Abstract
Microalgae constitute a diverse group of eukaryotic unicellular organisms that are of interest for pure and applied research. Owing to their natural synthesis of value-added natural products microalgae are emerging as a source of sustainable chemical compounds, proteins and metabolites, including but not limited to those that could replace compounds currently made from fossil fuels. For the model microalga, Chlamydomonas reinhardtii, this has prompted a period of rapid development so that this organism is poised for exploitation as an industrial biotechnology platform. The question now is how best to achieve this? Highly advanced industrial biotechnology systems using bacteria and yeasts were established in a classical metabolic engineering manner over several decades. However, the advent of advanced molecular tools and the rise of synthetic biology provide an opportunity to expedite the development of C. reinhardtii as an industrial biotechnology platform, avoiding the process of incremental improvement. In this review we describe the current status of genetic manipulation of C. reinhardtii for metabolic engineering. We then introduce several concepts that underpin synthetic biology, and show how generic parts are identified and used in a standard manner to achieve predictable outputs. Based on this we suggest that the development of C. reinhardtii as an industrial biotechnology platform can be achieved more efficiently through adoption of a synthetic biology approach.
Significance Statement
Chlamydomonas reinhardtii offers potential as a host for the production of high value compounds for industrial biotechnology. Synthetic biology provides a mechanism to generate generic, well characterised tools for application in the rational genetic manipulation of organisms: if synthetic biology principles were adopted for manipulation of C. reinhardtii, development of this microalga as an industrial biotechnology platform would be expedited.
Keywords: synthetic biology, industrial biotechnology, Chlamydomonas reinhardtii, metabolic engineering, rational design, transgene expression
Microalgae Present Unique Features for Applied Research and Biotechnology
Microalgae are a polyphyletic group of unicellular eukaryotic organisms that can occupy both aquatic and terrestrial environments, adopting photosynthetic, heterotrophic or mixotrophic lifestyles. The enormous diversity in the algal lineages is due to their long evolutionary history (Dorrell and Smith, 2011). For instance the green algae, which are ancestral to land plants, diverged 1300 million years ago from the stramenopiles (Yoon et al., 2004), a group that encompasses several microalgal lineages of biotechnological relevance such as diatoms. As a result, microalgae exhibit wide variation in both cellular architecture and biosynthetic capacity, and thus present a number of general and unique features as research models and for commercial exploitation. Specifically, the unicellular physiology combined with rapid cell division and photosynthetic growth mean that microalgae can be more productive per unit land area than any plant system. In addition, using microalgae as feedstocks for low value, high volume products such as biofuels, provides the opportunity to avoid a number of environmental factors that currently affect biofuel production from crop plants (Chisti, 2007). These include issues surrounding land and (fresh)water usage, mono-culture, crop rotation, and input requirements: for example microalgae are able to sequester nitrogen and phosphorus from industrial waste streams, thus reducing the need for fertilisers. Nevertheless, many challenges remain to be overcome before it will be possible to cultivate algae on the very large scales needed for biofuel production (Scott et al., 2010; Klein-Marcuschamer et al., 2013). Instead, microalgae offer considerable potential for the production of low volume, high(er) value compounds, which are characteristically produced by industrial biotechnology (IB). Moreover, the photosynthetic lifestyle of microalgae means that they offer a more sustainable source of compounds compared to bacterial and yeast hosts used in conventional IB, since there is no need to provide fixed carbon.
Currently microalgae are exploited commercially for compounds that they make naturally, such as the pigments β-carotene and astaxanthin, and polymers alginate, carrageenan and agar for food products. Several research studies have attempted to enhance the production of value-added compounds, including bio-hydrogen from Chlamydomonas reinhardtii (Baltz et al., 2014; Xu et al., 2014), or omega-3 long chain fatty acids such as docosahexanoic acid (DHA) or eicosapentaenoic acid (EPA) from Phaeodactylum tricornutum. Although P. tricornutum naturally makes EPA and some DHA, levels are low and the ratio of the two fatty acids is suboptimal for some commercial applications. In addition, they are present mainly in membrane rather than storage lipids, making extraction inefficient. Hamilton et al. (2014) were able to increase levels of DHA in P. tricornutum eight-fold by heterologous expression of two fatty-acid modifying enzymes, a Δ5-elongase and an acyl-CoA-dependent Δ6-desaturase, from the green alga Ostreococcus tauri using a constitutive promoter. Another desirable feature of microalgae is their cellular complexity, which provides the opportunity to partition or compartmentalise biochemical reactions, something not possible in bacterial hosts. This can facilitate the provision of precursors, or provide intracellular sinks for target products. Indeed, there are already reports of the production of human therapeutic proteins including erythropoietin, interferon β proinsulin and immunoglobulin A in the chloroplast of C. reinhardtii (Rasala et al., 2010).
Whilst the many benefits afforded by microalgae are yet to be demonstrated at scale, these studies serve to raise the profile of microalgae research and development. The challenge now lies in the application of metabolic engineering strategies to enhance natural features of microalgae, making commercial exploitation economically viable (Klein-Marcuschamer et al., 2013). For example, in dense cultures required for industrial cultivation, light capture is limited. Reduction of the light-harvesting apparatus reduces light absorption per cell, increasing penetration and productivity within the culture (Polle et al., 2003; Cazzaniga et al., 2014). Other targets for optimisation include resource-use efficiency and auto-flocculation to facilitate harvesting. In parallel, to optimise and expand the level and range of novel products made by microalgae, it will be necessary to introduce new biosynthetic capabilities, remove competing pathways, and optimise the provision of precursor substrates and cofactors. Thus for microalgal IB to come of age, we need a suite of molecular biological tools with which to carry out sophisticated metabolic engineering.
For more than 20 years scientists have been able to redirect the metabolism of a host cell in a targeted manner through genetic manipulation, and this has been applied to across the tree of life, in both prokaryotes and eukaryotes, working with well characterised laboratory strains as well as novel environmental isolates. In the commercial sector, several organisms have been developed as robust IB hosts, including bacteria (Escherichia coli, Bacillus subtilis, Corynebacterium glutamicum, Lactobacillus spp.), and yeasts and fungi (Saccharomyces cerevisiae, Aspergillus spp.). These are used as production systems for compounds ranging from organic acids to pharmaceutical proteins (Goel et al., 2012; Buchholz and Collins, 2013). In spite of this success, metabolic engineering seldom demonstrates the design aspect that is so fundamental to engineering per se, since the development of bespoke solutions for each experimental system/product results in parallel development, and limits the transferability of constructs and knowledge. The advent of synthetic biology, whose principles involve a combination of standard parts, predictive modelling, and iterative design and testing, is just beginning to be exploited in the field of metabolic engineering, and holds out real promise for considerable advances in IB for production of compounds to reduce reliance on fossil fuels as feedstocks (Keasling, 2012; Yadav et al., 2012).
In this review, we present an overview of what has been achieved in terms of genetic manipulation of C. reinhardtii, and frame this in the context of bacterial and yeast model systems, drawing comparisons to illustrate current capabilities and limitations. We then discuss the relevance and timely nature of the development of synthetic biology concepts for metabolic engineering and how this represents a step change in capacity and predictive power. Finally we highlight how, through relatively minor changes, synthetic biology approaches might be applied to the development of C. reinhardtii as an IB platform, and also to enhance the microalgal research field more broadly. It is worth mentioning that other microalgae are being explored for similar applications, and we direct the reader to excellent reviews on the subject (Wijffels et al., 2013; Bellou et al., 2014; Klok et al., 2014; Chauton et al., 2015). In addition, several cyanobacterial species are being used for metabolic engineering, including for biofuels (Quintana et al., 2011; Berla et al., 2013; Wijffels et al., 2013). As prokaryotes they are not algae, but their simpler genetic system and ease of transformation can offer advantages. Conversely, they are less versatile than algae in terms of metabolic pathways.
Molecular Resources for C. reinhardtii
Chlamydomonas reinhardtii (Chlorophyta) is a photosynthetic biflagellate microalga that has been studied for >30 years as a model for basic and applied physiology and biochemistry (see articles in the rest of this issue). The number of resources available for the manipulation of C. reinhardtii has increased markedly over this period (Figure1). These include well defined protocols for growth, sexual propagation, and mutagenesis, as well as numerous published biochemical, analytical and reporter assays. Supporting research into various aspects of cell biology is the Chlamydomonas stock centre, a collection of >300 plasmids and >2700 strains (http://www.chlamy.org). Sequences of the nuclear, chloroplast and mitochondrial genomes have been completed (Merchant et al., 2007). The approximately 17 000 genes are well annotated, due to the substantial amount of expression data available, as well as the efforts of the community. The current nuclear genome database, version 5.5, has had most gaps filled and is 93% complete (Blaby et al., 2014). This knowledge base is made accessible by web based tools including JGI Phytozome genome browser (Goodstein et al., 2012), the functional genomic ChlamyCyc portal (May et al., 2009), KEGG metabolic pathway portal (Kanehisa, 2000), and the PlantGDB comparative genomics resource (Duvick et al., 2008). The complexity of cellular metabolism is made accessible in silico by the generation of genome-scale metabolic models (GSMMs). GSMMs present a snapshot of metabolism in a network format and allow the topology of system to be investigated. Several GSMMs for C. reinhardtii have been generated (Boyle and Morgan, 2009; Christian et al., 2009; Manichaikul et al., 2009; Chang et al., 2011; Cogne et al., 2011; Dal'Molin et al., 2011; Kliphuis et al., 2012), and the efficacy of such models has been reviewed recently (Reijnders et al., 2014).
Figure 1.
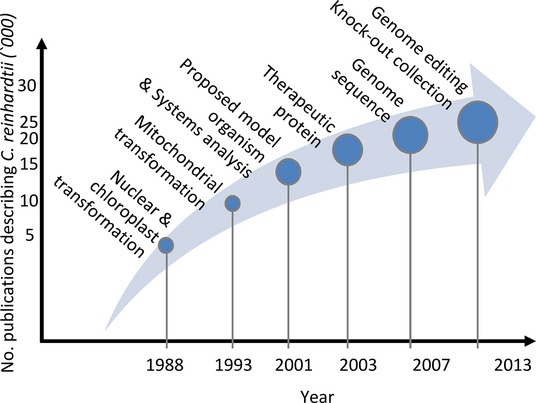
The rise of C. reinhardtii as a model system for molecular biology.Major breakthroughs include first nuclear and chloroplast transformations (Boynton et al., 1988; Blowers et al., 1989; Kindle et al., 1989), mitochondrial transformation (Randolph-Anderson et al., 1993), systems analysis by proteomics (Hippler et al., 2001), the proposal of C. reinhardtii as a model organism (Harris, 2001), the production of the first therapeutic recombinant protein in the chloroplast (Mayfield et al., 2003), the sequencing of the nuclear genome (Merchant et al., 2007) and the publication of genome editing techniques (Sizova et al., 2013; Gao et al., 2014; Jiang et al., 2014) and the creation of a knock-out collection (Zhang et al., 2014).
Protocols for transformation of the nucleus, chloroplast and mitochondria are available using biolistics (Debuchy et al., 1989), electroporation (Shimogawara et al., 1998), Agrobacterium (Kumar et al., 2004) or even simply vortexing with glass beads (Kindle, 1990). Facilitating nuclear transformation is the availability of several selectable markers, including auxotrophic markers, such as ARG7, which restores arginine prototrophy (Debuchy et al., 1989; Purton and Rochaix, 1995), as well as resistance genes, BLE (resistance against phleomycin; Stevens et al., 1996), APHVIII (paromomycin; Sizova et al., 1996), and HYG (hygromycin B; Berthold et al., 2002). Transformation of the chloroplast genome can be by homologous recombination, enabling targeted and specific manipulation, although because of the multiple copies of the chloroplast DNA, several rounds of selection are required to obtain homoplasmy (Day and Goldschmidt-Clermont, 2011). Nuclear transformation, on the other hand, is by random integration, and this means that multiple transformants must be screened to obtain stable lines expressing the transgene at the desired level (Debuchy et al., 1989). Important recent developments include the generation of a knock-out library consisting of >1600 single gene knockouts (Zhang et al., 2014) and the description of two strains, generated by random mutagenesis, which demonstrate enhanced protein expression due to an unknown mechanism that reduces the effects of transgene silencing (Neupert et al., 2009).
Complementing these resources are genomics studies of C. reinhardtii, of which there an increasing number (Table1), including several aimed at metabolic engineering. Matthew et al. (2009) analysed the metabolome of C. reinhardtii following sulfur depletion. This analysis demonstrated that between 0 and 24 h post sulfur deprivation, C. reinhardtii rapidly metabolised extra-cellular fixed carbon and synthesised the storage compounds starch and lipids in the form of triacylglycerides (TAGs). Over the next 96 h hydrogen production was achieved, peaking at 48 h post sulfur deprivation, while starch reserves were metabolised. However, amino acid and fatty-acid biosynthetic capacity and cellular stores remained intact, suggesting that the observed decline in hydrogen production was not due to metabolic depletion but instead because of the toxic accumulation of fermentation products, formate and ethanol. More recently, Lv et al. (2013) analysed the transcriptome of C. reinhardtii under exponential growth and lipid accumulation, identifying over 2500 genes that were upregulated. Of these 80% were assigned to functional categories which, as well as lipid biosynthesis, included several central metabolic pathways, suggesting an increase in general metabolism in response to the imposed stress. In addition, 41 transcription factors were differentially expressed under tested conditions. Building upon the genomic analyses are several GSMMs (Reijnders et al., 2014), which facilitate predictive attempts to harness the biosynthetic capacity of C. reinhardtii. Combined with a legacy of fundamental research and the current impetus placed on microalgae as a potentially sustainable source of energy, these modelling and other systems biology approaches are leading to an enormous increase in our understanding of C. reinhardtii, and efforts to manipulate it. At the same time, there has been a rise in the number of other microalgae with sequenced genomes and genomics tools (Figure S1 and Table S1); these data are leading to a much wider knowledge base of these organisms generally.
Table 1.
Examples of genomic studies completed for the analysis of specific physiological and biochemical characteristics of C. reinhardtii
| Study | Studies | Reference |
|---|---|---|
| Microarrays | Tandem repeat discovery | Zhao et al. (2014a) |
| Sexual production | Ning et al. (2013) | |
| Transcriptomics | Transcription factors | Riaño-Pachón et al. (2008) |
| Alternative splicing | Labadorf et al. (2010) | |
| Polyadenylation | Zhao et al. (2014b) | |
| Growth adaptation based on natural selection, serial dilution | Perrineau et al. (2014) | |
| TAG production improvement | Goodenough et al. (2014) | |
| Flagella regeneration | Stolc et al. (2005) | |
| CO2 effect | Fang et al. (2012) | |
| Proteomics | Basal body | Keller et al. (2005) |
| Mitochondria | van Lis et al. (2003) | |
| Thylakoids | Allmer et al. (2006) | |
| Metabolomics | ChlamyCyc database | May et al. (2009) |
| Partitioning oil and starch | Johnson and Alric (2013) | |
| S-deprived H2 production | Matthew et al. (2009) |
Advanced tools for manipulation of the C. reinhardtii nucleus
Efforts to express genes in the C. reinhardtii nucleus have been underpinned by the elucidation of mechanisms to regulate transgene expression. Examples include the use of promoters (Table2) that have been characterised either as constitutive (e.g. HSP70A/RBCS2; Schroda et al., 2000) or regulated in response to specific stimuli, such as light (PSAD; Fischer and Rochaix, 2001), copper (CYC6; Quinn and Merchant, 1995), nitrate (NIT1; Ohresser et al., 1997), and most recently vitamin B12 (METE; Helliwell et al., 2014). RNA-based elements have also been employed to augment transgene expression (Table2). Introns of the Rubisco Small Subunit 2 (RBCS2) have been used extensively since they were first shown to function as enhancer elements in a manner independent of orientation, and they are effective when positioned either upstream or downstream of the promoter (Lumbreras et al., 1998). Thiamine pyrophosphate riboswitches have been identified in the thiamine biosynthetic pathway of C. reinhardtii which cause alternative splicing on binding the ligand, (Croft et al., 2007). The THI4 riboswitch has subsequently been used to regulate expression of a nuclear encoded protein, NAC2, which in turn is required for chloroplast gene expression (Ramundo et al., 2013). Down-regulation of native C. reinhardtii genes has taken advantage of RNA interference, where binding of short fragments of RNA (microRNA or miRNA) complementary to specific mRNAs leads to cleavage and thus silencing of the target gene. C. reinhardtii encodes all the components of the RNAi pathway (Casas-Mollano et al., 2008), and techniques for artificial-micro-RNA (amiRNA) have been developed (Molnar et al., 2009; Zhao et al., 2009). In one report, combining amiRNA with the NIT1 promoter resulted in an inducible gene knockdown system (Schmollinger et al., 2010).
Table 2.
Regulatory elements available for C. reinhardtii nuclear gene expression. Categorised based upon functional properties, promoters, regulatory elements, functional peptides, colorimetric reporters and highly expressed strains
| Functional element | Name | Phytozyme gene ID | Property | Reference |
|---|---|---|---|---|
| Promoters | HSP70A | Cre08.g372100 (196 bp upstream to ATG) | Typically used as constitutive; expression can be enhanced by high light and temperature | Schroda et al. (2000) |
| RBCS2 or HSP70A/RBCS2 | Cre02.g120150 (180 bp upstream to ATG) | Strong constitutive (refer to HSP70A) | Lumbreras et al. (1998) | |
| PSAD | Cre05.g238332 (822 bp upstream to ATG) | Typically employed as strong constitutive; expression maybe enhanced by high light | Fischer and Rochaix (2001) | |
| CYC6 | Cre16.g651050 (127 bp upstream to ATG) | Metal (Cu) responsive | Quinn and Merchant (1995) | |
| NIT1 | Cre09.g410950 (282 bp upstream to ATG) | Ammonium responsive | Ohresser et al. (1997) | |
| ATX1 | Cre09.g392467 (532 bp upstream to ATG) | Iron (Fe) responsive | Fei and Deng (2007) | |
| CA1 | Cre05.g248400 (194 bp upstream to ATG) | CO2 responsive | Villand et al. (1997) | |
| SQD2 | Cre01.g038550 (75 bp upstream to ATG) | Phosphate (P) responsive | Iwai et al. (2014) | |
| CAMV 35S | – | Enhancer and minimal promoter only | Ruecker et al. (2008) | |
| METE | Cre03.g180750 (–574 to –89 bp from ATG) | Cobalamin (B12) suppression | Helliwell et al. (2014) | |
| Regulatory elements | RBCS2 intron 1, 2 and 3 | Cre02.g120150 | Enhance gene expression as intron in coding region or 5′UTR | Eichler-Stahlberg et al. (2009), Lumbreras et al. (1998) |
| THI4 | Cre04.g214150 (1414 bp 5′UTR) | 5′UTR, thiamine (B1) suppression | Croft et al. (2007), Moulin et al. (2013) | |
| RBCS2 | 32 aa | Rubisco small subunit 2 chloroplast transit peptide | León et al. (2007) | |
| Functional peptides | FD | 32 aa | Ferredoxin chloroplast targeting transit peptide | León et al. (2007) |
| ARS2 | Cre16.g671350 (63 bp 5′ end from ATG) | N-terminal secretion | Eichler-Stahlberg et al. (2009) | |
| 2 x simian virus 40 (SV40) | 20 aa | N- terminal nuclear target peptide | Rasala and Mayfield (2014) | |
| Foot-and-mouth disease virus (FMDV) 2A | 20 aa | Translational cleavage peptide | Rasala et al. (2012) | |
| Gaussia luciferase (gLuc) | 555 bp | Luciferase assay | Ruecker et al. (2008) | |
| Reporter genes | GFP, mCHERRY, EYFP, DsRED, tdTOMATO, VENUS | 708–1437 bp | Fluorescent protein | Rasala et al. (2013) |
| ARS | Cre16.g671400 (3012 bp cDNA) | Chromogenic assay | Davies et al. (1992) | |
| Highly expressed host strains | UVM4 and UVM11 | – | High expression host, transgene silencing supressed | Neupert et al. (2009) |
Several different reporter genes are available, both fluorescent and colorimetric, and in most instances these have been modified for expression in C. reinhardtii; the unusually high GC content of 65% (Merchant et al., 2007) means that most heterologous genes need to be codon optimised before they are expressed efficiently (e.g. Fuhrmann et al., 1999). Expressed proteins can be directed to different subcellular locations, or for secretion, using characterised targeting peptides, and tags for immunodetection or purification of expressed proteins have been developed (Eichler-Stahlberg et al., 2009). Eukaryotic cleavage peptides, such as viral 2A peptides (Rasala et al., 2012), have been demonstrated to work in C. reinhardtii, thus in principle allowing expression of two or more proteins from a single transgene construct. This would overcome potential difficulties in the limited number of promoters available, repeated use of which would be likely to increase the problems of gene silencing.
In the last 2 years, reports of advanced techniques for genome editing of C. reinhardtii have appeared. The first demonstration of genome editing was via zinc finger nucleases, which work by fusing a DNA binding domain from naturally occurring zinc finger nuclease with a DNA cleavage domain. Sizova et al. (2013) transformed C. reinhardtii with a heterologous non-functional APHVIII gene, interrupted with a 24-bp COP3 sequence, the target of the zinc finger nuclease. Subsequent transformation with the gene encoding the zinc finger nuclease, expressed from the HSP70A promoter, and a 120-bp sequence of DNA complementary to the non-functional region of APHVIII, followed by selection for paromomycin resistance, generated several colonies, indicating restoration of APHVIII gene function. No spontaneous resistance was apparent in the controls. The methodology was subsequently used to introduce targeted mutations into the native COP3 gene. The second example of genome editing employed Transcription Activator-Like Effectors (TALE) for specific activation of target genes (Gao et al., 2014). In this work, the endogenous arylsulfatase genes, ARS1 and ARS2, were targeted via their promoters. Nuclear expression of the respective TALE, from a HSP70A/RBCS2 promoter and RBCS2 terminator cassette increased the expression of each ARS at the level of both the transcript and protein, a fact confirmed by an assay of arylsulfatase activity. Finally the efficacy of the Clustered Regularly Interspersed Short Palindromic repeat (CRISPR) system was tested (Jiang et al., 2014). CRISPR associated protein 9 (CAS9) is an RNA-guided DNA nuclease that has been employed to introduce targeted mutations into eukaryotic genomes. No stable transformants of C. reinhardtii expressing the CAS9 protein could be recovered, suggesting that the protein was toxic (Jiang et al., 2014). As a result the authors employed transient expression in an attempt to orchestrate targeted mutagenesis of four independent gene targets, HYG, mutant green fluorescent protein (mGFP), Gaussia luciferase (gLUC) and peptidyl-prolyl cis-trans isomerase (FKB12). A PCR assay was employed to enrich for and detect the presence of Cas9 mediated mutations. Although the system demonstrated low efficiency, with only a single FKB12 mutant obtained, it provides a benchmark for future research in this area.
In spite of our ability to manipulate the genome of C. reinhardtii, few examples of actual metabolic engineering are available in the literature. Of these, engineered expression of native genes for diacylglycerol acyltransferases (DGATs) provides a good example of the current status. In this, three type-2 DGAT genes were expressed constitutively with the PSAD promoter and 3′UTR, and their impact on TAG biosynthesis and profile analysed (La Russa et al., 2012). Although expression varied due to random integration, DGAT mRNA levels were increased 1.7- to 29.1-fold compared to controls. In spite of this no significant increase in lipid biosynthesis was observed, possibly due to induction of lipid degradation pathways (Nguyen et al., 2011). A second example is provided by the attempt to engineer C. reinhardtii to make the keto-carotenoids, astaxanthin and canthaxanthin. Constitutive expression of a β-carotene ketolase gene (BKT1), derived from the green microalga Haematococcus pluvialis, in the C. reinhardtii nucleus using the RBCS2 promoter, combined with targeting to the chloroplast via RBCS2 or ferredoxin (FD) transit peptides (Table2) resulted in the undesired biosynthesis of 4-keto-lutein (León et al., 2007). Possible reasons for this failure include lack of substrate accessibility in the thylakoid membranes where the carotenoids are synthesised, and/or the inability of the heterologous β-ketolase to integrate into the carotenogenic enzyme complexes. For both this, and the example of expression of DGATs, optimisation would require many different genetic constructs to be tested, including down-regulation of endogenous pathways as well as expression of transgenes in the same cell line. The current tools available for manipulation of C. reinhardtii limit considerably the ability to carry out these combinatorial studies.
Engineering the C. reinhardtii Chloroplast
From an engineering perspective, the chloroplast provides a unique cellular compartment that harbours numerous essential processes, including those of biotechnological importance such as carotenoid and fatty-acid biosynthesis. The chloroplast genomes of more than 20 species are now accessible to genetic modification (Day and Goldschmidt-Clermont, 2011; Maliga, 2012). However, chloroplast engineering is routine for only three species, Nicotiana tabacum (Golds et al., 1993), N. benthamiana (Davarpanah et al., 2009), C. reinhardtii (Blowers et al., 1989). This further highlights the importance of C. reinhardtii as an IB platform. Working with the chloroplast provides a number of unique advantages, for example, the chloroplast genome is derived from a cyanobacterial genome that has been reduced by two orders of magnitude (Scharff and Bock, 2014). Its prokaryotic origin allows genes to be co-ordinated into operons, and to integrate DNA in a targeted manner via homologous recombination, so that mutant phenotypes can be complemented and resistance markers recycled to generate ‘marker-less’ strains (Fischer et al., 1996; Bateman and Purton, 2000) (Table3). Moreover, some established bacterial resources also work in the C. reinhardtii chloroplast, including the lac repressor system (Kato et al., 2007). This is useful as it has been reported that native promoters maybe compromised by tight regulation of essential photosynthetic processes. For example, the promoter and 5′UTR of the psbA gene, which encodes the D1 subunit of photosystem II, can only be used in a psbA-deficient background because of D1-dependent auto-repression (Rasala et al., 2011). In spite of this, some combinations of promoter and 5′UTR have been identified that achieve efficient transgene expression in photosynthetic backgrounds, such as the 16S rRNA promoter-atpA 5′UTR hybrid (Rasala et al., 2011), and there are several reports of expression of single proteins being produced within the C. reinhardtii chloroplast (Tissot-Lecuelle et al., 2014). However, examples of metabolic engineering are much fewer in number. The most relevant example is that of enhanced bio-hydrogen production. Wu et al. (2011) expressed codon optimised hemH and lba to enhance cellular respiration, reducing the dissolved oxygen concentration in the media, thereby causing an up regulation of hydrogenase genes hydA1 and hydA2. Nonetheless, as with examples in the nucleus, improvements obtained through chloroplast metabolic engineering are limited compared with what has been achieved in other biological platforms.
Table 3.
Regulatory elements available for the C. reinhardtii chloroplast. Chloroplast functional parts are classified as transcriptional leaders (promoter and 5′UTR), chloroplast-targeted peptides, selectable markers and destination sequences for homologous recombination
| Functional element | Name | Properties | Source |
|---|---|---|---|
| Transcriptional leaders | AtpA, psbD | Constitutive expression | Barnes et al. (2005) |
| psaA-exon1 | Strong expression | Michelet et al. (2011) | |
| Chloroplast-targeted peptides | psaD | 35 aa N-terminal | Fischer and Rochaix (2001) |
| rbcS2, FD | 32 aa N-terminal | León et al. (2007) | |
| Selectable marker | rrnS and rrnL point mutations | Spectomycin and erythromycin resistance | Kindle et al. (1991) |
| arg9 | Arginine complementation | Remacle et al. (2009) | |
| aadA | Spectinomycin resistance | Goldschmidt-Clermont (1991) | |
| aphA6 | Resistance kanamycin | Bateman and Purton (2000) | |
| GFP | Fluorescent reporter | Franklin et al. (2002) | |
| gusA | β-Glucuronidase activity | Sakamoto et al. (1993) | |
| atpB | 5-Fluorodeoxyuridine resistance | Kindle et al. (1991) | |
| coda | Cytosine deaminase sensitivity to 5-fluorocytosine | Young and Purton (2014) | |
| Destination sequences for homologous recombination | p322 | psbA and 16S rRNA | Manuell et al. (2007), Barnes et al. (2005) |
| patpint-cg11 (atpB-int) | Inverted repeat and atpB 3′UTR | Nickelsen et al. (1994) | |
| pLM7 (IR-int) | psbA and 5S/23S | Michelet et al. (2011) | |
| p72B | psbH and psbN | Bateman and Purton (2000) | |
| p71 | tscA and inverted repeat | Kindle et al. (1991) |
Harnessing Biological Systems for Technology
Industrial biotechnology employs biological systems, mainly microbes, for the production of commodities. Today these range from fuels, to platform chemicals, and many high-value products, including a vast range of therapeutics and pharmaceuticals. IB has its origins in fermentation practices for beer, wine and breadmaking, but as microbiology emerged as a science, the ability to control these technologies proceeded in parallel, and led to many important compounds being made this way (reviewed in Buchholz and Collins, 2013). A noteworthy example is penicillin from the fungus Penicillium notatum, as well as several other naturally occurring antibiotics from soil bacteria. The advent of genetic manipulation enabled the optimisation of biological production systems and enormously expanded the portfolio of compounds that could be produced by IB, including the ability to produce human insulin and growth hormone as recombinant proteins, avoiding the side effects of use of animal forms of the hormones (Vajo et al., 2001). Today, as well as pharmaceuticals, products from IB are an everyday feature of life, from vegetarian cheeses made with recombinant rennet, to plastics made from platform chemicals, to enzymes in washing powders (Goel et al., 2012).
The best known IB platforms are E. coli and S. cerevisiae, their utility being derived from decades of research that has progressed through various stages, ranging from the introduction of individual genes using standard expression systems, to the regulation of complex biochemical pathways using highly regulated genetic circuits (Figure2). Parallel to the development of tools for manipulation, the application of systems biology and GSMMs have enabled rational modification of discrete pathways (Yu et al., 2011; Chen and Zeng, 2013), whilst improvement of platform strains with respect to growth and productivity within the fermenter, as well as downstream processing, has led to optimised hosts for different bioprocesses (Goel et al., 2012). This progression is demonstrated by the work on the optimisation of E. coli strains expressing a gene encoding amorphodiene synthase from Artemisia annua, which catalyses the conversion of the isoprenoid intermediate farnesyl pyrophosphate into amorphodiene. Starting with a landmark metabolic engineering paper, Martin et al. (2003) introduced a yeast mevalonate pathway into E. coli to enhance production of the isoprenoid precursor, isopentenyl pyrophosphate. This increased amorphodiene biosynthesis 36-fold, to 24 μg mL−1. This strain was employed in several subsequent studies to achieve iterative improvements in target compound production by elucidating metabolic limitations in precursor biosynthesis and improving carbon flux towards the end products. First, expression of the mevalonate pathway gene 3-hydroxy-3-methyl-glutaryl-coenzyme A (HMG-CoA) reductase was modulated to improve precursor biosynthesis leading to a three-fold (ca. 70 μg mL−1) increase over the original strain (Pitera et al., 2007). Following this, mevalonate kinase and amorphadiene synthase were identified as rate limiting, and optimisation of promoter strength and gene copy number increased productivity seven-fold (ca. 160 μg mL−1) over the original construct (Anthony et al., 2009). This moved the metabolic bottleneck back to HMG-CoA reductase, and the introduction and optimisation of an alternative HMG-CoA reductase, derived from Delftia acidovorans, led to levels of 700 μg mL−1 amorphodiene (Ma et al., 2011). Most recently, Dahl et al. (2013) identified farnesyl pyrophosphate as a toxic intermediate and introduced dynamic control to regulate its synthesis and consumption. The application of systems biology methods identified promoters induced in response to intermediate accumulation, and these were employed to drive the expression of both the farnesyl pyrophosphate biosynthetic pathway and the amorphadiene biosynthetic genes, resulting in a doubling of the amorphadiene titre, to 1.6 mg mL−1. This approach is now routine in IB where iterative improvements enable optimisation of inputs and gene expression to maximise the economic viability of a bioprocess.
Figure 2.
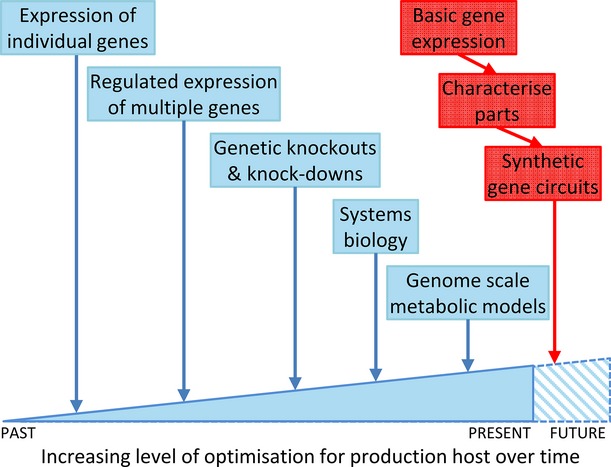
Schematic of host optimisation via metabolic engineering and synthetic biology.The chronologic development of a biological host via metabolic engineering (blue), including the recent advent of systems biology and genome-scale metabolic models. Synthetic biology (red) has the potential to expedite this process in new biological systems, including microalgae, providing an opportunity to proceed directly from basic knowledge and capacity to the generation of highly optimised productive hosts.
Tangential to these developments has been the rise of synthetic biology. Much of the current focus of this discipline is on the development and analysis of genetic circuits, emergent behaviour and minimal genomes (Juhas et al., 2011), as well as the generation of non-natural biological systems with an expanded genetic code (Neumann and Neumann-Staubitz, 2010). Nonetheless, the potential for synthetic biology to contribute to IB is significant (Khalil and Collins, 2010; Yadav et al., 2012).
Synthetic Biology Allows Rational Design of Biological Systems
Synthetic biology draws on principles first employed in the electronics field, and aims to create or (re)design complex biological circuits, networks or whole organisms in a rational way, combining computational models with assembly of standard or modularised components, to create useful outputs. Synthetic biology is characterised by an iterative workflow of continuous improvement, with generated knowledge informing subsequent rounds of design (shown schematically in Figure3a). The basis is the use of standard parts (Figure3b) for the design and engineering of novel biological systems, in this example DNA sequences such as promoters, terminators or introns. These parts or pathways may be described as orthogonal, that is they are transferrable between different cellular hosts with little/no impact on function (i.e. platform independent). They may be insulated, so they are unaffected by host cell metabolism, while an approach may be said to lead to abstraction, allowing design at higher levels of complexity because work builds upon well characterised inputs. This makes knowledge transferable, as a combination of elements that lead to a predetermined expression level of a transgene could be employed in subsequent studies to drive the expression of other transgenes. The aim is that, once sufficiently well characterised, the design process no longer needs to consider individual DNA parts. Instead these may be combined into expression cassettes or devices (Figure3c) with novel but predictable outputs. This will allow the field to progress more efficiently by building directly upon work that preceded it. Table4 provides a summary of other commonly used synthetic biology terms.
Figure 3.
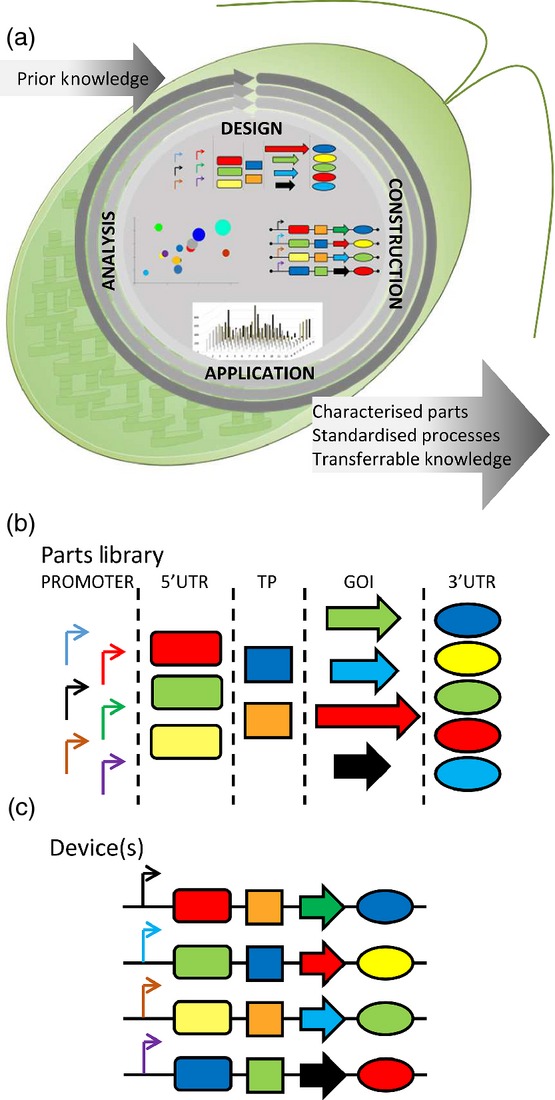
Schematic representation of a synthetic biology workflow.An example investigation may aim to increase metabolite x through heterologous expression of gene y. Prior to this knowledge must be generated to describe the function of individual parts, promoters, 5′UTRs, transit peptides (TP), (trans)genes of interest (GOI) and 3′UTRs, and/or introns to enhance mRNA stability. Achieved through progress from design through construction application and analysis (including in silico models), when completed in a standardised manner this represents a single iteration of the cyclic process (a). The completion of several rounds of characterisation generates the knowledge required assign functional parameters to individual (generic) parts. These parts can be considered in a discrete manner (b) providing the opportunity to deconstruction of complex modules and recombine the parts a rational manner to generate novel functional devices (c) that have a predictable output.
Table 4.
Summary of common synthetic biology terms
| Keyword | Description |
|---|---|
| Abstraction | Through standardisation of individual parts these may be combined into simple devices with predictable outputs, combination of these devices allows design at a higher level of complexity, i.e. circuits or systems. This is the concept of abstraction |
| Boolean logic gates | An engineering principle in which an output is dependent on one or more specific inputs, and where the output is 1 or 0, true or false, on or off |
| Chassis | The framework on which to base the standardised parts. Effectively, a host organism into which standard genetic constructs can be introduced easily |
| Device | A DNA construct incorporating required part(s) for desired expression of a transgene |
| Modularity | A design concept in which parts are considered as discrete elements. Characterised parts can be combined to create an expression cassette which, due to its predictable function, is considered a modular element in its own right, becoming a part |
| In-silico modelling | The application of informed metabolic models to inform experimental design and allow hypothesis-driven research into complex systems |
| Orthogonal | The function of a DNA part independently of: (i) cellular platform; and/or (ii) cellular context |
| Part(s) | DNA sequence(s) that encode a biological function, e.g. promoter, intron, 3′UTR, reporter gene |
| Standardisation | The application of uniformed strategies to assess, characterise or validate biological parts or processes. Examples include assembly protocols, reporter assays, or selection methods |
Synthetic biology in action – Escherichia coli
Escherichia coli is the best understood and most advanced biological platform available, demonstrating great utility for the development of ground-breaking molecular, genetic and biochemical techniques. As such it has been widely employed to pioneer synthetic biology (Cameron et al., 2014). For example, there is now a Registry of Standard Biological Parts, or Biobricks (Knight, 2003), the number of which now exceeds 12 000 (Vilanova and Porcar, 2014). Public access allows researchers to explore novel functionalities and applications, in return for contributing to the centralised knowledge database. This foundation provides the ability to engineer strains and create genetic circuits, such as the use of a two-component sensor and response system where genes were regulated in response to different light wavelengths (Olson et al., 2014). The system was composed of a histidine kinase with an N-terminal phytochrome domain sensor and a C-terminal bifunctional kinase-phosphatase signalling domain. Using biological information and modelling, the system was manipulated to regulate protein expression in response to different wavelengths and pulse profiles of light using an automated illumination program. The binary states demonstrated by this system provide a basis for future logic based gene circuits.
Escherichia coli synthetic biology has already advanced to population-scale regulation using synthetic networks. An example of this is the synchronisation of genetic clocks by post-translational events. Prindle et al. (2014) employed intracellular negative-feedback and a quorum-sensing oscillator in combination with proteins tagged for rapid protease catalysed degradation via the CLPXP signal. By coupling these processes the authors engineered a system in which the response delay was reduced by an order of magnitude over systems coupled at the level of transcription. Modification of the linker sequence length between the peptide and the degradation tag made the timing of the response tunable, offering the potential to create optimised expression profiles for synthetic gene circuits. As an illustration, a useful application of this for metabolic engineering would be regulation of key mevalonate biosynthesis genes in the amorphodiene strain discussed above.
Synthetic biology in action – Saccharomyces cerevisiae
The major chassis in eukaryotes is S. cerevisiae. This unicellular yeast benefits from numerous community resources that support fundamental research, including knock-out collections in which each protein-coding region has been systematically disrupted both individually (Dujon, 1998; Winzeler et al., 1999), and in pairs (Tong et al., 2001), allowing investigation of lethality and gene relationships. Aspects of S. cerevisiae research have moved to adopt the principles of synthetic biology. A pioneering example of this is the creation of an entirely artificial yeast chromosome (Annaluru et al., 2014), engineered to streamline and condense the encoded genetic information and allow re-organisation of coding regions on a massive scale. This paves the way for rapid laboratory-based evolution to generate novel phenotypes with biotechnological relevance. Further examples of how synthetic biology has been applied to S. cerevisiae include the development of novel synthetic inducible promoters, regulated by a ligand that does not influence cell physiology (i.e. is insulated), which are also tunable. McIsaac et al. (2014) constitutively expressed an artificial transcription factor (ATF) in a cell line harbouring a green fluorescent protein (GFP) reporter that was regulated via a synthetic promoter containing one or several binding sites for the ATF. The reporter was expressed in the presence of the inducer, β-estradiol, and modification to the number and position of the ATF binding sites provided tunable expression upon induction, with a dynamic range that spanned three orders of magnitude.
In another study, Yofe et al. (2014) investigated the ability of non-coding sequences to regulate gene expression in S. cerevisiae. Creating a library of 240 reporter lines, each containing a different intron within the reporter gene coding sequence, the authors observed variation in expression spanning 100-fold. These lines demonstrated robustness in expression level and a capacity for tunable gene expression, achieved through the modification of key intron motifs and/or the folding energy of the intron-exon boundary. Finally, to allow rational design, the authors developed a computational model that fitted the experimental performance of tested sequences.
To demonstrate the power of a combinatorial approach, the five genes of the uncharacterised violacein biosynthetic pathways were targeted. In this, five promoters which spanned nearly three orders of magnitude in expression level were employed to drive expression of the violacein biosynthetic genes from BBa_K274002, a plasmid from the Registry of Standard Biological Parts (Knight, 2003). A rapid PCR genotyping method combined with computational modelling to integrate experimental data enabled the prediction of pathway compositions that would minimise intermediate accumulation and/or maximise violacein biosynthesis. In doing so the authors demonstrated pathway optimisation without prior knowledge of absolute protein or metabolite levels, enzyme kinetics or thermodynamics, or even the order of the reactions (Lee et al., 2013).
Development of a C. reinhardtii Industrial Biotechnology Platform
The approaches presented above provide an overview of what is currently possible with well characterised IB platforms, and how a synthetic biology approach would augment both the range and amount of compounds that could be produced, and the rate of progress. For C. reinhardtii, although our ability to manipulate it genetically is advancing rapidly, many limitations remain for effective metabolic engineering. For example, fewer than 100 protein-coding genes from the C. reinhardtii genome have been functionally validated, compared with the approximately 6800 genes that have been characterised experimentally in Arabidopsis thaliana (Koornneef et al., 2004). Additionally, the number of tools to regulate transgene expression (Tables2 and 3) is small compared with standard IB hosts. There are also the well known complications of integration of transgenes into the nucleus via non-homologous end joining, and extensive gene silencing. A significant additional challenge is the fact that regulatory networks at the genetic, cellular, and metabolic levels in C. reinhardtii are almost completely unexplored, so that attempts at manipulation will be effectively empirical, whereas there are many studies of the interactome of E. coli (Juan et al., 2008) and S. cerevisiae (Ito et al., 2001). Although undoubtedly some of these knowledge gaps for C. reinhardtii will be filled in the near future (as discussed above), it should be noted that no biological chassis evolved to be optimal for scientific research, with extensive engineering required to create today's E. coli strains for routine protein expression (Goel et al., 2012; Buchholz and Collins, 2013). Thus rather than waiting for this to be true for C. reinhardtii, would it be possible to expedite progress by applying current leading concepts, techniques and technologies from synthetic biology? This might lead to quicker solutions to current research questions and, importantly, provide the opportunity to develop C. reinhardtii production systems that exceed the capacity of current metabolic engineering approaches. Adopting standardisation, modularity, and design/testing does not require drastic changes in approach, nor does it render all prior knowledge obsolete. Instead this progression can be achieved by subtle changes to minimise experimental variables and standardise workflows, creating a cycle of perpetual improvement.
To demonstrate this, let us consider an example of improved TAG biosynthesis for biodiesel production (Figure4). When cultured under diurnal conditions, i.e. with a light/dark photoperiod, C. reinhardtii will photosynthesise during the light, fixing carbon dioxide and storing any excess as starch. In the dark the accumulated starch is consumed to support cellular activity. When exposed to nutrient stress, such as nitrogen depletion, C. reinhardtii accumulates much higher amounts of storage molecules, principally starch and TAG (Merchant et al., 2012). Under conditions where the desired product is TAG, starch biosynthesis may be considered an undesired carbon sink. This principle was demonstrated through the analysis of mutants deficient in starch biosynthesis (Wang et al., 2009). However, starchless mutants are not able to be propagated under diurnal conditions because they lack the required starch to survive periods of darkness (Davey et al., 2014). To overcome this and optimise TAG production one may therefore consider a synthetic circuit that links nitrogen depletion and starch biosynthesis in a manner that would enhance carbon flux into TAG production only under nitrogen stress (shown schematically in Figure4a). Employing the workflow presented in Figure3, a regulatory system could be identified and used to regulate the expression of a starch biosynthesis gene in response to light or nitrogen depletion. Once suitable elements are identified data describing expression characteristics could be integrated into an in silico model to generate predicted outputs (Figure4b), and then tested to find the optimal combination of parts for the device. The next stage would be to engineer the system such that TAG production was induced independently of nitrogen depletion. Complementation of a starchless phenotype using this regulatory system would restore starch biosynthesis to that of a wild-type strain, with expression induced in response to light availability (input; light). Because the same system would be down-regulated in response to nitrogen stress (input; nitrogen depletion), this would over-ride the light-based regulation (OR gate logic). The designed circuit would, when exposed to nitrogen stress, suppress starch biosynthesis as native TAG production pathways are induced, producing more TAG because of increased fixed carbon availability.
Figure 4.
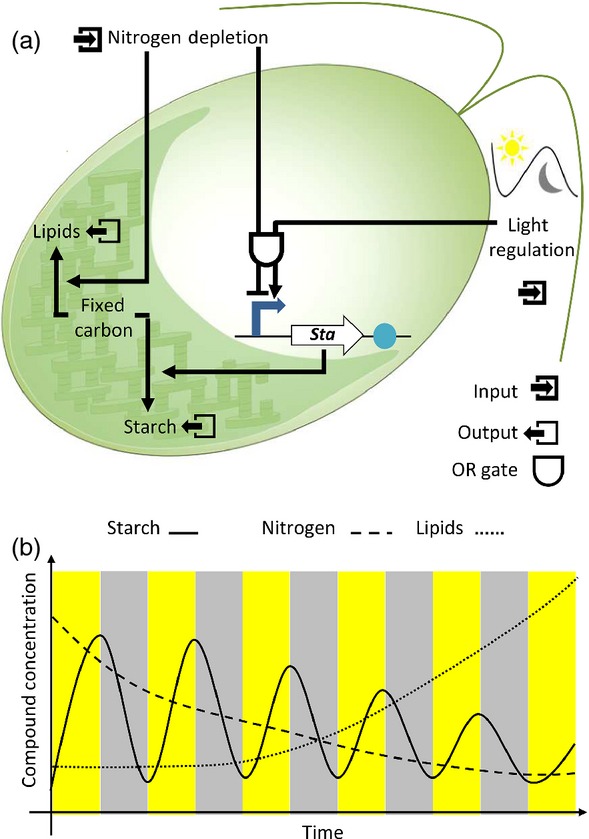
A schematic of synthetic biology applied to create a circuit for enhanced lipid production.(a) A representation of a synthetic gene circuit in a starchless mutant cell line. Starch biosynthesis is complemented by the STA gene, in a light-regulated manner (Light; input). This regulation is overruled under nitrogen stress (nitrogen; input) through an OR logic gate. Down-regulation of starch biosynthesis coincides with the induction of native TAG biosynthetic pathways, providing a larger substrate pool for increased TAG production.(b) A schematic model employed to correlate the relationship of starch, nitrogen and lipids with the inputs of light and cellular nitrogen concentration.
Conclusion
Microalgae are of interest to academic and commercial stakeholders due to their unique biology, which encompasses facets of bacterial and yeast systems, and their potential as a biotechnology platform. Although the microalgal research field is less well developed than for the former organisms, the model green alga, C. reinhardtii, is positioned for development as an IB platform. It is evident that the major limitation now lies in our capacity to engineer C. reinhardtii efficiently. By taking a synthetic biology approach, progress within the field should be accelerated.
One final point to make is that the diversity of microalgae means that C. reinhardtii does not provide all of the attributes assigned to ‘microalgae’. That said, its development as an IB platform would undoubtedly benefit the entire microalgal community, and pave the way for subsequent more efficient development of alternative microalgal hosts of biotechnological importance, including P. tricornutum, Thalassiosira pseudonana, O. tauri and Nannochloropsis spp. (Table S1). These will offer a wider range of products, and possibly other advantages such as homologous recombination, which has been reported for the latter two species. It is important to acknowledge that although E. coli and S. cerevisiae are the best known IB platforms, many other bacteria and yeasts are employed in million dollar biotechnology sectors for the production of value-added products. The time is right to consider extending this to microalgae to address the desire to improve the sustainability of these processes (Langevels et al., 2010).
Acknowledgments
The authors declare no conflict of interest. M.A.S and J.R were funded by the UK Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/I00680X/1, M.A.S was also funded by the European Commission 7th Framework Programme (FP7) project SPLASH (Sustainable PoLymers from Algae Sugars and Hydrocarbons), grant agreement number 311956. G.T.D.T.N was funded in part by Murray Edwards College and the Cambridge Philosophical Society. D.L. was funded by the Bill and Melinda Gates Foundation, and K.E.H was funded by BBSRC grant BB/I013164/1.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Algal-biotechnological advancements from a phylogenetic perspective.
Overview of microalgae that are transformable and/or have a published genome
References
- Allmer J, Naumann B, Markert C, Zhang M, Hippler M. Mass spectrometric genomic data mining: novel insights into bioenergetic pathways in Chlamydomonas reinhardtii. Proteomics. 2006;6:6207–6220. doi: 10.1002/pmic.200600208. [DOI] [PubMed] [Google Scholar]
- Annaluru N, Muller H, Mitchell LA, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony JR, Anthony LC, Nowroozi F, Kwon G, Newman JD, Keasling JD. Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4,11-diene. Metab. Eng. 2009;11:13–19. doi: 10.1016/j.ymben.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Baltz A, Dang K-V, Beyly A, Auroy P, Richaud P, Cournac L, Peltier G. Plastidial expression of type II NAD(P)H dehydrogenase increases the reducing state of plastoquinones and hydrogen photoproduction rate by the indirect pathway in Chlamydomonas reinhardtii. Plant Physiol. 2014;165:1344–1352. doi: 10.1104/pp.114.240432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D, Franklin S, Schultz J, Henry R, Brown E, Coragliotti A, Mayfield SP. Contribution of 5'- and 3'-untranslated regions of plastid mRNAs to the expression of Chlamydomonas reinhardtii chloroplast genes. Mol. Genet. Genomics. 2005;274:625–636. doi: 10.1007/s00438-005-0055-y. [DOI] [PubMed] [Google Scholar]
- Bateman JM, Purton S. Tools for chloroplast transformation in Chlamydomonas: expression vectors and a new dominant selectable marker. Mol. Gen. Genet. 2000;263:404–410. doi: 10.1007/s004380051184. [DOI] [PubMed] [Google Scholar]
- Bellou S, Baeshen MN, Elazzazy AM, Aggeli D, Sayegh F, Aggelis G. Microalgal lipids biochemistry and biotechnological perspectives. Biotechnol. Adv. 2014;32:1476–1493. doi: 10.1016/j.biotechadv.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Berla BM, Saha R, Immethun CM, Maranas CD, Moon TS, Pakrasi HB. Synthetic biology of cyanobacteria: unique challenges and opportunities. Front. Microbiol. 2013;4:246. doi: 10.3389/fmicb.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold P, Schmitt R, Mages W. An Engineered Streptomyces hygroscopicus aph 7” Gene Mediates Dominant Resistance against Hygromycin B in Chlamydomonas reinhardtii. Protist. 2002;153:401–412. doi: 10.1078/14344610260450136. [DOI] [PubMed] [Google Scholar]
- Blaby IK, Blaby-Haas CE, Tourasse N, et al. The Chlamydomonas genome project: a decade on. Trends Plant Sci. 2014;19:672–680. doi: 10.1016/j.tplants.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blowers AD, Bogorad L, Shark KB, Sanford JC. Studies on Chlamydomonas chloroplast transformation: foreign DNA can be stably maintained in the chromosome. Plant Cell. 1989;1:123–132. doi: 10.1105/tpc.1.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle N, Morgan J. Flux balance analysis of primary metabolism in Chlamydomonas reinhardtii. BMC Syst. Biol. 2009;3:4. doi: 10.1186/1752-0509-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J, Gillham N, Harris E, et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Buchholz K, Collins J. The roots – a short history of industrial microbiology and biotechnology. Appl. Microbiol. Biotechnol. 2013;97:3747–3762. doi: 10.1007/s00253-013-4768-2. [DOI] [PubMed] [Google Scholar]
- Cameron DE, Bashor CJ, Collins JJ. A brief history of synthetic biology. Nat. Rev. Microbiol. 2014;12:381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- Casas-Mollano JA, Rohr J, Kim E-J, Balassa E, van Dijk K, Cerutti H. Diversification of the core RNA interference machinery in Chlamydomonas reinhardtii and the role of DCL1 in transposon silencing. Genetics. 2008;179:69–81. doi: 10.1534/genetics.107.086546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzaniga S, Dall'Osto L, Szaub J, Scibilia L, Ballottari M, Purton S, Bassi R. Domestication of the green alga Chlorella sorokiniana: reduction of antenna size improves light-use efficiency in a photobioreactor. Biotechnol. Biofuel. 2014;157:7. doi: 10.1186/s13068-014-0157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang RL, Ghamsari L, Manichaikul A, et al. Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism. Mol. Syst. Biol. 2011;7:518. doi: 10.1038/msb.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauton MS, Reitan KI, Norsker NH, Tveterås R, Kleivdal HT. A techno-economic analysis of industrial production of marine microalgae as a source of EPA and DHA-rich raw material for aquafeed: research challenges and possibilities. Aquaculture. 2015;436:95–103. [Google Scholar]
- Chen Z, Zeng A-P. Protein design in systems metabolic engineering for industrial strain development. Biotechnol. J. 2013;8:523–533. doi: 10.1002/biot.201200238. [DOI] [PubMed] [Google Scholar]
- Chisti Y. Biodiesel from microalgae. Biotechnol. Adv. 2007;25:294–306. doi: 10.1016/j.biotechadv.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Christian N, May P, Kempa S, Handorf T, Ebenhöh O. An integrative approach towards completing genome-scale metabolic networks. Mol. Biosyst. 2009;5:1889–1903. doi: 10.1039/B915913b. [DOI] [PubMed] [Google Scholar]
- Cogne G, Rügen M, Bockmayr A, Titica M, Dussap CG, Cornet JF, Legrand J. A model-based method for investigating bioenergetic processes in autotrophically growing eukaryotic microalgae: application to the green algae Chlamydomonas reinhardtii. Biotechnol. Prog. 2011;27:631–640. doi: 10.1002/btpr.596. [DOI] [PubMed] [Google Scholar]
- Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proc. Natl Acad. Sci. USA. 2007;104:20770–20775. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl RH, Zhang F, Alonso-Gutierrez J, et al. Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotechnol. 2013;31:1039–1046. doi: 10.1038/nbt.2689. [DOI] [PubMed] [Google Scholar]
- Dal'Molin CG, Quek LE, Palfreyman RW, Nielsen LK. AlgaGEM–a genome-scale metabolic reconstruction of algae based on the Chlamydomonas reinhardtii genome. BMC Genom. 2011;12(Suppl. 4):S5. doi: 10.1186/1471-2164-12-S4-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davarpanah SJ, Jung SH, Kim YJ, Park Y-I, Min SR, Liu JR, Jeong WJ. Stable plastid transformation in Nicotiana benthamiana. J. Plant Biol. 2009;52:244–250. [Google Scholar]
- Davey MP, Horst I, Duong G-H, Tomsett EV, Litvinenko ACP, Howe CJ, Smith AG. Triacylglyceride production and autophagous responses in Chlamydomonas reinhardtii depend on resource allocation and carbon source. Eukaryot. Cell. 2014;13:392–400. doi: 10.1128/EC.00178-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Weeks DP, Grossman AR. Expression of the arylsulfatase gene from the f32-tubulin promoter in Chlamydomonas reinhardtii. Nucleic Acids Res. 1992;20:2959–2965. doi: 10.1093/nar/20.12.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A, Goldschmidt-Clermont M. The chloroplast transformation toolbox: selectable markers and marker removal. Plant Biotechnol. J. 2011;9:540–553. doi: 10.1111/j.1467-7652.2011.00604.x. [DOI] [PubMed] [Google Scholar]
- Debuchy R, Purton S, Rochaix JD. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 1989;8:2803–2809. doi: 10.1002/j.1460-2075.1989.tb08426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell RG, Smith AG. Do red and green make brown?: perspectives on plastid acquisitions within chromalveolates. Eukaryot. Cell. 2011;10:856–868. doi: 10.1128/EC.00326-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. European Functional Analysis Network (EUROFAN) and the functional analysis of the Saccharomyces cerevisiae genome. Electrophoresis. 1998;19:617–624. doi: 10.1002/elps.1150190427. [DOI] [PubMed] [Google Scholar]
- Duvick J, Fu A, Muppirala U, Sabharwal M, Wilkerson MD, Lawrence CJ, Lushbough C, Brendel V. PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res. 2008;36:D959–D965. doi: 10.1093/nar/gkm1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler-Stahlberg A, Weisheit W, Ruecker O, Heitzer M. Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta. 2009;229:873–883. doi: 10.1007/s00425-008-0879-x. [DOI] [PubMed] [Google Scholar]
- Fang W, Si Y, Douglass S, Casero D, Merchant SS, Pellegrini M, Ladunga I, Liu P, Spalding MH. Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1. Plant Cell. 2012;24:1876–1893. doi: 10.1105/tpc.112.097949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei X, Deng X. A novel Fe deficiency-responsive element (FeRE) regulates the expression of atx1 in Chlamydomonas reinhardtii. Plant Cell Physiol. 2007;48:1496–1503. doi: 10.1093/pcp/pcm110. [DOI] [PubMed] [Google Scholar]
- Fischer N, Rochaix JD. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genomics. 2001;265:888–894. doi: 10.1007/s004380100485. [DOI] [PubMed] [Google Scholar]
- Fischer N, Stampacchia O, Redding K, Rochaix J-D. Selectable marker recycling in the chloroplast. Mol. Gen. Genet. 1996;251:373–380. doi: 10.1007/BF02172529. [DOI] [PubMed] [Google Scholar]
- Franklin S, Ngo B, Efuet E, Mayfield SP, May SP. Development of a GFP reporter gene for Chlamydomonas reinhardtii chloroplast. Plant J. 2002;30:733–744. doi: 10.1046/j.1365-313x.2002.01319.x. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M, Oertel W, Hegemann P. A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 1999;19:353–361. doi: 10.1046/j.1365-313x.1999.00526.x. [DOI] [PubMed] [Google Scholar]
- Gao H, Wright DA, Li T, Wang Y, Horken K, Weeks DP, Yang B, Spalding MH. TALE activation of endogenous genes in Chlamydomonas reinhardtii. Algal Res. 2014;5:52–60. [Google Scholar]
- Goel A, Wortel MT, Molenaar D, Teusink B. Metabolic shifts: a fitness perspective for microbial cell factories. Biotechnol. Lett. 2012;34:2147–2160. doi: 10.1007/s10529-012-1038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golds T, Maliga P, Koop H-U. Stable plastid transformation in PEG-treated protoplasts of Nicotiana tabacum. Biotechnology. 1993;11:95–97. [Google Scholar]
- Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of Chlamydomonas. Nucleic Acids Res. 1991;19:4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Blaby I, Casero D, et al. The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot. Cell. 2014;13:591–613. doi: 10.1128/EC.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton ML, Haslam RP, Napier JA, Sayanova O. Metabolic engineering of Phaeodactylum tricornutum for the enhanced accumulation of omega-3 long chain polyunsaturated fatty acids. Metab. Eng. 2014;22:3–9. doi: 10.1016/j.ymben.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. Chlamydomonas as a model organism. Plant Biol. 2001;52:363–406. doi: 10.1146/annurev.arplant.52.1.363. [DOI] [PubMed] [Google Scholar]
- Helliwell KE, Scaife MA, Sasso S, Araujo APU, Purton S, Smith AG. Unraveling vitamin B12-responsive gene regulation in algae. Plant Physiol. 2014;165:388–397. doi: 10.1104/pp.113.234369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippler M, Klein J, Fink A, Allinger T, Hoerth P. Towards functional proteomics of membrane protein complexes: analysis of thylakoid membranes from Chlamydomonas reinhardtii. Plant J. 2001;28:595–606. doi: 10.1046/j.1365-313x.2001.01175.x. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai M, Ikeda K, Shimojima M, Ohta H. Enhancement of extraplastidic oil synthesis in Chlamydomonas reinhardtii using a type-2 diacylglycerol acyltransferase with a phosphorus starvation-inducible promoter. Plant Biotechnol. J. 2014;12:808–819. doi: 10.1111/pbi.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Brueggeman AJ, Horken KM, Plucinak TM, Weeks DP. Successful transient expression of Cas9/sgRNA genes in Chlamydomonas reinhardtii. Eukaryot. Cell. 2014;13:1465–1469. doi: 10.1128/EC.00213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Alric J. Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryot. Cell. 2013;12:776–793. doi: 10.1128/EC.00318-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan D, Pazos F, Valencia A. High-confidence prediction of global interactomes based on genome-wide coevolutionary networks. Proc. Natl Acad. Sci. USA. 2008;105:934–939. doi: 10.1073/pnas.0709671105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Eberl L, Glass JI. Essence of life: essential genes of minimal genomes. Trends Cell Biol. 2011;21:562–568. doi: 10.1016/j.tcb.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Marui T, Kasai S, Shinmyo A. Artificial control of transgene expression in Chlamydomonas reinhardtii chloroplast using the lac regulation system from Escherichia coli. J. Biosci. Bioeng. 2007;104:207–213. doi: 10.1263/jbb.104.207. [DOI] [PubMed] [Google Scholar]
- Keasling JD. Synthetic biology and the development of tools for metabolic engineering. Metab. Eng. 2012;14:189–195. doi: 10.1016/j.ymben.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Keller LC, Romijn EP, Zamora I, Yates JR, Marshall WF. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat. Rev. Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K, Schnell R, Fernandez E, Lefebvre P. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J. Cell Biol. 1989;109:2589–2601. doi: 10.1083/jcb.109.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL, Richards KL, Stern DB. Engineering the chloroplast genome: techniques and capabilities for chloroplast transformation in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA. 1991;88:1721–1725. doi: 10.1073/pnas.88.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Marcuschamer D, Chisti Y, Benemann JR, Lewis D. A matter of detail: assessing the true potential of microalgal biofuels. Biotechnol. Bioeng. 2013;110:2317–2322. doi: 10.1002/bit.24967. [DOI] [PubMed] [Google Scholar]
- Kliphuis AM, Klok AJ, Martens DE, Lamers PP, Janssen M, Wijffels RH. Metabolic modeling of Chlamydomonas reinhardtii: energy requirements for photoautotrophic growth and maintenance. J. Appl. Phycol. 2012;24:253–266. doi: 10.1007/s10811-011-9674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok AJ, Lamers PP, Martens DE, Draaisma RB, Wijffels RH. Edible oils from microalgae: insights in TAG accumulation. Trends Biotechnol. 2014;32:521–528. doi: 10.1016/j.tibtech.2014.07.004. [DOI] [PubMed] [Google Scholar]
- Knight T. 2003. Idempotent vector design for standard assembly of Biobricks. MIT Synthetic Biology Working Group Technical Reports http://hdl.handle.net/1721.1/21168.
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- Kumar SV, Misquitta RW, Reddy VS, Rao BJ, Rajam MV. Genetic transformation of the green alga – Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Sci. 2004;166:731–738. [Google Scholar]
- Labadorf A, Link A, Rogers MF, Thomas J, Reddy AS, Ben-Hur A. Genome-wide analysis of alternative splicing in Chlamydomonas reinhardtii. BMC Genom. 2010;11:114. doi: 10.1186/1471-2164-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevels JWA, Dixon J, Jaworski JF. Development perspectives of the biobased economy: a review. Crop Sci. 2010;50:142–151. [Google Scholar]
- Lee ME, Aswani A, Han AS, Tomlin CJ, Dueber JE. Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res. 2013;41:10668–10678. doi: 10.1093/nar/gkt809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- León R, Couso I, Fernández E. Metabolic engineering of ketocarotenoids biosynthesis in the unicellular microalga Chlamydomonas reinhardtii. J. Biotechnol. 2007;130:143–152. doi: 10.1016/j.jbiotec.2007.03.005. [DOI] [PubMed] [Google Scholar]
- van Lis R, Atteia A, Mendoza-Hernández G, González-Halphen D. Identification of novel mitochondrial protein components of Chlamydomonas reinhardtii. A proteomic approach. Plant Physiol. 2003;132:318–330. doi: 10.1104/pp.102.018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras V, Stevens DR, Purton S. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 1998;14:441–447. [Google Scholar]
- Lv H, Qu G, Qi X, Lu L, Tian C, Ma Y. Transcriptome analysis of Chlamydomonas reinhardtii during the process of lipid accumulation. Genomics. 2013;101:229–237. doi: 10.1016/j.ygeno.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Ma SM, Garcia DE, Redding-Johanson AM, et al. Optimization of a heterologous mevalonate pathway through the use of variant HMG-CoA reductases. Metab. Eng. 2011;13:588–597. doi: 10.1016/j.ymben.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Maliga P. Plastid Transformation in Flowering plants. In: Bock R, Knoop VE, editors. Genomics of Chloroplasts and Mitochondria. Vol. 35. the Netherlands: Springer; 2012. pp. 393–414. [Google Scholar]
- Manichaikul A, Ghamsari L, Hom EF, et al. Metabolic network analysis integrated with transcript verification for sequenced genomes. Nat. Methods. 2009;6:589–592. doi: 10.1038/nmeth.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuell AL, Beligni MV, Elder JH, Siefker DT, Tran M, Weber A, McDonald TL, Mayfield SP. Robust expression of a bioactive mammalian protein in Chlamydomonas chloroplast. Plant Biotechnol. J. 2007;5:402–412. doi: 10.1111/j.1467-7652.2007.00249.x. [DOI] [PubMed] [Google Scholar]
- Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- Matthew T, Zhou W, Rupprecht J, et al. The metabolome of Chlamydomonas reinhardtii following induction of anaerobic H2 production by sulfur depletion. J. Biol. Chem. 2009;284:23415–23425. doi: 10.1074/jbc.M109.003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P, Christian J-O, Kempa S, Walther D. ChlamyCyc: an integrative systems biology database and web-portal for Chlamydomonas reinhardtii. BMC Genom. 2009;10:209. doi: 10.1186/1471-2164-10-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield SP, Franklin SE, Lerner RA. Expression and assembly of a fully active antibody in algae. Proc. Natl Acad. Sci. USA. 2003;100:438–442. doi: 10.1073/pnas.0237108100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac RS, Gibney PA, Chandran SS, Benjamin KR, Botstein D. Synthetic biology tools for programming gene expression without nutritional perturbations in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:1–8. doi: 10.1093/nar/gkt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Kropat J, Liu B, Shaw J, Warakanont J. TAG, you're it Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr. Opin. Biotechnol. 2012;23:352–363. doi: 10.1016/j.copbio.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Michelet L, Lefebvre-Legendre L, Burr SE, Rochaix J-D, Goldschmidt-Clermont M. Enhanced chloroplast transgene expression in a nuclear mutant of Chlamydomonas. Plant Biotechnol. J. 2011;9:565–574. doi: 10.1111/j.1467-7652.2010.00564.x. [DOI] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe D. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 2009;58:165–174. doi: 10.1111/j.1365-313X.2008.03767.x. [DOI] [PubMed] [Google Scholar]
- Moulin M, Nguyen GTDT, Scaife MA, Smith AG, Fitzpatrick TB. Analysis of Chlamydomonas thiamin metabolism in vivo reveals riboswitch plasticity. Proc. Natl Acad. Sci. USA. 2013;110:14622–14627. doi: 10.1073/pnas.1307741110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Neumann-Staubitz P. Synthetic biology approaches in drug discovery and pharmaceutical biotechnology. Appl. Microbiol. Biotechnol. 2010;87:75–86. doi: 10.1007/s00253-010-2578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert J, Karcher D, Bock R. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 2009;57:1140–1150. doi: 10.1111/j.1365-313X.2008.03746.x. [DOI] [PubMed] [Google Scholar]
- Nguyen HM, Baudet M, Cuiné S, et al. Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics. 2011;11:4266–4273. doi: 10.1002/pmic.201100114. [DOI] [PubMed] [Google Scholar]
- Nickelsen J, Van Dillewijn J, Rahire M, Rochaix J. Determinants for stability of the chloroplast psbD RNA are located within its short leader region in Chlamydomonas reinhardtii. EMBO J. 1994;1:3182–3191. doi: 10.1002/j.1460-2075.1994.tb06617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning J, Otto TD, Pfander C, et al. Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev. 2013;27:1198–1215. doi: 10.1101/gad.212746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohresser M, Matagne RF, Loppes R. Expression of the arylsulphatase reporter gene under the control of the nit1 promoter in Chlamydomonas reinhardtii. Curr. Genet. 1997;31:264–271. doi: 10.1007/s002940050204. [DOI] [PubMed] [Google Scholar]
- Olson EJ, Hartsough LA, Landry BP, Shroff R, Tabor JJ. Characterizing bacterial gene circuit dynamics with optically programmed gene expression signals. Nat. Methods. 2014;11:449–455. doi: 10.1038/nmeth.2884. [DOI] [PubMed] [Google Scholar]
- Perrineau M-M, Gross J, Zelzion E, Price DC, Levitan O, Boyd J, Bhattacharya D. Using natural selection to explore the adaptive potential of Chlamydomonas reinhardtii. PLoS One. 2014;9:e92533. doi: 10.1371/journal.pone.0092533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitera DJ, Paddon CJ, Newman JD, Keasling JD. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab. Eng. 2007;9:193–207. doi: 10.1016/j.ymben.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Polle JEW, Kanakagiri S-D, Melis A. tla1, a DNA insertional transformant of the green alga Chlamydomonas reinhardtii with a truncated light-harvesting chlorophyll antenna size. Planta. 2003;217:49–59. doi: 10.1007/s00425-002-0968-1. [DOI] [PubMed] [Google Scholar]
- Prindle A, Selimkhanov J, Li H, Razinkov I, Tsimring LS, Hasty J. Rapid and tunable post-translational coupling of genetic circuits. Nature. 2014;508:387–391. doi: 10.1038/nature13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purton S, Rochaix JD. Characterisation of the ARG7 gene of Chlamydomonas reinhardtii and its application to nuclear transformation. Eur. J. Phycol. 1995;30:141–148. [Google Scholar]
- Quinn JM, Merchant S. Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell. 1995;7:623–628. doi: 10.1105/tpc.7.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana N, der Kooy FV, Van de Rhee MD, Voshol GP, Verpoorte R. Renewable energy from cyanobacteria: energy production optimization by metabolic pathway engineering. Appl. Microbiol. Biotechnol. 2011;91:471–490. doi: 10.1007/s00253-011-3394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramundo S, Rahire M, Schaad O, Rochaix J-D. Repression of essential chloroplast genes reveals new signaling pathways and regulatory feedback loops in Chlamydomonas. Plant Cell. 2013;25:167–186. doi: 10.1105/tpc.112.103051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph-Anderson BL, Boynton JE, Gillham NW, Harris EH, Johnson AM, Dorthu M-P, Matagne RF. Further characterization of the respiratory deficient dum-1 mutation of Chlamydomonas reinhardtii and its use as a recipient for mitochondrial transformation. Mol. Gen. Genet. 1993;236–236:235–244. doi: 10.1007/BF00277118. [DOI] [PubMed] [Google Scholar]
- Rasala BA, Mayfield SP. Photosynthetic biomanufacturing in green algae; production of recombinant proteins for industrial, nutritional, and medical uses. Photosynth. Res. 2014 doi: 10.1007/s11120-014-9994-7. DOI 10.1007/s11120-014-9994-7. [DOI] [PubMed] [Google Scholar]
- Rasala BA, Muto M, Lee PA, et al. Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol. J. 2010;8:719–733. doi: 10.1111/j.1467-7652.2010.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Muto M, Sullivan J, Mayfield SP. Improved heterologous protein expression in the chloroplast of Chlamydomonas reinhardtii through promoter and 5' untranslated region optimization. Plant Biotechnol. J. 2011;9:674–683. doi: 10.1111/j.1467-7652.2011.00620.x. [DOI] [PubMed] [Google Scholar]
- Rasala BA, Lee PA, Shen Z, Briggs SP, Mendez M, Mayfield SP. Robust expression and secretion of Xylanase1 in Chlamydomonas reinhardtii by fusion to a selection gene and processing with the FMDV 2A peptide. J. W. Stiller, ed. PLoS One. 2012;7:e43349. doi: 10.1371/journal.pone.0043349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Barrera DJ, Ng J, et al. Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 2013;74:545–556. doi: 10.1111/tpj.12165. [DOI] [PubMed] [Google Scholar]
- Reijnders MJMF, van Heck RGA, Lam CMC, Scaife MA, dos Santos VAPM, Smith AG, Schaap PJ. Green genes: bioinformatics and systems-biology innovations drive algal biotechnology. Trends Biotechnol. 2014;32:617–626. doi: 10.1016/j.tibtech.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Remacle C, Cline S, Boutaffala L, Gabilly S, Larosa V, Barbieri MR, Coosemans N, Hamel PP. The ARG9 gene encodes the plastid-resident N-acetyl ornithine aminotransferase in the green alga Chlamydomonas reinhardtii. Eukaryot. Cell. 2009;8:1460–1463. doi: 10.1128/EC.00108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaño-Pachón DM, Corrêa LGG, Trejos-Espinosa R, Mueller-Roeber B. Green transcription factors: a Chlamydomonas overview. Genetics. 2008;179:31–39. doi: 10.1534/genetics.107.086090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruecker O, Zillner K, Groebner-Ferreira R, Heitzer M. Gaussia-luciferase as a sensitive reporter gene for monitoring promoter activity in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genomics. 2008;280:153–162. doi: 10.1007/s00438-008-0352-3. [DOI] [PubMed] [Google Scholar]
- Russa MLa, Bogen C, Uhmeyer A, Doebbe A, Filippone E, Kruse O, Mussgnug JH. Functional analysis of three type-2 DGAT homologue genes for triacylglycerol production in the green microalga Chlamydomonas reinhardtii. J. Biotechnol. 2012;162:13–20. doi: 10.1016/j.jbiotec.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Kindle KL, Stern DB. In vivo analysis of Chlamydomonas chloroplast petD gene expression using stable transformation of beta-glucuronidase translational fusions. Proc. Natl Acad. Sci. USA. 1993;90:497–501. doi: 10.1073/pnas.90.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff LB, Bock R. Synthetic biology in plastids. Plant J. 2014;78:783–798. doi: 10.1111/tpj.12356. [DOI] [PubMed] [Google Scholar]
- Schmollinger S, Strenkert D, Schroda M. An inducible artificial microRNA system for Chlamydomonas reinhardtii; confirms a key role for heat shock factor 1 in regulating thermotolerance. Curr. Genet. 2010;56:383–389. doi: 10.1007/s00294-010-0304-4. [DOI] [PubMed] [Google Scholar]
- Schroda M, Blocker D, Beck C. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 2000;21:121–131. doi: 10.1046/j.1365-313x.2000.00652.x. [DOI] [PubMed] [Google Scholar]
- Scott SA, Davey MP, Dennis JS, Horst I, Howe CJ, Lea-Smith DJ, Smith AG. Biodiesel from algae: challenges and prospects. Curr. Opin. Biotechnol. 2010;21:277–286. doi: 10.1016/j.copbio.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Shimogawara K, Fujiwara S, Grossman A, Usuda H. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics. 1998;148:1821–1828. doi: 10.1093/genetics/148.4.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova IA, Lapina TV, Frolova ON, Alexandrova NN, Akopiants KE, Danilenko VN. Stable nuclear transformation of Chlamydomonas reinhardtii with a Streptomyces rimosus gene as the selective marker. Gene. 1996;181:13–18. doi: 10.1016/s0378-1119(96)00384-8. [DOI] [PubMed] [Google Scholar]
- Sizova I, Greiner A, Awasthi M, Kateriya S, Hegemann P. Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. Plant J. 2013;73:873–882. doi: 10.1111/tpj.12066. [DOI] [PubMed] [Google Scholar]
- Stevens DR, Rochaix JD, Purton S. The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 1996;251:23–30. doi: 10.1007/BF02174340. [DOI] [PubMed] [Google Scholar]
- Stolc V, Samanta MP, Tongprasit W, Marshall WF. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc. Natl Acad. Sci. USA. 2005;102:3703–3707. doi: 10.1073/pnas.0408358102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissot-Lecuelle G, Purton S, Dubald M, Goldschmidt-Clermont MP. Synthesis of recombinant products in the chloroplast. In: Theg SM, Wollman F-A, editors. Plastid Biology. New York: Springer; 2014. pp. 517–558. (eds. [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Vajo Z, Fawcett J, Duckworth WC. Recombinant DNA technology in the treatment of diabetes: insulin analogs. Endocr. Rev. 2001;22:706–717. doi: 10.1210/edrv.22.5.0442. [DOI] [PubMed] [Google Scholar]
- Vilanova C, Porcar M. iGEM 2.0 – refoundations for engineering biology. Nat. Biotechnol. 2014;32:420–424. doi: 10.1038/nbt.2899. [DOI] [PubMed] [Google Scholar]
- Villand P, Eriksson M, Samuelsson G. Carbon dioxide and light regulation of promoters controlling the expression of mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Biochem. J. 1997;327:51–57. doi: 10.1042/bj3270051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U. Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell. 2009;8:1856–1868. doi: 10.1128/EC.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijffels RH, Kruse O, Hellingwerf KJ. Potential of industrial biotechnology with cyanobacteria and eukaryotic microalgae. Curr. Opin. Biotechnol. 2013;24:405–413. doi: 10.1016/j.copbio.2013.04.004. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wu S, Xu L, Huang R, Wang Q. Improved biohydrogen production with an expression of codon-optimized hemH and lba genes in the chloroplast of Chlamydomonas reinhardtii. Bioresour. Technol. 2011;102:2610–2616. doi: 10.1016/j.biortech.2010.09.123. [DOI] [PubMed] [Google Scholar]
- Xu L, Wang Q, Wu S, Li D. Improvement of hydrogen yield of lba-transgenic Chlamydomonas reinhardtii caused by increasing respiration and impairing photosynthesis. Int. J. Hydrogen Energy. 2014;39:13347–13352. [Google Scholar]
- Yadav VG, De Mey M, Lim CG, Ajikumar PK, Stephanopoulos G. The future of metabolic engineering and synthetic biology: towards a systematic practice. Metab. Eng. 2012;14:233–241. doi: 10.1016/j.ymben.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yofe I, Zafrir Z, Blau R, Schuldiner M, Tuller T, Shapiro E, Ben-Yehezkel T. Accurate, model-based tuning of synthetic gene expression using introns in S. cerevisiae. G. P. Copenhaver, ed. PLoS Genet. 2014;10:e1004407. doi: 10.1371/journal.pgen.1004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- Young REB, Purton S. Cytosine deaminase as a negative selectable marker for the microalgal chloroplast: a strategy for the isolation of nuclear mutations that affect chloroplast gene expression. Plant J. 2014;80:915–925. doi: 10.1111/tpj.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Cao Y, Zou H, Xian M. Metabolic engineering of Escherichia coli for biotechnological production of high-value organic acids and alcohols. Appl. Microbiol. Biotechnol. 2011;89:573–583. doi: 10.1007/s00253-010-2970-z. [DOI] [PubMed] [Google Scholar]
- Zhang R, Patena W, Armbruster U, Gang SS, Blum SR, Jonikas MC. High-throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell. 2014;26:1398–1409. doi: 10.1105/tpc.114.124099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Wang W, Bai X, Qi Y. Gene silencing by artificial microRNAs in Chlamydomonas. Plant J. 2009;58:157–164. doi: 10.1111/j.1365-313X.2008.03758.x. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Guo C, Sutharzan S, Li P, Echt CS, Zhang J, Liang C. Genome-wide analysis of tandem repeats in plants and green algae. G3 (Bethesda) 2014a;4:67–78. doi: 10.1534/g3.113.008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Wu X, Raj Kumar PK, Dong M, Ji G, Li QQ, Liang C. Bioinformatics analysis of alternative polyadenylation in green alga Chlamydomonas reinhardtii using transcriptome sequences from three different sequencing platforms. G3 (Bethesda) 2014b;4:871–883. doi: 10.1534/g3.114.010249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Algal-biotechnological advancements from a phylogenetic perspective.
Overview of microalgae that are transformable and/or have a published genome


