Abstract
An understanding of how lipid droplets grow in the cell is important to current human health issues. Homotypic fusion of small lipid droplets to create larger ones is one proposed mechanism though the evidence for this process continues to be debated. By applying the technique of freeze-fracture electron microscopy to cells that have been stimulated to accumulate lipid droplets, we here present images which suggest that at least some large lipid droplets may indeed result from amalgamation of multiple smaller ones. These visual data add significantly to the notion that fusion contributes to lipid droplet growth.
Keywords: lipid droplet fusion freeze-fracture electron microscopy immunogold cytochemistry
The ever increasing incidence of obesity and atherosclerosis has highlighted the importance of understanding the function and dynamics of the lipid droplet, but our knowledge of how these structures grow remains rudimentary [1–7]. It is generally considered that the droplet is initially formed by accretion of lipid esters within and/or adjacent to the endoplasmic reticulum (ER) membrane [5–10]. Subsequent growth may be brought about by further accretion from adjacent ER membrane, cytoplasmic transport of lipid ester as soluble complexes to the droplet or direct accumulation at the droplet itself [5, 6, 10]. When cells accumulate lipid, the large numbers of small lipid droplets initially produced are subsequently replaced with fewer large ones, leading to the idea that homotypic fusion between droplets may also occur [5–7, 11, 12]. This mechanism, though frequently debated, remains controversial as clear-cut evidence for fusion events in situ has been difficult to obtain.
Possible fusion has been suggested from live cell fluorescence imaging but the low resolution, and other technical factors, such as vertical movement of structures out of the viewing plane, have prevented definitive conclusions to be reached [11]. Although thin-section electron microscopy has adequate resolution, extraction of lipids together with the surface phospholipid monolayers of closely apposed droplets during processing creates false fusion-like images. Freeze-fracture electron microscopy offers some advantages over other approaches [13, 14]. Although only static images are obtained, these are at high resolution from cells stabilized by the physical process of freezing, without recourse to chemical fixatives or solvents. Importantly, because the fracture plane is dictated by lipid organization, a wealth of structural detail within the droplet is revealed [9, 15].
Applying the freeze-fracture technique to THP-1 macrophages, abundant lipid droplets appear in the cytoplasm after 24 hrs’ incubation of the cells with acetylated low-density lipoprotein (LDL) (Fig. 1). Freeze fracture typically reveals the lipid droplets as closely associated but discrete spheroid or ovoid bodies, 0.5–1.0 μm in diameter, with a characteristic layered structure. Where the droplet is convexly or concavely fractured, a series of concentric layers is apparent, whereas cross fractures reveal less regular stacked layers in the droplet core. These features help define each droplet as an individual entity.
Figure 1.
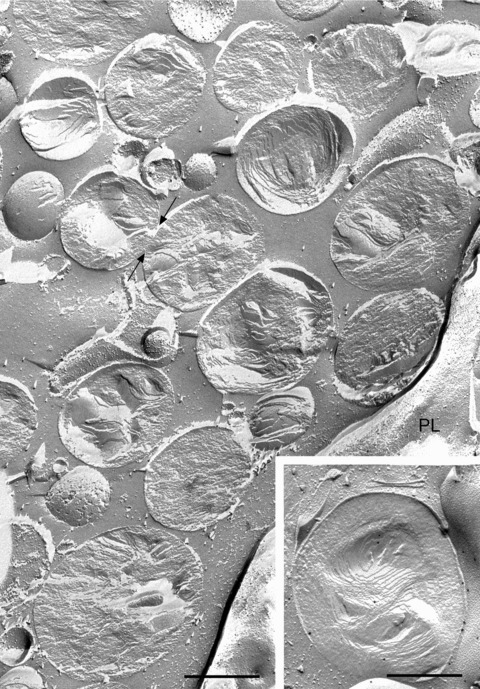
Freeze-fracture view of typical appearance of cytoplasmic lipid droplets in THP-1 macrophages. The cells were incubated with 50 μg/ml acetylated LDL for 24 hrs to stimulate lipid droplet accumulation. The fracture plane frequently follows along the plane of organized lipids to give convex or concave views of the droplet; as the fracture path skips back and forth between lipid layers of the droplet, an ‘onion-like’ morphology is revealed. Other lipid droplets are cross fractured, revealing a stack of lipid layers in the core. In either case, the boundary of each droplet is usually clearly defined, demarcating one droplet from the next. Only occasionally do side-by-side droplets show regions of continuity (arrows), raising the possibility of an ongoing fusion event. PL, plasma membrane. In some instances, two stacks of lipid layers, each approximately ovoid in overall shape, are found within a single droplet, creating the impression that two droplets have somehow combined to make one (inset). In this example, the droplet has been immunogold labelled for adipophilin. Bar: 0.5 μm.
Despite being so tightly packed that individual droplets appear to touch and even become flattened by mutual pressure at such contact regions, the boundaries of the individual droplets are normally quite distinct, suggesting a strong resistance to fusion (Fig. 1). Only occasionally is a possible site of continuity between the contents of apposed droplets seen. Whether such sites represent droplets caught in the act of fusion or the fracture path has simply failed to be deviated sufficiently to show the boundaries clearly at these sites, cannot be determined with certainty.
It is nevertheless striking that, from the appearances observed in freeze fracture, the larger lipid droplets frequently appear to be aggregates of smaller droplets. An example, in which a lipid droplet containing two distinct layered structures resembling those of separate droplets, is shown in the inset to Fig. 1. An exceptionally large droplet, containing multiple ovoid concentric structures, each reminiscent of those of individual droplets, is illustrated in Fig. 2.
Figure 2.
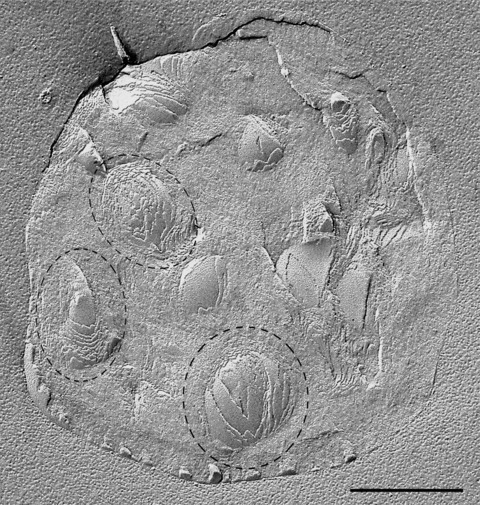
Large lipid droplet containing multiple small ovoid concentric layered structures which have the appearance of individual droplets (examples encircled with dashed lines). Such images suggest the amalgamation of many lipid droplets, each of which has retained the essential features of its individual structure as depicted in Fig. 1. Amalgamation of this nature implies that fusion events led to the formation of this droplet. Bar: 0.5 μm.
How are such structures formed? One possibility is that, as lipids are accreted into the enlarging droplet, they are internally organized into layered structures. However, if this were the case, it seems odd that multiple individual layered structures are formed rather than a single large one. The overwhelming impression created by these images is that numerous smaller lipid droplets have amalgamated to create a larger one. The ability of small lipid droplets to fuse would imply lipid fluidity with consequent intermixing of components; how their individual substructures would be maintained thus remains something of a puzzle. However, if such homotypic fusion between smaller lipid droplets does actually take place, then this could also explain how proteins that have traditionally been regarded as key components of the surface monolayer of the droplet are also detected in the core [15]. One of the theoretical objections to fusion as a mechanism of lipid droplet growth is that, owing to the reduction in overall surface area, an excess of surface monolayer would result that could not be accommodated via the mechanisms operating in membrane-bound vesicles [5, 6]. It could be suggested that some excess phospholipid monolayer becomes trapped in the droplet interior, in keeping with the localization of proteins there. In addition, however, the occasional observation of small vesicle-like blebs at the surface of large lipid droplets (Fig. 3) raises the intriguing possibility of a phospholipid shedding mechanism.
Figure 3.

Vesicular structures (v) at the periphery of a large lipid droplet. These might represent a mechanism for shedding excess phospholipid monolayer though how a stable bilayer vesicle could be created in such a situation is unclear. Alternatively, such structures may be involved in delivery to the droplet. In this example, the droplet has been double immunogold labelled for adipophilin (18 nm gold) and TIP 47 (12 nm gold). Bar: 0.2 μm.
Acknowledgments
We thank Karin Schlattmann, Christina Köppler and M. Opalka for competent and indispensable technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 492) and by the Seventh Framework Program of the EU-funded ‘Lipidomic Net’ (proposal number 202272).
References
- 1.Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40:325–438. doi: 10.1016/s0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- 2.Martinez-Botas J, Anderson JB, Tessier D, et al. Absence of perilipin results in leanness and reverses obesity in Lep (db/db) mice. Nat Genet. 2000;26:476–9. doi: 10.1038/82630. [DOI] [PubMed] [Google Scholar]
- 3.Mishra R, Eminacipator SN, Miller C, et al. Adipose differentiation related protein and regulators of lipid homeostasis identified by gene expression profiling in murine db/db diabetic kidney. Am J Physiol Renal Physiol. 2004;286:F913–21. doi: 10.1152/ajprenal.00323.2003. [DOI] [PubMed] [Google Scholar]
- 4.Larigauderie G, Furman C, Jaye M, et al. Adipophilin enhances lipid accumulation and prevents lipid efflux from THP-1 macrophages: potential role in atherogenesis. Arterioscler Thromb Vasc Biol. 2004;24:504–10. doi: 10.1161/01.ATV.0000115638.27381.97. [DOI] [PubMed] [Google Scholar]
- 5.Ohsaki Y, Cheng J, Suzuki M, et al. Biogenesis of cytoplasmic lipid droplets: from the lipid ester globule in the membrane to the visible structure. Biochim Biophys Acta. 2009;1791:399–407. doi: 10.1016/j.bbalip.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Fujimoto T, Ohsaki Y, Cheng J, et al. Lipid droplets: a classic organelle with new outfits. Histochem Cell Biol. 2008;130:263–79. doi: 10.1007/s00418-008-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiele C, Spandl J. Cell biology of lipid droplets. Curr Opin Cell Biol. 2008;20:378–85. doi: 10.1016/j.ceb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Brown DA. Lipid droplets: proteins floating on a pool of fat. Curr Biol. 2001;11:R446–9. doi: 10.1016/s0960-9822(01)00257-3. [DOI] [PubMed] [Google Scholar]
- 9.Robenek H, Hofnagel D, Buers I, et al. Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. J Cell Sci. 2006;119:4215–24. doi: 10.1242/jcs.03191. [DOI] [PubMed] [Google Scholar]
- 10.Buers I, Robenek H. Lorkowski S, et al. TIP 47, a lipid cargo protein involved in macrophage triglyceride metabolism. Arterioscler Thromb Vasc Biol. 2009;29:767–73. doi: 10.1161/ATVBAHA.108.182675. [DOI] [PubMed] [Google Scholar]
- 11.Nagayama M, Uchida T, Gohara K. Temporal and spatial variations of lipid droplets during adipocyte division and differentiation. J Lipid Res. 2007;48:9–18. doi: 10.1194/jlr.M600155-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Bostrom P, Andersson L, Rutberg M, et al. SNARE proteins mediate fusion netween cytosolic lipid droplets and are implicated in insulin sensitivity. Nat Cell Biol. 2007;9:1286–93. doi: 10.1038/ncb1648. [DOI] [PubMed] [Google Scholar]
- 13.Severs NJ. Freeze-fracture electron microscopy. Nat Protoc. 2007;2:547–76. doi: 10.1038/nprot.2007.55. [DOI] [PubMed] [Google Scholar]
- 14.Robenek H, Severs NJ. Recent advances in freeze-fracture electron microscopy: the replica immunolabeling technique. Biol Proc Online. 2008;10:9–19. doi: 10.1251/bpo138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robenek H, Buers I, Hofnagel O, et al. Compartmentalization of proteins in lipid droplet biogenesis. Biochim Biophys Acta. 2009;1791:408–18. doi: 10.1016/j.bbalip.2008.12.001. [DOI] [PubMed] [Google Scholar]


