Abstract
Anopheles atroparvus is one of the main malaria vectors of the Maculipennis group in Europe. Cytogenetic analysis based on salivary gland chromosomes has been used for taxonomic and population genetic studies of mosquitoes from this group. However, a high-resolution cytogenetic map that could be useful for physical genome mapping in An. atroparvus is still lacking. In the present study, a high-quality photomap for the polytene chromosomes from ovarian nurse cells of An. atroparvus was developed. Using fluorescent in situ hybridization, 10 genes from the 5 largest genomic supercontigs on the polytene chromosome were localized and 28% of the genome were anchored to the cytogenetic map. The study established chromosome arm homology between An. atroparvus and the major African malaria vector An. gambiae suggesting a whole-arm translocation between autosomes of these two species. The standard photomap for ovarian nurse cell chromosomes of An. atroparvus constructed will be useful for routine physical mapping. This map will assist in developing a fine-scale chromosome-based genome assembly for this species and will also facilitate comparative and evolutionary genomics studies in the genus Anopheles.
Keywords: malaria mosquito, cytogenetic map, genome assembly
Introduction
The malaria mosquito Anopheles atroparvus Van Thiel (Diptera, Culicidae) belongs to the subfamily Anophelinae, genus Anopheles, Maculipennis group and Maculipennis subgroup (Harbach, 2004). Species from the Maculipennis group are widely distributed throughout Eurasia and North America. The group comprises 13 palearctic members of which An. messeae Fall, An. sacharovi Favre, An. labranhiae Fall and An. atroparvus are currently considered as dominant vector species in Europe and Middle East (Sinka et al., 2010). The distribution of An. atroparvus in Europe ranges from Britain to Ukraine and the Caucasus region in Russia (Gornostaeva & Danilov, 2002; Sinka et al., 2010). Although malaria was eradicated in Europe in the mid 20th century, it has recently re-emerged in several European countries: Azerbaijan, Georgia, Kyrgyzstan, Tajikistan, Turkey and Uzbekistan (WHO, 2009). The global climate change raises concerns about a malaria re-establishment in Southern Europe (Sainz-Elipe et al., 2010) and revived interest in European malaria mosquitoes including An. atroparvus.
Due to its epidemiological importance and the availability of a well-established colony, An. atroparvus was one of the 16 species of Anopheles chosen for the genome sequencing project (Neafsey et al., 2013; Neafsey et al., 2014). The goal of this sequencing project was to understand the genetic basis of the difference in vectorial capacity between members of genus Anopheles and to establish a platform for comparative genomic analyses. The assembly of the An. atroparvus genome is currently available for analysis through VectorBase (http://www.vectorbase.org). The An. atroparvus genome assembly is 224 Mb in size and is represented by 1371 supercontigs. However, the development of a highly finished genome assembly for An. atroparvus, suitable for comparative genomic analyses, requires further anchoring, ordering and orienting the genomic supercontigs on chromosomes.
Two cytogenetic maps for the salivary gland polytene chromosomes of An. atroparvus have been developed: a drawn map (Kitzmiller et al., 1967) and a photomap (Stegnii & Kabanova, 1978). These maps have been used to establish a standard chromosome nomenclature for the Maculipennis subgroup and to reconstruct a chromosome phylogeny between the Palearctic members of Macilipennis group (Stegnii, 1981). The comparison of the chromosome banding patterns of different species discovered the presence of fixed overlapping inversions. These analyses also identified homosequential species, which have the same banding pattern of chromosomes but differ from each other by the amount and morphology of heterochromatin (Sharakhova et al., 1997). According to the chromosome phylogeny, homosequentional species An. atroparvus and An. labranhiae are basal members of the Maculipennis subgroup (Stegnii, 1981). This observation was confirmed by molecular phylogenetic analysis of enzymes (Bullini & Coluzzi, 1982) and the internal transcribe ribosomal spacer 2 (ITS2) (Marinucci et al., 1999; Kampen, 2005). However, an additional ITS2 comparison of mosquito samples from multiple populations in Iran revealed some contradictions with the original phylogeny (Djadid et al., 2007) and urged further validation. Molecular phylogenies usually reflect a high degree of genetic similarities related to the ancestral polymorphism and introgression, and they often contradict each other (Besansky et al., 1994; Besansky et al., 2003; White et al., 2011). The development of a robust species phylogeny requires a combination of the chromosome inversion data (Kamali et al., 2012) and molecular data (O'Loughlin et al., 2014) with known location on the physical genome map. The availability of the genomic sequences for the species of Anopheles provides an opportunity to perform whole-genome phylogenetic analyses. However, the absence of high-quality cytogenetic photomaps for some of the species remains an impediment to physical genome mapping.
Comparative chromosome analysis revealed striking differences among Palearctic members of the Maculipennis group in the amount and morphology of pericentromeric heterochromatin and in spatial organization of the chromatin inside the nucleus (Sharakhova et al., 1997; Stegniy, 1993). This observation led to a conclusion that reorganization of the three-dimensional chromatin structure is important for speciation in mosquitoes (Stegnii, 2006; Artemov et al., 2010). Microdissection and sequencing of heterochromatin from the centromeric area of chromosome 2 in An. atroparvus demonstrated the presence of genes and repetitive elements in this region (Grushko et al., 2004). Interspecific fluorescent in situ hybridization (FISH) of DNA clones from the microdissected region revealed rapid evolution of heterochromatic sequences among members of the Maculipennis group (Grushko et al., 2009). Availability of the high-quality cytogenetic photomap for An. atroparvus, a basal species of the Maculipennis group, will stimulate further investigation of heterochromatin evolution and speciation in this group of mosquitoes.
The present study aims to construct the first cytogenetic photomap for the polytene chromosomes from ovarian nurse cells of An. atroparvus. The map will use phase-contrast digital images of unstained chromosomes.
Materials and methods
Mosquito strain and ovary preservations
Anopheles atroparvus mosquitoes were obtained from a laboratory colony hosted in the Tomsk State University, Russia. Adult females were fed on guinea pigs. Approximately 24 h post blood feeding, ovaries were pulled out and fixed in Carnoy’s solution (3 ethanol : 1 glacial acetic acid by volume). Ovaries were preserved in fixative solution from 24 h up to one month at −20°C.
Chromosome preparation
For one preparation of ovarian nurse cell chromosomes, a single ovary from one pair was taken. Ovaries were held for 5 min, maturated and squashed in a drop of 50% propionic acid. The quality of the preparation was checked under an AxioImager A1 microscope (Carl Zeiss, OPTEC Company. Siberian Office, Novosibirsk, RF). High-quality preparations were then frozen in liquid nitrogen. Preparations were dehydrated in a series of ethanol (50%, 70%, 90% and 100%) and air dried.
Chromosome map development
Chromosome images were observed using an AxioImager A1 microscope with an attached CCD camera MRc5 in phase contrast using AxioVision rel. 4.7.1 software (OPTEC Company. Siberian Office, Novosibirsk, RF). Images were combined, straightened, shaped and cropped using AdobePhotoshop CS2 (George et al., 2010; Artemov & Stegnii, 2011). Chromosome nomenclature was adopted from previously published chromosome maps for salivary glands of An. atroparvus (Stegnii & Kabanova, 1978) and ovarian nurse cell chromosomes of An. messeae (Stegnii & Sharakhova, 1991).
Fluorescent in situ hybridization
For the probe preparation, gene-specific primers were designed to amplify unique exon sequences from the beginning and end of each of 5 supercontigs using the Primer3 software (v.0.4.0) (Rozen & Skaletsky, 2000). Primer design was based on gene annotations from An. atroparvus genome assembly AatrE1 (Table 3) available at VectorBase (http://www.vectorbase.org). PCR was performed using 2X Immomix DNA polymerase (Bioline USA Inc., MA, USA) and a standard Immomix amplification protocol. Amplified fragments were labeled by nick-translation with Cy3 and Cy5 fluorescent dyes (GE Health Care, UK Ltd., Buckinghamshire, UK). FISH was performed using previously described standard protocol (Sharakhova et al., 2006).
Table 3.
List of primer sequences used for PCR amplification of DNA probes
| Supercontig accession # |
Gene accession # |
Forward primer | Reverse primer |
|---|---|---|---|
| KI421882 | AATE008453 | GTCGAATGCGCAGTCTGTAA | GCCAGGATCACTCCAATGTT |
| AATE010796 | CGCGTACATGTCATCGAAAC | TGAATTCGCTGTACAACACG | |
| KI421883 | AATE005670 | GAAGAACCAAACCACCCAGA | TACTGGAACAGTGGCTCGTG |
| AATE008545 | GCTACAAGATGCAGGGCTTC | TGGTCGAGCTGGAAGAAGTT | |
| KI421886 | AATE001693 | GTGTCATCCCACGAGGACTT | AGCGCTCCTCGTACTGTGTT |
| AATE008759 | CTCCAAGGTGGACAAATCGT | CGACCGTGATGTAGATGTGC | |
| KI421887 | AATE011741 | GAGATTACGCGGACGAAGAG | GATCTTGGTGGGTGTTTGCT |
| AATE006315 | CCGAGAGTGAGGAGAAAACG | TTGCTGATCTAGCCCGTACC | |
| KI421898 | AATE018661 | GCGAGGAGCTGATACAGACC | TTCTTCTTGCGCTCCTTGAT |
| AATE018218 | GTTCCCGGACGAACAGTTTA | CAGCTTGAAGTGGTGGATGA |
Results
Cytogenetic map for Anopheles atroparvus
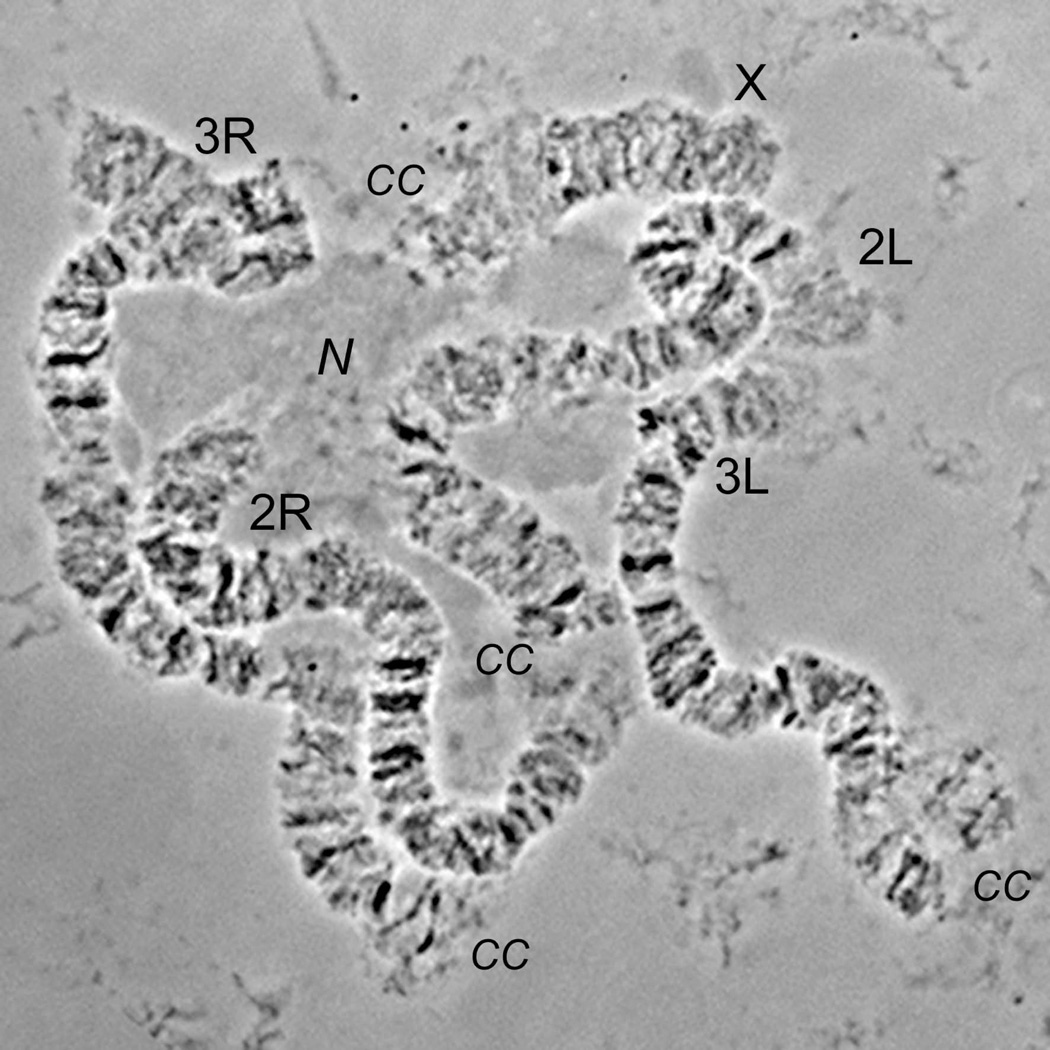
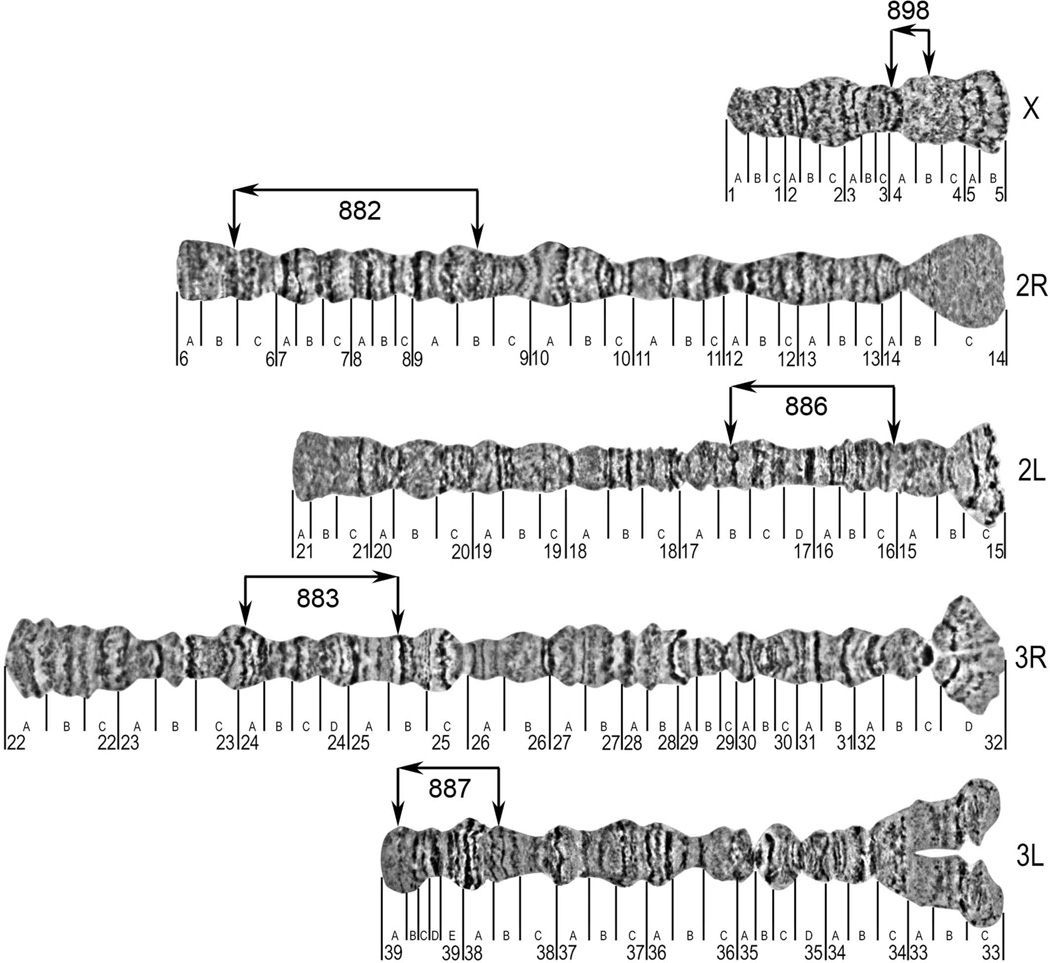
Similar to other mosquitoes (Rai, 1963), An. atroparvus has 3 pairs of chromosomes (Kitzmiller et al., 1967). Polytene chromosome complement from the nurse cells is represented by the smallest sex chromosome X, intermediate chromosome 2 with arms almost equal in length and the largest chromosome 3, which has 3R arm slightly longer than 3L arm (Table 1, Figure 1). Arm 3R is the longest among the others. Chromosomes of ovarian nurse cells do not form a chromocentre, and pericentromeric areas of each chromosome form independent attachments to the nuclear envelope (Sharakhova et al., 1999; Stegniy, 1993). A photomap for the polytene chromosomes was constructed from ovarian nurse cells of An. atroparvus (Figure 2). Chromosomes were straightened using AdobePhotoshop and divided into 33 numbered divisions and 118 lettered subdivisions. The division borders and nomenclature were adopted from previously developed salivary gland chromosome map of An. atroparvus (Stegnii & Kabanova, 1978) and another map for the polytene chromosome from ovarian nurse cells of An. messeae (Stegnii & Sharakhova, 1991).
Table 1.
The measurements of An. atroparvus ovarian nurse cells polytene chromosomes
| Chromosome | X | 2 | 3 |
| Average length (µm) | 210 | 1135 | 1200 |
| Relative length (%) | 8.3 | 44.4 | 47.3 |
| Centromere position (%) | Not applicable | 46 | 38.4 |
Figure 1. Chromosome complement of ovarian nurse cells of An. atroparvus.
Chromosome arms are shown as X, 2R, 2L, 3R, 3L. Pericentromeric regions and nucleolus are indicated as CC and N respectively.
Figure 2. A standard cytogenetic photomap of ovarian nurse cell chromosomes of An. atroparvus.
The lines below the chromosomes indicate the boundaries of numbered and lettered divisions and subdivisions respectively. The locations of DNA probes from the start and the end of the genomic supercontigs are indicated by vertical arrows above the chromosomes. The orientations of the genomic supercontigs are shown by horizontal arrows above the chromosomes.
There are easily identifiable robust landmarks for the An. atroparvus chromosome arms. The sex chromosome X has a slightly flared light telomere and a large flared light centromeric end terminated by three diffuse bands in subdivisions 4C-5B. Some of these bands often become invisible because of their diffuse morphology and granulated mesh-like structure. Arms of chromosome 2 can be easily distinguished from each other by the morphology of their telomeres. The telomere end of 2R is dark and consists of series of distinct bands in subdivisions 6A-B. In contrast, the telomere end of the 2L arm was flared, light and had a dark thin band in region 21C. Arms of chromosome 2 are connected to each by diffuse-light granulated areas in regions 14BC-15BC, which has two dark diffuse bands on 2L arm in subdivision 15C. Two sets of wide dark bands in regions 10A-B can be considered as an additional landmark for the 2R arm. Arms of chromosome 3 are usually not connected to each other in squashed preparations due to the strong attachment to the nuclear envelope. Pericentromeric region of the 3R arm had a dark band in subdivision 32C. The homologous chromosomes are often unpaired in subdivision 32D. An additional landmark for the 3R arm was a set of three dark bands in the middle of the arm in regions 25A-C. Arm 3L was characterized by a long asynaptic area of homologous in pericentromeric subdivisions 33A-C. Region 35B with two adjacent dark bands--dot-like and half-circle-like, “a bird eye,”--can be considered as additional landmark for the 3L arm (Stegnii & Kabanova, 1978) (Figure 2).
Physical mapping of the An. atroparvus genome
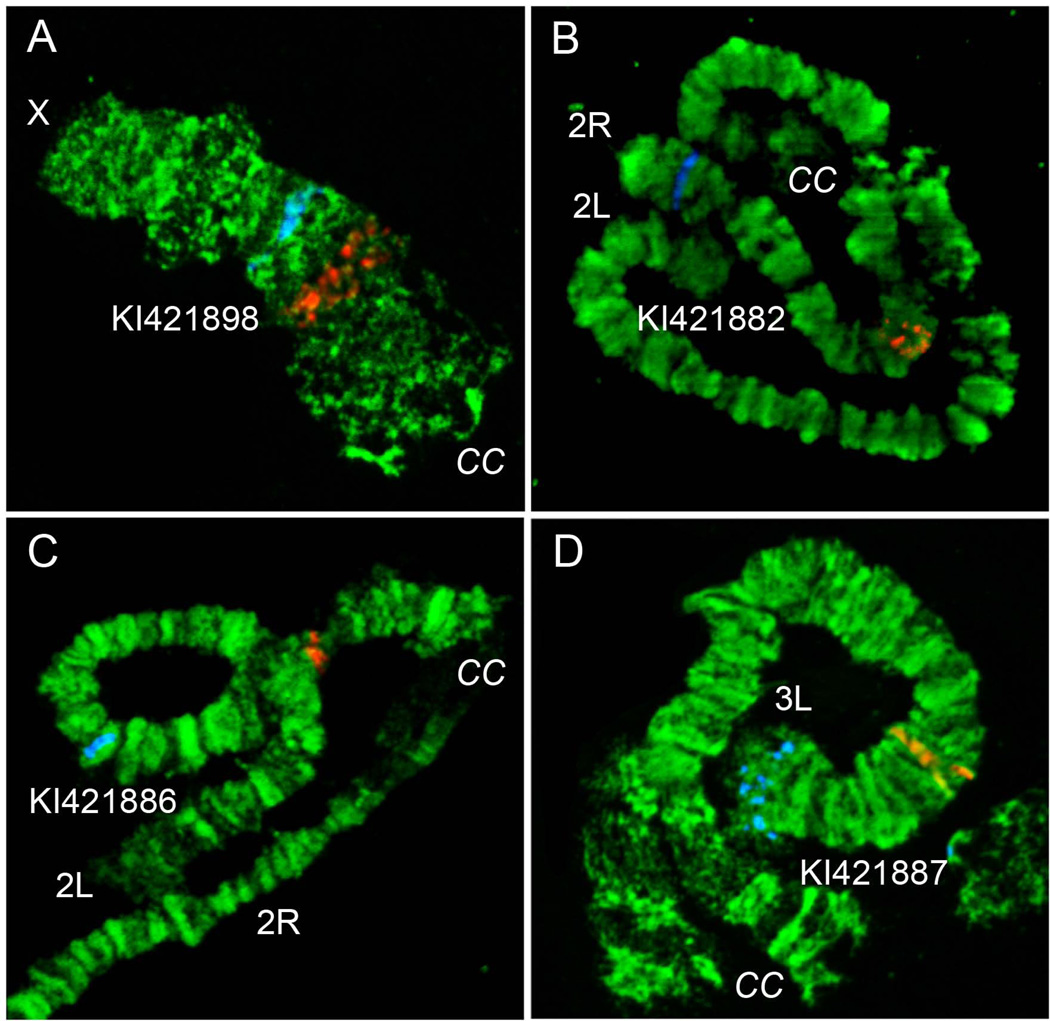
The utility of a cytogenetic map developed by the present study for An. atroparvus was validated by the physical mapping of 10 PCR-amplified DNA probes labeled with Cy3 and Cy5 dUTPs. For the mapping, the four largest supercontigs from autosomes and additional supercontigs were selected from the sex chromosome X (Table 2). Supercontigs were chosen based on the arm homology to An. gambiae established using Basic Local Alignment Search Tool (BLAST) in the VectorBase (https://www.vectorbase.org). One probe from the start of the supercontigs and another probe from the end of the supercontigs were labeled using red and blue fluorescent dyes, respectively (Figure 3). The PCR probes were designed to amplify the unique sequences from gene exons (Table 2). All probes produced clear unique signals and were successfully mapped to the chromosomes based on the banding patterns (Figure 3). Probes from the same genomic supercontigs were found on the same chromosome arm indicating no mistakes in sequence assembly. The largest genomic supercontig KI421882 of 20 Mb was mapped to the 2R arm in regions 6B-9B. This supercontig covered almost 9 lettered subdivisions on the 2R chromosome arm. The smallest supercontig KI421898 of 4 Mb was placed to the chromosome X in divisions 4A-4B. A good quality An. atroparvus genome assembly allowed assignment and orientation of 63 Mb equal to 28% of the genome to the chromosomes by physical mapping of 5 genomic supercontigs.
Table 2.
List of supercontigs/genes mapped to the chromosome positions
| Supercontig accession# |
Supercontig size (bp) |
Probe/gene | Gene start-end positions |
Chromosome location |
|---|---|---|---|---|
| KI421882 | 20238125 | AATE008453 | 66762-76445 | 2R:9B |
| AATE010796 | 16318761-16325046 | 2R:6B | ||
| KI421883 | 16623535 | AATE005670 | 1516240-1518054 | 3R:24A |
| AATE008545 | 13695270-13697405 | 3R:25B | ||
| KI421886 | 11808074 | AATE001693 | 35088-46700 | 2L:16C |
| AATE008759 | 11195642-11201764 | 2L:17B | ||
| KI421887 | 10405585 | AATE011741 | 966864-974650 | 3L:38B |
| AATE006315 | 9676291-9681308 | 3L:39A | ||
| KI421898 | 4025992 | AATE018661 | 2294-6247 | X:4B |
| AATE018218 | 3538365-3539771 | X:4A |
Figure 3. Examples of FISH on chromosomes.
The position of red and blue signal demonstrate the beginning and the end of the genomic supercontigs KI421898 (A), KI421882 (B), KI421886 (C) and KI421887 (D) on chromosomes X, 2R, 2L and 3L, respectively.
Arm homology between An. atroparvus and An. gambiae
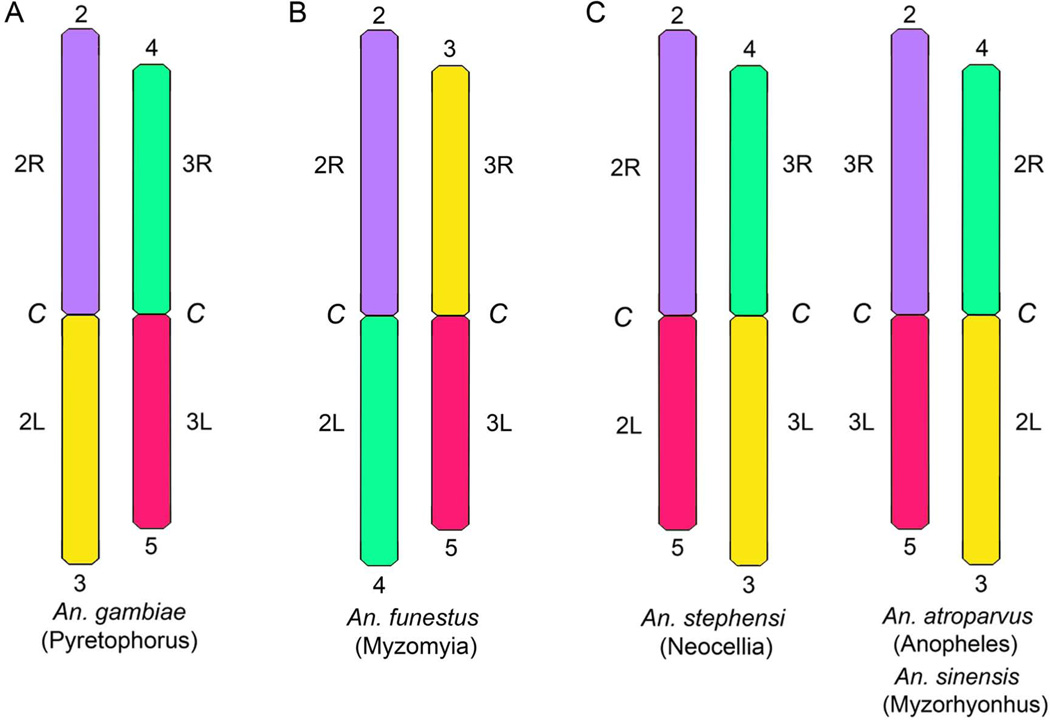
The arm homology between An. atroparvus and An. gambiae was established using a BLAST tool in the VectorBase (https://www.vectorbase.org). The genomic supercontig KI421883 from the longest autosome 3R arm of An. atroparvus was found homologous to the longest arm 2R in An. gambiae. Supercontig KI421882 from 2R arm of An. atroparvus was mapped in silico to the 3R arm of An. gambiae. Supercontigs from 2L (KI421886) and 3L (KI421887) arms of An. atroparvus were found in 2L and 3L of An. gambiae, respectively. Probes from the supercontig KI421898 homologous to the X chromosome in An. gambiae hybridized also to the X chromosome in An. atroparvus. These results suggest a whole-arm translocation between autosomes of these two species. Thus, An. atroparvus has arm association typical to that found in two other Asian species from genus Anopheles, An. stephensi (Sharakhova et al., 2010) and An. sinensis (Lianga et al., 2014) (Figure 4).
Figure 4. Chromosome arm homology between An. gambiae (A), An. funestus (B) and An. stephensi, An. atroparvus, An. sinensis (C).
Homologous arms are indicated by the numbers above and below chromosomes. Notations for chromosome arms are shown on sides of the chromosomes. C - pericentromeric regions. Names of series are shown in parentheses below the species names.
Discussion
A standard cytogenetic photomap of the ovarian nurse cell polytene chromosomes for the dominant European malaria vector An. atroparvus was developed. This map is one from the series of cytogenetic maps constructed for species from the genus Anopheles during the last decade (Sharakhov et al., 2001; Sharakhova et al., 2006; George et al., 2010; Sharakhova et al., 2011; Lianga et al., 2014). The original photomap for An. atroparvus was developed using chromosomes stained with orsein (Stegnii & Kabanova, 1978), and its utilization for the physical mapping purposes was difficult. Herein, phase contrast images of unstained chromosomes taken with digital technology were used. This approach allowed a clear banding pattern of the chromosomes to be obtained. To make the pattern recognition and mapping convincing, chromosomes were completely straightened using AdobePhotoshop. Each of the 10 PCR-amplified probes of genes from the An. atroparvus genome assembly was assigned to a band or interband position on the chromosome map facilitating the precise physical mapping of 5 genomic supercontigs (Figures 2 and 3). Because of the good quality of the genome assembly for An. atroparvus, 28% of the genome was placed to the chromosomes. In the present study, a much higher efficiency of physical mapping was obtained using the already assembled genome compared with when physical mapping was performed before the genome assembly became available. For example, 88% of the An. gambiae genome was placed to chromosomes based on 2000 BAC clones in situ hybridizations (Holt et al., 2002). Many of these clones hybridized to the same positions on the chromosomes.
Arm homology was demonstrated between An. atroparvus and An. gambiae; the major malaria vector in Africa. Similar to other studies, a whole-arm translocation between autosomes of these two species was found. This type of chromosome rearrangement is common in the genus Anopheles (Sharakhova et al., 2013). To compare chromosome arrangements in different species, a system of autosome arm notation proposed by Green and Hunt was adopted (Green & Hunt, 1980). Accordingly, the autosome arms in An. gambiae were named as follows: 2R = 2, 2L = 4, 3R = 3 and 3L = 5. Three different arrangements of chromosome arm association are possible: 1) (2+3) (4+5); 2) (2+4) (3+5) and 3) (2+5) (3+4). The first type of arm association has been described for the African mosquitoes of the An. gambiae complex from the ubgenus Cellia (Figure 4A). The second arm association has been discovered in An. funestus, another African mosquito from the subgenus Cellia (Green & Hunt, 1980). Interestingly, the same arm association has also been found in the American mosquito An. albimanus from the subgenus Nyssorynchus (Figure 4B) (Cornel & Collins, 2000). The present study demonstrated that An. atroparvus has the third type of arm association (2+5) (3+4) (Figure 4C). The same arm association has been described in Asian malaria vectors: An. stephensi from the subgenus Cellia (Sharakhova et al., 2006) and An. sinensis from the subgenus Anopheles (Lianga et al., 2014), and also in an African mosquito An. nili from the subgenus Cellia (Sharakhova et al., 2011). It is unclear if these rearrangements reflect phylogenetic relationships between the species or if they had happened multiple times in evolution of the genus Anopheles. Physical mapping of additional species may help to solve this problem.
Further application of the cytogenetic map constructed by this study for An. atroparvus, the dominant malaria vector in Europe, will help to develop a high-quality genome assembly for this species. The availability of the chromosome-based genome maps for multiple species of Anopheles will facilitate comparative genomic studies and will improve our understanding of phylogenetic relationship in this group of mosquitoes.
Acknowledgements
Authors thank AGC Consortium for the opportunity to use the genome assembly for An. atroparvus before the publication (Neafsey et al., 2014). This work was supported by National Institutes of Health grant 1R21AI099528-01A1 (to I.V.S).
Footnotes
Author’s declaration of interests
No competing interests have been declared.
References
- Artemov GN, Anan'ina TV, Fisenko O, Stegnii VN. Analysis of the spatial organization of the XL chromosome attachment site in nurse cell nuclei of the malaria mosquito Anopheles atroparvus. Genetika. 2010;46:1181–1184. [PubMed] [Google Scholar]
- Artemov GN, Stegnii VN. Molecular genetic analysis of the X-chromosomal nuclear envelope attachment region in nurse cells of the malaria mosquitoes Anopheles messeae Fall. Genetika. 2011;47:1307–1314. [PubMed] [Google Scholar]
- Besansky NJ, Krzywinski J, Lehmann T, Simard F, Kern M, Mukabayire O, et al. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc Natl Acad Sci U S A. 2003;100:10818–10823. doi: 10.1073/pnas.1434337100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky NJ, Powell JR, Caccone A, Hamm DM, Scott JA, Collins FH. Molecular phylogeny of the Anopheles gambiae complex suggests genetic introgression between principal malaria vectors. Proc Natl Acad Sci U S A. 1994;91:6885–6888. doi: 10.1073/pnas.91.15.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullini L, Coluzzi M. Evolutionary and taxonomic inferences of electrophoretic studies in mosquitoes. In: Steiner WWM, Tabachnick WJ, Rai KS, Narang S, editors. Recent developments in the genetics of insect disease vectors. Champaign, Illinois: Stipes Publishing Company; 1982. [Google Scholar]
- Cornel AJ, Collins FH. Maintenance of chromosome arm integrity between two Anopheles mosquito subgenera. J Hered. 2000;91:364–370. doi: 10.1093/jhered/91.5.364. [DOI] [PubMed] [Google Scholar]
- Djadid ND, Gholizadeh S, Tafsiri E, Romi R, Gordeev M, Zakeri S. Molecular identification of Palearctic members of Anopheles maculipennis in northern Iran. Malar J. 2007;6:6. doi: 10.1186/1475-2875-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George P, Sharakhova MV, Sharakhov IV. High-resolution cytogenetic map for the African malaria vector Anopheles gambiae. Insect Mol Biol. 2010;19:675–682. doi: 10.1111/j.1365-2583.2010.01025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornostaeva RM, Danilov AV. On ranges of the malaria mosquitoes (Diptera: Culicidae: Anopheles) of the Maculipennis complex on the territory of Russia. Parazitologiia. 2002;36:33–47. [PubMed] [Google Scholar]
- Green C, Hunt R. Interpretation of variation in ovarian polytene chromosomes of Anopheles funestus Giles, A. parensis Gillies, and A. aruni? Genetica. 1980;51:187–195. [Google Scholar]
- Grushko OG, Sharakhova MV, Shevchenko AI, Karagodin DA, Karamysheva TV, Rubtsov NB, et al. Characterization and comparative analysis of DNA from the pericentric heterochromatin of chromosome 2 of Anopheles atroparvus V. Tiel (Culicidae, Diptera) Genetika. 2004;40:1325–1335. [PubMed] [Google Scholar]
- Grushko OG, Sharakhova MV, Stegnii VN, Sharakhov IV. Molecular organization of heterochromatin in malaria mosquitoes of the Anopheles maculipennis subgroup. Gene. 2009;448:192–197. doi: 10.1016/j.gene.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Harbach RE. The classification of genus Anopheles (Diptera: Culicidae): a working hypothesis of phylogenetic relationships. Bull Entomol Res. 2004;94:537–553. doi: 10.1079/ber2004321. [DOI] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Kamali M, Xia A, Tu Z, Sharakhov IV. A new chromosomal phylogeny supports the repeated origin of vectorial capacity in malaria mosquitoes of the Anopheles gambiae complex. PLoS Pathog. 2012;8:e1002960. doi: 10.1371/journal.ppat.1002960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampen H. The ITS2 ribosomal DNA of Anopheles beklemishevi and further remarks on the phylogenetic relationships within the Anopheles maculipennis group of species (Diptera: Culicidae) Parasitol Res. 2005;97:118–128. doi: 10.1007/s00436-005-1393-8. [DOI] [PubMed] [Google Scholar]
- Kitzmiller JB, Frizzi G, Baker RH. Evolution and speciation within the Maculipennis Complex of the genus Anopheles. In: Wright JW, Pal R, editors. Genetics of insect vectors of disease. Amsterdam-London-New York: Elsevier Publishing Company; 1967. pp. 151–210. [Google Scholar]
- Lianga J, Sharakhova MV, Lan Q, Sharakhov IV, Xia A. A standard cytogenetic map for Anopheles sinensis and chromosome arm homology between subgenera Anopheles and Cellia. Medical and Veterinary Entomology. 2014 doi: 10.1111/mve.12048. In print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinucci M, Romi R, Mancini P, Di Luca M, Severini C. Phylogenetic relationships of seven palearctic members of the maculipennis complex inferred from ITS2 sequence analysis. Insect Mol Biol. 1999;8:469–480. doi: 10.1046/j.1365-2583.1999.00140.x. [DOI] [PubMed] [Google Scholar]
- Neafsey DE, Christophides GK, Collins FH, Emrich SJ, Fontaine MC, Gelbart W, et al. The evolution of the Anopheles 16 genomes project. G3 (Bethesda) 2013;3:1191–1194. doi: 10.1534/g3.113.006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, et al. Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science. 2014 doi: 10.1126/science.1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Loughlin SM, Magesa S, Mbogo C, Mosha F, Midega J, Lomas S, et al. Genomic Analyses of Three Malaria Vectors Reveals Extensive Shared Polymorphism but Contrasting Population Histories. Mol Biol Evol. 2014;31:889–902. doi: 10.1093/molbev/msu040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai KS. A comparative study of mosquito karyotypes. Ann ent Soc Am. 1963;56:160–170. [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sainz-Elipe S, Latorre JM, Escosa R, Masia M, Fuentes MV, Mas-Coma S, et al. Malaria resurgence risk in southern Europe: climate assessment in an historically endemic area of rice fields at the Mediterranean shore of Spain. Malar J. 2010;9:221. doi: 10.1186/1475-2875-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhov IV, Sharakhova MV, Mbogo CM, Koekemoer LL, Yan G. Linear and spatial organization of polytene chromosomes of the African malaria mosquito Anopheles funestus. Genetics. 2001;159:211–218. doi: 10.1093/genetics/159.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhova MV, Antonio-Nkondjio C, Xia A, Ndo C, Awono-Ambene P, Simard F, et al. Cytogenetic map for Anopheles nili: application for population genetics and comparative physical mapping. Infect Genet Evol. 2011;11:746–754. doi: 10.1016/j.meegid.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhova MV, Braginets OP, Stegnii VN. Spatial organization of polytene chromosomes in ovarian trophocyte nuclei of the malaria mosquito Anopheles labranchiae Fall. Tsitologiia. 1999;41:226–229. [PubMed] [Google Scholar]
- Sharakhova MV, Peery A, Antonio-Nkondjio C, Xia A, Ndo C, Awono-Ambene P, et al. Cytogenetic analysis of Anopheles ovengensis revealed high structural divergence of chromosomes in the Anopheles nili group. Infect Genet Evol. 2013;16:341–348. doi: 10.1016/j.meegid.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhova MV, Stegnii VN, Braginets OP. Interspecies differences in the ovarian trophocyte precentromere heterochromatin structure and evolution of the malaria mosquito complex Anopheles maculipennis. Genetika. 1997;33:1640–1648. [PubMed] [Google Scholar]
- Sharakhova MV, Xia A, McAlister SI, Sharakhov IV. A standard cytogenetic photomap for the mosquito Anopheles stephensi (Diptera: Culicidae): application for physical mapping. J Med Entomol. 2006;43:861–866. doi: 10.1603/0022-2585(2006)43[861:ascpft]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Sharakhova MV, Xia A, Tu Z, Shouche YS, Unger MF, Sharakhov IV. A physical map for an Asian malaria mosquito, Anopheles stephensi. Am J Trop Med Hyg. 2010;83:1023–1027. doi: 10.4269/ajtmh.2010.10-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinka ME, Bangs MJ, Manguin S, Coetzee M, Mbogo CM, Hemingway J, et al. The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionomic precis. Parasit Vectors. 2010;3:117. doi: 10.1186/1756-3305-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegnii VN. Genetic basis of evolution in malaria mosquitoes Anopheles from Maculipennis complex. Communications 1: Chromosome phylogeny relationships Russian Journal of Zoology. 1981;60:69–70. [Google Scholar]
- Stegnii VN. Evolutionary significance of chromosome architecture for epigenetic control of eukaryote development and phylogeny. Genetika. 2006;42:1215–1224. [PubMed] [Google Scholar]
- Stegnii VN, Kabanova VM. Chromosome analysis of Anopheles atroparvus and A. maculipennis (Diptera, Culicidae) Russian Journal of Zoology. 1978;57:613–619. [Google Scholar]
- Stegnii VN, Sharakhova MV. Systemic reorganization of the architechtonics of polytene chromosomes in onto- and phylogenesis of malaria mosquitoes. Structural features regional of chromosomal adhesion to the nuclear membrane. Genetika. 1991;27:828–835. [PubMed] [Google Scholar]
- Stegniy VN. Architectonics of genome, system mutation and evolution. Novosobirsk: Novosibirsk University Pub; 1993. [Google Scholar]
- [Accessed 2014 October 24];Vector Base [ http://www.vectorbase.org]
- White BJ, Lawniczak MK, Cheng C, Coulibaly MB, Wilson MD, Sagnon N, et al. Adaptive divergence between incipient species of Anopheles gambiae increases resistance to Plasmodium. Proc Natl Acad Sci U S A. 2011;108:244–249. doi: 10.1073/pnas.1013648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. World malaria report 2009. Geneva: World Health Organization; 2009. [Google Scholar]