Abstract
Milk fat globule-EGF factor 8 (MFG-E8) is expressed by macrophages and plays an important role in attenuating inflammation and maintaining tissue homeostasis. Previously, we and others found that LPS inhibits MFG-E8 gene expression in macrophages. Here, we characterized the 5′-flanking region of the mouse MFG-E8 gene. To functionally analyze the upstream regulatory region of the MFG-E8 gene, a series of luciferase reporter gene constructs containing deleted or mutated regulatory elements were prepared. Using the luciferase assay, we revealed that Sp1 binding motifs within the proximal promoter region were necessary for full activity of the MFG-E8 promoter, whereas AP-1 like binding sequence at −372 played a role in governing the promoter activity at a homeostatic level. With chromatin immunoprecipitation assay, we showed that Sp1 and c-Jun physically interact with the MFG-E8 promoter region in vivo. In addition, Sp1 was found to regulate the MFG-E8 promoter activity positively and c-Jun negatively. Furthermore, we demonstrated that LPS inhibited MFG-E8 promoter activity via targeting Sp1 and AP-1-like motifs in the 5′-flanking region. Collectively, our data indicate that Sp1 and AP-1-related factors are involved in the regulation of MFG-E8 gene transcription by targeting their binding sites in the 5′-flanking region under physiological and inflammatory states.
Keywords: Inflammation, Gene regulation, Transcription factors, Macrophages
INTRODUCTION
Milk fat globule-EGF factor 8 (MFG-E8) is a glycoprotein derived from macrophages (Reviewed in [Matsuda et al., 2011]). It binds several cell surface molecules such as phosphatidylserine and integrins αvβ3/αvβ5 [Andersen et al., 1997; Hanayama et al., 2002; Taylor et al., 1997; Thery et al., 1999]. Previously, we and others [Atabai et al., 2009; Aziz et al., 2009; Bu et al., 2007; Cui et al., 2010; Hanayama et al., 2002; Hanayama et al., 2004; Wu et al., 2010] have shown that MFG-E8 mediates critical processes involved in anti-inflammation, repair and remodeling of tissues, clearance of apoptotic cells, removal of collagen, neovascularization, and epithelial restitution. It has been demonstrated that MFG-E8 inhibits neutrophil infiltration in acute lung injury via modulation of CXCR2 [Aziz et al., 2012]. MFG-E8 has been found to block lipopolysaccharide-induced TNF-α production in macrophages via STAT3-mediated SOCS3 activation [Aziz et al., 2011]. Furthermore, evidence has suggested that MFG-E8 protects against acute inflammatory injury in numerous organs such as lung, brain, and intestines [Ajakaiye et al., 2012; Aziz et al., 2012; Cheyuo et al., 2012]. Recently, MFG-E8 has been found to regulate osteoclast homeostasis and inflammatory bone loss as well as angiogenesis in cutaneous wound healing [Abe et al., 2014; Uchiyama et al., 2014]. It inhibits inflammasome-induced IL-1β production and limits postischemic cerebral injury [Deroide et al., 2013]. Together, these studies suggest that MFG-E8 plays an important role in mediating diverse homeostatic functions, innate immunity, and tissue protection.
MFG-E8 gene expression is altered in various pathophysiological states. Down-regulation of MFG-E8 has been shown in acute inflammatory conditions such as sepsis, ischemia/reperfusion, and acute alcohol exposure [Bu et al., 2007; Miksa et al., 2006; Wang et al., 2013]. On the other hand, MFG-E8 level was found to be increased during the development of tissue fibrosis and obesity [Atabai et al., 2009; Khalifeh-Soltani et al., 2014]. Furthermore, it has been demonstrated that inflammatory mediators such as lipopolysaccharide (LPS) and reactive oxygen species directly inhibit MFG-E8 gene expression in macrophages [Miksa et al., 2006; Wang et al., 2013]. In addition, Komura et al. reported that MFG-E8 production is down-regulated in sepsis via a CD14 dependent fashion [Komura et al., 2009]. Lack of MFG-E8 may contribute to the pathogenesis of lupus-like autoimmune disorders [Asano et al., 2004; Hanayama et al., 2004; Peng and Elkon, 2011]. However, details of molecular mechanisms underlying the regulation of MFG-E8 gene expression by inflammatory mediators are still not clear.
In the present study, we first characterized the 5′-regulatory region of the mouse MFG-E8 gene and identified the functional role of the interaction between c-Jun and Sp1 in the regulation of MFG-E8 gene promoter activity. Subsequently, we investigated the molecular mechanism by which LPS represses the promoter activity in the MFG-E8 gene. We found that LPS down-regulates MFG-E8 gene expression by targeting Sp1 and AP-1-like motifs in the 5′-flanking region of the MFG-E8 gene. Together, these studies provided evidence that AP-1 and Sp1 are key transcription factors for the regulation of the MFG-E8 gene expression under physiological and pathophysiological conditions.
MATERIALS AND METHODS
Reagents
All cell culture media were obtained from American Type Culture Collection (ATCC, Manassas, VA). Heat-inactivated fetal bovine serum was purchased from Life Technologies (Grand Island, NY). LPS from Escherichia coli 0111:B4, chemicals, mouse mAb against mouse β-actin (catalog # A5441), and molecular biology reagents were purchased from Sigma-Aldrich (St. Louis, MO). RNeasy RNA extraction kit and real-time PCR reagents were purchased from QIAGEN (Valencia, CA). Quick Change Lightning Site-Directed Mutagenesis kit was purchased from Agilent Technologies (La Jolla, CA). iScript™ cDNA synthesis kit was purchased from Bio-Rad (Hercules, CA). All primers and oligonucleotides were synthesized by Integrated DNA Technology (Coralville, IA). Goat polyclonal antibody against murine MFG-E8 (catalog # AF2805) was purchased from R&D Systems (Minneapolis, MN). Horseradish peroxidase (HRP) -conjugated anti-goat or anti-mouse IgG polyclonal antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc (West Grove, PA). Pierce ECL 2 Western Blotting Substrate System was purchased from Thermo Scientific (Southfield, MI). Antibodies against Sp1 and c-Jun were purchased from Active Motif (Carlsbad, CA). The pCMV-c-Jun expression plasmid that encodes the wild-type human c-Jun protein and the empty mammalian expression vector driven by CMV promoter (i.e. pCMV) were generous gifts from Dr. Michael Birrer in NCI at NIH [Brown et al., 1993]. The pCMV-Sp1, a Sp1 expression plasmid (catalog # 12097) was purchased from Addgene (Cambridge, MA).
Cell culture
RAW 264.7 cells (a murine macrophage-like cell line) were purchased from ATCC. The cells were cultured in a water-saturated atmosphere with 5% CO2 at 37°C in Dulbecco’s modified Eagle’s minimum essential medium supplemented with 10% heat-inactivated fetal bovine serum, 1% nonessential amino acids, 100 U/ml penicillin, and 100 μg/ml streptomycin. Passages of 10–18 were used for experiments.
Isolation of peritoneal macrophages
Murine peritoneal macrophages were isolated using a standard protocol in our lab [Bu et al., 2006; Wang et al., 2013]. The procedure was conducted according to the Materials and Methods approved by the Institutional Animal Care and Use Committee. Briefly, C57BL/6 mice (8 weeks old, Jackson Laboratory) were intraperitoneally injected with 1.5 ml of 3% (w/v) thioglycollate (Sigma) for macrophages elicitation. After 4 days, mice were sacrificed with inhalation of CO2 and used for isolation of peritoneal macrophages. To this end, the peritoneum was lavaged with cold serum-free DMEM for several times. The exudate cells were washed, plated at 1 × 106 cells/ml on 6-well plates in DMEM with 10% FBS and antibiotics, and incubated for 2 h at 37°C in a humidified air atmosphere containing 5% CO2. At the end, non-adherent cells were removed by washing plates with culture medium. Adherent cells were then cultured in DMEM containing FBS (10%) overnight and used for experiments.
RNA extraction and quantitative real-time RT-PCR
RNA was isolated from 1 × 106 RAW 264.7 cells using RNeasy RNA extraction kit and reverse-transcribed using iScript™ cDNA synthesis kit according to the manufacturer’s protocol. Quantitative real-time PCR for measuring MFG-E8 gene transcripts was performed using a standard protocol as previously described [Wang et al., 2013]. Sequences of forward (F) and reverse (R) primers for real-time PCR were murine MFG-E8F 5′-ATCTACTGCCTCTGCCCTGA-3′, murine MFG-E8R 5′-CCAGACATTTGGCATCATTG-3′, murine 18S rRNAF 5′-TGCCCTATCAACTTTCGATG-3′, and murine 18S rRNAR 5′-GATGTGGTAGCCGTTTCTCA-3′. The fold change in expression levels of MFG-E8 mRNA in each treatment group were calculated from the threshold cycle (Ct) as 2−ΔΔCT using 18S rRNA as the internal reference. The ΔΔCT value is defined as the Ct difference between the normalized amount of the candidate gene studied and the normalized amount of internal control.
Protein extraction and Western blotting
Total cellular proteins were isolated from RAW 264.7 cells using a protocol as previously described [Wang et al., 2013]. Protein extracts (15 μg) were fractionated in NuPAGE® 4–12% Bis-Tris Gels (precast polyacrylamide gels supplied by Invitrogen) followed by electrophoretically transferred to PVDF membranes. Goat polyclonal antibody against murine MFG-E8 (1:800) was used to detect MFG-E8 protein in blots. HRP-conjugated anti-goat IgG polyclonal antibody (1:3,000) was used as the secondary antibody. After washing with PBS-T, membranes were treated with solutions supplied in the Pierce ECL 2 Western Blotting Substrate System, scanned with Typhoon 7000 (GE Healthcare, Piscataway, NJ), and analyzed with Image Quant™ TL7.0 software (IQ, GE Healthcare). Then, blots were stripped and reprobed with a mouse mAb against β-actin (1/10,000) followed by incubation with a HRP-conjugated anti-mouse IgG (dilution 1:5,000), development with the Pierce ECL 2 Western Blotting Substrate System, scanning with Typhoon 7000, and analyzing with Image Quant™ TL7.0 software as described above.
Plasmid constructs and 5′-sequential deletion
Table 1 describes the sequences of the primers used. Using murine genomic DNA as a template, we prepared a 590-bp DNA fragment by a high fidelity PCR with primer A and primer Z that contained a restriction enzyme site of MluI and XhoI respectively. The amplified DNA fragment contains the murine MFG-E8 gene 5′-upstream region between −444 nt and +146 nt. The PCR-amplified product was run on a 1.5% agarose gel, purified by QIAquick gel extraction kit from QIAGEN, and digested with MluI and XhoI. The digested fragment was purified and subsequently subcloned into MluI and XhoI sites located upstream of the promoterless luciferase gene in the pGL3-basic vector (Promega, Madison, WI) to generate the pGL3-444/+146 plasmid construct. The same Materials and Methods was used for preparing a deletion panel of 5′-flanking MFG-E8 promoter constructs including pGL3-324/+146, pGL3-94/+146, pGL3-19/+146, and pGL3+27/+146. The primer pairs used for generating these constructs were B/Z for pGL3-324/+146, C/Z for pGL3-94/+146, D/Z for pGL3-19/+146, and E/Z for pGL3+27/+146. All constructs were sequenced to verify the correct orientation and sequence of the inserts. The sequence of the MFG-E8 gene promoter in constructs was confirmed by blasting with the sequence of NCBI Gene ID 17304.
Table I.
List of PCR primers used for preparing luciferase promoter constructs
| Primer name | Primer sequence |
|---|---|
| A | 5′-AGCCCAGACTAGTCTGGAACTC-3′ |
| B | 5′-ACTCCTGGATTATTTTATTTTATTT-3′ |
| C | 5′-AGCCCTCCCTCTTCCCAC-3′ |
| D | 5′-GGCCAGTGGGCGGAGCT-3′ |
| E | 5′-GGAGCGGACGCAGGAACT-3′ |
| Z | 5′-GAGGCGCAGAGTAGCATG-3′ |
Site-directed mutagenesis
Quick Change Lightning Site-Directed Mutagenesis System (Agilent Technologies) was used to introduce mutations in the MFG-E8 promoter constructs. The protocol supplied by the manufacturer was followed. The primer pairs used to generate AP-1-like motif mutants were 5′-TGGAACTGTCTGATCCTCCCGCTCCAGCTTCCTAACAGTTGGG-3′ (sense) and 5′-CCCAACTGTTAGGAAGCTGGAGCGGGAGGATCAGACAGTTCCA-3′ (antisense). The primer pairs used to generate Sp1 motif mutants were (1) 5′-CCGGGGCTGTACGGTTCTGAGCCCTTGGGCCA-3′ (sense) and 5′-TGGCCCAAGGGCTCAGAACCGTACAGCCCCGG-3′ (antisense), and (2) 5′-GCCCTTGGGCCAGTGTTCTGAGCTGAGGCGCCTG-3′ (sense) and 5′-CAGGCGCCTCAGCTCAGAACACTGGCCCAAGGGC-3′ (antisense). The underlined nucleotides indicate mutated elements. Primers including the mutation sites were designed with QuickChange Primer Design Program from Agilent Technologies (La Jolla, CA). All constructs containing mutated sites were confirmed by sequencing.
Transient transfection and luciferase reporter assay
A previously described standard protocol was used [Wang et al., 2013]. Briefly, RAW 264.7 cells and antibiotic-free culture medium were used for experiments. For analysis of MFG-E8 promoter activity, cells were plated at 0.4 × 105 cells per well in 24-well plates 1 day prior transfection. Then, they were cotransfected with 0.1 μg of pRL-null reporter plasmid and 0.4 μg of pGL3 luciferase reporter constructs that fused with serial 5′ deletion fragments of the MFG-E8 promoter using Lipofectamine 2000 transfection reagent (Life Technologies) and following the manufacturer’s protocol. Twenty-four hours after transfections, cells were treated with or without LPS (100 ng/ml) for an additional 18 h. Then, cells were processed for measuring luciferase activity using Dual-Luciferase Reporter Assay System (Promega) and following a protocol provided by the manufacturer. Wallac Victor2 1420 Multilabel Counter (PerkinElmer, Waltham, MA) was used for measuring levels of bioluminescence generated in the assay. Levels of the firefly luciferase activity represented the murine MFG-E8 promoter activity, whereas the Renilla luciferase activity was used for normalization of the transfection efficiency. The luciferase activity of each construct was compared with that of the promoterless pGL3 basic vector.
Chromatin immunoprecipitation (ChIP) assay
The assay was performed using ChIP-IT™ Express (Active Motif, CA). In brief, 0.54 ml of 37% formaldehyde was added to RAW 264.7 cells in a 15-cm cell culture dish containing 20 ml of culture medium. Cells were incubated on a shaking platform for 10 min at room temperature for cross-linking of DNA to proteins. Treated cells were then washed with Glycine Stop-Fix Solution and followed by fragmentation of chromatin DNA with sonication and immunoprecipitation of targeted protein-chromatin DNA complexes with an antibody against Sp1 or c-Jun using a protocol for ChIP assay provided by the manufacturer. Then, the isolated protein-chromatin DNA complexes were heated at 65°C for 2.5 h for reverse crosslinking, followed by deproteinization with proteinase K. Finally, MFG-E8 promoter bound to Sp1 or c-Jun were detected using PCR with the following sets of primers: Sp1-F 5′-AATTTCCCCCTGTCCAGTCTAT-3′ and Sp1-R 5′-CTCCTCTCACTCCGGAATAAATC-3′ for the −87 to +31 region; AP-1F 5′-AGGAAACAGGGTCCCATTCT-3′ and AP-1R 5′-AGCATGCTGGAGACCCTAGA-3′ for the −466 to −266 region. The non-specific primers (NSP) used for the post-ChIP PCR were NSP-F 5′-TCCCAAGTGCTGGGATTAAGG-3′ and NSP-R: 5′-AACTCTCAGCTCCTCCTGCACC -3′. The NSP primers detected a region located at about 2.5 kb from −466 nt upstream of the MFG-E8 promoter. The PCR products were analyzed with 2% agarose gel.
Preparation of nuclear extracts
Nuclear extracts from cultured RAW 264.7 cells were prepared using a nuclear protein kit (Millipore, Billerica, MA). The protocol provided by the manufacturer was followed. Protein concentration was measured by the Bradford assay method (BioRad, Hercules, CA). Nuclear extracts were stored in liquid nitrogen till using.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using the Gel Shift Assay System (Promega, Madison, WI) as described before [Chen et al., 2000; De Plaen et al., 1998]. Briefly, double-stranded oligonucleotide probe was prepared by annealing the synthetic oligonucleotides containing −43 nt to +7 nt of the mouse MFG-E8 promoter (5′-GGGGCTGTACGGGGCGGAGCCCTTGGGCCAGTGGGCGGAGCTGAGGCGCC-3′ and 5′-GGCGCCTCAGCTCCGCCCACTGGCCCAAGGGCTCCGCCCCGTACAGCCCC-3′) and was labeled with [γ-32P]ATP (3,000 Ci/mmol, 10 mCi/ml) using T4 polynucleotide kinase. Sequences corresponding to the putative Sp1 binding motifs are underlined. For DNA-protein binding reactions, 5 μg of nuclear extract proteins were incubated in 15 μl of reaction mixture containing 4% glycerol, 1 mM MgCl2, 0.5 mM dithiothreitol, 0.5 mM EDTA, 50 mM NaCl, 10 mM Tris.HCl, pH7.5, and 2.0 μg PolydIdC with or without 125-fold molar excess of an appropriate unlabeled double-stranded DNA element (Figure 5A) at room temperature for 15 min, followed by addition of the 32P-labeled oligonucleotide probe (1.75 pmol/reaction) and incubation at room temperature for 20 min. For supershift experiments, 1 μg of an antibody against Sp1 was added to the completed binding reaction and the mixture was incubated for an additional 10 min at 4°C. Finally, all reaction mixtures were resolved by electrophoresis on a 6% nondenaturing polyacrymide gel at 120 V in 1 x Tris-glycine-EDTA buffer. The gel was dried and analyzed using a phosphorimaging system.
Figure 5.
Identification of the binding of Sp1 protein to putative Sp1 elements in the proximal promoter region of the MFG-E8 gene with EMSA. (A) Oligonucleotide sequences used for EMSA. Only top strands are shown. They correspond to the murine MFG-E8 promoter sequence from − 43 to + 7. Underlined sequences indicate putative Sp1 elements. Lowercase letters in each oligonucleotide sequence indicate the positions of the base substitutions compared to the wild-type sequence. WT, oligonucleotide with wild-type sequence; MUT, oligonucleotide containing the desired mutations within the Sp1 element; Rep, oligonucleotide containing replacement mutated segment in the entire sequence of Sp1 binding motif; NSC, non-specific competitor. (B) To determine the relative affinity of the oligonucleotides of the MFG-E8 proximal promoter region to Sp1 protein, nuclear proteins were extracted from RAW 264.7 cells. EMSA/Supershift assay was performed as described under “Materials and Methods”. (C) To confirm the specificity of the binding affinity of putative Sp1 motifs in the MFG-E8 proximal promoter region, EMSA was performed as described under “Materials and Methods”. In the assay, a 125-fold molar excess of unlabeled double-stranded WT or mutagenized oligonucleotides were used as competitors as indicated in the figure.
Statistical analysis
All experiments were performed at least twice, and most were performed three or more times with similar results. Data were tested by one-way ANOVA and, when appropriate, post hoc Tukey-Kramer multiple comparison tests. Differences were considered significant at P < 0.05. The results were reported as means ± SEM.
RESULTS
1. LPS represses MFG-E8 gene expression in murine macrophages
First, we examine the effect of inflammation on MFG-E8 gene expression in primary mouse peritoneal macrophages. Briefly, primary mouse peritoneal macrophages were stimulated with LPS (100 ng/ml) or incubated with culture medium alone. After 24 hrs, cells were harvested, total RNAs was isolated, and the MFG-E8 gene transcription was measured using real-time RT-PCR. As shown in Figure 1A, LPS treatment markedly inhibited MFG-E8 gene transcription in primary murine macrophages.
Figure 1.
LPS inhibits MFG-E8 gene expression in macrophages. Primary mouse peritoneal macrophages (A) or RAW 264.7 macrophage cells (B and C) were treated with DMEM containing LPS or medium alone (control for 6 and 24 h) as indicated in the figure. At the end of treatments, cells were harvested and processed for extraction of total cellular RNA and protein respectively. Levels of MFG-E8 transcripts were measured by qRT-PCR (A and B), whereas MFG-E8 protein was assessed by western blot analysis (C). Reported values were mean changes vs. control ± SEM (n = 6). **: P < 0.01. The upper panel in C is a representative western blot result and the lower panel in C represents MFG-E8 protein expression as measured by densitometric analysis of immunoblot data. Bars represent mean signal intensity relative to control group. The levels of β-actin gene expression were used for normalizing equal protein loading in experiments. Uncropped images of the Western blot membranes used in this figure are shown in Supplementary Figure 1.
Furthermore, we examined MFG-E8 gene expression in response to LPS in RAW 264.7 cells (a murine macrophage-like cell line). The MFG-E8 mRNA and protein were determined by quantitative real-time RT-PCR and western blotting respectively. We found that MFG-E8 mRNA expression was profoundly decreased 6 h after LPS treatment in a dose-dependent fashion (Fig. 1B). This change persisted 24 h after LPS challenge. Also, the level of MFG-E8 protein was persistently decreased by more than 74% (P<0.01) after LPS treatment (Fig. 1C). Together, the data suggest that LPS directly suppresses MFG-E8 gene expression in macrophages. As the response to LPS stimulation observed in RAW 264.7 cells was similar to the one observed in primary mouse macrophages, suggesting a common mechanism involved in both cell types, and to minimize the animal burden, we chose to use RAW 264.7 cells for the rest of our studies.
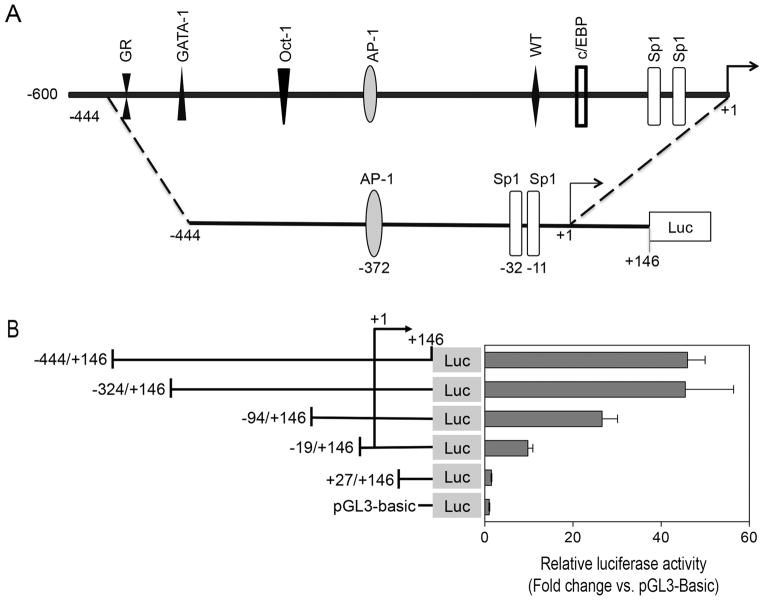
2. Characterization of the 5′-flanking region of the murine MFG-E8 gene promoter
To examine the molecular mechanism underlying repression of the MFG-E8 gene expression by LPS, we characterized the promoter of the murine MFG-E8 gene. Previous studies have suggested that the 5′-regulatory region of the mouse MFG-E8 gene may be present in up to 3 kb upstream of the transcription start site [Aziz et al., 2008; Okuyama et al., 2008]. Through in silico analysis, we predicted that the proximal promoter region of the murine MFG-E8 gene (NCBI Gene ID 17304) is present in the 5′-upstream region extending to a nucleotide position about −450 nt from the transcription initiation site (Fig. 2A). To map the promoter transcriptional activity, we cloned the MFG-E8 5′-upstream sequence (−444 to +146 nt) into a pGL3-basic reporter plasmid and further prepared a series of luciferase reporter constructs representing 5′ deletions of the MFG-E8 promoter (Fig. 2B). These constructs were transiently transfected into RAW 264.7 cells for luciferase assay. As shown in Fig. 2B, transfection of pGL3-444/+146 construct into RAW 264.7 cells resulted in a 46-fold increase in luciferase activity over the promoterless pGL3 basic construct, suggesting that the promoter-like regulatory region lies between −444 and +146. Removal of 5′ sequence up to position −324 did not affect luciferase activity, but deletions beyond this position through −94 resulted in approximate 40% reduction of luciferase activity. Further deleting the 5′-flanking region of the MFG-E8 promoter to −19 caused additional loss of luciferase activity, suggesting the presence of transcriptional enhancers especially between −324 and −19. In addition, it was observed that cells transfected with luciferase reporters containing the nucleotide sequences between −94 to +146 as well as −19 to +146 maintained 60% and 23% luciferase activity respectively. No luciferase activity was detected when deletion was extended to +27. These results suggest that the regulatory elements necessary for maintaining basal transcription of MFG-E8 gene are present between the proximal −94 and the transcription start site of the promoter region.
Figure 2.
Deletion analysis of the 5′-upstream region of the mouse MFG-E8 promoter. (A) Schematic representation of the MFG-E8 gene promoter. The upper panel shows the location of putative transcriptional response elements in the promoter region, whereas the lower panel illustrates the location of Sp1 and AP-1-like elements in the luciferase reporter constructs. (B) Functional and mapping analysis of the mouse MFG-E8 gene promoter under basal conditions by transfection assay. RAW 264.7 cells in each group were cotransfected with pRL-null plasmid (internal control for normalizing the transfection efficiency) and one of a serial deletion constructs shown schematically in the left panel. The position of each construct relative to the transcription start site (+1) and the positions of AP-1-like and Sp1 elements within the reporter contracts are described in the panel A. Cell lysates were prepared 48 h after transfection and tested for luciferase activity as described under “Materials and Methods”. Data represent the mean relative luciferase activity ± SEM from a representative experiment. Similar results were found in 3 independent experiments.
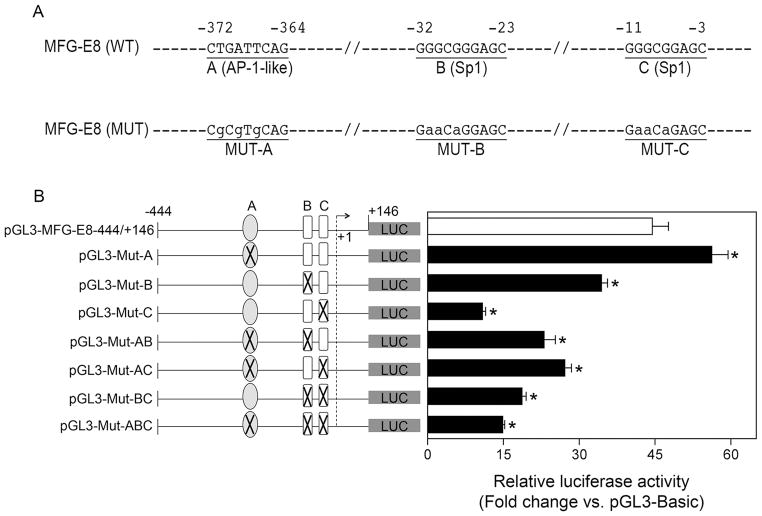
3. Identification of critical motifs maintaining basal activity of the mouse MFG-E8 gene promoter
To determine which transcription factor binding sites that are critical for maintaining the promoter activity of the mouse MFG-E8 gene, we searched for known transcription-factor binding-consensus sequences in the nucleotide sequences between −444 and +146 nt using the Transfast program (Gesellschaft für Biotechnologische, Braunschweig, Germany). This search revealed that the MFG-E8 promoter region contains multiple putative transcription-factor binding motifs (Fig. 2A). However, the sequence of the 5′-flanking region of the gene contains no putative NF-κB consensus binding sites.
We particularly focused on determining whether AP-1-like binding site (at −372 nt) and canonical Sp1 binding sites (at −32 and −11) were functional elements because their role in the transcriptional regulation of the MFG-E8 gene expression has not been elucidated yet. To do so, we introduced point mutations into site A, B, and/or C in the pGL3-444/+146 as indicated in Fig. 3A and 3B (left panel). These sites represent AP-1 or Sp1 binding motifs in the promoter region. The luciferase reporter gene assay indicated that mutation of AP-1-like binding site (i.e. site A) resulted in significant increase in luciferase expression (Fig. 3B, right panel). In contrast, mutation of either site B or C or both B and C sites that are Sp1 binding motifs markedly reduced the basal activity of the MFG-E8 promoter. In addition, using a ChIP assay, we found that c-Jun (a factor in AP-1 complex) and Sp1 physically interact with the MFG-E8 promoter region in vivo (Fig. 4). Together, these data suggest that Sp1 elements within the proximal sequence upstream of the transcriptional initiation site are key motifs for maintaining the basal activity of the MFG-E8 promoter function, whereas AP-1 binding sequence plays a role in governing the promoter activity at a homeostatic level.
Figure 3.
Functional analysis of the MFG-E8 gene promoter with site-directed mutagenesis. (A) Location and site-directed mutagenesis of putative AP-1-like and Sp1 binding sites in the MFG-E8 gene promoter. WT indicates wild-type; MUT indicates mutagenized candidate sites. Mutagenized bases are indicated by lowercase letters. Candidate AP-1-like and Sp1 sites are underlined. (B) Effect of site-directed mutagenesis of putative AP-1-like and Sp1 binding sites on the MFG-E8 promoter activities. The characteristics of MFG-E8 promoter-luciferase reporter constructs used for the experiments are shown on the left panel, whereas the data of luciferase assay for the promoter activity are shown on the right panel. Circles named with A indicate the location of the AP-1-like binding motif. Rectangles named with B and C indicate the locations of the Sp1 binding motifs. The transcriptional start site is indicated as a broken line. The rectangles or circles are marked with an X when a site-replaced mutation in one or more positions is present. RAW 264.7 cells were co-transfected with one of pGL3 luciferase reporter constructs indicated in the figure and pRL-null plasmid (used as an internal control to normalize the transfection efficiency). After 48 h of culture, cells were harvested and cell lysates were assayed for activities of firefly and Renilla luciferase as described under “Materials and Methods”. The firefly luciferase measurements were normalized to Renilla luciferase activity within a given sample. Data represent the mean ± SEM from a representative experiment. Similar results were found in 2 independent experiments. *: P<0.05 vs. the pGL3-MFG-E8-444/+146 control.
Figure 4.

Sp1 and c-Jun bind to the MFG-E8 promoter in vivo. To determine whether Sp1 and c-Jun bind to the MFG-E8 promoter in vivo, ChIP assay was performed. Naïve RAW 264.7 cells were briefly treated with formaldehyde to cross-link proteins to the chromatin DNA as described under “Materials and Methods”. Formaldehyde-treated cells were lysed and sonicated. Immunoprecipitations were then performed using an antibody against Sp1 or c-Jun (AP-1-like). A portion of material not subjected to immunoprecipitation was used as control for the amount of DNA added to each immunoprecipitate (Input). Chromatin DNA-protein complex pulled-down by the antibodies were processed for PCR using primer pairs of P1/P2 or P3/P4 overlapping regions −466 to −266 nt and −87 to +31 nt of the MFG-E8 gene (panel A). NSP indicates non-specific primer set. PCR products were size-fractionized with 2% agarose gel. Results shown in panel B are from a representative experiment. Similar results were found in three independent experiments.
To further confirm the physical association between Sp1 and the identified regions, EMSA experiments were performed using a 32P-labeled double-stranded oligonucleotide probe, namely, Sp1-WT. This probe includes nucleotide sequences present between −43 to +7 nt of the MFG-E8 gene proximal promoter (Fig. 5A), which contains two Sp1 binding motif sequences. Using nuclear protein extracted from naïve RAW 264.7 cells, 32P-labeled Sp1-WT probe was found to form a strong complex with nuclear proteins (Fig. 5B, lane 2). Furthermore, supershift assay confirmed that the Sp1-WT probe indeed bound with Sp1 protein in nuclear proteins of naïve RAW 264.7 cells (Fig. 5B, lane 5). In addition, the cold double-stranded Sp1-WT oligonucleotide but not non-specific competitor (NSC) was found to compete with 32P-labeled Sp1-WT probe binding to nuclear extract (Fig. 5B, lanes 3 and 4), indicating specific binding of the Sp1-WT probe to the targeted nuclear proteins. To determine which Sp1 binding motif in the cold double-stranded Sp1-WT oligonucleotide is a functional element, we prepared two groups of mutated Sp1-WT double-stranded oligonucleotides as described in Fig. 5A. Among them, oligonucleotides in Group I have one intact and one mutated Sp1 motifs, whereas oligonucleotides in Group II have mutations or alterations on both Sp1 motifs. We the performed an EMSA experiment to examine whether these mutated oligonucleotides still have any competitor-like activity. It was found that all oligonucleotides in the Group I can function as a competitor to inhibit the binding of 32P-labeled Sp1-WT probe to nuclear extract proteins (Fig. 5C). In contrast, the oligonucleotides in Group II showed no effect. Taken together, the data suggested that both Sp1 elements contributed to the affinity of the Sp1-WT oligonucleotides to the nuclear extract proteins.
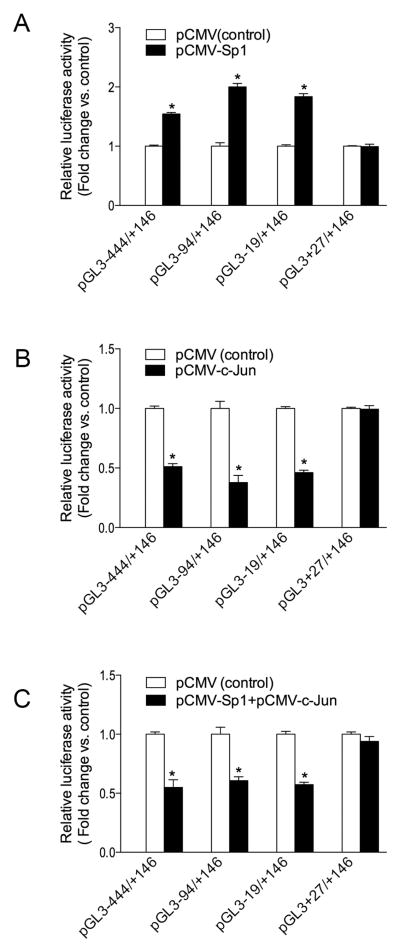
4. Sp1 and c-Jun modulate MFG-E8 promoter activity
In this experiment, we determined the role of Sp1 and c-Jun in the regulation of the MFG-E8 promoter activity using an approach consisting in the ectopic expression of these proteins in RAW 264.7 cells that were co-transfected with pGL3 luciferase reporters fused with the full length or 5′ truncated fragments of the mouse MFG-E8 gene promoter. Luciferase assay showed that the activity of MFG-E8 promoter was significantly increased with all constructs except with pGL3+27/+146 when Sp1 was overexpressed (Fig. 6A). In contrast, the promoter activity was markedly inhibited with all constructs except with pGL3+27/+146 when c-Jun was overexpressed (Fig. 6B). To our surprise, ectopic expression of c-Jun also inhibited luciferase activity in 5′ truncated fragments of the MFG-E8 promoter constructs that lack the AP-1-like binding motif (i.e. constructs −94/+146 and −19/+146 in Fig. 6B). In addition, overexpression of c-Jun also attenuated the effect of ectopic expression of Sp1 on MFG-E8 promoter activity (Fig. 6C). Together, the data further suggest that Sp1 activates the MFG-E8 promoter through targeting the Sp1 sequences in the promoter region, whereas c-Jun negatively regulates the MFG-E8 promoter activity via AP-1-like binding motif dependent and independent mechanisms.
Figure 6.
Role of the ectopic expression of Sp1 and c-Jun on the MFG-E8 promoter activity. RAW 264.7 cells were co-transfected with the plasmids indicated in the figure using the method described in “Materials and Methods”. Each pGL3 luciferase reporter construct contains an appropriate 5′ truncated fragment of the mouse MFG-E8 gene promoter as labeled in each group. pCMV-c-Jun and pCMV-Sp1 are expression plasmids that encode the wild-type human c-Jun and Sp1 proteins. pCMV is an empty mammalian expression vector driven by the CMV promoter and was used as transfection control. After 48 h of transfection, cells were harvested and cell lysates were processed for measuring luciferase activities as described in Fig. 3 legend. The effect of ectopic expression of Sp1, c-Jun, and Sp1 plus c-Jun on the promoter activity is shown in the panels A, B, and C respectively. Values are shown as fold change of luciferase activity relative to pCMV transfection controls. Data represent the mean ± SEM from a representative experiment. Similar results were found in 2 independent experiments. *P<0.05 vs. the control.
5. Characterization of the LPS-targeted functional motifs on the MFG-E8 promoter
In this experiment, we first co-transfected RAW 264.7 cells with pGL3 plasmid constructs containing either −444/+146, −94/+146, −19/+146, and +27/+146 sequences (as described in Fig. 2) and pRL-null plasmid. pGL3-444/+146 is only one to contain an AP-1 binding motif. The pGL3-444/+146 and pGL3-94/+146 have two Sp1 binding motifs, whereas the pGL3-19/+146 contains one Sp1 binding motif. The pGL3+27/+146 was used as a negative control as it lacks the MFG-E8 promoter activity and has no binding motif to either AP-1 or Sp1. Twenty-four hours after transfection, cells were treated with LPS (100 ng/ml) or vehicle (i.e. control) for 24 h and luciferase assay was performed with a standard protocol as previously described [Wang et al., 2013]. We found that the relative luciferase activity was reduced by 38.7 ± 1.1 % in pGL3-444/+146-transfected cells and 23.4 ± 0.47% in pGL3-94/+146-transfected cells 24 h after LPS treatment (P<0.05) compared to control cells (Fig. 7A). In addition, LPS treatment also resulted in a slight but significant reduction of the luciferase expression in pGL3-19/+146-transfected cells. These results indicate that the 5′-flanking region of the MFG-E8 promoter comprises LPS-responsive sequences. Furthermore, AP-1-like and Sp1 binding motifs on the MFG-E8 promoter appeared to be critical elements targeted by LPS.
Figure 7.
LPS inhibits MFG-E8 expression through targeting Sp1 and AP-1-like motifs in the MFG-E8 gene promoter. RAW 264.7 cells were transfected with pGL3 luciferase reporter construct as described under “Materials and Methods”. The pGL3 plasmids containing a full length (i.e. pGL3-444/+146) and 5′ truncated fragments of the MFG-E8 promoter were used for the experiments in panel A, whereas pGL3 plasmid constructs harboring mutated MFG-E8 promoter illustrated in Fig. 3B were used for experiments in panel B. Twenty-four hours after transfection, cells were treated with LPS or medium alone (control) for 24 h. Then, cells were harvested and lysed. Cell lysates were assayed for luciferase activity as described under “Materials and Methods”. Values are shown as fold change of luciferase activity relative to controls. Data represent the mean ± SEM from a representative experiment. Similar results were found in 2 independent experiments. *P<0.05 vs. the control.
To further evaluate the importance of AP-1-like and Sp1 binding motifs in response to LPS, RAW 264.7 cells were transfected with a set of pGL3-444/+146 constructs that have point mutations in the AP-1-like and/or Sp1 elements as indicated in Fig. 3B. The wild-type promoter (pGL3-444/+146) was used as a positive control. As shown in Fig. 7B, the effect of LPS on the inhibition of relative luciferase activity remained in cells transfected with a construct containing the promoter region with mutations at the AP-1-like motif (i.e. site A), a single Sp1 motif (i.e. site B or C), or AP-1-like plus the 1st Sp1 motif (i.e. site A plus site B). In contrast, LPS did not repress the luciferase activity in cells transfected with constructs containing mutations of all Sp1 (i.e. site B and site C) or AP-1-like plus the 2nd Sp1 elements (i.e. site A and site C). Collectively, these data suggest that LPS inhibits the MFG-E8 promoter activity via targeting the Sp1 and AP-1-like motifs in the 5′-flanking region of the murine MFG-E8 gene.
DISCUSSION
We previously showed that the macrophages of intestinal lamina propria express the MFG-E8 protein [Bu et al., 2007]. Recently, we reported that the 5′-flanking region of the murine MFG-E8 gene has a promoter-like activity that is affected by inflammatory stimulations [Wang et al., 2013]. In addition, the expression of the MFG-E8 gene has been shown to be transcriptional regulated by a number of stimuli [Bu et al., 2007; Komura et al., 2009; Miksa et al., 2006; Wang et al., 2013]. However, little is known about the mechanisms underlying the transcriptional control of the MFG-E8 gene in macrophages under physiological conditions and during inflammation. In the present study, we confirmed that the expression of MFG-E8 is down-regulated by LPS. We further characterized the regulatory elements in the promoter region of the MFG-E8 gene which are necessary to respond to inflammation and found that they contain consensus DNA sequences of a number of transcription factor binding motifs. Among them, Sp1 and AP-1-like sites were found to be critical regulatory elements for maintaining the constitutive expression of the MFG-E8 gene and for its repression by inflammatory stimulations in macrophages.
Of particular interest is the identification of two Sp1 binding sites located at the proximal end of the promoter region of the mouse MFG-E8 gene. By deletion and mutation analyses, we showed that Sp1 sites are critical elements that maintain the basal activity of the promoter. In addition, we found that the sequences of the Sp1 elements are highly conserved between mouse and human MFG-E8 genes (data not shown), suggesting that the role of the Sp1 binding motifs in regulating the MFG-E8 gene expression seems to be conserved between the two species. Furthermore, we demonstrated that ectopic expression of Sp1 increases the MFG-E8 promoter activity in a Sp1 binding motif dependent manner, suggesting that the interaction between the transcription factor Sp1 and its binding sites in the promoter region plays a critical role in MFG-E8 gene expression. Indeed, Sp1 is a ubiquitously expressed transcription factor that is involved in maintaining the basal transcription of many genes. A growing body of evidence shows that Sp1 governs gene transcription through direct harboring Sp1-binding GC box sites in the promoter of target genes [Wierstra, 2008]. In addition, Sp1 has been shown to functionally interact with other transcription factors that bind to their respective response elements in the regulatory region of responsive genes [Wierstra, 2008]. With ChIP and EMSA assays, we confirmed that the putative Sp1 elements in the promoter region of MFG-E8 gene constantly interact with the transcription factor Sp1. Together, our data suggest that basal promoter activity of the MFG-E8 gene is maintained by the transcription factor Sp1 under physiological conditions.
In addition, our study provides two pieces of evidence indicating that the AP-1-like site at −372 is a functional motif that interacts with c-Jun for transcriptional regulation of MFG-E8 gene expression. First, we showed that mutation of this AP-1-like sequence resulted in an increase in the promoter activity of the MFG-E8 gene. Secondly, the in vivo interaction between this AP-1-like sequence in the promoter and the transcription factor c-Jun is revealed by ChIP assay. In addition, overexpression of c-Jun resulted in the repression of the MFG-E8 promoter activity. Collectively, our data suggest that c-Jun is involved in the transcriptional regulation of the MFG-E8 gene via a complex mechanism: Under physiological conditions, endogenous c-Jun is bound to the AP-1-like motif at −372, and this interaction plays an important role in adjusting the basal level of expression of the gene. In contrast, when the level of cellular c-Jun increases under pathophysiological conditions, this transcription factor inhibits the MFG-E8 promoter activity through an AP-1 binding site independent manner.
c-Jun has been found to be up-regulated in response to inflammation [Raivich, 2008] and to participate in the suppression of the promoter activity of several inflammatory genes [Afonso et al., 2006; Bois-Joyeux et al., 1995; Drosatos et al., 2007; Farrow et al., 1996; Franklin et al., 1995; Hadzopoulou-Cladaras et al., 1998; Matsuguchi et al., 2003; Wang et al., 2000]. c-Jun physically interacts with Sp1 [Chen and Chang, 2000; Kardassis et al., 1999; Wu et al., 2003], and this interaction has been shown to either enhance or repress the expression of target genes [Chen and Chang, 2000; Kardassis et al., 1999; Yuan et al., 2010]. When c-Jun was ectopically expressed to mimic an increase in cellular c-Jun as found in inflammation, we found that the up-regulation of c-Jun represses the MFG-E8 promoter activity via mechanisms that were both dependent and independent of the AP-1 binding site at −372 nt. In addition, co-transfection of c-Jun with Sp1 attenuated Sp1-mediated enhancement of the MFG-E8 promoter activity, further suggesting that c-Jun is a potent inhibitor of the MFG-E8 gene expression. Thus, it appears that functional interaction between c-Jun and Sp1 are responsible for suppression of the MFG-E8 transcriptional activity under inflammatory conditions. However, the exact mechanism by which c-Jun impairs Sp1-controlled MFG-E8 gene expression remains to be explored.
Furthermore, we analyzed regulatory elements including AP-1 and Sp1 binding motifs in the promoter region to see which ones have critical function in mediating LPS effect on the inhibition of the MFG-E8 gene expression. Using deletion and mutation approaches, we provide evidence that an AP-1-like sequence (−372 to −364 nt) and Sp1 sequences (−32 to −23 nt and −11 to −3 nt) are necessary for the inhibition of transcription of the murine MFG-E8 gene expression by LPS. The functional assays indicate that an AP-1-like element at −372 nt confers a potent motif that mediates the inhibitory effect of LPS. However, to achieve full inhibition by LPS, AP-1-like and Sp1 elements are both needed to be present. These results suggest that LPS inhibits MFG-E8 gene expression via a signaling pathway that involves the cooperation between the AP-1 and Sp1 transcription factors.
It is unclear how AP-1 mediates the LPS inhibitory effects on MFG-E8 gene expression. AP-1 is a family of proteins which contain multiple regulatory factors such as Jun (c-Jun, JunB, JunD) and Fos (c-Fos, FosB, Fra-1 and Fra2) which form complexes with different regulatory functions [Shaulian and Karin, 2002; Wagner and Eferl, 2005]. LPS has been found to up-regulate expression of c-jun and c-fos genes as well as to induce AP-1 binding activity [Dokter et al., 1993; Hambleton et al., 1996]. AP-1 can activate or repress many different genes [Shaulian and Karin, 2002; Wagner and Eferl, 2005]. AP-1 has been shown to mediate repression of gene expression by LPS and TNF [Afonso et al., 2006; Gafencu et al., 2007]. c-Jun is a critical component of the AP-1 protein complex. Previous data revealed that c-Jun is activated during inflammation [Raivich, 2008]. It participates in the suppression of the promoter activity of several inflammatory genes [Afonso et al., 2006; Bois-Joyeux et al., 1995; Drosatos et al., 2007; Farrow et al., 1996; Franklin et al., 1995; Hadzopoulou-Cladaras et al., 1998; Matsuguchi et al., 2003; Wang et al., 2000]. In the present study, we found that (1) an AP-1-like binding site is required for LPS to repress the activity of the MFG-E8 gene promoter; and (2) over-expression of c-Jun results in the down-regulation of the MFG-E8 gene promoter activity. Thus, it is likely that LPS down-regulates MFG-E8 promoter via the AP-1 signaling pathway.
In the present study, we also revealed two additional notable features of the MFG-E8 gene promoter with deletion and mutagenesis assays. First, we found that the deletion of the AP-1-contained DNA fragment (−444 to −324 nt) has no effect on the constitutive activation of the promoter (Figure 2B), whereas mutation of the AP-1 binding site at −372 nt significantly increased the promoter activity (Figure 3B). Such a difference may be explained by the fact that the nucleotide sequences between −444 and −324 nt may pose a “functional unit” containing not only the AP-1 binding site but also an enhancer-like element that is repressed by AP-1. This would be why removal of the entire functional unit by deleting the region between −444 and −324 nt would show no effect on the promoter activity. Secondly, the deletion analysis demonstrated that the construct containing 324 bp upstream of the transcription start site showed full basal promoter activity (Figure 2B). The promoter activity was reduced with the stepwise deletion, which included the removal of putative regulatory elements, such as Sp1, c/EBP, and WT (Figure 2A and B). In the present study, we focused on Sp1 sites to determine their contributions to the basal promoter activity of MFG-E8 and their role in LPS-induced suppression of MFG-E8 gene expression. However, it seems other regulatory sequences within this region may play a role on constitutive activation of MFG-E8 promoter. Taken together, it is of interest to investigate the exact role of other undefined regulatory elements in regulation of MFG-E8 gene expression in futures.
Finally, there are several limitations that should be considered in the interpretation of this study. The main limitation is that we are using only LPS as a stimulus to assess inflammation-induced MFG-E8 gene regulation and utilizing only macrophage-like cells as a cell model in the present study. Evidence showed that MFG-E8 is expressed in numerous cells such as macrophages, dendritic cells, and osteoclasts [Abe et al., 2014; Hanayama et al., 2002; Jinushi et al., 2007]. Previous studies suggest that MFG-E8 gene expression is regulated by multiple factors such as fractakine, alcohol, reactive oxygen species, and LPS [Komura et al., 2009; Leonardi-Essmann et al., 2005; Wang et al., 2013]. Although we defined the molecular mechanism underlying regulation of MFG-E8 promoter activity in macrophages by LPS, it remains unknown (1) whether this regulation is cell-type dependent; and (2) how other inflammatory mediators regulate MFG-E8 gene expression. Another limitation is that we only examined the 5′-promoter region-associated transcriptional regulation of the MFG-E8 gene expression in physiological and inflammation conditions. Evidence has shown that not only transcriptional events but also post-transcriptional processes are critical mechanisms underlying regulation of gene expression. In the present study, we have found that the reduction in MFG-E8 protein levels is notably more dramatic than the change of MFG-E8 mRNA (Figure 1), suggesting that post-transcriptional regulation may play a role. Thus, it would be interesting to examine whether and how RNA stability, the process of RNA turnover, and mechanisms of RNA post-transcriptional regulation are involved in regulation of MFG-E8 gene expression in futures.
In summary, we characterized the promoter region of the mouse MFG-E8 gene. Results from the present study suggest that the transcriptional regulation of MFG-E8 gene is strictly controlled under homeostatic conditions as well as during inflammation. The interaction among transcription factors Sp1, c-Jun, and their binding motifs in the promoter region plays a critical role in the transcriptional regulation of the MFG-E8 gene expression. MFG-E8 has been shown to attenuate inflammation and regulate intestinal epithelial homeostasis. In this study, we clearly show how LPS targets the MFG-E8 gene, leading to these functional consequences. However, further research is needed to understand the exact signaling pathway by which inflammation targets Sp1, c-Jun and inflammation responsive motifs in the promoter of the MFG-E8 gene to alter its expression.
Supplementary Material
Acknowledgments
Grant sponsor: National Institutes of Health; Grant numbers: R01DK064240 (to XDT), R21AA020494 (to XDT), and R01HD060876 (To IDP). Grant sponsor: US Department of Veterans Affairs; Grant number: I01BX001690 (to XDT).
The authors thank Dr. Michael Birrer for providing the pCMV/c-Jun expression plasmid.
Non-standard abbreviations
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
- HRP
horseradish peroxidase
- MFG-E8
milk fat globule-EGF factor 8
- NSC
non-specific competitor
- NSP
non-specific primer
Footnotes
CONFLICT OF INTEREST
All authors declare that they have no conflict of interests to disclose.
References
- Abe T, Shin J, Hosur K, Udey MC, Chavakis T, Hajishengallis G. Regulation of Osteoclast Homeostasis and Inflammatory Bone Loss by MFG-E8. J Immunol. 2014;193:1383–1391. doi: 10.4049/jimmunol.1400970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso V, Santos G, Collin P, Khatib AM, Mitrovic DR, Lomri N, Leitman DC, Lomri A. Tumor necrosis factor-alpha down-regulates human Cu/Zn superoxide dismutase 1 promoter via JNK/AP-1 signaling pathway. Free Radic Biol Med. 2006;41:709–721. doi: 10.1016/j.freeradbiomed.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Ajakaiye MA, Jacob A, Wu R, Yang WL, Nicastro J, Coppa GF, Wang P. Recombinant human MFG-E8 attenuates intestinal injury and mortality in severe whole body irradiation in rats. PLoS ONE. 2012;7:e46540. doi: 10.1371/journal.pone.0046540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen MH, Berglund L, Rasmussen JT, Petersen TE. Bovine PAS-6/7 binds alpha v beta 5 integrins and anionic phospholipids through two domains. Biochemistry. 1997;36:5441–5446. doi: 10.1021/bi963119m. [DOI] [PubMed] [Google Scholar]
- Asano K, Miwa M, Miwa K, Hanayama R, Nagase H, Nagata S, Tanaka M. Masking of phosphatidylserine inhibits apoptotic cell engulfment and induces autoantibody production in mice. J Exp Med. 2004;200:459–467. doi: 10.1084/jem.20040342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atabai K, Jame S, Azhar N, Kuo A, Lam M, McKleroy W, Dehart G, Rahman S, Xia DD, Melton AC, Wolters P, Emson CL, Turner SM, Werb Z, Sheppard D. Mfge8 diminishes the severity of tissue fibrosis in mice by binding and targeting collagen for uptake by macrophages. J Clin Invest. 2009;119:3713–3722. doi: 10.1172/JCI40053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz M, Jacob A, Matsuda A, Wu R, Zhou M, Dong W, Yang WL, Wang P. Pre-treatment of recombinant mouse MFG-E8 downregulates LPS-induced TNF-alpha production in macrophages via STAT3-mediated SOCS3 activation. PLoS ONE. 2011;6:e27685. doi: 10.1371/journal.pone.0027685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz M, Matsuda A, Yang WL, Jacob A, Wang P. Milk fat globule-epidermal growth factor-factor 8 attenuates neutrophil infiltration in acute lung injury via modulation of CXCR2. J Immunol. 2012;189:393–402. doi: 10.4049/jimmunol.1200262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz MM, Ishihara S, Mishima Y, Oshima N, Moriyama I, Yuki T, Kadowaki Y, Rumi MA, Amano Y, Kinoshita Y. MFG-E8 attenuates intestinal inflammation in murine experimental colitis by modulating osteopontin-dependent alphavbeta3 integrin signaling. J Immunol. 2009;182:7222–7232. doi: 10.4049/jimmunol.0803711. [DOI] [PubMed] [Google Scholar]
- Aziz MM, Ishihara S, Rumi MA, Mishima Y, Oshima N, Kadota C, Moriyama I, Li YY, Rahman FB, Otani A, Oka A, Ishimura N, Kadowaki Y, Amano Y, Kinoshita Y. Prolactin induces MFG-E8 production in macrophages via transcription factor C/EBPbeta-dependent pathway. Apoptosis. 2008;13:609–620. doi: 10.1007/s10495-008-0201-1. [DOI] [PubMed] [Google Scholar]
- Bois-Joyeux B, Denissenko M, Thomassin H, Guesdon S, Ikonomova R, Bernuau D, Feldmann G, Danan JL. The c-jun proto-oncogene down-regulates the rat alpha-fetoprotein promoter in HepG2 hepatoma cells without binding to DNA. J Biol Chem. 1995;270:10204–10211. doi: 10.1074/jbc.270.17.10204. [DOI] [PubMed] [Google Scholar]
- Brown PH, Alani R, Preis LH, Szabo E, Birrer MJ. Suppression of oncogene-induced transformation by a deletion mutant of c-jun. Oncogene. 1993;8:877–886. [PubMed] [Google Scholar]
- Bu HF, Wang X, Zhu YQ, Williams RY, Hsueh W, Zheng X, Rozenfeld RA, Zuo XL, Tan XD. Lysozyme-modified probiotic components protect rats against polymicrobial sepsis: role of macrophages and cathelicidin-related innate immunity. J Immunol. 2006;177:8767–8776. doi: 10.4049/jimmunol.177.12.8767. [DOI] [PubMed] [Google Scholar]
- Bu HF, Zuo XL, Wang X, Ensslin MA, Koti V, Hsueh W, Raymond AS, Shur BD, Tan XD. Milk fat globule-EGF factor 8/lactadherin plays a crucial role in maintenance and repair of murine intestinal epithelium. J Clin Invest. 2007;117:3673–3683. doi: 10.1172/JCI31841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BK, Chang WC. Functional interaction between c-Jun and promoter factor Sp1 in epidermal growth factor-induced gene expression of human 12(S)-lipoxygenase. Proc Natl Acad Sci. 2000;97:10406–10411. doi: 10.1073/pnas.180321497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Lu Y, De Plaen IG, Wang LY, Tan XD. Transcription factor NF-kappaB signals antianoikic function of trefoil factor 3 on intestinal epithelial cells. Biochem Biophys Res Commun. 2000;274:576–582. doi: 10.1006/bbrc.2000.3176. [DOI] [PubMed] [Google Scholar]
- Cheyuo C, Jacob A, Wu R, Zhou M, Qi L, Dong W, Ji Y, Chaung WW, Wang H, Nicastro J, Coppa GF, Wang P. Recombinant human MFG-E8 attenuates cerebral ischemic injury: its role in anti-inflammation and anti-apoptosis. Neuropharmacology. 2012;62:890–900. doi: 10.1016/j.neuropharm.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui T, Miksa M, Wu R, Komura H, Zhou M, Dong W, Wang Z, Higuchi S, Chaung W, Blau SA, Marini CP, Ravikumar TS, Wang P. MFG-E8 Attenuates Acute Lung Injury in Mice after Intestinal Ischemia and Reperfusion. Am J Respir Crit Care Med. 2010 Nov 5; doi: 10.1164/rccm.200804-625OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Plaen IG, Tan XD, Chang H, Qu XW, Liu QP, Hsueh W. Intestinal NF-kappaB is activated, mainly as p50 homodimers, by platelet-activating factor. Biochim Biophys Acta. 1998;1392:185–192. doi: 10.1016/s0005-2760(98)00024-1. [DOI] [PubMed] [Google Scholar]
- Deroide N, Li X, Lerouet D, Van VE, Baker L, Harrison J, Poittevin M, Masters L, Nih L, Margaill I, Iwakura Y, Ryffel B, Pocard M, Tedgui A, Kubis N, Mallat Z. MFGE8 inhibits inflammasome-induced IL-1beta production and limits postischemic cerebral injury. J Clin Invest. 2013;123:1176–1181. doi: 10.1172/JCI65167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokter WH, Esselink MT, Halie MR, Vellenga E. Interleukin-4 inhibits the lipopolysaccharide-induced expression of c-jun and c-fos messenger RNA and activator protein-1 binding activity in human monocytes. Blood. 1993;81:337–343. [PubMed] [Google Scholar]
- Drosatos K, Sanoudou D, Kypreos KE, Kardassis D, Zannis VI. A dominant negative form of the transcription factor c-Jun affects genes that have opposing effects on lipid homeostasis in mice. J Biol Chem. 2007;282:19556–19564. doi: 10.1074/jbc.M700986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow KN, Manning N, Schaufele F, Gutierrez-Hartmann A. The c-Jun delta-domain inhibits neuroendocrine promoter activity in a DNA sequence- and pituitary-specific manner. J Biol Chem. 1996;271:17139–17146. doi: 10.1074/jbc.271.29.17139. [DOI] [PubMed] [Google Scholar]
- Franklin CC, McCulloch AV, Kraft AS. In vitro association between the Jun protein family and the general transcription factors, TBP and TFIIB. Biochem J. 1995;305 (Pt 3):967–974. doi: 10.1042/bj3050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafencu AV, Robciuc MR, Fuior E, Zannis VI, Kardassis D, Simionescu M. Inflammatory signaling pathways regulating ApoE gene expression in macrophages. J Biol Chem. 2007;282:21776–21785. doi: 10.1074/jbc.M611422200. [DOI] [PubMed] [Google Scholar]
- Hadzopoulou-Cladaras M, Lavrentiadou SN, Zannis VI, Kardassis D. Transactivation of the ApoCIII promoter by ATF-2 and repression by members of the Jun family. Biochemistry. 1998;37:14078–14087. doi: 10.1021/bi9804176. [DOI] [PubMed] [Google Scholar]
- Hambleton J, Weinstein SL, Lem L, DeFranco AL. Activation of c-Jun N-terminal kinase in bacterial lipopolysaccharide-stimulated macrophages. Proc Natl Acad Sci. 1996;93:2774–2778. doi: 10.1073/pnas.93.7.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S. Identification of a factor that links apoptotic cells to phagocytes. Nature. 2002;417:182–187. doi: 10.1038/417182a. [DOI] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miyasaka K, Aozasa K, Koike M, Uchiyama Y, Nagata S. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science. 2004;304:1147–1150. doi: 10.1126/science.1094359. [DOI] [PubMed] [Google Scholar]
- Jinushi M, Nakazaki Y, Dougan M, Carrasco DR, Mihm M, Dranoff G. MFG-E8-mediated uptake of apoptotic cells by APCs links the pro- and antiinflammatory activities of GM-CSF. J Clin Invest. 2007;117:1902–1913. doi: 10.1172/JCI30966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardassis D, Papakosta P, Pardali K, Moustakas A. c-Jun transactivates the promoter of the human p21(WAF1/Cip1) gene by acting as a superactivator of the ubiquitous transcription factor Sp1. J Biol Chem. 1999;274:29572–29581. doi: 10.1074/jbc.274.41.29572. [DOI] [PubMed] [Google Scholar]
- Khalifeh-Soltani A, McKleroy W, Sakuma S, Cheung YY, Tharp K, Qiu Y, Turner SM, Chawla A, Stahl A, Atabai K. Mfge8 promotes obesity by mediating the uptake of dietary fats and serum fatty acids. Nat Med. 2014;20:175–183. doi: 10.1038/nm.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komura H, Miksa M, Wu R, Goyert SM, Wang P. Milk fat globule epidermal growth factor-factor VIII is down-regulated in sepsis via the lipopolysaccharide-CD14 pathway. J Immunol. 2009;182:581–587. doi: 10.4049/jimmunol.182.1.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi-Essmann F, Emig M, Kitamura Y, Spanagel R, Gebicke-Haerter PJ. Fractalkine-upregulated milk-fat globule EGF factor-8 protein in cultured rat microglia. J Neuroimmunol. 2005;160:92–101. doi: 10.1016/j.jneuroim.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Matsuda A, Jacob A, Wu R, Zhou M, Nicastro JM, Coppa GF, Wang P. Milk fat globule-EGF factor VIII in sepsis and ischemia-reperfusion injury. Mol Med. 2011;17:126–133. doi: 10.2119/molmed.2010.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuguchi T, Masuda A, Sugimoto K, Nagai Y, Yoshikai Y. JNK-interacting protein 3 associates with Toll-like receptor 4 and is involved in LPS-mediated JNK activation. EMBO J. 2003;22:4455–4464. doi: 10.1093/emboj/cdg438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miksa M, Wu R, Dong W, Das P, Yang D, Wang P. Dendritic cell-derived exosomes containing milk fat globule epidermal growth factor-factor VIII attenuate proinflammatory responses in sepsis. Shock. 2006;25:586–593. doi: 10.1097/01.shk.0000209533.22941.d0. [DOI] [PubMed] [Google Scholar]
- Okuyama T, Kurata S, Tomimori Y, Fukunishi N, Sato S, Osada M, Tsukinoki K, Jin HF, Yamashita A, Ito M, Kobayashi S, Hata RI, Ikawa Y, Katoh I. p63(TP63) elicits strong trans-activation of the MFG-E8/lactadherin/BA46 gene through interactions between the TA and DeltaN isoforms. Oncogene. 2008;27:308–317. doi: 10.1038/sj.onc.1210646. [DOI] [PubMed] [Google Scholar]
- Peng Y, Elkon KB. Autoimmunity in MFG-E8-deficient mice is associated with altered trafficking and enhanced cross-presentation of apoptotic cell antigens. J Clin Invest. 2011;121:2221–2241. doi: 10.1172/JCI43254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivich G. c-Jun expression, activation and function in neural cell death, inflammation and repair. J Neurochem. 2008;107:898–906. doi: 10.1111/j.1471-4159.2008.05684.x. [DOI] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- Taylor MR, Couto JR, Scallan CD, Ceriani RL, Peterson JA. Lactadherin (formerly BA46), a membrane-associated glycoprotein expressed in human milk and breast carcinomas, promotes Arg-Gly-Asp (RGD)-dependent cell adhesion. DNA Cell Biol. 1997;16:861–869. doi: 10.1089/dna.1997.16.861. [DOI] [PubMed] [Google Scholar]
- Thery C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J Cell Biol. 1999;147:599–610. doi: 10.1083/jcb.147.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama A, Yamada K, Ogino S, Yokoyama Y, Takeuchi Y, Udey MC, Ishikawa O, Motegi S. MFG-E8 Regulates Angiogenesis in Cutaneous Wound Healing. Am J Pathol. 2014;184:1981–1990. doi: 10.1016/j.ajpath.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol Rev. 2005;208:126–140. doi: 10.1111/j.0105-2896.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- Wang CH, Tsao YP, Chen HJ, Chen HL, Wang HW, Chen SL. Transcriptional repression of p21((Waf1/Cip1/Sdi1)) gene by c-jun through Sp1 site. Biochem Biophys Res Commun. 2000;270:303–310. doi: 10.1006/bbrc.2000.2422. [DOI] [PubMed] [Google Scholar]
- Wang X, Bu HF, Zhong W, Asai A, Zhou Z, Tan XD. MFG-E8 and HMGB1 Are Involved in the Mechanism Underlying Alcohol-Induced Impairment of Macrophage Efferocytosis. Mol Med. 2013;19:170–182. doi: 10.2119/molmed.2012.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra I. Sp1: emerging roles--beyond constitutive activation of TATA-less housekeeping genes. Biochem Biophys Res Commun. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- Wu R, Chaung WW, Zhou M, Ji Y, Dong W, Wang Z, Qiang X, Wang P. Milk fat globule EGF factor 8 attenuates sepsis-induced apoptosis and organ injury in alcohol-intoxicated rats. Alcohol Clin Exp Res. 2010;34:1625–1633. doi: 10.1111/j.1530-0277.2010.01248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhang X, Zehner ZE. c-Jun and the dominant-negative mutant, TAM67, induce vimentin gene expression by interacting with the activator Sp1. Oncogene. 2003;22:8891–8901. doi: 10.1038/sj.onc.1206898. [DOI] [PubMed] [Google Scholar]
- Yuan H, Young CY, Tian Y, Liu Z, Zhang M, Lou H. Suppression of the androgen receptor function by quercetin through protein-protein interactions of Sp1, c-Jun, and the androgen receptor in human prostate cancer cells. Mol Cell Biochem. 2010;339:253–262. doi: 10.1007/s11010-010-0388-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.