Abstract
Hepatic cancer is one of the most lethal cancers worldwide. Here, we report that the expression of Ca2+/calmodulin-dependent protein kinase kinase 2 (CaMKK2) is significantly up-regulated in hepatocellular carcinoma (HCC) and negatively correlated with HCC patient survival. The CaMKK2 protein is highly expressed in all eight hepatic cancer cell lines evaluated and is markedly up-regulated relative to normal primary hepatocytes. Loss of CaMKK2 function is sufficient to inhibit liver cancer cell growth, and the growth defect resulting from loss of CaMKK2 can be rescued by ectopic expression of wild-type CaMKK2 but not by kinase-inactive mutants. Cellular ablation of CaMKK2 using RNA interference yields a gene signature that correlates with improvement in HCC patient survival, and ablation or pharmacological inhibition of CaMKK2 with STO-609 impairs tumorigenicity of liver cancer cells in vivo. Moreover, CaMKK2 expression is up-regulated in a time-dependent manner in a carcinogen-induced HCC mouse model, and STO-609 treatment regresses hepatic tumor burden in this model. Mechanistically, CaMKK2 signals through Ca2+/calmodulin-dependent protein kinase 4 (CaMKIV) to control liver cancer cell growth. Further analysis revealed that CaMKK2 serves as a scaffold to assemble CaMKIV with key components of the mammalian target of rapamycin/ribosomal protein S6 kinase, 70 kDa, pathway and thereby stimulate protein synthesis through protein phosphorylation.
Conclusion
The CaMKK2/CaMKIV relay is an upstream regulator of the oncogenic mammalian target of rapamycin/ribosomal protein S6 kinase, 70 kDa, pathway, and the importance of this CaMKK2/CaM-KIV axis in HCC growth is confirmed by the potent growth inhibitory effects of genetically or pharmacologically decreasing CaMKK2 activity; collectively, these findings suggest that CaMKK2 and CaMKIV may represent potential targets for hepatic cancer.
Hepatocellular carcinoma (HCC) constitutes nearly 80% of all hepatic cancers and represents a significant health risk. While the global incidence of HCC largely stems from hepatitis B and C infections or aflatoxin exposure,1 the continuing rise of HCC in the United States directly correlates with the increase in adulthood obesity.2 In fact, HCC is now the third leading cause of cancer-related death worldwide due largely to inadequate early diagnosis coupled with a limited number of effective therapies.3 Perhaps more alarming is that while the incidence of hepatic cancer continues to rise, there has been a steady decline in the 5-year survival rate for HCC patients over the past three decades.4 These observations create an immediate demand to understand the molecular defects responsible for HCC in order to identify suitable biomarkers and functionally characterize candidate-signaling pathways that can be exploited for therapeutic intervention.
Alterations of Ca2+ signaling have been causally linked to the transition toward obesity and are involved in the subsequent inflammatory events that respond to and propagate obesity. Central to this Ca2+ response is Ca2+/calmodulin (CaM)–dependent protein kinase kinase 2 (CaMKK2), which is an important regulator of energy balance, cellular differentiation, and inflammation.5 Mice that are Camkk2−/− display resistance to high-fat diet–induced obesity and liver steatosis, which are potent risk factors for HCC.6 Further supporting these findings, Camkk2−/− mice also show protection against lipopolysaccharide-induced fulminant hepatitis, which closely recapitulates the DNA damage, inflammation, and aberrant cell growth observed during the onset of hepatic cancer.7 Collectively, these findings suggest that CaMKK2 may play a central role in liver cancer development and progression.
A member of the CaMK family, CaMKK2 phosphorylates Ca2+/CaM-dependent protein kinase 1 (CaMKI), Ca2+/CaM-dependent protein kinase 4 (CaMKIV), and adenosine monophosphate–activated protein kinase (AMPK) in response to an increase of intracellular Ca2+ (Fig. 1A).5 Regulation of CaMKI and/or CaMKIV by CaMKK2 regulates cell cycle progression, cell motility, survival, and gene transcription.8 Also, CaMKK2 impacts energy homeostasis in the hypothalamus, adipocyte differentiation, macrophage functions, as well as lipid and carbohydrate metabolism in the liver.5 Whereas others have reported that CaMKK2 is up-regulated by androgen signaling in prostate cancer,9 the mechanistic significance is poorly understood and no information exists on the function of CaMKK2 in hepatic cancer.
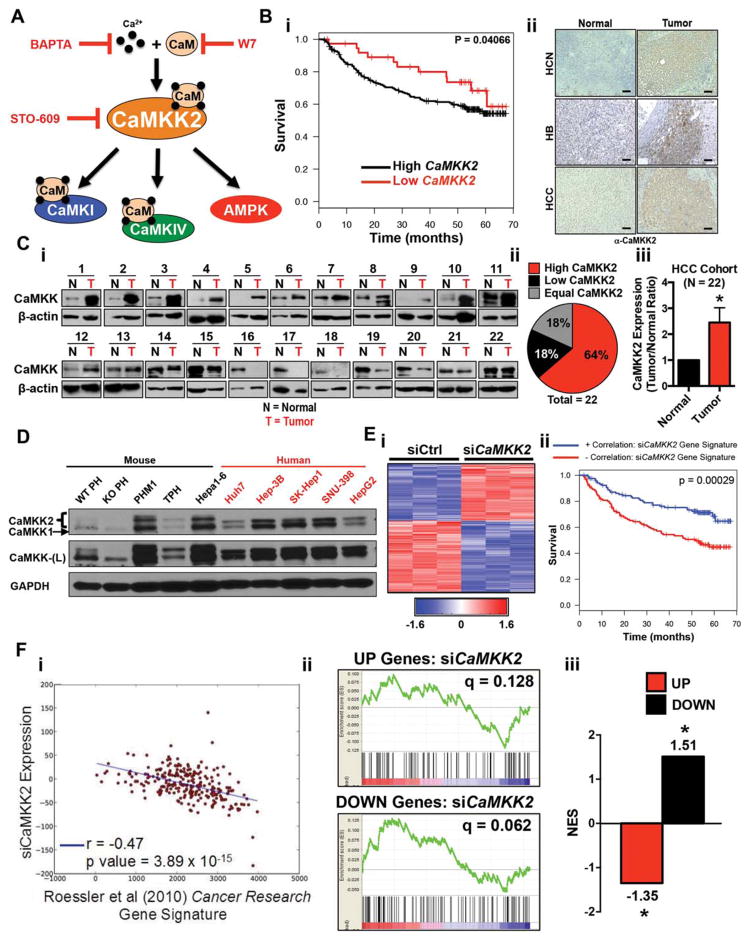
Fig. 1.
Expression of CaMKK2 is up-regulated in HCC and inversely correlates with patient survival. (A) Schematic of the CaMK signaling pathway and pharmacological inhibitors that target each component. (Bi) Kaplan-Meier disease-free survival curve of liver cancer patients (n=247) with high (black line) versus low (red line) Camkk2 expression. (Bii) Immunohistochemical staining of CaMKK2 in matched normal and tumor human liver cancer samples: (left) normal liver samples, (right) liver tumor samples. Scale bar=10 μm. (C) Comparison of CaMKK2 protein expression in tumor tissues and matched adjacent normal tissues from 22 HCC patients. (Ci) Immunoblot of CaMKK2 in normal and tumor tissues. (Cii) Percentage breakdown of immunoblot data with high (red, 64%), low (black, 18%), or equal (gray, 18%) CaMKK2 protein expression in tumor compared to normal tissue. (Ciii) Normalized densitometric quantification of CaMKK2 protein expression in matched tumor versus normal liver tissue. (D) Immunoblot analysis of CaMKK2 protein expression in WT and Camkk2−/− primary hepatocytes and eight liver cancer cell lines. (Ei) Hierarchical clustering of microarray data from PHM1 cells treated with either siControl or siCamkk2. (Eii) Kaplan-Meier survival plot of liver cancer patients (n=247) comparing a positive correlation (blue lines) with the siCamkk2 gene signature or negative correlation (red lines) with the siCamkk2 gene signature. (Fi) Pearson correlation of siCaMKK2 gene signature in PHM1 cells compared to the Roessler et al.14 gene signature (r=−0.47; P value=3.89×10−15). (Fii) Comparison of the up- and down-regulated gene signatures in liver cancer patients with those impacted by silencing Camkk2 by siRNA using GSEA. (Fiii) Histogram of normalized enrichment score from GSEA comparison in Fii. Abbreviations: BAPTA, 1,2,-bis(o-aminophenoxy)ethane-N,N,-N′,N′-tetra-acetic acid; HB, hepatoblastoma; HCN, hepatocellular neoplasia; L, long exposure; N, normal liver; NES, normalized enrichment score; si, small interfering; T, liver tumor.
Herein, we demonstrate that CaMKK2 is overexpressed in HCC patient biopsies. Consistent with these findings, we have identified critical roles for CaMKK2 in the regulation of hepatic cancer cell growth. Our data reveal that CaMKK2 functions through CaMKIV to regulate the mammalian target of rapamycin (mTOR)/ribosomal protein S6 kinase, 70 kDa (S6K), pathway, which is essential for cancer cell growth and protein synthesis.10,11 In fact, aberrant mTOR/S6K pathway activation in HCC has been reported to be as high as 41%,12 thus identifying this pathway as an independent predictor for hepatic cancer recurrence.13 Our results show that CaMKK2 serves a scaffolding role that integrates its Ca2+-responsive kinase activity to control protein translation, which is required for optimal liver cancer cell growth. Moreover, this study suggests that CaMKK2 inhibitors could have therapeutic potential against liver cancer.
Materials and Methods
Colony Formation Assay
Liver cancer cells were seeded at 1000/mL/well into 12-well plates. Colonies were formed for 14 days (7 days for PHM1). Medium and inhibitors were refreshed every 3 days. Cells were fixed with 4% paraformaldehyde, and colonies were stained with 0.025% crystal violet. The crystal violet stain was eluted with 60% isopropanol and quantified at an optical density of 550 nm.
Pulse Chase
Cells were serum-fasted overnight, and 10% serum containing 5 μM puromycin was added back to the medium for 2 hours. Protein synthesis was detected by immunoblot with anti-puromycin antibody. Total protein was determined by Coomassie staining.
2-Deoxy-2-(18F)Fluoro-D -Glucose Positron Emission Tomographic Imaging
The small animal Inveon System with multimodality computed tomography (CT) and docked positron emission tomography (PET) was used for in vivo μPET/CT. The CT parameters were set at 80 kV, 500 μA, and 290 ms exposure time at each of the 180 rotation steps over 360° at low magnification. Mice received 150 μCi of 2-deoxy-2-(18F)fluoro-D-glucose in 0.1-cc by tail vein injection 1 hour prior to PET/CT imaging. Scanning by PET employed an acquisition time of 5 minutes. The CT images were reconstructed using a Feldkamp conebeam algorithm. The system utilizes a two-dimensional filtered back-projection for reconstruction of PET images.
Diethylnitrosamine-Induced Hepatic Tumorigenesis
Male wild-type (WT) mice (n=10) were i.p.- injected with diethylnitrosamine (DEN; 25 mg/kg in dimethyl sulfoxide) at postnatal day 15. Tumor progression was monitored for 9 months postinjection, and livers were isolated and homogenates made for immunoblot analysis. For STO-609 treatment, a baseline measurement of hepatic tumor burden in WT mice (n=8) 6 months postinjection was established by PET/CT imaging of 2-deoxy-2-(18F)fluoro-D-glucose. Each group of mice received either vehicle (10% dimethyl sulfoxide in phosphate-buffered saline) or STO-609 by i.p. injection (30 μg/kg body weight) twice per week for 4 weeks. Following 4 weeks of treatment, a second round of PET/CT imaging was performed to determine final tumor burden. Mice were sacrificed, and livers were isolated for immunoblot analysis.
Results
CaMKK2 Expression Is Up-Regulated in HCC and Inversely Correlates With Patient Survival
In response to Ca2+/CaM binding, CaMKK2 phosphorylates and activates CaMKI, CaMKIV, and AMPK (Fig. 1A). To examine the relationship of CaMKK2 expression to patient survival in human liver cancer, transcriptome profiles of 247 human HCC patients from a publicly available microarray data set14,15 were stratified into patients with high versus low CaMKK2, and those bins were used to generate a Kaplan-Meier survival plot (Fig. 1Bi). The clinical follow-up data of the HCC patient cohort demonstrate that high CaMKK2 expression correlates with poor disease-free survival (Fig. 1Bi, black line). These expression data are consistent with immunohistochemical staining of CaMKK2 in tumor tissue sections from liver cancer biopsies (hepatocellular neoplasia, hepatoblastoma, HCC), which show stronger CaMKK2 staining in the tumor regions than in adjacent normal tissue (Fig. 1Bii). In a separate cohort of 22 HCC patient samples, CaMKK2 protein was up-regulated in tumor compared to adjacent normal tissue (Fig. 1Ci) as 64% (14/22) of the patient-derived tumors display increased CaMKK2 protein selectively (Fig. 1Cii), and the level of overexpression was approximately 2.5 times greater than in the adjacent normal tissue (Fig. 1Ciii).
To examine CaMKK2 expression in a relevant cellular context, we evaluated CaMKK2 protein levels in normal primary hepatocytes isolated from WT and Camkk2−/− mice alongside eight liver cancer cell lines of human or mouse origin. All tumor cell lines examined display significantly increased CaMKK2 expression compared to normal hepatocytes (Fig. 1D). To determine the extent to which the CaMKK2 responsive gene signature in liver cancer cells correlates with human cases of HCC, we knocked down Camkk2 in PHM1 cells by small interfering RNA (siRNA) and performed a microarray analysis. Hierarchical clustering of the transcriptome profiles revealed a number of genes whose expression increased or decreased upon Camkk2 silencing (Fig. 1Ei). Gene ontology analyses comparing the siCamkk2 gene signature with HCC microarray data sets identified 15 common pathways (Supporting Fig. S1D,E). We investigated the association of the CaMKK2 gene signature with survival and recurrence data using the same HCC human cohort described in Fig. 1Bi. We found that the gene signature produced by silencing Camkk2 conferred a marked improvement in survival (Fig. 1Eii) and recurrence (Supporting Fig. S1C) of HCC patients. Correlation of the siCamkk2 gene signature with the HCC tumor progression signature over the gene transcriptome profiles from Roessler et al.14,15 revealed a strong negative correlation (r=−0.47, P=3.89 × 10−15), further verifying the protective effect of attenuating CaMKK2 signaling on HCC patient survival (Fig. 1Fi). To determine the directionality of gene changes resulting from Camkk2 knockdown with the HCC microarray data, we performed Gene Set Enrichment Analysis (GSEA). These data highlight that genes with low expression in HCC display higher expression upon Camkk2 silencing (Fig. 1Fii–iii). Conversely, genes with high expression in HCC show a negative normalized enrichment score upon Camkk2 silencing, indicating an inverse correlation (Fig. 1Fii–iii) that is consistent with the findings observed in Fig. 1Eii and Fi, respectively.
Loss of CaMKK2 Activity Attenuates Liver Cancer Cell Growth In Vitro
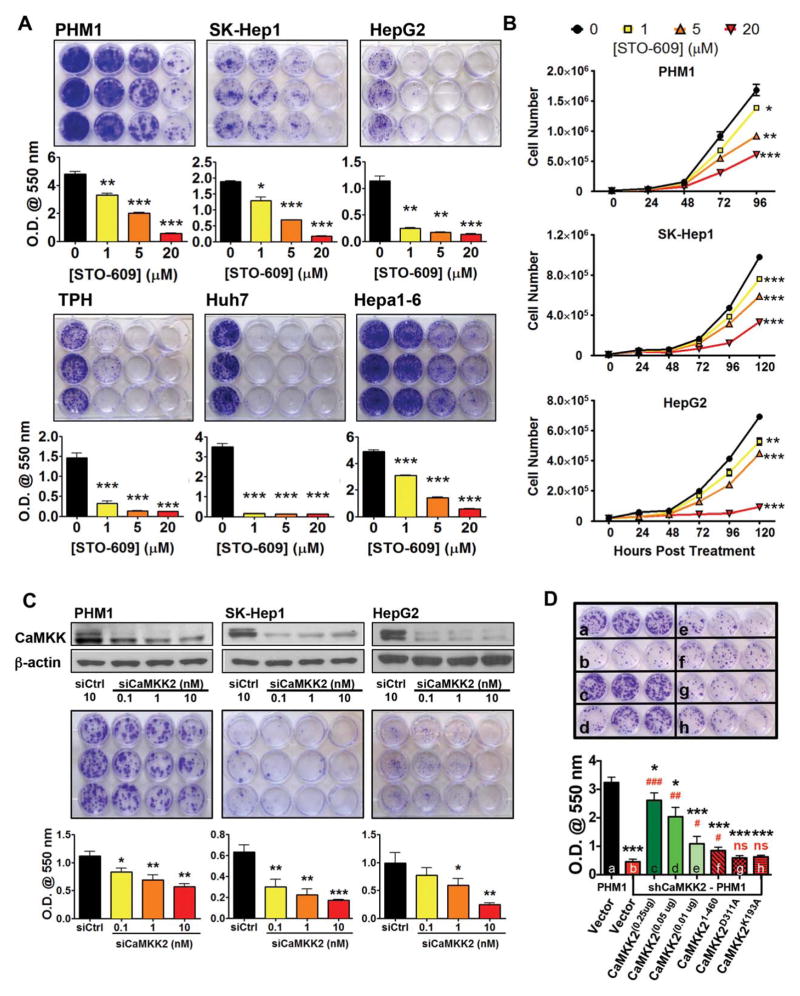
The overexpression of CaMKK2 observed in cellular models of hepatic cancer suggests that up-regulation of CaMKK2 confers a growth advantage to the cancer cell. To evaluate this possibility, we performed colony formation assays on six liver cancer cell lines treated with a selective CaMKK2 inhibitor, STO-609.16 Consistently, STO-609 attenuated colony formation in a dose-dependent manner (Fig. 2A). Proliferation assays performed over 5 days in the presence of increasing concentrations of STO-609 confirmed these results (Fig. 2B). We knocked down CaMKK2 expression by siRNA in three different liver cell lines (PHM1, SK-Hep1, and HepG2) to ascertain whether the STO-609-mediated growth attenuation results from CaMKK2 inhibition. We found that the dose-dependent loss of CaMKK2 impairs liver cancer cell growth in a manner similar to STO-609 treatment, suggesting the effects of the inhibitor are primarily mediated by attenuating CaMKK2 action (Fig. 2C). Impressively, loss or inhibition of CaMKK2 is sufficient to inhibit growth of PHM1 cells (green fluorescent protein [GFP]-c-Myc– transformed/p53−/− liver progenitor cells), demonstrating the effectiveness of CaMKK2 inhibition to impair aggressive cancer cell proliferation.
Fig. 2.
Loss of CaMKK2 activity attenuates liver cancer cell growth in vitro. (A) (Top panels) Crystal violet staining of colony formation assays of six liver cancer cell lines with increasing doses of STO-609. (Bottom panels) Measurement of extracted crystal violet stain at an optical density of 550 nm. (B) Proliferation assays of PHM1, SK-Hep1, and HepG2 liver cancer cells treated with increasing doses of STO-609. (C) Cell lines PHM1, SK-Hep1, and HepG2 were transfected with increasing doses of Camkk2 siRNA and assayed for colony formation potential. (Top panels) Immunoblot analysis of CaMKK2 and β-actin. (Middle panels) Crystal violet staining of colony formation assays upon Camkk2 siRNA knockdown. (Bottom panels) Measurement of extracted crystal violet stain at an optical density of 550 nm. (D) In PHM1 cells, CaMKK2 was stably knocked down using shRNA, and colony formation was performed upon transient transfection with increasing doses of WT CaMKK2 or kinase-inactive CaMKK2 mutants. (Top panel) Crystal violet staining of colony formation. (Bottom panels) Measurement of extracted crystal violet stain at an optical density of 550 nm. Data are graphed as the mean±standard error of the mean. Statistical comparisons: * versus a, # versus b. Shown are the representative replicates of at least three independent experiments. *P<0.05, **P<0.01, ***P<0.001, #P<0.05, ##P<0.01, ###P<0.001. Abbreviation: O.D., optical density.
Next, we generated three independent clones of PHM1 cells with stable knockdown of CaMKK2 by short hairpin RNA (shRNA). Consistent with the cell proliferation inhibition observed with STO-609 treatment or siRNA knockdown of Camkk2, PHM1 cells with stable loss of CaMKK2 displayed decreased growth in both colony formation and proliferation assays (Supporting Fig. S2B,C). To determine whether the proliferation block resulting from loss of CaMKK2 can be rescued by exogenous expression of CaMKK2, we re-introduced CaMKK2 into established Camkk2 shRNA PHM1 cells and found this was sufficient to restore colony-forming potential in a dose-dependent manner (Fig. 2D; Supporting Fig. S2D). Importantly, re-expression of CaMKK2 harboring selective inactivation mutations in the kinase domain (D311A, K192A) failed to rescue liver cancer cell growth (Fig. 2D; Supporting Fig. S2D).
CaMKK2 Coordinates Protein Synthesis by Regulating mTOR/S6K Activity
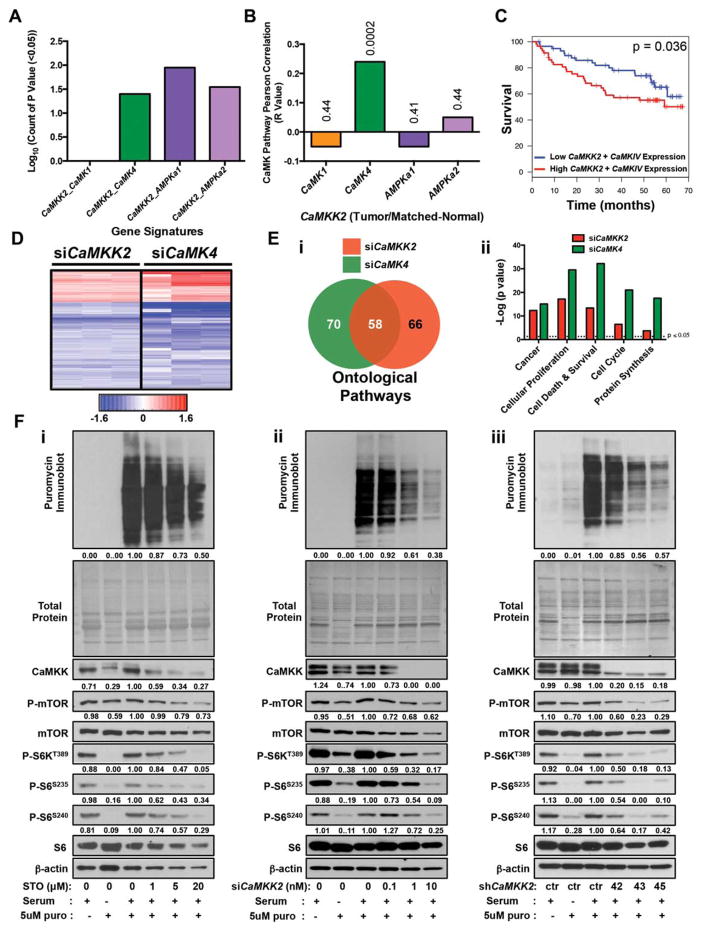
The robust effect of CaMKK2 action on liver cancer cell proliferation prompted us to investigate the mechanistic nature of this molecular phenotype. We began by stratifying the HCC patient survival data on the basis of various CaMKK2 gene signatures (i.e., CaMKK2_CaMK1, CaMKK2_CaMK4, CaMKK2_AMPKa1, CaMKK2_ AMPKa2). These pathway component signatures were analyzed at 148 threshold cutoffs, and the number of statistically significant correlations was determined and graphed (Fig. 3A; Supporting Table S1).17 A significant association was shown between CaMKK2 and survival at numerous thresholds, similar to that of CaMKIV and AMPKa1/a2, but not CaMKI (Fig. 3A). We next analyzed the correlation of Camkk2 gene expression with that of Camki, Camkiv, Ampka1, and Ampka2 in the human HCC microarray data set. Interestingly, we found that only Camkiv positively and significantly correlated with Camkk2 expression (r=0.24, P=0.0002), suggesting a specific functional relationship between these two components of the CaMK signaling pathway in the context of hepatic cancer (Fig. 3B; Supporting Fig. S3A). To examine the importance of Camkk2 and Camkiv expression to patient survival, HCC microarray data sets were first stratified into patients with high versus low Camkiv, and those bins were used to generate Kaplan-Meier plots. The survival and recurrence data demonstrate that, similar to observations made for Camkk2 expression in Fig. 1B, high Camkiv expression also correlates with increased recurrence and poor survival (Supporting Fig. S3B,C, red lines). Importantly, patients with high Camkk2/Camkiv correlated with poor survival and increased HCC recurrence, suggesting that this combined signature can serve as a predictor of liver cancer outcome (Fig. 3C; Supporting Fig. S3D). To understand the downstream actions of CaMKK2 and CaMKIV in liver cancer cells, we knocked down Camkiv in PHM1 cells by siRNA, performed microarray analyses, and compared the siCamkiv gene signature to that of siCamkk2 from Fig. 1Ei. Hierarchical clustering of the transcriptomic profiles revealed that the expression of a number of common genes increases or decreases upon Camkk2 or Camkiv silencing (Fig. 3D). Gene ontology was performed using the CaMKK2 and CaMKIV gene signatures to identify candidate pathways that overlap (Fig. 3E). We identified a cassette of putative overlapping cellular processes that might explain this cellular phenotype. First, to test the effect of CaMKK2 on cellular processes that influence cell growth, we performed cell cycle analysis of PHM1 cells in the presence of STO-609 or upon selective knockdown using siRNA against Camkk2 (Supporting Fig. S3E,F). These data demonstrate that inhibition or acute deletion of CaMKK2 had negligible effects on cell cycle progression. Next, we examined whether inhibition of CaMKK2 activated apoptosis. We found no change in cleaved caspase 3 in PHM1 cells with increasing doses of STO-609, suggesting that loss of CaMKK2 activity fails to induce apoptosis (Supporting Fig. S3G).
Fig. 3.
CaMKK2 correlates with CaMKIV and coordinates protein synthesis by regulating mTOR/S6K activity. (A) The HCC patient survival data were stratified on the basis of various CaMKK2 gene signatures (i.e., CaMKK2_CaMK1, CaMKK2_CaMK4, CaMKK2_AMPKa1, CaMKK2_AMPKa2). These pathway component signatures were analyzed at 148 different threshold cutoffs (see Supporting Table S1), and the statistically significant correlations (P<0.05) observed were counted and graphed on a log scale. (B) Graphical representation of Pearson correlations of Camkk2 gene expression with that of Camki, Camkiv, Ampka1, and Ampka2 in the human HCC microarray data set.14,15 Only Camkiv positively and significantly correlated with Camkk2 expression (r=0.24, P=0.0002). (C) Kaplan-Meier survival plot of liver cancer patients (n=247) comparing low Camkk2+Camkiv expression (blue lines) with high Camkk2+Camkiv expression (red lines). (D) Hierarchical clustering comparison of microarray data from PHM1 cells treated with either siCamkk2 (from Fig. 1F) or siCamkiv. (Ei) Ingenuity pathway analysis was used to directionally compare microarray data from (D). A Venn diagram shows the overlap of ontological pathways from common gene signatures resulting from siRNA knockdown of Camkk2 (red) and Camkiv (green) in PHM1 cells. (Eii) Graphical summary of candidate overlapping ontological pathways from (Ei). Data are graphed as the −log of the P value for each ontological pathway. Dotted line represents the statistical cutoff for P values <0.05. (F) PHM1 cells in the presence of increasing doses of STO-609 (i), siCamkk2 (ii), or shCamkk2 (iii) were serum-fasted overnight and protein synthesis was monitored by readdition of 10% serum and 5 μM puromycin for 2 h. (Top panels) Puromycin incorporation was analyzed by immunoblot using a puromycin-selective antibody. (Middle panels) Following the immunoblot, the nitrocellulose membrane was stained with Coomassie to examine total protein loading. (Bottom panels) Immunoblot analysis of PHM1 cells for various components of the mTOR/S6K pathway. Densitometry is provided below each immunoblot panel to emphasize significant changes. Shown are representative replicates of at least three independent experiments.
Numerous studies report that changes in Ca2+/CaM signaling influence protein synthesis, although the underlying mechanism that connects these events remains unknown.18,19 To test whether inhibition of CaMKK2 impacts protein translation as suggested by our microarray analyses, we assessed protein synthesis through the treatment of PHM1 cells with increasing doses of STO-609 followed by immunoblot analysis of the regulatory components of the mTOR/S6K pathway. In these experiments, protein synthesis was paused by overnight serum starvation and reinitiated upon refeeding. Once resumed, protein synthesis was monitored by quantifying puromycin incorporation.20 We found that STO-609 markedly decreases protein synthesis in a dose-dependent manner that mirrors the effects observed on S6K/S6 phosphorylation (Fig. 3Fi). Similarly, siCamkk2 attenuates protein synthesis, confirming the inhibitory effect of STO-609 (Fig. 3Fii). Finally, stable knockdown of Camkk2 with shRNA recapitulates the inhibition of protein synthesis resulting from STO-609 or siRNA, highlighting this essential cellular process as a primary mechanism by which loss of CaMKK2 attenuates liver cancer cell growth (Fig. 3Fiii). We found that phosphorylation of p70S6K is decreased upon STO-609 treatment in a dose-dependent manner, as is phosphorylation of its substrate S6 (Fig. 3Fi). Silencing Camkk2 by siRNA resulted in a dose-dependent decrease in phosphorylation of S6K and S6 (Fig. 3Fii). Similar observations were made in PHM1 cells with stable knockdown of CaMKK2 (Fig. 3Fiii). We also examined the effect of pharmacological inhibition or acute ablation of CaMKK2 on the mTOR/S6K pathway in cycling PHM1 cells and observed a robust decrease in S6K and S6 phosphorylation (Supporting Fig. S3J–L). We found similar results in SK-Hep1, a human liver cancer cell line (Supporting Fig. S3H,I), revealing that control of protein synthesis is a fundamental function of CaMKK2 in multiple liver cancer cell lines. Although loss of CaMKK2 expression or activity results in a clear reduction of S6K/S6 phosphorylation, the phosphorylation of mTOR seems to be marginally affected, suggesting that the role of CaMKK2 in S6K signaling could be downstream of mTOR or to facilitate the interaction between mTOR and S6K (Supporting Fig. S3J–L).
CaMKK2 Signals Through CaMKIV to Regulate Liver Cancer Cell Growth
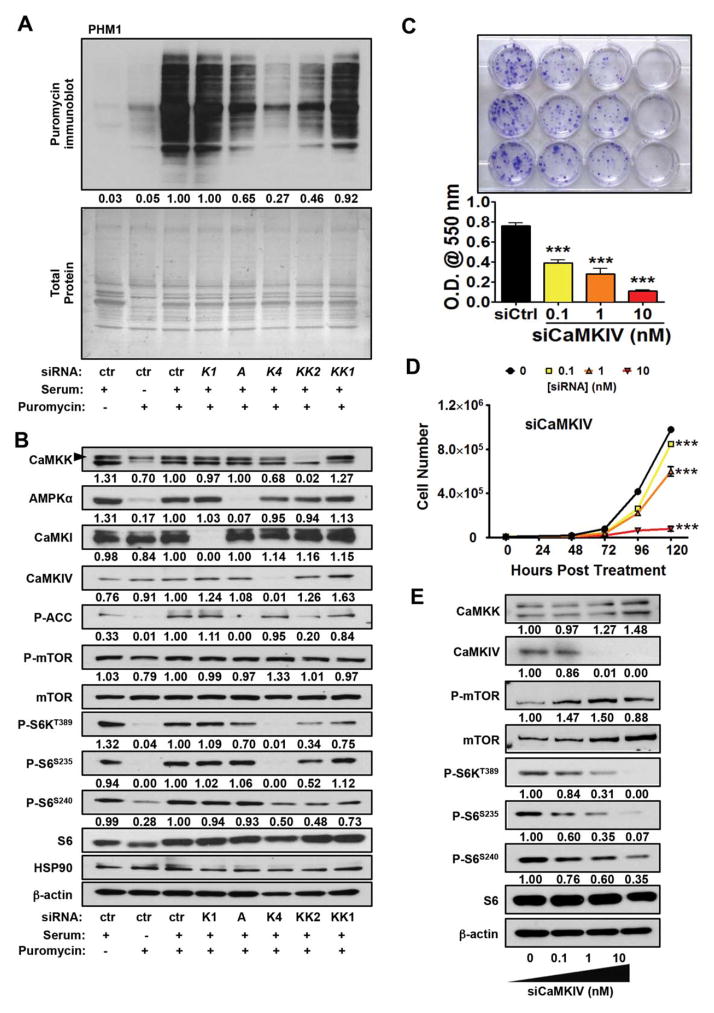
The fact that CaMKK2 activity is necessary to maintain S6K/S6 phosphorylation and optimal protein translation combined with strong correlative data suggesting a functional interplay between CaMKK2 and CaMKIV prompted us to investigate the effects of CaMKIV on protein translation. We compared the effects of acute CaMKK2 ablation to loss of AMPK, CaMKI or CaMKIV on protein synthesis in PHM1. Consistent with our bioinformatics analyses, we found that loss of either CaMKK2 or CaMKIV confers the most significant inhibitory effect on protein synthesis (Fig. 4A). Similar observations were made in SK-Hep1 cells (Supporting Fig. S4A). Analysis of components of the mTOR/S6K pathway by immunoblotting indicates that, consistent with their effect on protein synthesis, knockdown of CaMKK2 or CaMKIV markedly impairs S6K/S6 activity (Fig. 4B). On the contrary, knockdown of AMPK demonstrated a minimal effect on protein synthesis with no impact on S6K/S6 signaling, while loss of CaMKI activity failed to influence either of these indices (Fig. 4A,B), further highlighting the selectivity of CaMKK2 signaling through CaMKIV for controlling protein translation.
Fig. 4.
CaMKK2 functions through CaMKIV to regulate liver cancer cell growth. (A) PHM1 cells were transfected with Control (ctr), Camki (K1), Ampka (A), Camkiv (K4), Camkk2 (KK2), or Camkk1 (KK1) siRNA. Cells were serum-fasted overnight, and protein synthesis was monitored by adding back 10% serum and 5 μM puromycin. Immunoblot of puromycin incorporation and Coomassie staining for total protein are represented. (B) Immunoblot analysis of protein knockdown from (A) and components of the mTOR/S6K pathway are shown. (C) (Top panel) Crystal violet staining of colony formation assays of PHM1 liver cancer cells treated with increasing concentrations (0 to 10 nM) of Camkiv siRNA. (Bottom panel) Measurement of extracted crystal violet stain at an optical density of 550 nm. (D) Proliferation assays of PHM1 cells treated with increasing concentrations (0 to 10 nM) of Camkiv siRNA measured every day for 5 days. (E) Immunoblot analysis of components of the mTOR/S6K pathway in PHM1 cells transfected with increasing doses of Camkiv siRNA. Shown are representative replicates of at least three independent experiments. Data are graphed as the mean±standard error of the mean. *P<0.05, **P<0.01, ***P<0.001. Densitometry is provided below each immunoblot panel to emphasize significant changes. Abbreviation: O.D., optical density.
Given that loss of CaMKIV impairs protein synthesis, we tested the effect of acute CaMKIV ablation on liver cancer cell growth. Treatment of PHM1 cells with increasing doses of siCamkiv results in a marked decrease in colony formation and proliferation (Fig. 4C,D), and immunoblot analysis of the mTOR/S6K pathway indicates that knockdown of CaMKIV attenuates S6K/S6 activity in a manner similar to that of CaMKK2 (Fig. 4E). In addition, we observed that CaMKIV knockdown in SK-Hep1 cells perturbed cell proliferation and S6K/S6 signaling (Supporting Fig. S4B,D). We next assessed whether overexpression of CaMKIV was sufficient to rescue the growth inhibitory effects upon loss of CaMKK2. Colony formation assays were performed using control PHM1 cells or those with stable knockdown of CaMKK2, which were transfected with either WT CaMKIV or a constitutively active CaMKIV (CaMKIV1-317). These data show that overexpression of WT CaMKIV, in the absence of CaMKK2, is insufficient to rescue cell growth (Supporting Fig. S4E). Moreover, overexpression of CaMKIV1-317 only partially rescues the growth inhibition brought on by loss of CaMKK2 (Supporting Fig. S4E). These data argue that CaMKK2 is ratelimiting and likely required for CaMKIV actions on mTOR/S6K signaling.
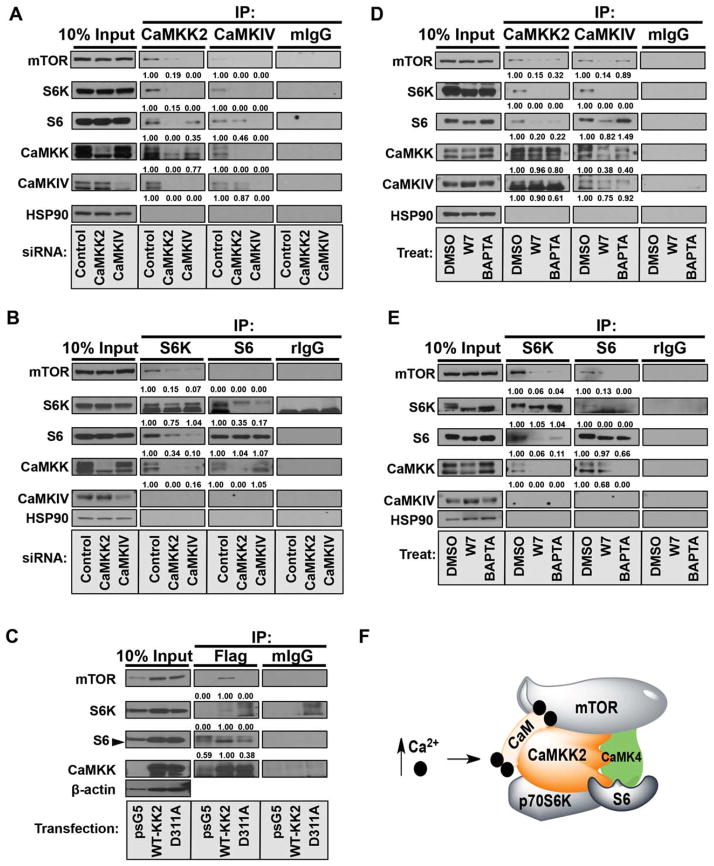
CaMKK2 Complexes With Core Components of the mTOR/S6K Pathway
Because CaMKIV is a well-established substrate of CaMKK221 and this signaling axis was found to regulate protein synthesis, we hypothesized that CaMKK2/CaMKIV may complex with components of the mTOR/S6K pathway. To test this hypothesis, reciprocal coimmunoprecipitations were performed using PHM1 cells in which either Camkk2 or Camkiv was silenced by siRNA. First, either endogenous CaMKK2 or CaMKIV was immunoprecipitated and analyzed for interaction of components of the mTOR/S6K pathway (Fig. 5A). These data demonstrate that CaMKK2 has a stronger interaction with mTOR than does CaMKIV, but silencing CaMKIV markedly disrupts the interaction of mTOR with CaMKK2. A similar observation was made of the interaction with S6K, which shows a stronger association with CaMKK2, yet silencing CaMKIV markedly disrupted this interaction. Together, these data suggest that CaMKK2 interacts with the core mTOR/S6K complex and that CaMKIV is integral to the stability of this complex. Next, we performed immunoprecipitations for S6K or S6 from PHM1 cells in which either Camkk2 or Camkiv was silenced. These data confirm the interaction of mTOR, S6K, and S6 with CaMKK2 and CaMKIV and demonstrate that loss of CaMKK2 abolishes the interaction of S6K with mTOR and S6 (Fig. 5B). Loss of CaMKIV has a similar impact on the constituency of the mTOR/S6K complex. These findings highlight the importance of CaMKK2 and CaMKIV to the integrity of the mTOR/S6K complex and suggest a possible mechanism by which signaling through this axis could regulate protein synthesis.
Fig. 5.
Both CaMKK2 and CaMKIV complex with core components of the mTOR/S6K pathway. (A) Endogenous immunoprecipitation with antibodies specific for CaMKK2, CaMKIV, or mouse immunoglobulin G from PHM1 cells treated with control siRNA, siCamkk2, or siCamkiv, respectively. Immunoprecipitation material was immunoblotted for components of the mTOR/S6K pathway as indicated. (B) Endogenous immunoprecipitation with antibodies specific for S6K, S6, or rabbit immunoglobulin G from PHM1 cells treated with control siRNA, siCamkk2, or siCamkiv, respectively. Immunoprecipitation material was immunoblotted for components of the mTOR/S6K pathway as indicated. (C) Immunoprecipitation from stable shCamkk2-treated PHM1 cells ectopically expressing either full-length CaMKK2 or a catalytic mutant of CaMKK2 (CaMKK2D311A) with antibodies specific for Flag or mouse immunoglobulin G. Immunoprecipitation material was immunoblotted for components of the mTOR/S6K pathway as indicated. (D) Endogenous immunoprecipitation with antibodies specific for CaMKK2, CaMKIV, or mouse immunoglobulin G from PHM1 cells treated with either W7 (CaM antagonist) or BAPTA (Ca2+ chelator). Immunoprecipitation material was immunoblotted for components of the mTOR/S6K pathway as indicated. (E) Endogenous immunoprecipitation with antibodies specific for S6K, S6, or rabbit immunoglobulin G from PHM1 cells treated with either W7 (CaM antagonist) or BAPTA (Ca2+ chelator). Immunoprecipitation material was immunoblotted for components of the mTOR/S6K pathway as indicated. For all immunoprecipitations, 10% of input lysate used for each immunoprecipitation was immunoblotted as a loading control. Densitometry is provided below each immunoblot panel to emphasize significant changes. (F) Proposed model for the organization of Ca2+/CaM stimulation of CaMKK2 and CaMKIV with components of the mTOR/S6K complex. Abbreviations: BAPTA, 1,2,-bis(o-aminophenoxy)ethane-N,N,-N′,N′-tetra-acetic acid; DMSO, dimethyl sulfoxide; IP, immunoprecipitation; mIgG, mouse immunoglobulin G.
To determine if the kinase activity of CaMKK2 is required for its interaction with the mTOR/S6K complex, shCaMKK2-stable PHM1 cells were used to immunoprecipitate ectopically expressed full-length CaMKK2 or a catalytically inactive CaMKK2 mutant (CaMKK2D311A) and analyzed for interaction with components of the mTOR/S6K pathway (Fig. 5C). Immunoprecipitation of ectopically expressed CaMKK2 showed an interaction with mTOR, S6K, as well as S6; and these interactions seem to require the kinase activity of CaMKK2 as a kinase mutant of CaMKK2 is not sufficient to pull down any of the mTOR pathway components (Fig. 5C). To interrogate whether other known substrates of CaMKK2 are involved in this complex, we performed immunoprecipitations of CaMKK2 in PHM1 cells in which Camkk1, Camkk2, Camki, or Ampk was silenced by siRNA. These data confirm the importance of CaMKK2 and CaMKIV in the mTOR/S6K complex, whereas loss of CaMKK1, CaMKI, or AMPK failed to influence this complex (Supporting Fig. S5).
To examine whether Ca2+ and/or CaM signaling is required for the ability of CaMKK2 to complex with the mTOR/S6K machinery, we performed endogenous coreciprocal immunoprecipitations in PHM1 cells treated with either vehicle, W7 (CaM antagonist), or BAPTA (1,2,-bis(o-aminophenoxy)ethane-N,N,-N′,N′- tetra-acetic acid) (Ca2+ chelator). Immunoprecipitation of either CaMKK2 or CaMKIV clearly shows an interaction with components of the mTOR/S6K pathway (Fig. 5D). Treatment with W7 or BAPTA significantly attenuates this complex, suggesting a dependency of Ca2+/CaM on the CaMKK2/CaMKIV/mTOR/S6K complex. Additionally, immunoprecipitation of either S6K or S6 again demonstrates that disruption of Ca2+/CaM signaling is sufficient to blunt the interaction of CaMKK2 with components of the mTOR translation apparatus, highlighting the scaffolding role of CaMKK2 (Fig. 5E). Taken together, these data suggest that CaMKK2 may function in conjunction with CaMKIV as a signaling platform for coordinating accessibility to components of the protein translation regulatory machinery as depicted by the model in Fig. 5F.
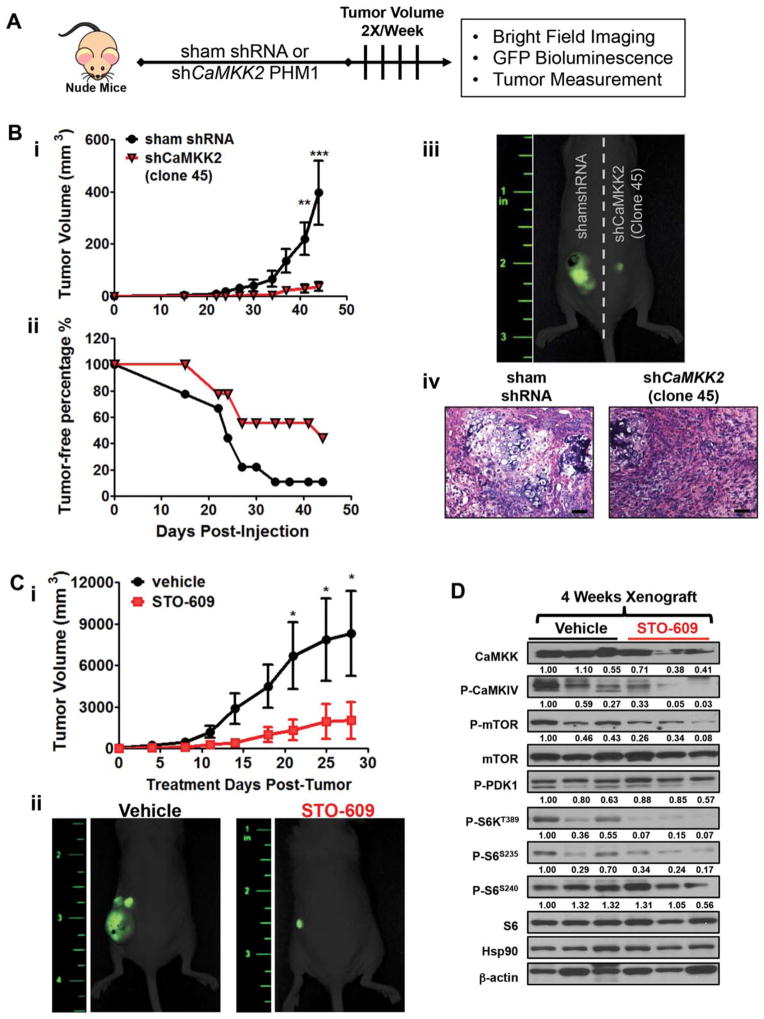
Loss of CaMKK2 Activity Attenuates Liver Cancer Cell Growth In Vivo
We have demonstrated that inhibition or loss of CaMKK2 is sufficient to impair liver cancer cell growth in vitro. To validate these observations in vivo, control PHM1 cells and three independent clones of PHM1 cells with stable knockdown of Camkk2 were injected subcutaneously into nude mice, respectively (Fig. 6A). We measured the tumor outgrowth resulting from control PHM1 cells or those with stable loss of CaMKK2 (shCamkk2) and found that ablation of Camkk2 markedly impaired tumor outgrowth compared to control PHM1 cells (Fig. 6Bi; Supporting Fig. S6A–C). Additionally, visible tumors appeared much later when Camkk2 was ablated (Fig. 6Bii). At the conclusion of the experiment, mice were imaged by GFP fluorescence to visualize PHM1-induced tumors (Fig. 6Biii; Supporting Fig. S6A–C). These data support the tumor measurement data by showing that stable knockdown of CaMKK2 markedly dampens the GFP signal arising from PHM1 cell outgrowth. Hematoxylin and eosin staining of tumor sections from control cells and cells stably expressing shCamkk2 confirmed the inhibitory effect of CaMKK2 knockdown by revealing more organized and less vascularized tumor architecture (Fig. 6Biv; Supporting Fig. S6A–C).
Fig. 6.
Loss of CaMKK2 activity attenuates liver cancer cell growth in vivo. (A) Schematic representation of PHM1 tumor outgrowth model conducted in nude mice; sham shRNA PHM1 cells were injected into the left flank, while PHM1 cells with stable knockdown of Camkk2 were injected into the right flank. (Bi) Longitudinal measurement of tumor outgrowth between control PHM1 cells and PHM1 cells treated with shCamkk2 (n=9). (Bii) Tumor-free percentage of mice from (Bi). (Biii) Images using GFP fluorescence of endpoint tumor outgrowth to detect tumors formed in mice in (Bi). (Biv) Hematoxylin and eosin–stained, paraffin-embedded sections of tumor biopsies from livers of nude mice injected with pLKO control PHM1 cells (left) or shCamkk2-treated PHM1 cells (right). Scale bars=10 μm. (C) Control PHM1 cells (2 × 106/100 μL) were subcutaneously injected into nude mice, and tumor volume was allowed to develop to 3 mm3. Tumor-bearing mice were i.p.- injected with either vehicle (10% dimethyl sulfoxide in phosphate-buffered saline) or STO-609 (30 μg/kg body weight) twice per week for 4 weeks (n=8/treatment group). (Ci) Longitudinal measurement of tumor volume in response to vehicle or STO-609 treatment. (Cii) Images using GFP fluorescence of endpoint tumor outgrowth to detect tumors formed by mice in (Ci). (D) Representative immunoblot analysis of CaMKK2 targets and components of the mTOR/S6K pathway in tumors from (C). Data are graphed as the mean±standard error of the mean. *P<0.05, **P<0.01, ***P<0.001. Densitometry is provided below each immunoblot panel to emphasize significant changes.
Given that stable knockdown of CaMKK2 in cultured liver cells attenuates their growth in vitro, it is not surprising that these cells also lack the ability to initiate tumor outgrowth in vivo. To test the potency of CaMKK2 inhibition for reducing tumor growth in vivo, we injected nude mice with PHM1 cells, allowed tumors to develop (3 mm3), and then treated the tumor-bearing mice longitudinally with either vehicle or STO-609 twice per week for 4 weeks. The STO-609 treatment significantly repressed tumor outgrowth compared to vehicle, as evaluated by the tumor growth curve and GFP fluorescence (Fig. 6Ci,ii). At the end of the 4-week treatment, the tumor mass was excised and used to generate protein lysate for immunoblot analysis of the mTOR/S6K pathway components. Consistent with our in vitro data, inhibition of CaMKK2 with STO-609 blocked the activation of CaMKIV and blunted the mTOR/S6K pathway (Fig. 6D). These data strongly suggest that CaMKK2 activity is required for aggressive liver cancer cell growth in vivo.
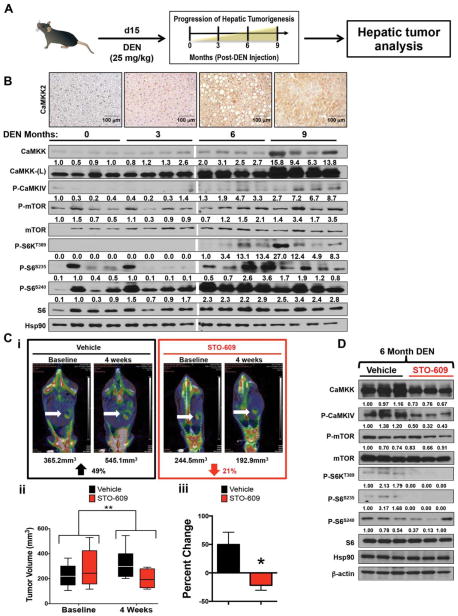
Loss of CaMKK2 Has Beneficial Effects on DEN-Induced Tumorigenesis
Our data reveal that CaMKK2 function is essential for the proliferation of liver cancer cells and that its high expression directly correlates with poor survival in HCC patients. To determine if CaMKK2 inhibition has therapeutic efficacy in attenuating hepatic tumorigenesis, we utilized the DEN-induced mouse model for hepatic cancer that closely resembles the progression and molecular etiology of human HCC22 (Fig. 7A). First, we evaluated CaMKK2 protein expression in WT mice following a time course of DEN treatment. Consistent with the data from matched tumor versus normal HCC samples (Fig. 1Bii) and the analysis of hepatic cancer cell lines (Fig. 1D), CaMKK2 protein levels increase over the course of hepatic tumor progression (Fig. 7B). Analysis of Camkk2 transcript from these same DEN-induced liver cancer samples revealed an increase in Camkk2 expression (Supporting Fig. S7A), which is consistent with the elevated Camkk2 expression observed in microarray data from HCC patients (Fig. 1B). Furthermore, the enhanced CaMKK2 expression observed during tumorigenesis appears to have functional consequences as we also found similar increases in the phosphorylation of CaMKIV (Fig. 7B). Moreover, elevated CaMKK2 seems to coincide with increased protein translation as demonstrated by elevated mTOR/S6K pathway activation (Fig. 7B).
Fig. 7.
Loss of CaMKK2 has beneficial effects on the DEN-induced HCC model. (A) Schematic representation of the DEN-induced HCC model. (B) Immunohistochemical staining of CaMKK2 and immunoblot analysis of CaMKK2 targets and components of the mTOR/S6K pathway in the liver samples of WT mice 0, 3, 6, and 9 months post-DEN injection (n=4 for each time point). (C) Wild-type mice were injected with DEN. (Ci) Six months postinjection, mice were monitored by PET/CT imaging to establish a baseline tumor burden. Tumor-bearing mice were i.p.-injected with either vehicle (10% dimethyl sulfoxide in phosphate-buffered saline) or STO-609 (30 μg/kg body weight) twice per week for 4 weeks. After 4 weeks, mice were monitored again by PET/CT imaging to quantify changes in tumor burden. (Cii) Quantitation of tumor volume for selected mice with equivalent average tumor volume prior to administering either vehicle or STO-609 treatment. (Ciii) Comparison of percentage change of tumor burden between vehicle and STO-609 treatment groups (n=6 for vehicle cohort and n=5 for STO-609 cohort). (D) Immunoblot analysis of CaMKK2 targets and components of the mTOR/S6K pathway in the tumor samples isolated from (C) (n=3 for each treatment group). Data are graphed as the mean±standard error of the mean. *P<0.05, **P<0.01. Densitometry is provided below each immunoblot panel to emphasize significant changes.
To test the therapeutic efficacy of CaMKK2 inhibition, DEN-induced tumorigenesis was initiated and allowed to proceed for 6 months in WT mice, followed by longitudinal treatment with either vehicle or STO-609. We used PET/CT to monitor 2-deoxy-2-(18F)fluoro-D-glucose tumor uptake to establish a baseline hepatic tumor burden prior to drug treatment. Vehicle or STO-609 was administered i.p. twice per week for 4 weeks, after which the treated mice were imaged again by PET/CT. Mice treated with STO-609 for 4 weeks displayed approximately a 21% reduction in tumor burden, while tumors in vehicle-treated mice increased nearly 50% over the same 4-week period (Fig. 7Ci–iii). Consistent with our in vitro and xenograft data, inhibition of CaMKK2 with STO-609 in mice with endogenous liver tumors blocked the activation of CaMKIV and blunted mTOR/S6K signaling (Fig. 7D).
Discussion
Driven by a limited number of therapeutic options and the lack of prognostic markers for early detection of hepatic cancer, the incidence and death rates of HCC have nearly quadrupled in the United States over the last 30 years. These disturbing statistics emphasize the importance and urgency of understanding the mechanisms that underlie the onset and progression of liver cancer.23 Work from our laboratory has shown the importance of CaMKK2 for controlling appetite, energy balance, inflammation, and cellular differentiation.5 In the present study, we demonstrate that CaMKK2 is essential for liver cancer cell growth using both in vitro and in vivo models. Impressively, all eight liver cancer cell lines examined display elevated CaMKK2 expression and activity, underscoring the importance of this signaling node for optimal cell growth. CaMKK2 is one of the most upstream kinases in the Ca2+/CaM kinase cascade.24 As such, it is exciting to discover that inhibition or knockdown of CaMKK2 efficiently attenuates liver cancer cell proliferation even though these cells express extremely high levels of potent oncogenes such as SRC-1, SRC-3, and β-catenin (data not shown), all of which are important for promoting hepatic tumorigenesis. Strong evidence supporting a fundamental role of CaMKK2 for achieving optimal proliferation is that its activity is essential for the growth of PHM1 cells, which are very aggressive p53-null/Myc-transformed liver cancer cells.25 This cell-based conclusion is further supported by the fact that ablation or inhibition of CaMKK2 greatly attenuates tumor growth in vivo. Therefore, it is not surprising to observe an inverse relationship between Camkk2 expression and HCC patient survival. Intriguingly, we demonstrated that untransformed primary murine embryonic fibroblasts and preadipocytes generated from littermate WT or Camkk2−/− mice have identical proliferation rates, suggesting that amplifying CaMKK2 expression/activity is one mechanism by which aberrant cancer cells can achieve a proliferation advantage.6 Consistently, we found that STO-609 treatment of primary hepatocytes does not influence viability, further suggesting that CaMKK2 action in cancer cells is distinct from that of normal cells (data not shown). Therefore, these findings highlight CaMKK2 as an attractive candidate for potential therapeutic intervention of HCC.
The progression toward aberrant cell growth in cancer can be achieved by activating proliferation pathways and/or attenuating those that control apoptosis. Thus, we investigated the effects of CaMKK2 inhibition on cell cycle progression and apoptosis. Interestingly, CaMKK2 inhibition failed to significantly impact cell cycle progression (Supporting Fig. S3E,F) or apoptosis (Supporting Fig. S3G). Nevertheless, based on the striking effects of CaMKK2 inhibition/ablation, we predicted that CaMKK2 must control some fundamental process required for cancer cell growth. Indeed, our literature search and hypothesis-driven approaches suggest that CaMKK2 activity might be necessary for protein synthesis, a process sufficient to confer a growth advantage to the cancer cell. These approaches were supported by a variety of bioinformatic analyses that suggested a strong correlation between CaMKK2 and CaMKIV. Our microarray analysis of CaMKK2 and CaMKIV function in liver cancer cells provided additional validation as pathways for cell growth and protein synthesis were among the candidates. As confirmation of our hypothesis, in response to the same experimental conditions, STO-609 treatment fails to trigger apoptosis but dramatically attenuates the phosphorylation and activation of S6K and S6 (Supporting Fig. S3G). Furthermore, ablation of Camkk2 confirms its role in the regulation of S6K and S6. Our pulse-chase experiments strongly suggest that a primary biological outcome of CaMKK2 action is to promote protein synthesis. Indeed, the mTOR/S6K/S6 protein synthesis pathway is a well-known oncogenic signaling cascade routinely hijacked by cancer cells.11 The robust increase in CaMKK2 protein compared to the modest change in Camkk2 mRNA in liver cancer cell lines, patient samples, and our DEN tumor model suggests that the primary mechanism responsible for this effect is likely post-transcriptional or perhaps translational. Numerous attempts have been made to block the protein translation pathway for cancer therapy, but the requirement for protein synthesis in normal cells hinders their clinical potential. However, the fact that CaMKK2 has restricted expression, coupled with the availability of one selective inhibitor (STO-609), creates the potential to exploit CaMKK2 inhibition for cancer treatment. The feasibility of this idea is supported by our data demonstrating that inhibition of CaMKK2 is sufficient to drastically attenuate the mTOR/S6K pathway, which is critical for cancer cell growth.
Interestingly, although protein synthesis is clearly repressed when CaMKK2 activity is inhibited, the phosphorylation of mTOR is modestly changed when CaMKK2 expression or activity is blunted, suggesting that CaMKK2 may regulate S6K/S6 in an indirect manner. One alternate mechanism for how CaMKK2 might regulate S6K signaling is through PDK1.26 However, PDK1 phosphorylation remains unchanged when CaMKK2 activity is lost (Supporting Fig. S3JL), suggesting that auxiliary kinase pathways downstream of CaMKK2 impact S6K. Indeed, our use of siRNAs against AMPK, CaMKI, and CaMKIV demonstrates that while all three CaMKK2 targets affect liver cancer cell growth (data not shown), CaMKIV was the predominant target of CaMKK2 that impacts liver cancer cell protein synthesis. In fact, inhibition of CaMKIV often showed similar efficacy as inhibiting or ablating CaMKK2 in attenuating liver cancer cell growth, although overexpression of CaMKIV in the absence of CaMKK2 was insufficient to rescue this effect. In fact, although a recent report suggests a functional interplay between CaMKIV and mTOR, our findings provide the first direct evidence demonstrating a coordinated role for CaMKK2 and CaMKIV in the regulation of protein synthesis.27
Signaling of Ca2+ is a well-acknowledged determinant for controlling translation, but the precise mechanism by which Ca2+ regulates this pathway has remained undefined.28,29 Our data suggest that CaMKK2 and CaMKIV provide a platform for the recruitment and coordinated regulation of the mTOR/S6K complex to control protein synthesis. These mechanistic findings combined with the correlation of high Camkk2/Camkiv expression with poor HCC prognosis raise the exciting possibility that combinatorial therapies targeting CaMKK2 and CaMKIV might offer a promising treatment strategy for hepatic cancer. Although the mechanism(s) that promotes elevated CaMKK2 expression in liver cancer remains unknown, analysis of available TCGA-LIHC data shows no copy number increase or selective mutations in Camkk2 (data not shown). However, Camkk2 is an androgen-responsive target gene in prostate cancer9 and, given the importance of androgens in liver cancer, opens new areas of research to determine if hepatic androgen regulation of Camkk2 may provide a partial explanation for the gender bias of liver cancer in men.
Supplementary Material
Acknowledgments
We thank Naomi Gonzales and Thomas Ribar (Duke University) for technical assistance. We thank Chundong Yu (Xiamen University) for HCC samples. The Pathology Core provided histological resources. We acknowledge the joint participation by the Adrienne Helis Malvin Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with Baylor College of Medicine and the CaMKK2 Inhibitors for Therapeutic Treatment of Hepatic Cancer program.
Supported by the National Institutes of Health (GM033976 to A.R.M.).
Abbreviations
- AMPK
adenosine monophosphate–activated protein kinase
- CaM
calmodulin
- CaMKI
Ca2+/calmodulin-dependent protein kinase 1
- CaMKIV
Ca2+/calmodulin-dependent protein kinase 4
- CaMKK2
Ca2+/calmodulin-dependent protein kinase kinase 2
- DEN
diethylnitrosamine
- GFP
green fluorescent protein
- GSEA
gene set enrichment analysis
- HCC
hepatocellular carcinoma
- mTOR
mammalian target of rapamycin
- PET/CT
positron emission tomography/computed tomography
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- S6K
ribosomal protein S6 kinase, 70 kDa
- WT
wild type
Footnotes
Potential conflict of interest: Nothing to report.
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep.27832/suppinfo.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Alzahrani B, Iseli TJ, Hebbard LW. Non-viral causes of liver cancer: does obesity led inflammation play a role? Cancer Lett. 2014;345:223–229. doi: 10.1016/j.canlet.2013.08.036. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 4.Edwards BK, Noone AM, Mariotto AB, Simard EP, Boscoe FP, Henley SJ, et al. Annual report to the nation on the status of cancer, 1975–2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer. Cancer. 2014;120:1290–1314. doi: 10.1002/cncr.28509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Racioppi L, Means AR. Calcium/calmodulin-dependent protein kinase kinase 2: roles in signaling and pathophysiology. J Biol Chem. 2012;287:31658–31665. doi: 10.1074/jbc.R112.356485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin F, Ribar TJ, Means AR. The Ca2+/calmodulin-dependent protein kinase kinase, CaMKK2, inhibits preadipocyte differentiation. Endocrinology. 2011;152:3668–3679. doi: 10.1210/en.2011-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Racioppi L, Noeldner PK, Lin F, Arvai S, Means AR. Calcium/calmodulin-dependent protein kinase kinase 2 regulates macrophage-mediated inflammatory responses. J Biol Chem. 2012;287:11579–11591. doi: 10.1074/jbc.M111.336032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hook SS, Means AR. Ca2+/CaM-dependent kinases: from activation to function. Annu Rev Pharmacol Toxicol. 2001;41:471–505. doi: 10.1146/annurev.pharmtox.41.1.471. [DOI] [PubMed] [Google Scholar]
- 9.Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazli L, et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011;30:2719–2733. doi: 10.1038/emboj.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahin F, Kannangai R, Adegbola O, Wang J, Su G, Torbenson M. mTOR and P70 S6 kinase expression in primary liver neoplasms. Clin Cancer Res. 2004;10:8421–8425. doi: 10.1158/1078-0432.CCR-04-0941. [DOI] [PubMed] [Google Scholar]
- 13.Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972–1983. doi: 10.1053/j.gastro.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–966. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tokumitsu H, Inuzuka H, Ishikawa Y, Ikeda M, Saji I, Kobayashi R. STO-609, a specific inhibitor of the Ca2+/calmodulin-dependent protein kinase kinase. J Biol Chem. 2002;277:15813–15818. doi: 10.1074/jbc.M201075200. [DOI] [PubMed] [Google Scholar]
- 17.Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S, et al. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PloS One. 2012;7:e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf-Brandstetter C, Roessler S, Storch S, Hempel U, Gbureck U, Nies B, et al. Physicochemical and cell biological characterization of PMMA bone cements modified with additives to increase bioactivity. J Biomed Mater Res B Appl Biomater. 2013;101:599–609. doi: 10.1002/jbm.b.32862. [DOI] [PubMed] [Google Scholar]
- 19.Oishi N, Kumar MR, Roessler S, Ji J, Forgues M, Budhu A, et al. Transcriptomic profiling reveals hepatic stem-like gene signatures and interplay of miR-200c and epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma. Hepatology. 2012;56:1792–1803. doi: 10.1002/hep.25890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodman CA, Hornberger TA. Measuring protein synthesis with SUn-SET: a valid alternative to traditional techniques? Exerc Sport Sci Rev. 2013;41:107–115. doi: 10.1097/JES.0b013e3182798a95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selbert MA, Anderson KA, Huang QH, Goldstein EG, Means AR, Edelman AM. Phosphorylation and activation of Ca2+-calmodulin-dependent protein kinase IV by Ca2+-calmodulin-dependent protein kinase Ia kinase. Phosphorylation of threonine 196 is essential for activation. J Biol Chem. 1995;270:17616–17621. doi: 10.1074/jbc.270.29.17616. [DOI] [PubMed] [Google Scholar]
- 22.Williams GM, Iatropoulos MJ, Jeffrey AM. Mechanistic basis for non-linearities and thresholds in rat liver carcinogenesis by the DNA-reactive carcinogens 2-acetylaminofluorene and diethylnitrosamine. Toxicol Pathol. 2000;28:388–395. doi: 10.1177/019262330002800306. [DOI] [PubMed] [Google Scholar]
- 23.Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27:1485–1491. doi: 10.1200/JCO.2008.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson KA, Means RL, Huang QH, Kemp BE, Goldstein EG, Selbert MA, et al. Components of a calmodulin-dependent protein kinase cascade. Molecular cloning, functional characterization and cellular localization of Ca2+/calmodulin-dependent protein kinase kinase beta. J Biol Chem. 1998;273:31880–31889. doi: 10.1074/jbc.273.48.31880. [DOI] [PubMed] [Google Scholar]
- 25.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pullen N, Dennis PB, Andjelkovic M, Dufner A, Kozma SC, Hemmings BA, et al. Phosphorylation and activation of p70s6k by PDK1. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 27.Koga T, Hedrich CM, Mizui M, Yoshida N, Otomo K, Lieberman LA, et al. CaMK4-dependent activation of AKT/mTOR and CREM-alpha underlies autoimmunity-associated Th17 imbalance. J Clin Invest. 2014;124:2234–2245. doi: 10.1172/JCI73411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brostrom CO, Brostrom MA. Calcium-dependent regulation of protein synthesis in intact mammalian cells. Annu Rev Physiol. 1990;52:577–590. doi: 10.1146/annurev.ph.52.030190.003045. [DOI] [PubMed] [Google Scholar]
- 29.Brostrom MA, Brostrom CO. Calcium dynamics and endoplasmic reticular function in the regulation of protein synthesis: implications for cell growth and adaptability. Cell Calcium. 2003;34:345–363. doi: 10.1016/s0143-4160(03)00127-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.