.28a,b | 7.21 ± 0.92 | 25.02 ± 2.83 | 11.03 ± 1.38 |
| SGC7901/PrPC | 7.14 ± 0.79a | 6.21 ± 0.64a | 7.89 ± 1.12a | 2.46 ± 0.39a | 2.93 ± 0.23a | 6.89 ± 0.83 | 24.85 ± 2.57 | 10.84 ± 1.33 |
| SGC7901/pcDNA3.1 | 0.47 ± 0.05 | 0.23 ± 0.03 | 0.45 ± 0.04 | 0.38 ± 0.04 | 0.34 ± 0.07 | 8.74 ± 1.08 | 23.96 ± 2.36 | 12.75 ± 1.52 |
P < 0.05 comparing with SGC7901/pcDNA3.1;
P > 0.05 comparing SGC7901/PrPC.
By flow cytometry, the intracellular accumulation and retention of ADR were examined. As shown in Table 4, fluorescence intensity in SGC7901/PrPC (1-OPRD) and SGC7901/PrPC were significantly lower than the control cells, SGC7901/pcDNA3.1 (P < 0.05), while SGC7901/PrPC (1-OPRD) and SGC7901/PrPC cells had no significant difference in fluorescence (P > 0.05).
Table 4.
Fluorescence intensity of intracellular ADR in transfected cells
| Mean fluorescence intensity | |||
|---|---|---|---|
| SGC7901/PrPC (1-OPRD) | SGC7901/PrPC | SGC7901/pcDNA3.1 | |
| Accumulation | 9.32 ± 1.18a,b | 10.07 ± 1.07a | 17.25 ± 0.84 |
| Retention | 0.38 ± 0.07a,b | 0.35 ± 0.05a | 0.21 ± 0.08 |
P < 0.05 comparing with SGC7901/pcDNA3.1;
P > 0.05 comparing SGC7901/PrPC.
Overexpression of PrPC (1-OPRD) enhanced tumour cell proliferation and cellular transformation
MTT assay, plate and soft agar colony formation assays were done to examine the effects of PrPC (1-OPRD) and PrPC on cell proliferation and cellular transformation in vitro. Compared with empty vector-transfected cells and parental cells, PrPC (1-OPRD) and PrPC-transfected cells showed significantly increased rate of cell proliferation as shown by MTT assay. Meanwhile, SGC7901/PrPC (1-OPRD) exhibited more increase on cell proliferation than that of SGC7901/PrPC (Fig. 3A).
Figure 3.
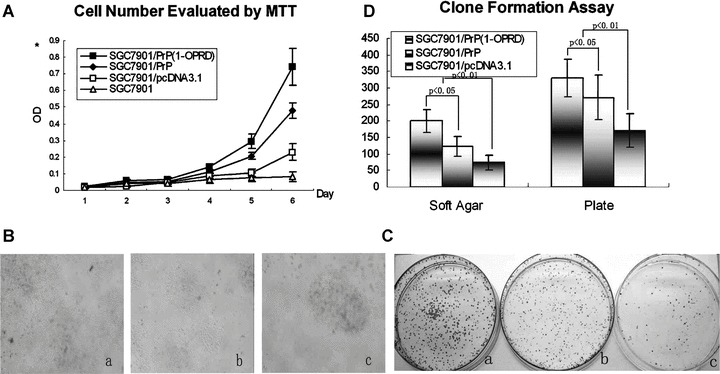
Effects of PrPC (1-ORPD) on cell proliferation of gastric cancer cells. (A) Detection of the cell growth rate in vitro. Cell number was evaluated by the absorbance at 490 nm in MTT assay at the indicated time. The value shown is the mean of three determinations. (B) Detection of the clone formation in soft agar. Cell were placed in media containing soft agar and incubated for 20 days. The number of foci > 100 μm was counted. (a) SGC7901/PrPC (1-OPRD); (b) SGC7901/PrPC and (c) SGC7901/pcDNA3.1. (C) Detection of the clone formation in plate. Cell were placed in media containing plate and incubated for 20 days. The number of foci > 100 μm was counted. (a) SGC7901/PrPC (1-OPRD); (b) SGC7901/PrPC and (c) SGC7901/pcDNA3.1. (D) Detection of clone formation. Vertical bars represent mean ± S.D. from at least three separate experiments, each conducted in triplicate.
Anchorage-independent growth is one of the important characteristics of in vitro tumour growth; therefore, we examined whether up-regulation of PrPC (1-OPRD) expression could promote SGC7901 cells growth in soft agar and plate. As shown in Fig. 3B and C, PrPC (1-OPRD) and PrPC transfection resulted in increased accumulation of growth of SGC7901 in colony formation assay, with average increase rates of 170.4% and 65.1% in soft agar and 93.3% and 58.6% in plate assay, respectively (Fig. 3D).
PrPC (1-OPRD) promoted G0-/G1- to S-phase transition in the cell cycle
To explore the possible roles of PrPC (1-OPRD) and PrPC in controlling cell proliferation, we examined the cell cycles of transfected cells by FACS three times. The mean PI of SGC7901/PrPC (1-OPRD) (0.584 ± 0.032) was higher than that of SGC7901/PrPC(0.449 ± 0.017), even higher than that of SGC7901/pcDNA3.1 (0.373 ± 0.023) and SGC7901 (0.360 ± 0.009) (P < 0.05). As reported previously [12], PrPC could promote gastric cancer cells transition from G0-/G1- to S-phase. PrPC (1-OPRD) had more effect on the cell proliferation from G0-/G1- to S-phase, and therefore contributed to the enhanced proliferation rate (Fig. 4A).
Figure 4.
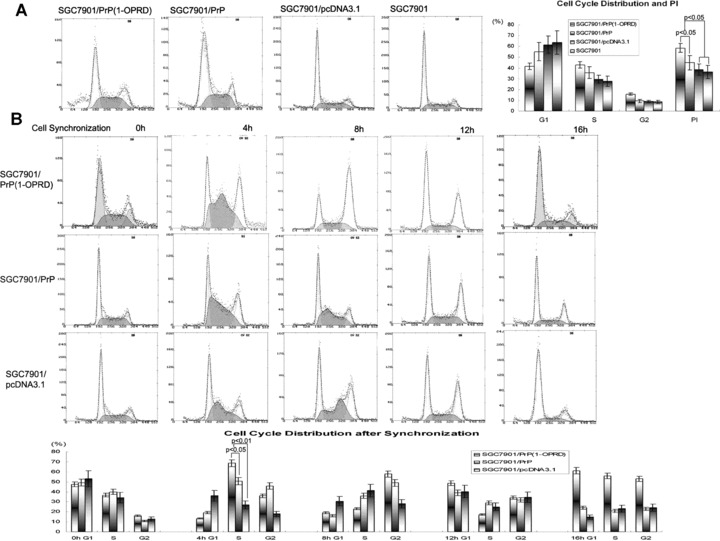
Effects of PrPC (1-ORPD) on cell cycle distribution in gastric cancer cells. (A) Cell cycle distribution in transfected cells. The cells were detergent extracted, stained with propidium iodide, and analysed by flow cytometry. The PI were calculated: PI = (S+G2)/(G1+S+G2). (B) Cell cycle synchronization. Effect of PrPC (1-OPRD) on cell cycle by arresting cells at G1/S boundary by adding thymidine (2 mM) at 0 hrs. Four hours after releasing, the cells began entering into S-phase. The average releasing rate of cells from G1- to S-phase in SGC7901/PrPC (1-OPRD) was higher than that of SGC7901/PrPC, even higher than that of vector control cells. Eight hours later after releasing, most cells had gone into G2-phase. By 16 hrs, cells entered a new cell cycle.
We further analysed the effects of PrPC (1-OPRD) and PrPC on cell cycles by synchronization. It was found that after cell cycle blocking by addition of thymidine (2 mM), cells were mostly arrested in G1-phase. Four hours after releasing, the cells began entering into S-phase. The average releasing rate of cells from G1- to S-phase in SGC7901/PrPC (1-OPRD) was 67.4%, which was higher than that of SGC7901/PrPC (45.7%), and significantly higher than that of vector control cells (34.6%) (P < 0.05). Eight hours later after releasing, most cells had gone into G2-phase. By 16 hrs, cells entered a new cell cycle (Fig. 4B).
CyclinD3 was involved in proliferation and G1/S transition of gastric cancer cells regulated by PrPC (1-OPRD)
To identify the molecules which were specifically regulated by PrPC (1-OPRD) and were responsible for the effect caused by PrPC (1-OPRD) in gastric cancer cells, gene array was used to screen for target molecules. Of the 22,575 sequences represented on the microarray, there were 3,798 genes whose expression was significantly altered by PrPC expression compared with emptor vector transfected cells [17]. Between SGC7901/PrPC (1-OPRD) and SGC7901/PrPC, there were 2337 genes whose expression was significantly altered (Fig. 5A). Figure 5B showed, as a scatter plot, the distribution of all the genes in the cDNA microarrays according to their expressions with respect to SGC7901/PrPC, nearly half were up-regulated and half are down-regulated. However, among them, seven genes closely related to G1-/S-phase transition CCND3 (NM_001760), CCNH (NM_001239), CCNG1 (NM_004060), CDK7 (NM_001799), CDC2 (NM_001786), CKS2 (NM_001827), MNAT1 (NM_002431) were found to be up-regulated in PrPC (1-OPRD)-transfected cells.
Figure 5.
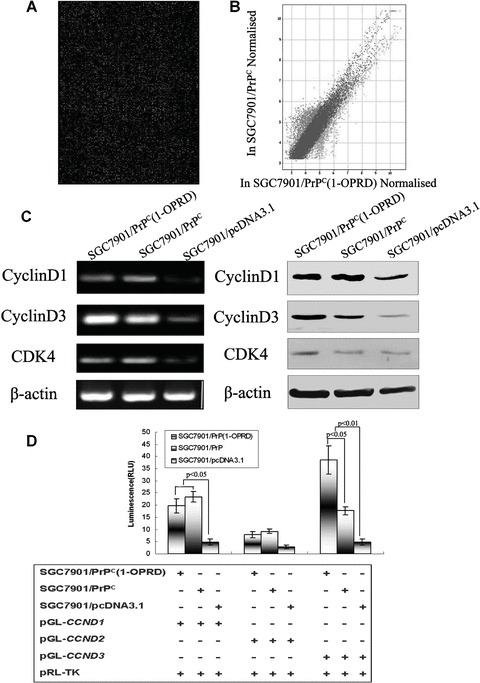
The inducible effect of PrPC (1-OPRD) on cyclinD. (A) Expression intensity of SGC7901/PrPC (1-OPRD) cells versus SGC7901/PrPC cells based on microarray data from all 22,575 human cDNA chip. cDNA amplicons from cells labelled with Cy5 and Cy3. Microarray hybridization was conducted as described in the ‘Materials and methods’. (B) Scatter plot, in logarithmic scales, of signal intensities representing the gene expression profiles of SGC7901/PrPC versus SGC7901/PrPC (1-OPRD). Green and red colours represent genes whose expression levels were significantly up and down-regulated in SGC7901/PrPC (1-OPRD), respectively. Blue spots show insignificant expression in both cells. (C) RT-PCR and Western blot analysis of the PrPC (1-OPRD), PrPC and vector transfectants. β-actin was used as a loading control. (D) Relative luciferase activity of cyclinD promoters in SGC7901 cells co-transfected with PrPC (1-OPRD), PrPC or empty vector were evaluated by dual luciferase reporter assay.
Confirmed by previous gene array results [18], cyclinD was found to be up-regulated in PrPC transfected cells, which was a key molecule in G1-/S-phase transition regulation. Therefore, we examined the expression of cyclinD in gastric cancer cells after PrPC (1-OPRD) and PrPC transfection. Increased transcription of cyclinD3 in PrPC (1-OPRD) was confirmed by RT-PCR. Similarly, overexpression of PrPC (1-OPRD) was found to increase the protein expression of cyclinD3 in SGC7901 cells by Western blotting (Fig. 5C). For PrPC (1-ORPD), it would specifically up-regulate cyclinD3 to promote cell transition from G1 to S.
To investigate the possible mechanisms involved in the regulation of cyclinD by PrPC (1-OPRD) and PrPC, dual-luciferase-reporter assay was performed. Luciferase reporter plasmids pGL3-CCND1, pGL3-CCND2 and pGL3-CCND3, containing cyclinD1, cyclinD2 and cyclinD3 promoters, respectively, were transiently transfected into SGC7901/PrPC (1-ORPD), SGC7901/PrPC and SGC7901/pcDNA3.1 together with pRL-TK. As seen in Fig. 5D, the intensity of luciferase luminescence in SGC7901/PrPC (1-ORPD) cells co-transfected with either pGL3-CCND3 was 2.18-fold higher than that of SGC7901/PrPC and 7.96-fold higher than that of SGC7901/pcDNA3.1, respectively, indicating that PrPC (1-ORPD) triggered transactivation of cyclinD3 to promote gastric cancer cell transition from G0-/G1- to S-phase.
Discussion
In the present study, we present the first evidence that forced overexpression of mutation form of PrPC, PrPC (1-OPRD) could promote the proliferation of gastric cancer cells and facilitate G1- to S-phase transition in the cell cycle. PrPC (1-OPRD) could also inhibit apoptosis and promote adhesion, invasion and MDR in gastric cancer cells. However, it exhibited no significant difference with full length PrPC on apoptosis, adhesion, invasion or MDR effects. Further experiments indicated that PrPC (1-OPRD) could trigger the transactivation of cyclinD3 to promote the cell transition and proliferation. To our knowledge, this is the first report on the function of mutation form of PrPC, PrPC (1-OPRD) and gastric cancer.
Approximately 10–15% of the human prion disease is inherited and more than 20 pathogenic mutations have been found, especially on the octapeptide repeat region (aa51–90). The N-terminal fragment of PrPC is a flexible region, which is higher conserved among different species [19]. The octapeptide repeat region was believed to be a copper binding site and mediated PrP-dependent activation of superoxide dismutase [20]. Prion proteins with mutations have altered N-terminal conformation and increased ligand binding activity, then more susceptible to oxidative attack [21]. Deletion of one Gly-Pro rich octapeptide repeat from the N-terminal of PrP was also seen in some species [22]. A survey in China found the PrPC (1-OPRD) mutation in four Hui(2.0%) and one Han (0.5%) in Chinese population [23]. 1-OPRD allele frequencies are 0.5% in Western Europeans [24], 0.54% in Italians [25], 0.45% in Germans [26] and 1.0% in Turkish [27]. In neural system, this mutation form was generally thought to be a non-pathogenic polymorphism. However, our previous work found that the PrPC (1-OPRD) existed in four of six kinds of gastric cancer cell lines detected and the mutation frequency was higher in gastric cancer cases [4].
For the high frequency mutation of PrPC (1-OPRD) in gastric cancer, two hypotheses were given accordingly: if PrPC (1-OPRD) is related to cell survival; or if PrPC (1-OPRD) can promote gastric cancer development. Though cancer DNA could show allelic imbalance (including loss of heterozygosity and gene amplification or multiplication) and cancer cell lines might have scrambled genomes (like irregular and variable chromosome numbers), it is really interesting that only this mutation of 1-OPRD but not others no others (like insertion of octapeptide repeats which is often seen in nervous system disease caused by PrP) existed in gastric cancer.
In the present study, the results of flow cytometry detection with annexin V/PI staining or immunofluorescence microscopy detection with Hoechest 3325/PI staining both suggested that PrPC (1-OPRD) could inhibit the apoptosis of gastric cancer cells. However, the effect between PrPC and PrPC (1-OPRD) showed no significant difference. Cell adhesion and invasion assays proved that PrPC (1-OPRD) could promote adhesion and invasion of gastric cancer cells, which was similar with the effect of PrPC. MTT assay and intracellular accumulation and retention of ADR were used to study the effect of PrPC (1-OPRD) on anticancer drugs, which mediated the MDR of ADR, VCR, VP-16, 5-FU and CDDP as PrPC. Taken together, PrPC (1-OPRD) had the same effect as PrPC on gastric cancer cell apoptosis, metastasis and MDR.
However, MTT assay and clone formation assay both suggested that forced PrPC (1-OPRD) overexpression could more significantly promote the proliferation of gastric cancer cells SGC7901. PrPC synthesis in T98G cells was previously demonstrated to be dependent on G1-phase of the cell cycle [28] and PrPC was found to promote gastric cancer cell transition from G1- to S-phase [12]. FACS and synchronization with thymidine (2 mM) in this study further proved that PrPC (1-OPRD) also promoted SGC7901 cells progression by regulating of the G1-/S-phase cell cycle. D-type cyclins played an important role in G1/S cell cycle transition [29]. Between PrPC (1-OPRD) and PrPC, seven genes closely related to G1-/S-phase transition CCND3 (NM_001760), CCNH (NM_001239), CCNG1 (NM_004060), CDK7 (NM_001799), CDC2 (NM_001786), CKS2 (NM_001827), MNAT1 (NM_002431) were found to be up-regulated in PrPC (1-OPRD)-transfected cells. The cyclinD3 was further proved to be up-regulated by PrPC (1-OPRD) at both mRNA and protein level, through transcriptional activation. Generally speaking, PrPC (1-OPRD) might stimulate cells proliferation by accelerating the transcription of cyclinD3 besides cyclinD1 and thus facilitating cell cycle transition from G1- to S-phase.
An interesting finding was that cells transfected with the plasmids containing PrPC deleted of the whole octapeptide repeat region (Δ51–90) would significantly inhibit the cell proliferation-promoting effect of PrPC[14]. While forced expression of PrPC (1-OPRD), deleted one octapeptide repeat, had more stimulating effect on the growth of gastric cancer cells. The same phenomenon also occurred in gastric cancer cell line AGS (human gastric adenocarcinoma cell line) (originally expressed full length of PrPC) and MKN28 (originally homozygotes expressed PrPC[1-OPRD]). Octapeptide repeat region was reported normally comprised one nonapeptide (R1) followed by four octapeptides (R2, R2, R3, R4), which had the same amino acid sequence but could be distinguished by variations in DNA sequence [30]. After sequencing and carefully analysis, PrPC (1-OPRD) was found to be deleted a functional octapeptide (R4) in gastric cancer cells. The different effects just fell in accord with previous report that different mutations altered N-terminal conformation of PrPC[21] and then might mediate different functions in gastric cancer. However, the relationship between the structure and function needed our further study.
In conclusion, this study demonstrated that overexpression of PrPC (1-OPRD) might promote the proliferation of gastric cancer cells at least partially through transcriptional activation of cyclinD3, besides cyclinD1 to accelerate the G1-/S-phase transition. The promoting proliferation effect of PrPC (1-OPRD) was more than that of full length PrPC. However, they showed no difference on apoptosis, adhesion, invasion or MDR effects of gastric cancer cells. It confirmed one of our hypotheses that PrPC (1-OPRD) could promote gastric cancer development. Nevertheless, detailed work on the relationship between this mutation form and function of PrPC in gastric cancer cells are still waiting to be investigated.
Conflict of interest
No conflicts of interest exist.
Acknowledgments
We thank Richard G. Pestell and Chenguang Wang (Lombardi Comprehensive Cancer Center and Department of Oncology, Georgetown University Medical Center, USA) for the cyclinD1 promoter plasmid. We are also grateful to Bo Huang for help in luciferase activity analysis, professors Xin Wang and Jie Liu for good suggestions during experiment, and technician Taidong Qiao, Zheng Chen, Baojun Chen and Baohua Song for their excellent technical assistance. This study was supported in part by grants from National Science Foundation of China (No. 30572134, 30872965, 30900551) and the National Basic Research Program of China (2009CB521703, 2010CB529300).
References
- 1.Yu S, Yin S, Li C, et al. Aggregation of prion protein with insertion mutations is proportional to the number of inserts. Biochem J. 2007;403:343–51. doi: 10.1042/BJ20061592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croes EA, Theuns J, Houwing-Duistermaat JJ, et al. Octapeptide repeat insertions in the prion protein gene and early onset dementia. J Neurol Neurosurg Psychiatry. 2004;75:1166–70. doi: 10.1136/jnnp.2003.020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu SL, Jin L, Sy MS, et al. Polymorphisms of the PRNP gene in Chinese populations and the identification of a novel insertion mutation. Eur J Hum Genet. 2004;12:867–70. doi: 10.1038/sj.ejhg.5201245. [DOI] [PubMed] [Google Scholar]
- 4.Liang J, Wang JB, Pan YL, et al. High frequency occurrence of 1-OPRD variant of PRNP gene in gastric cancer cell lines and Chinese population with gastric cancer. Cell Biol Int. 2006;30:920–3. doi: 10.1016/j.cellbi.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 5.Manuelidis L, Yu ZX, Banquero N, et al. Cells infected with scrapie and Creutzfeldt-Jakob disease agents produce intracellular 25-nm virus-like particles. Proc Natl Acad Sci USA. 2007;104:1965–70. doi: 10.1073/pnas.0610999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aguzzi A, Polymenidow M. Mammalian prion biology. One century of evolving concepts. Cell. 2004;116:313–27. doi: 10.1016/s0092-8674(03)01031-6. [DOI] [PubMed] [Google Scholar]
- 7.Yin S, Yu S, Li C, et al. Prion proteins with insertion mutations have altered N-terminal conformation, increased ligand-binding activity and are more susceptible to oxidative attack. J Biol Chem. 2006;281:10698–705. doi: 10.1074/jbc.M511819200. [DOI] [PubMed] [Google Scholar]
- 8.Mitteregger G, Vosko M, Krebs B, et al. The role of the octarepeat region in neuroprotective function of the cellular prion protein. Brain Pathol. 2007;17:174–83. doi: 10.1111/j.1750-3639.2007.00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roucou X, LeBlanc AC. Cellular prion protein neuroprotective function: implications in prion diseases. J Mol Med. 2005;83:3–11. doi: 10.1007/s00109-004-0605-5. [DOI] [PubMed] [Google Scholar]
- 10.Liang J, Pan YL, Ning XX, et al. Overexpression of PrPC and its antiapoptosis function in gastric cancer. Tumour Biol. 2006;27:84–91. doi: 10.1159/000092488. [DOI] [PubMed] [Google Scholar]
- 11.Liang J, Ge FL, Lu YY, et al. Role of PrPC related to apoptosis. EXCLI J. 2006;5:11–24. [Google Scholar]
- 12.Liang J, Pan Y, Zhang D, et al. Cellular prion protein promotes proliferation and G1(S transition of human gastric cancer cells SGC7901 and AGS. FASEB J. 2007;21:2247–56. doi: 10.1096/fj.06-7799com. [DOI] [PubMed] [Google Scholar]
- 13.Pan Y, Zhao L, Liang J, et al. Cellular prion protein promotes invasion and metastasis of gastric cancer. FASEB J. 2006;20:1886–8. doi: 10.1096/fj.06-6138fje. [DOI] [PubMed] [Google Scholar]
- 14.Du J, Pan Y, Shi Y, et al. Overexpression and significance of prion protein in gastric cancer and multidrug-resistant gastric carcinoma cell line SGC7901(ADR) Int J Cancer. 2005;113:213–20. doi: 10.1002/ijc.20570. [DOI] [PubMed] [Google Scholar]
- 15.Liang J, Bai F, Luo G, et al. Hypoxia induced overexpression of PrP(C) in gastric cancer cell lines. Cancer Biol Ther. 2007;6:769–74. doi: 10.4161/cbt.6.5.4001. [DOI] [PubMed] [Google Scholar]
- 16.Liang J, Luo GH, Ning XX, et al. Differential expression of calcium-related genes in gastric cancer cells transfected with cellular prion protein. Biochem Cell Biol. 2007;85:375–83. doi: 10.1139/o07-052. [DOI] [PubMed] [Google Scholar]
- 17.Brooks AR, Shiffman D, Chan CS, et al. Functional analysis of the human cyclin D2 and cyclin D3 promoters. J Biol Chem. 1996;271:9090–9. doi: 10.1074/jbc.271.15.9090. [DOI] [PubMed] [Google Scholar]
- 18.Jun-ichi S, Yasuo K, Shigeru K. Gene expression profile in prion protein-deficient fibroblasts in culture. Am J Pathol. 2000;157:59–68. doi: 10.1016/s0002-9440(10)64517-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wopfner F, Weidenhofer G, Schneider R, et al. Analysis of 27 mammalian Schneider and 9 avian PrPs reveals high conservation of flexible regions of the prion protein. J Mol Biol. 1999;289:1163–78. doi: 10.1006/jmbi.1999.2831. [DOI] [PubMed] [Google Scholar]
- 20.Sakudo A, Lee DC, Nishimura T, et al. Octapeptide repeat region and N-terminal half of hydrophobic region of prion protein (PrP) mediate PrP-dependent activation of superoxide dismutase. Biochem Biophys Res Commun. 2005;326:600–6. doi: 10.1016/j.bbrc.2004.11.092. [DOI] [PubMed] [Google Scholar]
- 21.Yin S, Yu S, Li C, et al. Prion proteins with insertion mutations have altered N-terminal conformation and increased ligand binding activity and are more susceptible to oxidative attack. J Biol Chem. 2006;281:10698–705. doi: 10.1074/jbc.M511819200. [DOI] [PubMed] [Google Scholar]
- 22.Schatzl HM, Da Costa M, Taylor L, et al. Prion protein gene variation among primates. J Mol Biol. 1995;245:362–74. doi: 10.1006/jmbi.1994.0030. [DOI] [PubMed] [Google Scholar]
- 23.Yu SL, Jin L, Sy MS, et al. Polymorphisms of the PRNP gene in Chinese populations and the identification of a novel insertion mutation. Eur J Hum Genet. 2004;12:867–70. doi: 10.1038/sj.ejhg.5201245. [DOI] [PubMed] [Google Scholar]
- 24.Palmer MS, Mahal SP, Campbell TA, et al. Deletions in the prion protein gene are not associated with CJD. Hum Mol Genet. 1993;2:541–4. doi: 10.1093/hmg/2.5.541. [DOI] [PubMed] [Google Scholar]
- 25.Salvatore M, Genuardi M, Petraroli R, et al. Polymorphisms of the prion protein gene in Italian patients with Creutzfeldt - Jakob disease. Hum Genet. 1994;94:375–9. doi: 10.1007/BF00201596. [DOI] [PubMed] [Google Scholar]
- 26.Windl O, Giese A, Schulz-Schaeffer W, et al. Molecular genetics of human prion diseases in Germany. Hum Genet. 1999;105:244–52. doi: 10.1007/s004399900124. [DOI] [PubMed] [Google Scholar]
- 27.Erginel-Unaltuna N, Peoc’h K, Komurcu E, et al. Distribution of the M129V polymorphism of the prion protein gene in a Turkish population suggests a high risk for Creutzfeldt - Jakob disease. Eur J Hum Genet. 2001;9:965–8. doi: 10.1038/sj.ejhg.5200754. [DOI] [PubMed] [Google Scholar]
- 28.Kikuchi Y, Kakeya T, Yamazaki T, et al. G1-dependent prion protein expression in human glioblastoma cell line T98G. Biol Pharm Bull. 2002;25:728–33. doi: 10.1248/bpb.25.728. [DOI] [PubMed] [Google Scholar]
- 29.Arber N, Hibshoosh H, Moss SF, et al. Increased expression of cyclin D1 is an early event in multistage colorectal carcinogenesis. Gastroenterology. 1996;110:669–74. doi: 10.1053/gast.1996.v110.pm8608874. [DOI] [PubMed] [Google Scholar]
- 30.Yanagihara C, Yasuda M, Maeda K, et al. Rapidly progressive dementia syndrome associated with a novel four extra repeat mutation in the prion protein gene. J Neurol Neurosurg Psychiatry. 2002;72:788–91. doi: 10.1136/jnnp.72.6.788. [DOI] [PMC free article] [PubMed] [Google Scholar]


