Abstract
Embryonic stem cell (ESC) cultures display a heterogeneous gene expression profile, ranging from a pristine naïve pluripotent state to a primed epiblast state. Addition of inhibitors of GSK3β and MEK (so-called 2i conditions) pushes ESC cultures toward a more homogeneous naïve pluripotent state, but the molecular underpinnings of this naïve transition are not completely understood. Here, we demonstrate that DAZL, an RNA-binding protein known to play a key role in germ-cell development, marks a subpopulation of ESCs that is actively transitioning toward naïve pluripotency. Moreover, DAZL plays an essential role in the active reprogramming of cytosine methylation. We demonstrate that DAZL associates with mRNA of Tet1, a catalyst of 5-hydroxylation of methyl-cytosine, and enhances Tet1 mRNA translation. Overexpression of DAZL in heterogeneous ESC cultures results in elevated TET1 protein levels as well as increased global hydroxymethylation. Conversely, null mutation of Dazl severely stunts 2i-mediated TET1 induction and hydroxymethylation. Our results provide insight into the regulation of the acquisition of naïve pluripotency and demonstrate that DAZL enhances TET1-mediated cytosine hydroxymethylation in ESCs that are actively reprogramming to a pluripotent ground state.
Keywords: 2i conditions, Dazl, DNA hydroxymethylation, naïve pluripotency, TET1
Introduction
Embryonic stem cells (ESCs) are derived from the inner cell mass (ICM) of pre-implantation blastocyst embryos 1. ESCs possess broad developmental potential and are able to generate every cell type in the developing embryo. Cells in the blastocyst ICM as well as in ESCs at the single-cell level display a heterogeneous gene expression profile, while in serum-cultured ESCs, heterogeneity takes more extreme forms and stretches well outside the developmental boundaries of the blastocyst embryo 2,3. In fact, the expression of genes naturally associated with germ-cell development is a hallmark property of murine embryonic stem cells and sets them apart from other pluripotent stem cell types such as the epiblast stem cells (EpiSCs) derived from post-implantation epiblast embryos 4,5. To date, the significance of this germ-cell profile for ESC biology remains elusive.
A so-called “2i inhibitor cocktail”, consisting of a MEK and GSK3β inhibitor, facilitates ESC derivation and maintenance 6. 2i culture conditions enhance ESC homogeneity by suppressing lineage differentiation 7 and induce genome-wide DNA demethylation in murine ESCs 8,9,10. Consequently, ESCs in 2i conditions are epigenetically and transcriptionally more similar to naïve cells in the blastocyst ICM from which these cells have been derived.
We explored the molecular changes that accompany the transition to this naïve pluripotent state. To this end, we used several ESC lines expressing fluorescent reporter genes for naïve pluripotency, including Nanog, Stella, and Dazl. We demonstrate that DAZL, an RNA-binding protein and a marker for late PGC development, is expressed in 5–10% of serum-cultured ESCs and induced to approximately 80% during 2i culture. We explored the significance of DAZL in ESC biology and observed that DAZL is also expressed in vivo in a subpopulation of cells in the blastocyst ICM. Under serum culture conditions, DAZL-positive ESCs are transcriptionally more similar to ESCs cultured in 2i and also exhibit high levels of 5-hydroxymethylation, whereas 5-hydroxymethylation is low in DAZL-negative ESCs. 5-hydroxymethylation results from the hydroxylation of methylated cytosine residues by TET1 or TET2 enzymes and is an important step in the opening of heterochromatic regions 11. We observed that, upon 2i induction, DAZL-positive ESCs transition faster to a homogeneous naïve pluripotent state than their DAZL-negative counterparts. Finally, we observed that DAZL is an essential component of TET1-dependent DNA demethylation during reprogramming and in the absence of Dazl expression, the induction of TET1 enzymes is impaired. We found that DAZL functions as a translational enhancer of Tet1 mRNA molecules, which are complexed with Dazl protein in mESCs. Indeed, overexpression of Dazl results in an increase in TET1 protein levels and high 5-hydroxymethylation.
Our findings shed important light on the mechanism by which ES cells transition to a naïve pluripotent state, and demonstrate that Dazl plays an essential role in active TET1-mediated global DNA demethylation.
Results and Discussion
DAZL is heterogeneously expressed in mESCs and induced by 2i culture conditions
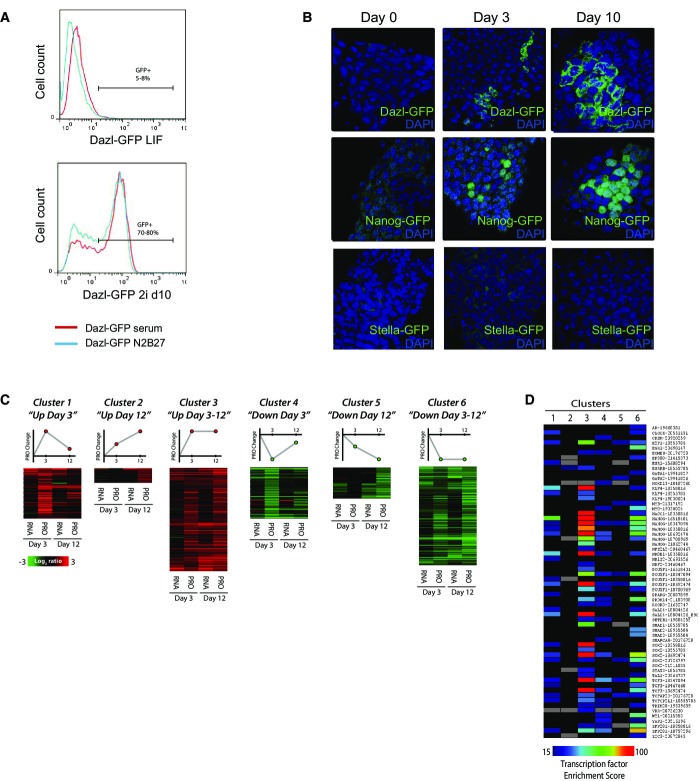
To study the role of DAZL in murine ESCs, we used Dazl-GFP reporter ESC lines derived from Dazl-GFP transgenic mice 12. This Dazl-GFP reporter line faithfully recapitulates DAZL expression as GFP levels correlate with the amount of mRNA molecules found in individual cells (Appendix Fig S1A). DAZL was previously shown to be expressed at the start of PGC migration toward the future gonads and Dazl RNA expression has been used as a specific marker of naïve pluripotent stem cells in murine ESCs 2,4,5,13. However, to date, its role in ESC biology remains unknown. The Dazl-GFP transgene is heterogeneously expressed in 5–8% of mESCs in LIF/MEF/serum and N2B27/LIF culture conditions (Fig1A). Upon FACS separation of Dazl-GFP-positive from the Dazl-GFP-negative cells, the sorted cells re-establish the original equilibrium within a few days (Appendix Fig S1B). A similar heterogeneous equilibrium has been reported for other ESC genes such as Stella, Nanog, and Rex1 2,3,14. Suppression of ESC differentiation by a combination of an ERK and a GSK3β inhibitor, so-called 2i conditions, promotes a more homogeneous state of ESC self-renewal 15,16,17.
Figure 1.
- FACS analysis showing Dazl-GFP expression in serum- or N2B27-cultured ESCs in LIF (upper panel) and 10 days in 2i + LIF (lower panel).
- Expression profiles of germ-cell reporter ESCs Dazl-GFP, Nanog-GFP, and Stella-GFP at day 0, 3, and 10 after 2i induction analyzed by fluorescent microscopy.
- mESCs were profiled for mRNA (RNA) and protein (PRO) expression at day 3 and 12 after addition of 2i inhibitors to the culture media. We focused on those genes that demonstrated a minimum twofold change in protein levels at one of the time points (day 3 and/or day 12). mRNA and protein expression profiles were clustered based on the expression pattern of the genes and their corresponding proteins. n = 2 biological replicates/time point.
- Gene members of each cluster were analyzed with the X2K analysis tool. Heat map output demonstrates 2i-induced genes are highly enriched for downstream targets of the pluripotency network.
We analyzed the effect of 2i addition to the culture media on our Dazl-GFP reporter cells as well as on Nanog-GFP and Stella-GFP reporter ESCs 18,19. As reported previously, Nanog is expressed in 80–90% of ESCs in serum culture conditions 14, and while 2i induction does not profoundly change the total percentage of Nanog+ cells, we did observe the emergence of an additional Nanog-bright population in accordance with a recent paper of Miyanari and Torres-Padilla (Fig1B, middle panel, Appendix Fig S1C) 16. During early 2i conversion, we also noticed an increase in the percentage of Stella-GFP-positive cells, as reported previously 20, but we observed that Stella expression wanes upon prolonged 2i culture, and by day 10, the expression of this marker is almost completely abrogated (Fig1B, lower panel, Appendix Fig S1C). Stella is known to be a specific marker of nascent PGCs around E7.5 of post-implantation development 21. The expression of this marker in serum-cultured ESCs demonstrates that the heterogeneous gene expression profile observed in ESCs stretches beyond the developmental boundaries of the blastocyst ICM from which these cells are derived, and 2i culture appears to limit this promiscuous gene expression. We were therefore surprised to find that 2i culture conditions increased the expression of Dazl-GFP, a marker reported to be expressed at even later stages of germ-cell development, to approximately 80% after 10 days (Fig1A and B, Appendix Fig S1C).
To further explore the temporal molecular changes that accompany the transition of heterogeneous ESC cultures to 2i-induced naïve pluripotency, we analyzed mRNA and protein levels of Dazl-GFP ESCs grown in conventional culture medium and subsequently cultured for 3 and 12 days in 2i conditions by microarray and mass spectrometry. In total, 4,368 genes were identified at both protein and mRNA levels (Table EV1). We focused on genes that demonstrated at least a twofold change in protein levels at one of the time points. We identified 563 genes with corresponding changes in mRNA and protein levels. GO analysis of these genes revealed a role in broad cellular functions, including enrichment for metabolic processes in 2i culture conditions, which is in agreement with a recent publication analyzing RNA expression in 2i-induced cells (Appendix Fig S1D) 15. However, none of these processes point to a clear role in stem cell pluripotency.
We subdivided the differentially expressed genes into six clusters based on their expression pattern and identified corresponding upstream transcription factors with X2K software (Fig1C and D, Tables EV2 and EV3) 22. Interestingly, genes in cluster 3, which contains genes that are sustainably upregulated in 2i culture conditions, reveal a significant enrichment for key transcriptional regulators of pluripotency, Oct4, Sox2, Nanog, and Klf4 (Fig1D). As such, these factors appear to reduce heterogeneity of genes that are important for the maintenance of the naïve pluripotent state, thereby making the pluripotency network more robust. In contrast, we find that a series of germ-cell-related genes, including Stella, Lefty1, and Tcfap2c, are transiently expressed at day 3 of 2i induction, and almost absent upon sustained 2i culture. The heterogeneous expression of these in serum-cultured ESCs falls outside the developmental boundary of the blastocyst ICM and therefore appears to result from serum-mediated signals that drive differentiation toward the post-implantation epiblast 23.
The only PGC-specific gene that was continuously upregulated in 2i conditions was Dazl, thought to be a marker for late PGC development 21, which is expressed in approximately 80% of the ESCs after 2i induction. While the importance of Dazl PGC development is known, its expression appeared out of context in ESCs.
DAZL is expressed in late blastocyst embryos
To further examine the role of DAZL in ESC biology, we explored the expression of DAZL during pre-implantation development. Dazl-GFP embryos were isolated at the morula stage and cultured in KSOM for 96 h. While we did not observe Dazl-GFP expression at the morula and early blastocyst stages, late blastocyst cultures revealed a subpopulation of Dazl-GFP-positive cells in the ICM (Fig2A). Late blastocyst Dazl-GFP embryos flushed at E4.5 demonstrated a similar GFP expression in the ICM confirming that the Dazl-GFP expression was not the result of the in vitro maturation of the embryos (Appendix Fig S2A). To validate that the observed Dazl-GFP expression was not the result of aberrant expression of the transgenic reporter, we performed single-molecule RNA FISH on wild-type blastocyst embryos to visualize the expression of Dazl and Oct4 mRNA 24. As shown in Fig2B, Dazl mRNA is co-expressed with Oct4 in a subpopulation of cells in the blastocyst ICM, demonstrating that Dazl expression is restricted to pluripotent cells in the early embryo. We therefore conclude that DAZL expression in cultured ESCs reflects the expression of this gene in the late pre-implantation blastocyst.
Figure 2.
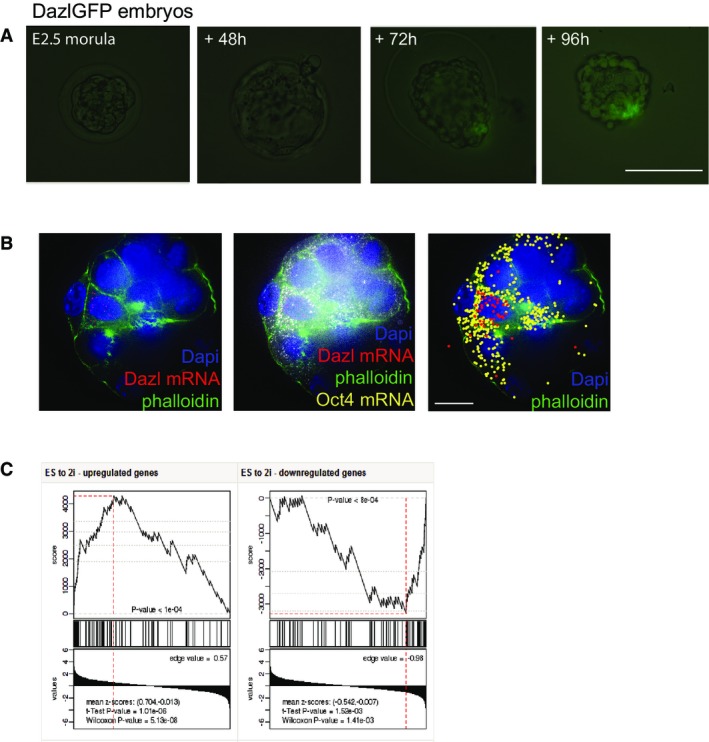
- DazlGFP embryo cultured in vitro in KSOM from E2.5 morula stage to late blastocyst stage. Scale bar, 100 μm.
- Single-molecule FISH experiment showing single Dazl and Oct4 mRNA molecules in E3.5 blastocyst embryos. Right panel, artificial visualization of single mRNA molecules. Scale bar, 50 μm.
- The top panel of this GSEA plot shows a rank-sum-based score, which is calculated depending on the correlation of the differentially expressed genes in DazlGFP ESCs in 2i conditions with its expression in embryos cultured in 2i conditions vs. control embryos. The enrichment score reflects the degree to which the examined gene set is over-represented at the extremes. The middle panel shows vertical lines corresponding to the rank of the genes in the examined gene set. Here, most of the black lines are clustered to the left meaning that the genes in DazlGFP 2i dataset are among the most upregulated in the embryo 2i measurements. The bottom panel shows the actual expression difference scores from the embryos cultured in 2i conditions vs. control embryos. The overall P-value (shown on the top) is determined based on the value of the rank-sum statistics (top-plot) at its extreme point (marked by a vertical red line).
Embryos grown to the blastocyst stage in the presence of 2i inhibitors express high uniform levels of OCT4 and NANOG in the ICM, while the hypoblast is absent 25. To explore how 2i culture conditions affect gene expression in developing blastocysts just prior to ESC derivation, morula-stage embryos were cultured with or without 2i inhibitors until the late blastocyst stage. mRNA sequencing was performed on single blastocyst embryos. At the single gene level, we observed similar changes to those we observed when ESCs are cultured in 2i conditions, including an increase in the expression of Dazl, Klf8, and Id3 (Appendix Fig S2B), suggesting that the transcriptional changes observed upon 2i culture of ESCs are similar to those observed during 2i expansion of the naïve pluripotent cell population in the blastocyst ICM.
Indeed, this was confirmed when we compared the changes in gene expression induced by 2i culture in ESCs and blastocysts at the global level by Gene Set Enrichment Analysis (GSEA) 26. Figure2C demonstrates that the majority of gene expression changes observed in ESCs cultured in 2i conditions show similar changes in expression levels in the 2i embryos compared to controls (Fig2C, Appendix Fig S2B). These data demonstrate that blastocyst embryos in 2i conditions, as well as in ESCs cultured in 2i conditions, reflect similar biological states.
DAZL marks actively reprogramming ESCs
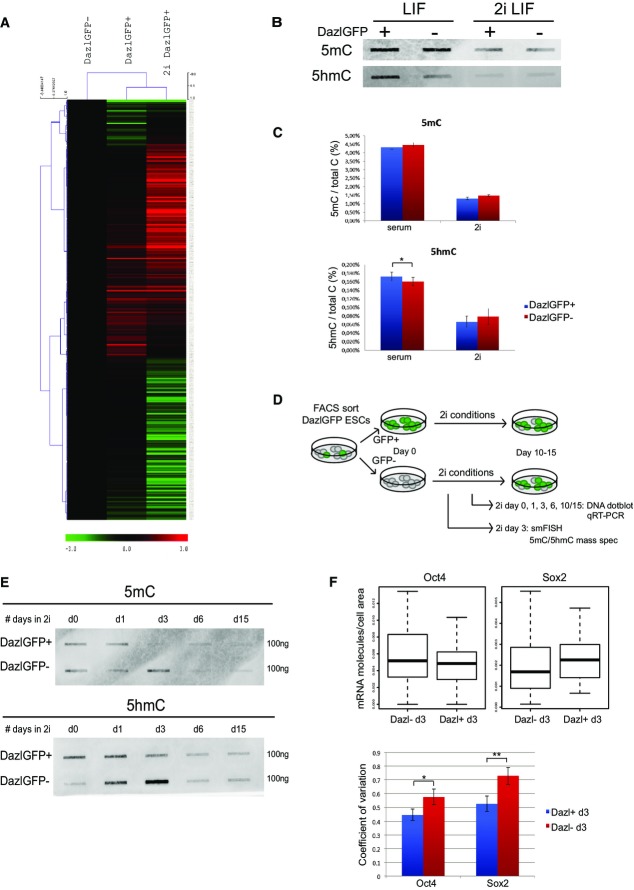
The experiments above demonstrate that DAZL is expressed in the ICM of late, pre-implantation blastocyst embryos, as well as in ESCs. Furthermore, 2i-induced expansion of the pluripotent epiblast enhances DAZL expression in pre-implantation embryos and induces the percentage of Dazl-positive cells in ESC cultures. Together, these data suggest that Dazl marks unique naïve cells within the heterogeneous ESC population akin to the cells in the naïve ICM. Indeed, comparison of DAZL-negative and DAZL-positive cells cultured in serum with DAZL-positive cells cultured in 2i by proteomic analysis shows that Dazl-positive cells in serum cluster together with DAZL-positive cells in 2i (Fig3A). ESCs cultured in 2i conditions represent homogeneous cell populations, and indeed, we find that Dazl-GFP-positive and Dazl-GFP-negative cells cultured in 2i are transcriptionally similar (Appendix Fig S3A) 10,27,28. This means that DAZL-positive cells in serum already display a gene expression pattern that is more related to ESCs in 2i conditions than to the 90–95% Dazl-negative cells in conventional serum culture.
Figure 3.
- Proteomic analysis of genes expressed in Dazl-negative ESCs, Dazl-positive ESCs, and Dazl-positive ESCs cultured for 12 days in 2i conditions.
- Dot-blot analysis for 5meC and 5hmeC in Dazl-positive and Dazl-negative ESCs FACS-sorted from serum-cultured cells or sorted from ESCs cultured in 2i + LIF for 12 days.
- Mass spectrometric measurement of global 5mC and 5hmC levels in DazlGFP+ and DazlGFP− ESCs in serum as well as in 2i conditions. P-values were calculated using paired t-test. *P < 0.05. Error bars indicate s.e.m. values of ≥ 3 biological replicates.
- Schematic overview of experimental setup: DazlGFP+ ESCs and DazlGFP− ESCs are FACS-sorted from serum-cultured ESCs followed by culture of each population in 2i conditions. Different time points during the transition to a naïve pluripotent state were analyzed for gene expression and DNA methylation.
- Dot-blot analysis for 5meC and 5hmeC in Dazl-positive and Dazl-negative cells FACS-sorted at day 0 followed by culture in 2i + LIF conditions for several days.
- The upper panel shows a boxplot with the distribution of single Oct4 and Sox2 mRNA molecules per cell area in > 50 individual ESCs that were sorted for Dazl-GFP+ and Dazl-GFP− followed by 3 days of culture in 2i conditions. The lower panel shows the coefficient of variation (or normalized variance) of the single Oct4 and Sox2 transcripts within each cell population. *P < 0.05, **P < 0.001. Error bars indicate s.d. values of 100,000 times in silico random sampling of 50 cells per condition.
One of the hallmark events that occur during pre-implantation embryonic development is genome-wide demethylation. DNA methylation of CpG dinucleotides (5mC) in mammalian cells is associated with gene silencing. The maintenance methyltransferase DNMT1 is responsible for copying these patterns during DNA replication and DNMT3a and DNMT3b set up de novo DNA methylation during development (reviewed by Bagci and Fisher 29). DNA demethylation can occur via active and passive mechanisms. Passive replication-dependent loss of DNA methylation is achieved by downregulation of DNMT1 or NP95 or their exclusion from the nucleus resulting in the dilution of global DNA methylation during subsequent cell proliferation 30. Active DNA demethylation is regulated by TET (Tet1-3) enzymes that catalyze the conversion of 5-methylcytosine to 5-hydroxymethylcytosine (5hmC). 5hmC can subsequently be actively demethylated by its oxidation into 5-formylcytosine (5fC) and 5-carboxymethylcytosine (5caC) or passively by replication-dependent passive demethylation since 5hmC is not recognized by DNMT1 and NP95 31,32,33.
Genome-wide DNA demethylation occurs in early embryos up to the blastocyst stage and during primordial germ-cell development and is associated with pluripotency 34,35. However, ESCs are highly methylated compared to the ICM of blastocyst embryos 35. In accordance with our microarray and proteomics data, it was demonstrated that DNMT3s are downregulated in ESCs in 2i conditions, resulting in DNA demethylation making them more reminiscent of the ICM 8,35,36.
Since our observed changes in DAZL expression in ESCs switched to 2i conditions coincided with the timing of changes in global DNA methylation, we explored the methylation state of DAZL-positive cells in serum-cultured ESCs. To investigate the DNA methylation status of Dazl-GFP-positive and Dazl-GFP-negative cells in serum and 2i conditions, we performed dot-blot analysis as well as mass spectrometric measurement of global 5mC and 5hmC levels on genomic DNA isolated from these cells. As expected based on previous reports, 5meC levels are globally decreased in 2i-cultured cells, while no clear difference is observed between DAZL-positive and DAZL-negative ESCs in these conditions (Fig3B and C) 9,37. Interestingly, DAZL-positive ESCs in serum are highly hydroxy-methylated compared to DAZL-negative ESCs in serum and they also express Tet1 and Tet2 genes at significantly higher levels (4- and 2-fold difference respectively) (Fig3B and C, Appendix Fig S3A). TET-mediated conversion of 5meC to 5hmeC can result in both active and passive loss of DNA methylation 32,33. While mice deficient in both Tet1 and Tet2 can develop postnatally, they display many epigenetic abnormalities and increased 5meC levels highlighting the importance of TET proteins in regulating cytosine methylation 38.
The observation that protein expression levels of DAZL-positive cells in serum cluster with those in 2i conditions, together with high basal level of 5hmeC in DAZL-positive ESCs, suggests they would convert more quickly to a fully naïve pluripotent state than DAZL-negative cells. To investigate this, we FACS-sorted Dazl-GFP-positive and Dazl-GFP-negative ESCs and analyzed 5hmeC levels over time during transition to a naïve state in 2i conditions (Fig3D). As shown in Fig3E, Dazl-GFP-positive cells already show a loss of 5meC after 3 days in 2i comparable to levels of ESCs that were cultured in 2i conditions for several passages (Fig3E, Appendix Fig S3B). In contrast, Dazl-GFP-negative cells show higher methylation and hydroxymethylation levels at this time point in a similar time frame as has been reported for other ESC lines converted to 2i culture conditions (Fig3E, Appendix Fig S3B) 8,9. However, while Dazl-GFP-positive ESCs in serum display higher global expression levels of naïve genes, during their transition to a naïve state in 2i conditions, gene expression changes follow similar patterns in Dazl-GFP-positive and Dazl-GFP-negative starting populations (Appendix Fig S3C).
While global expression levels of the core pluripotency factors Oct4 and Sox2 are unchanged in serum vs. 2i culture conditions, a more homogeneous gene expression pattern is induced in the latter 15,17,39. To test whether mRNA transcripts are more equally distributed among the cells in DAZL-positive cells cultured in 2i for 3 days than in DAZL-negative cells at day 3 in 2i, we performed single-molecule RNA FISH for Oct4 and Sox2 in these cells (Appendix Fig S3D). As shown in Fig3F, the distribution of Oct4 as well as Sox2 mRNA transcripts is smaller in the DAZL-positive cells at day 3 in 2i conditions meaning that they are more homogeneously expressed among the cells. Indeed, the coefficients of variation (cv) in DAZL-negative cells for Oct4 as well as Sox2 are significantly different from the cv in Dazl-positive cells cultured for 3 days in 2i conditions (Fig3F, lower panel). Thus, DAZL marks a subpopulation of ESCs that is more akin to 2i-induced naïve pluripotent stem cells, as measured by their global gene expression level, their more homogeneous expression of core pluripotency factors, and their enhanced levels of TET hydroxylases and 5-hydroxymethylation, resulting in more rapid genome demethylation upon 2i induction.
DAZL is required for TET1-mediated hydroxymethylation
Finally, we explored whether DAZL, aside from being a marker for more naïve ESCs within the culture, also had a functional role in the establishment of the naïve pluripotent state. To this end, we established an ESC line in which DAZL can be induced by doxycycline (Dox). We tested whether the overexpression of Dazl was sufficient to induce naïve pluripotency in ESCs cultured in serum conditions at different time points. Indeed, not only Dazl transcript levels increased following Dazl overexpression with doxycycline, but also other genes associated with naïve pluripotency such as Prdm14, Rex1, and Tfcp2 l1 (Fig4A). While Tet1 and Tet2 transcript levels remain unchanged, Dazl overexpression does induce significant higher levels of hydroxymethylation (Fig4A and B). Global DNA methylation does not decrease, which is in accordance with the observation that de novo methyltransferases are not downregulated (Fig4A, Appendix Fig S4A). Similar results have been published recently for the induction of hydroxymethylation by vitamin C, which leads to DNA demethylation at germline genes, but global DNA methylation remains unchanged 40.
Figure 4.
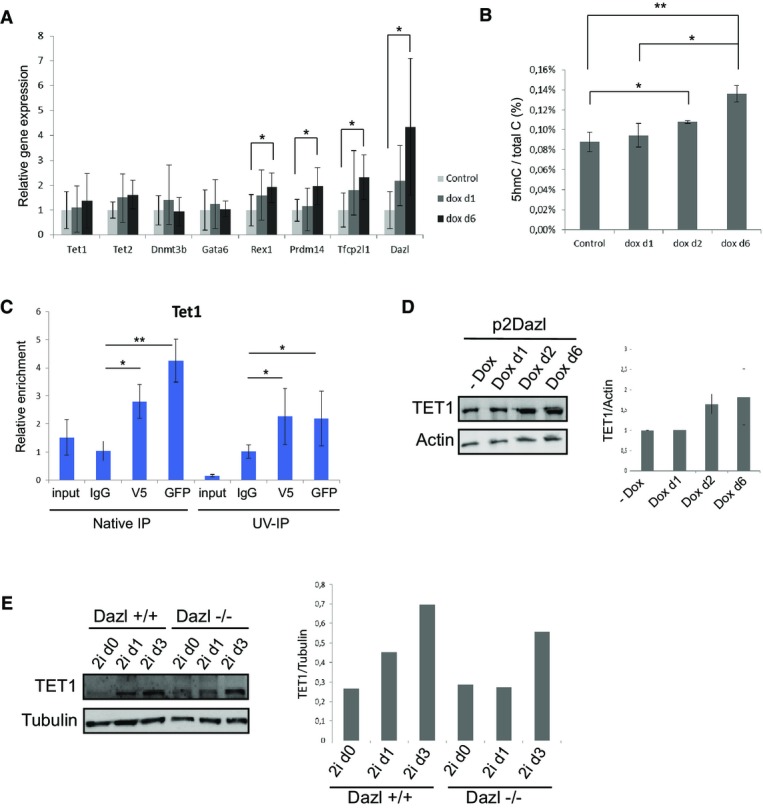
- qRT–PCR results for genes involved in DNA methylation (Dnmt3b) and hydroxymethylation (Tet1 and Tet2), primed pluripotency (Gata6) and naïve pluripotency (Rex1, Prdm14, Tfcp2 l1 and Dazl) upon Dazl overexpression at d1 and d6 after treatment of ESCs with doxycycline.
- Mass spectrometric quantification of global 5hmC levels upon overexpression of Dazl in ESCs at d0, d1, d2, and d6.
- qRT–PCR results for Tet1 in both native as well as UV-cross-linked RNA-IP experiments in Dazl-GFP mESCs. Antibodies against GFP and V5 were used to immunoprecipitate Dazl, and normal mouse IgG was used as a negative control.
- Western blot for TET1 protein levels upon overexpression of Dazl in ESCs at d0, d1, d2, and d6. Relative enrichment of TET1 in different samples was quantified using ImageJ on Western blots of 3 biological replicates.
- Western blot for TET1 protein levels in Dazl+/+ and Dazl−/− ESCs upon conversion to 2i culture conditions. Relative enrichment of TET1 in different samples was quantified using ImageJ.
Data information: P-values were calculated using Student's t-test. *P < 0.05, **P < 0.001. Error bars indicate s.d. values of > 3 biological replicates.
Vitamin C activates TET proteins to oxidize 5-methylcytosine to 5-hydroxymethylcytosine. Since DAZL does not have an antioxidant role like vitamin C, we wondered by which mechanism DAZL can induce hydroxymethylation. In the adult testis, DAZL has been shown to stabilize associated mRNA targets thereby functioning as an enhancer of mRNA translation 41,42. We hypothesized that DAZL could have a role in stabilization of Tet genes causing the observed increase in hydroxymethylation. To investigate whether Tet1 and Tet2 are mRNA targets of DAZL, we performed RNA immunoprecipitation experiments followed by qRT–PCR. We found that mRNAs associated with Dazl-GFP-V5 are enriched for Tet1 transcripts compared to IgG control RNA-IP in UV-cross-linked IP experiments as well as in native IPs, but not for Tet2 (Fig4C, Appendix Fig S4B). These results suggest that DAZL could have a direct effect on the stabilization or promotion of translation of Tet1 genes. Indeed, while the expression of Tet1 and Tet2 genes does not change upon Dazl overexpression (Appendix Fig S4E), elevated TET1 protein levels are observed in ESCs from 2 days onwards after the induction of Dazl with doxycycline, indicating that DAZL supports more efficient translation (Fig4D).
The transition of serum-cultured ESCs to a naïve pluripotent state in 2i conditions is accompanied by a transient increase in hydroxymethylation and Tet gene expression during the first 3 days after switching culture conditions 8. To test whether DAZL is essential for the acquisition of a naïve pluripotent state, we examined the effect of loss of Dazl expression on DNA methylation and hydroxymethylation levels upon 2i-induced transition to a naïve pluripotent state. We cultured Dazl−/− ESCs, Dazl+/− ESCs, and Dazl+/+ ESCs in 2i conditions for several days and investigated DNA methylation and gene expression levels at different time points 43. Interestingly, 5-hydroxymethylation does not increase after 24 h in two different Dazl knockout ESC lines that we studied, whereas wild-type ESCs from the same genetic background do show this increase in accordance with previous reports (Appendix Fig S4C) 8,36. DNA dot-blot analysis also shows that global 5-hydroxymethylation is detected at lower levels in Dazl knockout ESCs as compared to Dazl heterozygous ESCs in 2i culture (Appendix Fig S4D). However, no significant difference is observed in the transcription of Tet1 and Tet2 genes during the first days upon conversion to 2i culture conditions between Dazl−/−, Dazl+/−, and Dazl+/+ ESCs (Appendix Fig S4E). To investigate whether DAZL is necessary for enhancing Tet1 translation during the transition to a naïve pluripotent state, we performed Western blot to analyze the changes in TET1 protein levels in Dazl−/− and Dazl+/+ ESCs during the first 3 days in 2i conditions. As a matter of fact, Fig4E shows that TET1 protein is expressed at high levels after 24 h and 3 days in 2i medium in Dazl+/+ ESCs, while TET1 induction is slower and at lower levels in Dazl−/− ESCs explaining the delayed hydroxymethylation as observed in Appendix Fig S4C (Fig4E, Appendix Fig S4C).
Despite low 5hmC levels, Dazl knockout ESCs do reach a demethylated state upon prolonged 2i culture (Appendix Fig S4D). We hypothesized that, in the absence of Dazl, mESCs mainly undergo passive replication-dependent demethylation through downregulation of DNMTs 31,32. Indeed, qRT–PCR analysis of the expression of DNA methyltransferases demonstrates a significant downregulation of Dnmt3a and Dnmt3b upon conversion to 2i culture conditions in Dazl−/−, Dazl+/−, and Dazl+/+ ESCs. However, the downregulation of Dnmt3b is greater in Dazl−/− and Dazl+/− ESCs compared to wild-type cells, and additionally, they also show a significant downregulation of the maintenance methyltransferase Dnmt1 (Appendix Fig S4F). Thus, while cytosine demethylation occurs mainly through a different route in 2i-cultured Dazl−/− ESCs, the cells ultimately reach a demethylated state as well.
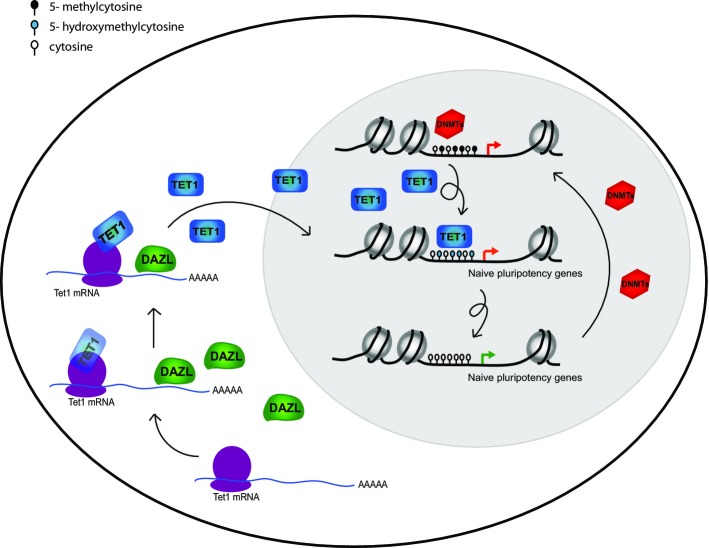
Together, our results demonstrate that DAZL marks actively reprogramming cells in ESC cultures. Furthermore, DAZL plays an active role in this process by associating with Tet1 mRNA and regulating its translation, thereby controlling the oxidization of 5mC to 5hmC (Fig5). Overexpression of DAZL is sufficient for inducing continuously higher levels of 5hmC and TET protein expression. Previous studies have reported that TET1 binding is enriched near the transcriptional start site of germline- and naïve pluripotency genes 40. Indeed, these genes are upregulated upon sustained ectopic expression of Dazl in ESCs 40,44. DAZL may therefore facilitate rapid and active transition of ESCs to a more naïve state, by enhancing TET1 protein levels and promoting hydroxymethylation of promoters of the pluripotent network. Indeed, in the absence of Dazl expression, TET1 protein expression and 5-hydroxymethylation are aberrantly regulated upon 2i induction. Our findings highlight the essential role of DAZL in the regulation of hydroxymethylation. Since Dazl is known to play an essential role in germ-cell differentiation, it would be interesting to further investigate the role of DAZL in DNA demethylation during primordial germ-cell development.
Figure 5.
DAZL is required for TET1-mediated hydroxymethylation to a more naïve pluripotent state
Model showing the mechanisms by which DAZL can induce hydroxymethylation in serum-cultured ESCs. DAZL associates with Tet1 mRNA and enhances its translation. Increased Tet1 levels oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC). This can be followed by both active and passive demethylation resulting in the expression of germline and naïve pluripotency genes since TET1 is enriched at the promoters of these genes. DNMTs can subsequently induce DNA methylation again.
Materials and Methods
Cell culture
Mouse embryonic stem cells were cultured on γ-irradiated feeder MEFs in DMEM containing 15% FBS or serum-free B27N2 medium both supplemented with leukemia inhibitory factor (LIF) 6. For the 2i experiments, 1 μM MEK inhibitor PD0325901 (Axon Medchem) and 5 μM GSK3β inhibitor Kenpaullone (Tocris) were used. Dazl-GFP ESCs were generated as described 12. Dazl−/− and Dazl+/− ESCs were isolated from blastocysts from Dazl heterozygous matings 43.
Inducible Dazl ESCs were generating by cloning Dazl cDNA into the p2Lox-V5 vector as described previously 45.
Microarray analysis, immunofluorescence microscopy, qRT–PCR gene expression analysis, Western blotting, and RNA immunoprecipitation
Microarray analysis, immunofluorescence microscopy, qRT–PCR gene expression analysis, Western blotting, and RNA immunoprecipitation were performed as described 12. Full experimental procedures are found in the Appendix Supplementary Methods. The NCBI Gene Expression Omnibus accession numbers for the microarray analysis data in this paper are: GSE69055, GSE69356 and GSE69357.
Mass spectrometry-based proteomics analysis
LC/MS analysis of mC and hmC in the genomic DNA
All oligonucleotides, nucleosides (dC, 5mdC), ammonium acetate, and LC/MS-grade acetonitrile were from Sigma-Aldrich. 5hmdC was purchased from Berry & Associates, Inc. (Dexter, MI).15N3-dCTP and 15N3-dC were from Silantes, GmbH (Munich, Germany). 2H3-5mdC was from TRC, Inc (Toronto, Canada). 15N3-5hmdC was self-synthesized by a series of in vitro reactions using ngTet1. In brief, the PCR-synthesized oligonucleotide with the incorporated 15N3-dC was MSP1-digested and PAGE-purified to remove the non-labeled primer regions, then incubated with M.Sss1 methyl transferase to convert all 15N3-dC into 15N3-mdC, and then incubated with ngTet1 to further oxidize it to 15N3-hmdC. Finally, the oligonucleotide was digested to nucleosides and 15N3-hmdC was HPLC-purified. The ngTet1 plasmid DNA was a kind donation of Dr. Cheng. 46. All solutions were prepared using Millipore quality water (Barnstead GenPure xCAD Plus, Thermo Scientific). Genomic DNA was isolated after RNase A (Fermentas) treatment of lysed cells by DNeasy blood & tissue kit (Qiagen) according to the manufacturer's instructions followed by ethanol precipitation using ammonium acetate as salt or using phenol–chloroform DNA isolation followed by ethanol precipitation using ammonium acetate. About 1 μg of DNA was degraded to nucleosides with 0.003 U nuclease P1 (Roche), 0.01 U snake venom phosphodiesterase (Worthington), and 0.1 U alkaline phosphatase (Fermentas) 47. Separation of the nucleosides from the digested DNA samples was performed with an Agilent 1290 UHPLC system equipped with ReproSil 100 C18 column (3 μm, 4.6 × 150 mm, Jasco GmbH, Groß-Umstadt, Germany) with a gradient of 5 mM ammonium acetate (pH 7) and acetonitrile. Quantitative MS/MS analysis was done with an Agilent 6490 triple quadruple mass spectrometer coupled with stable isotope dilution, as described 48,49,50. For specific experimental instrument setting, see Tables EV5 and EV6.
Single-molecule FISH
Single-molecule FISH on E3.5 embryos and ESCs was performed and analyzed as previously described 51.
RNA sequencing of blastocyst embryos
This procedure is based on the protocol of Tang et al 52. Differential expression was analyzed using the Bayesian method according to the protocol of Kharchenko and colleagues 53.
More detailed Materials and Methods are provided in the Appendix Supplementary Methods.
Acknowledgments
We thank Sarah Opitz for critically reading the manuscript, Nune Schelling for technical support, and Lucas Kaaij for help with statistical analysis. We also thank Laura Prickett-Rice and Kat Folz-Donahue at the HSCI FACS facility at Mass. General Hospital; Stefan van der Elst at the Hubrecht Institute FACS facility for help with cell sorting; Anko de Graaf of the Hubrecht Institute imaging facility; and David Egan of the Cell Screening facility at the UMCU. MW is supported by a grant from the Netherlands Institute for Regenerative Medicine. JM and AJRH acknowledge support by the Netherlands Proteomics Centre, embedded in the Netherlands Genomics Initiative. This work was funded in part by The Netherlands Organization for Scientific Research (NWO, project 91796323).
Author contributions
MW and NG conceived work and wrote the manuscript, and MW, H-HC, and NG designed the experiments and analyzed the data. MW and H-HC performed the majority of the experiments. JM and NM analyzed the proteomics data. MM performed LC/MS of 5 m and 5hm in genomic DNA. LK and JPJ performed smFISH in blastocyst embryos. MA generated inducible ES cell lines. EK assisted in blastocyst immonostainings. LS and PVK performed and analyzed RNA sequencing of blastocyst embryos. MG and HvdV helped with embryo analysis. CN supervised the LC/MS for DNA methylation. AvO supervised the smFISH in blastocysts. AJRH supervised the proteomics analysis. NG supervised all aspects of the project.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Appendix
Table EV1
Table EV2
Table EV3
Table EV4
Table EV5
Table EV6
Review Process File
References
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 4.Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, de Sousa C, Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 5.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 6.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wray J, Kalkan T, Smith AG. The ground state of pluripotency. Biochem Soc Trans. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- 8.Ficz G, Hore TA, Santos F, Lee HJ, Dean W, Arand J, Krueger F, Oxley D, Paul YL, Walter J, et al. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leitch HG, McEwen KR, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich JG, Smith A, et al. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaji M, Ueda J, Hayashi K, Ohta H, Yabuta Y, Kurimoto K, Nakato R, Yamada Y, Shirahige K, Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen HH, Welling M, Bloch DB, Munoz J, Mientjes E, Chen X, Tramp C, Wu J, Yabuuchi A, Chou YF, et al. DAZL limits pluripotency, differentiation, and apoptosis in developing primordial germ cells. Stem Cell Rep. 2014;3:892–904. doi: 10.1016/j.stemcr.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Y, Page DC. Dazl deficiency leads to embryonic arrest of germ cell development in XY C57BL/6 mice. Dev Biol. 2005;288:309–316. doi: 10.1016/j.ydbio.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 14.Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 15.Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyanari Y, Torres-Padilla ME. Control of ground-state pluripotency by allelic regulation of Nanog. Nature. 2012;483:470–473. doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- 17.Wray J, Kalkan T, Gomez-Lopez S, Eckardt D, Cook A, Kemler R, Smith A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Payer B, Chuva de Sousa Lopes SM, Barton SC, Lee C, Saitou M, Surani MA. Generation of stella-GFP transgenic mice: a novel tool to study germ cell development. Genesis. 2006;44:75–83. doi: 10.1002/gene.20187. [DOI] [PubMed] [Google Scholar]
- 19.Hatano SY, Tada M, Kimura H, Yamaguchi S, Kono T, Nakano T, Suemori H, Nakatsuji N, Tada T. Pluripotential competence of cells associated with Nanog activity. Mech Dev. 2005;122:67–79. doi: 10.1016/j.mod.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- 22.Chen EY, Xu H, Gordonov S, Lim MP, Perkins MH, Ma'ayan A. Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics. 2013;28:105–111. doi: 10.1093/bioinformatics/btr625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zwaka TP, Thomson JA. A germ cell origin of embryonic stem cells? Development. 2005;132:227–233. doi: 10.1242/dev.01586. [DOI] [PubMed] [Google Scholar]
- 24.Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, Songyang Z, Thomson JA, Zwaka TP. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nichols J, Silva J, Roode M, Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martello G, Bertone P, Smith A. Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. EMBO J. 2013;32:2561–2574. doi: 10.1038/emboj.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye S, Li P, Tong C, Ying QL. Embryonic stem cell self-renewal pathways converge on the transcription factor Tfcp2l1. EMBO J. 2013;32:2548–2560. doi: 10.1038/emboj.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bagci H, Fisher AG. DNA demethylation in pluripotency and reprogramming: the role of tet proteins and cell division. Cell Stem Cell. 2013;13:265–269. doi: 10.1016/j.stem.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Habibi E, Brinkman AB, Arand J, Kroeze LI, Kerstens HH, Matarese F, Lepikhov K, Gut M, Brun-Heath I, Hubner NC, et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, Powell BE, Cheng AW, Faull KF, Lyko F, et al. Loss of tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell. 2014;29:102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grun D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- 40.Blaschke K, Ebata KT, Karimi MM, Zepeda-Martinez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds N, Collier B, Bingham V, Gray NK, Cooke HJ. Translation of the synaptonemal complex component Sycp3 is enhanced in vivo by the germ cell specific regulator Dazl. RNA. 2007;13:974–981. doi: 10.1261/rna.465507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds N, Collier B, Maratou K, Bingham V, Speed RM, Taggart M, Semple CA, Gray NK, Cooke HJ. Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells. Hum Mol Genet. 2005;14:3899–3909. doi: 10.1093/hmg/ddi414. [DOI] [PubMed] [Google Scholar]
- 43.Ruggiu M, Speed R, Taggart M, McKay SJ, Kilanowski F, Saunders P, Dorin J, Cooke HJ. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 44.Hackett JA, Dietmann S, Murakami K, Down TA, Leitch HG, Surani MA. Synergistic mechanisms of DNA demethylation during transition to ground-state pluripotency. Stem Cell Rep. 2013;1:518–531. doi: 10.1016/j.stemcr.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazzoni EO, Mahony S, Iacovino M, Morrison CA, Mountoufaris G, Closser M, Whyte WA, Young RA, Kyba M, Gifford DK, et al. Embryonic stem cell-based mapping of developmental transcriptional programs. Nat Methods. 2011;8:1056–1058. doi: 10.1038/nmeth.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hashimoto H, Pais JE, Zhang X, Saleh L, Fu ZQ, Dai N, Correa IR, Jr, Zheng Y, Cheng X. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature. 2014;506:391–395. doi: 10.1038/nature12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kellner S, Ochel A, Thuring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M. Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucleic Acids Res. 2014;42:e142. doi: 10.1093/nar/gku733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu S, Wang J, Su Y, Guerrero C, Zeng Y, Mitra D, Brooks PJ, Fisher DE, Song H, Wang Y. Quantitative assessment of Tet-induced oxidation products of 5-methylcytosine in cellular and tissue DNA. Nucleic Acids Res. 2013;41:6421–6429. doi: 10.1093/nar/gkt360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfaffeneder T, Spada F, Wagner M, Brandmayr C, Laube SK, Eisen D, Truss M, Steinbacher J, Hackner B, Kotljarova O, et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat Chem Biol. 2014;10:574–581. doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- 50.Tsuji M, Matsunaga H, Jinno D, Tsukamoto H, Suzuki N, Tomioka Y. A validated quantitative liquid chromatography-tandem quadrupole mass spectrometry method for monitoring isotopologues to evaluate global modified cytosine ratios in genomic DNA. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;953–954:38–47. doi: 10.1016/j.jchromb.2014.01.050. [DOI] [PubMed] [Google Scholar]
- 51.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K, Surani MA. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc. 2010;5:516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Table EV1
Table EV2
Table EV3
Table EV4
Table EV5
Table EV6
Review Process File