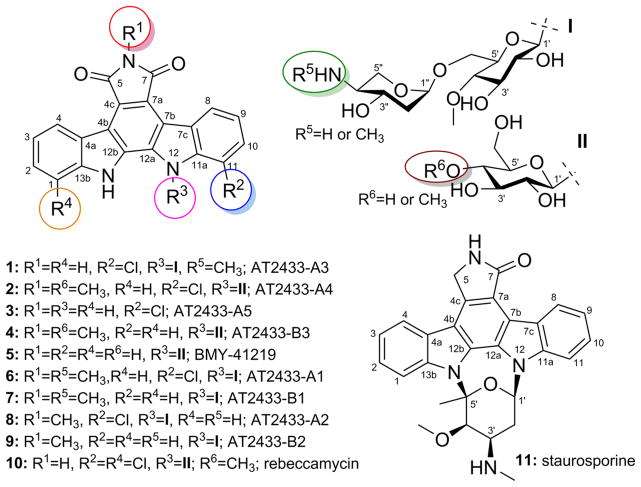
Abstract
Actinomadura melliaura ATCC 39691, a strain isolated from a soil sample collected in Bristol Cove, California, is a known producer of the disaccharide-substituted AT2433 indolocarbazoles (6–9). Reinvestigation of this strain using new media conditions led to >40-fold improvement in the production of previously reported AT2433 metabolites and the isolation and structure elucidation of the four new analogues, AT2433-A3, A4, A5, and B3 (1–4). The availability of this broader set of compounds enabled a subsequent small antibacterial/fungal/cancer SAR study that revealed disaccharyl substitution, N-6 methylation, and C-11 chlorination as key modulators of bioactivity. The slightly improved anticancer potency of the newly reported N-6-desmethyl 1 (compared to 6) contrasts extensive SAR of monoglycosylated rebeccamycin-type topoisomerase I inhibitors where N-6 alkylation has contributed to improved potency and ADME. Complete 2D NMR assignments for the known metabolite BMY-41219 (5) and 13C NMR spectroscopic data for the known analogue AT2433-B1 (7) are also provided for the first time.
Indolocarbazoles (exemplified by staurosporine, rebeccamycin, and AT2433; Figure 1) are actinomycete-derived alkaloids and potent inhibitors of topoisomerase I and kinases relevant to anticancer, antitubercular, antimalarial, and antiviral lead development.1–4 Of the indolocarbazoles that have advanced to the clinic, midostaurin (PKC412) recently completed a successful phase II trial for AML (Acute Myelogenous Leukemia) and is currently in phase II evaluation for metastatic melanoma, lestauritinib (CEP-701) is currently in clinical phase II/III evaluation for treating FLT3-ITD AML, and CT327 (a PEGylated formulation of the natural product K252a) is currently in phase IIb for treating psoriasis.5,6 While many indolocarbazole analogues have been synthesized,2,7–13 it is noteworthy that those that have advanced furthest clinically are either a known natural product (CT327) or subtle variations thereof (including the N-alkylated becatecarin or N-acylated PKC412). The vast majority of the >90 microbial indolocarbazole natural products exist as glycosides.14,15 Yet, of these, only four are disaccharide-substituted indolocarbazoles (AT2433 A1, B1, A2, and B2; Figure 1, 6–9, respectively).16,17 While the fundamental mechanism of the disaccharide-substituted indolocarbazole cytotoxicity is poorly understood, the terminal amino sugar has been implicated in both their unique mode of DNA binding and putative topisomerase-independent mechanism of action.18–20 As part of an effort to identify unique reagents to advance indolocarbazole biosynthetic studies,3,8,21–36 herein we describe the isolation, structure elucidation, and biological activity of four new naturally occurring indolocarbazoles (AT2433-A3, A4, A5, and B3; 1–4) along with three known counterparts, BMY-41219 (5), AT2433-A1 (6), and AT2433-B1 (7), from Actinomadura melliaura ATCC 39691. A comparison of cancer cell line cytotoxicities revealed the attached sugars as critical to bioactivity, where the disaccharide-substituted metabolites (1, 6, and 7) were found to be more potent than their monosaccharide-substituted congeners (2, 4, and 5). Chlorination of the indolocarbazole core was also found to be important to bioactivity, particularly in the context of antitubercular, antifungal, and Gram-positive antibacterial assays.
Figure 1.
Chemical structures of indolopyrrolocarbazoles 1–11.
RESULTS AND DISCUSSION
Disaccharide-substituted 6 and 7 represent the major metabolites of A. melliaura ATCC 39691, which also produces other related minor metabolites including the aminopentose N-desmethyl analogues 8 and 9 (Figure 1).16,17 In an effort to access these, and potentially additional indolocarbazole analogues as reagents for biosynthetic studies, the strain was fermented in A-medium and the extracts were analyzed by LC-ESIMS. Consistent with previous studies, this preliminary analysis revealed 6 and 7 as the predominate products along with several additional metabolites that displayed indolopyrrolocarbazole UV and MS-fragmentation signatures. Subsequent purification of compounds from a large-scale fermentation extract using progressive chromatography led to the isolation of four new naturally occurring indolocarbazoles [AT2433-A3 (1; 4.3 mg), AT2433-A4 (2; 3.0 mg), AT2433-A5 (3; 1.7 mg), and AT2433-B3 (4; 3.3 mg)] along with the three known previously reported indolocarbazoles [BMY-41219 (5; 5.9 mg),37 AT2433-A1 (6, 1.75 g),16,17 and AT2433-B1 (7, 1.2 g)16,17] (Table 1, 5Figures 1–3 and S76). Of this latter set, the full NMR assignments for BMY-41219 (; Table S2, Figures S2 and S80) and 13C NMR spectroscopic data for AT2433-B1 (7; Table S2) are reported here for the first time. It is also noteworthy that the production levels in A-medium reported herein of compounds AT2433-A1 (6) and AT2433-B1 (7) were substantially higher than from previously reported conditions (41-fold and 92-fold, respectively).17 Additional compounds isolated included harman (also known as 1-methyl-β-carboline, 2.0 mg),38–43 5′-methylthioadenosine (2.0 mg),44,45 riboflavin (4.3 mg), and lumichrome (1.7 mg)44,46 (see Experimental Section and Supporting Information Figures S76–S78).
Table 1.
13C (100 MHz) and 1H (500 MHz) NMR Data of Compounds 1–4a
| position | AT2433-A3 (1)b,c
|
AT2433-A4 (2)b,d
|
AT2433-A5 (3)b,d
|
AT2433-B3 (4)b,c
|
||||
|---|---|---|---|---|---|---|---|---|
| δC, type | δH, m (J in Hz) | δC, type | δH, m (J in Hz) | δC, type | δH, m, (J in Hz) | δC, type | δH, m (J in Hz) | |
| 1 | 112.7, CH | 7.72, d (8.0) | 112.2, CH | 7.75, d (8.0) | 112.2, CH | 7.82, dd (8.0, 1.0) | 113.1, CH | 7.69, d (7.5) |
| 2 | 129.2, CH | 7.64, t (7.0) | 127.6, CH | 7.61, td (7.5, 1.5) | 127.1, CH | 7.56, td (8.0, 1.0) | 128.0, CH | 7.53, td (8.0, 1.0) |
| 3 | 122.4, CH | 7.32, t (7.0) | 120.7, CH | 7.40, t (7.0) | 121.3, CH | 7.36, td (8.0, 1.0) | 121.6, CH | 7.32, td (8.0, 1.0) |
| 4 | 126.8, CH | 8.99, d (8.0) | 124.5, CH | 9.12, d (8.0) | 124.2, CH | 8.95, d (8.0) | 126.2, CH | 9.13, d (8.0) |
| 4a | 121.4, C | 121.0, C | 119.8, C | 123.5, C* | ||||
| 4b | 119.8, C | 117.5, C | 115.9, C | 119.6, C* | ||||
| 4c | 124.4, C | 118.2, C | 118.9, C | 119.7, C | ||||
| 5 | 172.2, C | 169.3, C | 169.7, C | 171.3, C* | ||||
| 6-CH3 | 23.9, CH3 | 3.20, s | 23.6, CH3 | 3.11, s | 24.0, CH3 | 3.24, s | ||
| 7 | 172.2, C | 169.5, C | 169.7, C | 171.3, C* | ||||
| 7a | 123.4, C | 121.9, C | 120.5, C | 119.7, C | ||||
| 7b | 121.4, C | 119.4, C | 115.5, C | 119.6, C | ||||
| 7c | 127.5, C | 125.5, C | 123.1, C | 123.5, C* | ||||
| 8 | 125.9, CH | 9.20, d (8.0) | 123.8, CH | 9.27, dd (8.0, 1.0) | 123.3, CH | 8.87, d (8.0) | 126.4, CH | 9.23, d (8.0) |
| 9 | 123.8, CH | 7.30, t (8.0) | 122.5, CH | 7.42, t (8.0) | 121.3, CH | 7.34, t (7.5) | 122.0, CH | 7.34, t (8.0) |
| 10 | 131.0, CH | 7.52, d (7.5) | 129.6, CH | 7.65, dd (7.5, 1.0) | 126.0, CH | 7.62, dd (7.5, 1.0) | 128.3, CH | 7.56, td (8.5, 1.5) |
| 11 | 118.1, C | 116.5, C | 115.8, C | 112.1, CH | 7.81, d (8.5) | |||
| 11a | 139.9, C | 138.2, C | 137.1, C | 142.9, C | ||||
| 12 | 11.95, brs | |||||||
| 12a | 131.8, C | 129.7, C | 128.8, C | 130.8, C | ||||
| 12b | 131.4, C | 130.3, C | 129.0, C | 131.6, C | ||||
| 13 | 11.81, brs | 11.64, brs | ||||||
| 13a | 141.9, C | 140.8, C | 140.1, C | 141.9, C | ||||
| 1′ | 86.5, CH | 7.01, d (9.5) | 83.8, CH | 6.90, d (9.0) | 86.3, CH | 6.18, d (8.5) | ||
| 2′ | 73.8, CH | 3.82, m | 72.1, CH | 3.41, m | 73.7, CH | 3.84, t (8.0) | ||
| 2′-OH | 4.94, d (5.5) | |||||||
| 3′ | 79.5, CH | 3.68, m | 76.6, CH | 3.51, m | 79.1, CH | 3.98, t (9.5) | ||
| 3′-OH | 5.28, d (6.5) | |||||||
| 4′ | 80.3, CH | 3.68, m | 77.0, CH | 3.67, t (9.5) | 79.6, CH | 3.85, t (9.5) | ||
| 4′-OCH3 | 61.5, CH3 | 3.74, s | 60.1, CH3 | 3.61, s | 61.4, CH3 | 3.80, s | ||
| 5′ | 79.5, CH | 4.06, m | 77.6, CH | 3.90, d (10.0) | 75.0, CH | 4.06, m | ||
| 6′ | 67.5, CH2 | 4.41, d (10.0) 4.08, m |
58.7, CH2 | 4.00, brd (2.0) | 60.4, CH2 | 4.12, d (11.5) 4.08, dd (11.5, 2.0) |
||
| 6′-OH | 6.34, t (4.0) | |||||||
| 1″ | 100.4, CH | 5.28, brs | ||||||
| 2″ | 38.7, CH2 | 2.54, m 2.03, m |
||||||
| 3″ | 65.4, CH | 4.02, m | ||||||
| 4″ | 61.5, CH | 3.05, m | ||||||
| 4″-NHCH3 | 31.6, CH3 | 2.51, s | ||||||
| 5″ | 59.0, CH2 | 4.04, m 3.84, m |
||||||
Assignments supported by 2D HSQC and HMBC experiments.
See Supporting Information for the NMR spectra.
CD3OD.
DMSO-d6.
Obtained from the HMBC spectrum.
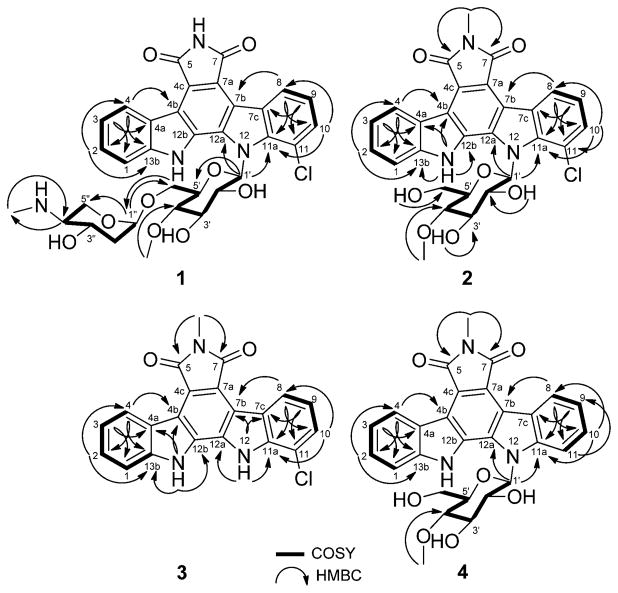
Figure 3.
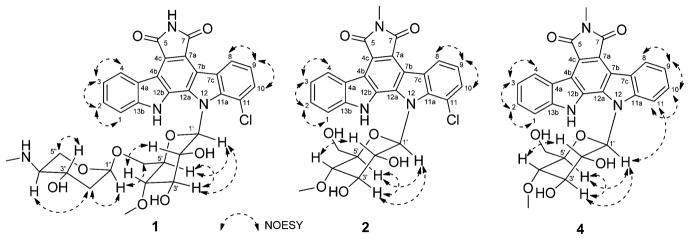
Key NOESY correlations for indolocarbazoles 1, 2, and 4.
Structure Elucidation
The physicochemical properties of compounds 1–7 are summarized in the Experimental Section. Compound 1 was isolated as a yellow solid (4.3 mg) using progressive chromatography (Figure S76). The UV spectrum of 1 was similar to that of AT2433-A1 (6) with indolocarbazole UV absorption signatures at 396, 316, 290, and 236 nm (Figure S1). The molecular formula of 1 was deduced as C33H33ClN4O9 from HRESIMS and 1H and 13C NMR. Compared to 6, the Δm/z = 14 difference observed in 1 implicated the loss of a methyl group. The 1H and 13C NMR spectra of 1 (Table 1) and 6 (Table S2, Figure S81) in CD3OD revealed both to share a common disaccharide-substituted indolopyrrolocarbazole core, where, compared to 6, compound 1 lacked the N-6 methyl. Confirmation of the common N-12 4″-amino-4″-N-methyl-2″,4″-dideoxy-α-L-xylose-(1″→6′)-4′-O-methyl-β-D-N-glucosyl moiety was confirmed through the cumulative analyses of 1H–1H-COSY and HMBC spectroscopic data (Figure 2). Specifically, the 3J HMBC cross-peaks observed from H-1″ to CH2-6′ (δ 67.5) and from H2-6′ to C-1 (δ 100.4) were consistent with the attachment of the 4″-amino-4″-N-methyl-2″,4″-dideoxy-α-L-xylose moiety at the 6′-position of indole 4′-O-methyl-β-D-N-glucoside. The corresponding chemical shift of the C-1′ anomeric carbon (δC 86.5) and the 3J HMBC correlation observed from H-1′ to the quaternary carbons at δ 139.9 (C-11a) and δ 131.8 (C-12a) confirmed the N-12 glycosyl attachment. All of the remaining HMBC correlations (Figure 2) and NMR data (Table 1) were in full agreement with structure 1. The relative configurations at the sterocenters of disaccharide sugar residue were indirectly established through the analyses of NOESY correlations (Figure 3), coupling constants, and comparison to those of 6 (Figure S81, Table S2). Thus, HRMS and cumulative 1H, 13C, 1H–1H-COSY, TOCSY (Supporting Information, Figure S2), HSQC, HMBC, and NOESY spectral data revealed compound 1 as a new analogue of the monochlorinated AT2433-A series, and 1 was thereby designated as AT2433-A3.
Figure 2.
1H–1H–COSY and selected HMBC correlations for indolocarbazoles 1–4.
Compound 2 was obtained as a yellow solid (3.0 mg, Figure S76) and displayed common indolocarbazole UV–vis (Figure S1) and physicochemical properties. The molecular formula of 2 was deduced as C28H24ClN3O7 based on the molecular ion peak observed at m/z 550.1381 in the HRESIMS spectrum, where the 129 amu difference from 6 implicated the absence of the terminal pentose. Consistent with this, no pentosyl proton or carbon signals in the 1H/13C NMR/HSQC spectra of 2 (Table 1) were found. Further COSY, TOCSY (Figure S2), HMBC, and NOESY correlations were in full agreement with compound 2 (Figures 2 and 3) as a new analogue of the monochlorinated AT2433-A series, and 2 was thereby designated as AT2433-A4. Importantly, 2 differs from the prototype dichlorinated monosaccharide-substituted rebeccamycins (Figure 1, 10) via the additional N-6 methyl and lack of the second C-1 chlorine.
Compound 3 was obtained as a yellow solid (1.7 mg, Figure S76) and also displayed common indolocarbazole UV–vis (Figure S1) and physicochemical properties. The molecular formula of 3 was confirmed as C21H12ClN3O2, where the 176 amu difference from 2 suggested the absence of the N-12 4′-O-methyl-β-D-N-glucosyl moiety. Consistent with this, the 1H NMR spectrum of 3 in DMSO-d6 (Table 1) displayed two broad indole-NH signals at δ 11.95 and 11.64. In addition, no corresponding glucosyl proton or carbon signals in the 1H/13C NMR/HSQC spectra of 3 (Table 1) were observed. Further COSY, TOCSY (Figure S2), HMBC, and NOESY correlations were in full agreement with compound 3 (Figures 2 and 3) as a new analogue of the monochlorinated AT2433-A series, and 3 was thereby designated as AT2433-A5.
Compound 4 was also obtained as a yellow solid (3.3 mg, Figure S76) and displayed common indolocarbazole UV–vis (Figure S1) and physicochemical properties. The molecular formula of 4 was confirmed as C28H25N3O7 on the basis of HRESIMS, where the 35 amu difference from 2 suggested the absence of the C-11 chlorine. The observed additional C-11 proton signal at δ 7.81 (d, J = 8.5 Hz), along with full 1D and 2D NMR (Table 1, 4Figures 2, 3, and S2), provided further support for this distinguishing feature. Thus, compound , as a new analogue of the deschlorinated AT2433-B series, was designated as AT2433-B3. It should be noted that while synthetic 4 was previously reported as a selective topoisomerase I inhibitor,10,47–50 the discovery of 4 as a natural product and the corresponding full NMR assignments for 4 (Figures 2 and 3; Table 1) are reported here for the first time.
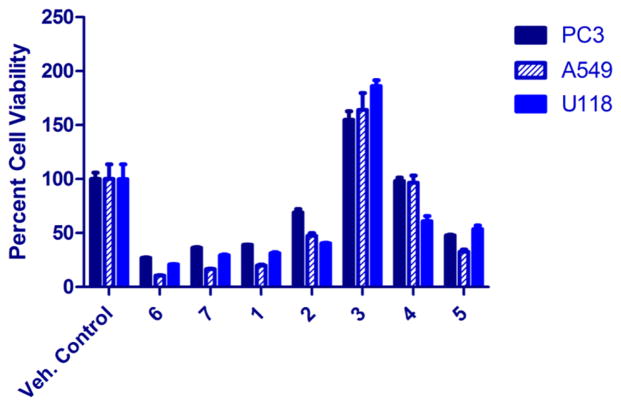
Including AT2433-A3 (1), -A4 (2), -A5 (3), and -B3 (4) reported herein, the indolopyrrolocarbazoles make up 74 of the 94 naturally occurring microbial indolocarbazoles, only five of which contain disaccharyl substitutions (the new 1 along with previously reported 6–9).14,15 Indolocarbazoles including staurosporines,51–60 K-252 derivatives,61–63 rebeccamycins,64,65 RK-1409B,66 RK-286 C and D,67,68 tjipanazoles,69 TAN-999S and TAN-1030A analogues,54,70 fradcarbazoles,71 indocarba-zostatins,72–75 ZHD-0501,76 fluoroindolocarbazoles,77 holy-rines,78 MLR-52,79 and BE-13793C80,81 have been reported to have promising antibacterial, antifungal, antitumor, and neuroprotective activities. Thus, compounds 1–7 were tested against five bacterial strains (Staphylococcus aureus ATCC 6538, Micrococcus luteus NRRL B-287, Mycobacterium smegmatis ATCC 14468, Salmonella enterica ATCC 10708, and Escherichia coli NRRL B-3708), one fungal strain (Saccharomyces cerevisiae ATCC 204508), and three human cancer cell lines (PC-3, prostate; A549, lung; and U118, brain). As highlighted in Table 2, the disaccharyl-substituted indolocarbazoles were the most active, where AT2433-A1 (6) displayed the best overall antibacterial/fungal activities ranging from 1 μM (0.7 μg/mL, Staph. aureus) to 60 μM (40 μg/mL, M. smegmatis). Within this context, the removal of the N-6 methyl (1) or the C-11 chlorine (7) led to similar reductions in overall potency, where the differences in activity against S cerevisiae and M. smegmatis were the most dramatic. Similar trends were observed in the single-dose in vitro anticancer study (Figure 4) with a disaccharyl (1, 6, and 7) > mono (2, 4, and 5) > indolocarbazole aglycon (3) (Table 3 and Supporting Information, Table S1 and Figure S79). To more accurately assess potency differences, the most active new analogue from the single-dose study (1) was compared to the corresponding most active known comparator AT2433-A1 (6) in a full-dose study (Table 3). Both compounds displayed notable potencies (IC50’s of 46–106 nM), where 1 was found to be slightly more active than 6 (2-fold) against the PC3 prostate cancer cell line. While 3 appears to stimulate growth in this assessment, this dose–response is consistent with hormesis, a biphasic response with apparent stimulation and inhibition zones observed common to a variety of different drugs, antibodies, and radiation.82
Table 2.
In Vitro Antimicrobial Activities of AT2433-A3 (1), AT2433-B3 (4), AT2433-A1 (6), and AT2433-B1 (7)
| organism | compounda
|
|||
|---|---|---|---|---|
| 1 | 4 | 6 | 7 | |
| Staphylococcus aureus ATCC 6538 | 7.5 (5) | 7.5 (4) | 1 (0.7) | 7.5 (5) |
| Micrococcus luteus NRRL B-287 | 7.5 (5) | 15 (7.7) | 7.5 (5) | 7.5 (5) |
| Saccharomyces cerevisiae ATCC 204508 | >60 (>40) | >60 (>31) | 7.5 (5) | >60 (>40) |
| Mycobacterium smegmatis ATCC 14468 | 120 (80) | >120 (>62) | 60 (40) | 120 (80) |
MIC in μM (μg/mL) values are based on three independent replicates. Compounds 2, 3, and 5 were inactive against all strains up to 60 μM (120 μm for M. smegmatis). Compounds 1–7 were inactive against Salmonella enterica ATCC 10708 and Escherichia coli NRRL B-3708 up to 60 μM. Kanamycin and ampicillin (S. aureus, M. luteus, S. enterica, and E. coli), spectinomycin and rifampicin (M. smegmatis), and amphotericin B (S. cerevisiae) were used as positive controls.
Figure 4.
Viability of PC3 (prostate), A549 (lung), and U118 (brain) human cancer cell lines at 10 μM treatment for compounds 1–7 after 72 h (see Supporting Information, Table S1).
Table 3.
Cancer Cell Line Inhibition by AT2433-A3 (1) and AT2433-A1 (6)a
| compound | cell line [(IC50 (nM)]
|
||
|---|---|---|---|
| PC3 | A549 | U118 | |
| AT2433-A3 (1) | 45.8 | 69.6 | 47.3 |
| AT2433-A1 (6) | 106.2 | 76.4 | 56.8 |
See Supporting Information, Figure S79. AT2433-A1, as a known compound, was also considered as a positive control.
In summary, A. melliaura ATCC 39691 is a known producer of the indolocarbazole AT2433 series (6–9). Reinvestigation of this strain using new growth conditions led to a dramatic improvement in production of three known indolocarbazoles (5–7) and the discovery of four new naturally occurring indolopyrrolocarbazoles (1–4). The availability of this broader set of compounds enabled a subsequent small antibacterial/ fungal/cancer structure–activity relationship (SAR), which revealed disaccharyl substitution, N-6 methylation, and C-11 cholorination as key modulators of activity. The slightly improved anticancer potency of N-6-desmethyl 1 (compared to 6) contrasts the extensive SAR of the rebeccamycin-type topoisomerase I inhibitors that have advanced to clinical evaluation (as exemplified by becatecarin,83,84 edotecarin,85 or NB-50686), where N-6 alkylation has been a key to improvements in potency and ADME (absorption, distribution, metabolism, and excretion).87 While this subtle distinction could be consistent with the putative uncharacterized top-isomerase-independent mechanism of action for the disacchar-yl-substituted AT2433 metabolites, further SAR and mechanistic study is required.
EXPERIMENTAL SECTION
General Experimental Procedures
UV spectra were recorded on an Ultrospec 8000 spectrometer (GE, Pittsburgh, PA, USA). NMR spectra were measured using Varian Vnmr 500 (1H, 500 MHz; 13C, 125.7 MHz) and Vnmr 400 (1H, 399.8 MHz; 13C, 100.5 MHz) spectrometers, where δ-values were referenced to respective solvent signals [CD3OD, δH 3.31 ppm, δC 49.15 ppm; DMSO-d6, δH 2.50 ppm, δC 39.51 ppm]. High-resolution electrospray ionization (ESI) mass spectra were recorded on a Thermo Scientific (Rockford, IL, USA) Q Exactive (Orbitrap mass spectrometer) via direct infusion at 3 μL/min. Full-scan mass spectra were recorded in positive (3.8 kV) and negative (3.8 kV) ion modes (capillary temperature: 225 °C; nominal resolution: 140 000). HPLC-MS analyses were accomplished using a Waters (Waters Corp., Milford, MA, USA) 2695 LC module (Waters Symmetry Anal. C18, 4.6 × 250 mm, 5 μm; solvent A: H2O–0.1% formic acid, solvent B: CH3CN–0.1% formic acid; flow rate: 0.5 mL min−1; 0–4 min, 10% B; 4–22 min, 10%–100% B; 22–27 min, 100% B; 27–29 min, 100%–10% B; 29–35 min, 10% B). HPLC analyses were performed on an Agilent 1260 system equipped with a photodiode array (PDA) detector and a Phenomenex C18 column (250 × 4.6 mm, 5 μm; Phenomenex, Torrance, CA, USA) (solvent A: H2O–0.1% TFA; solvent B: CH3CN; flow rate: 1.0 mL min−1; 0–35 min, 5%–100% B; 35–40 min, 100% B; 40–41 min, 100%–5% B; 41–45 min, 5% B). Semipreparative HPLC was accomplished using Phenomenex on a Varian (Varian, Palo Alto, CA, USA) ProStar model 210 equipped with a PDA detector and a gradient elution profile (C18 column, 10 × 250 mm, 5 μm; solvent A: 0.05% TFA–H2O, solvent B: CH3CN; flow rate: 5.0 mL min−1; 0–2 min, 25% B; 2–15 min, 25%–100% B; 15–17 min, 100% B; 17–18 min, 100%–25% B; 18–19 min, 25% B). All solvents used were of ACS grade and purchased from the Pharmco-AAPER (Brookfield, CT, USA). Rf values were measured on Polygram SIL G/UV254 (Macherey-Nagel & Co., Dueren, Germany). C18-functionalized silica gel (40–63 μm) was purchased from Material Harvest Ltd. (Cambridge, United Kingdom). Amberlite XAD16N resin (20–60 mesh) was purchased from Sigma-Aldrich (Saint Louis, MO, USA). Size exclusion chromatography was performed on Sephadex LH-20 (25–100 μm, GE Healthcare, Piscataway, NJ, USA). Staphylococcus aureus, Mycobacterium smegmatis, Salmonella enterica, and Saccharomyces cerevisiae strains were obtained from ATCC (Manassas, VA, USA); Micrococcus luteus and Escherichia coli were obtained from NRRL (Peoria, IL, USA). PC3, A549, and U118 cells were obtained from ATCC. All other reagents used were reagent grade and purchased from Sigma-Aldrich.
Media, Fermentation, Extraction, Isolation, and Purification
Actinomadura melliaura ATCC 39691 was cultivated on M2-agar plates [glucose (4.0 g), malt extract (10.0 g), yeast extract (4.0 g), and agar (15.0 g) dissolved in 1 L of H2O (pH 7.2) and sterilized by autoclaving for 33 min at 121 °C] at 28 °C for 3 days. To prepare the seed culture, a small piece of grown agar plate was used to inoculate one 250 mL baffled flask, containing 100 mL of A-medium [glucose (10.0 g), yeast extract (5.0 g), soluble starch (20.0 g), peptone (5.0 g), NaCl (4.0 g), K2HPO4 (0.5 g), MgSO4·7H2O (0.5 g), and calcium carbonate (2 g) dissolved in 1 L of H2O (pH 7.0) and sterilized by autoclaving for 33 min at 121 °C], and the culture was grown at 28 °C with shaking (210 rpm) for 2 days. An aliquot of seed culture (1 mL) was subsequently used to inoculate 80 250 mL baffled flasks, each containing 100 mL of A-medium. Fermentation was continued at 28 °C with shaking (210 rpm) for 6 days. The obtained yellow culture broth was centrifuged and filtered over Celite, and the supernatant was mixed with XAD-16 (4%) resin overnight, followed by filtration. The recovered resin was washed with H2O (3 × 1200 mL) and then extracted with MeOH (3 × 800 mL). The MeOH extract was subsequently evaporated in vacuo at 38 °C to afford 7.10 g of yellow, oily crude extract. The biomass (mycelium) was extracted with MeOH (3 × 1200 mL), which was then evaporated in vacuo at 38 °C to yield 44.35 g of yellow, oily crude extract. Both extracts revealed the same sets of metabolites based upon HPLC and TLC analysis and were therefore combined to afford a total of 51.45 g of crude extract that served as the source material for the following workup and isolation procedure (Supporting Information, Figure S76).
The obtained crude extract (51.45 g) was dissolved in 50% MeOH–H2O (100 mL) and fractionated with a gradient of H2O/0–100% CH3CN, followed by 100% MeOH using RP-18 column chromatography (column 5 × 45 cm, 150 g) to provide the following fractions: 1.2 L 0% CH3CN, fractions F1A–F1F (200 mL each); 0.6 L 10% CH3CN, fractions F2A–F2C (200 mL each); 0.6 L 20% CH3CN, fractions F3A–F3C (200 mL each); 0.6 L 30% CH3CN, fractions F4A–F4C (200 mL each); 0.6 L 40% CH3CN, fractions F5A–F5C (200 mL each); 0.6 L 50% CH3CN, fractions F6A–F6C (200 mL each); 0.6 L 60% CH3CN, fractions F7A–F7C (200 mL each); 0.6 L 70% CH3CN, fractions F8A–F8C (200 mL each); 0.6 L 80% CH3CN, fractions F9A–F9C (200 mL each); 0.6 L 90% CH3CN, fractions F10A–F10C (200 mL each); 1.5 L 100% CH3CN, fractions F11A–F11E (300 mL each); 1.5 L 100% CH3OH, fractions F11F–F11K (250 mL each); 0.8 L 0.1% TFA CH3OH, fraction F12A. All fractions were analyzed by TLC and HPLC as a basis for the subsequent final purification steps described in the next paragraph.
As highlighted in Figure S76, fractions F4B–F5A (0.6 g) were combined and subsequently purified by preparative TLC (CH2Cl2–MeOH, 9:1), Sephadex LH-20 (CH2Cl2–MeOH, 6:4; column 1 × 40 cm), and semipreparative HPLC to afford harman (1.3 mg), riboflavin (4.3 mg), and 5′-methylthioadenosine (2.0 mg). Similarly, fractions F5C–F6C (0.4 g) were combined and purified by Sephadex LH-20 (CH2Cl2–MeOH, 6:4; 2.5 × 50 cm) and semipreparative HPLC to yield lumichrome (1.7 mg). Treatment of the combined fractions F7A–F7C (70 mg) using preparative TLC (CH2Cl2–MeOH, 9:1), followed by Sephadex LH-20 (CH2Cl2–MeOH, 6:4; column 1 × 40 cm), afforded BMY-41219 (5; 5.9 mg). Purification of combined fractions F9B–F10C (0.45 g) by preparative TLC and semipreparative HPLC gave AT2433-A4 (2, 3.0 mg). Semipreparative HPLC purification of fraction F11A (150 mg) afforded AT2433-A3 (1, 2.6 mg) and AT2433-B1 (7, 45.0 mg), while resolution of combined fractions F11B–F11E (0.4 g) via Sephadex LH-20 (CH2Cl2–MeOH, 6:4; column 2.5 × 50 cm) and semipreparative HPLC provided AT2433-A3 (1, 1.7 mg), AT2433-A5 (3, 1.7 mg), AT2433-B3 (4, 3.3 mg), AT2433-B1 (7, 51.9 mg), and harman (0.7 mg). Finally, purification of combined fractions F11H–11K (0.7 g) using Sephadex LH-20 (CH2Cl2–MeOH, 6:4; column 2.5 × 50 cm) gave AT2433-A1 (6, 450 mg), while the major fractions F11F (1.1 g) and F11G (1.3 g) were composed of AT2433-B1 (7) and AT2433-A1 (6), respectively (each ≥95% purity), based on HPLC/MS analysis.
AT2433-A3 (1): yellow solid; UV/vis (MeOH) λmax (log ε) 236 (4.17), 290 (3.99), 316 (4.26), 396 (3.25) nm; 1H NMR (CD3OD, 500 MHz) and 13C NMR (CD3OD, 100 MHz), see Table 1; (+)-APCI-MS m/z 665 [M + H]+; (+)-HRESIMS m/z 665.2008 [M + H]+ (calcd for C33H34ClN4O9, 665.2008); (−)-HRESIMS m/z 663.1846 [M – H] − (calcd for C33H32ClN4O9, 663.1863).
AT2433-A4 (2): yellow solid; UV/vis (MeOH) λmax (log ε) 238 (3.97), 290 (3.84), 319 (3.96), 402 (2.93) nm; 1H NMR (DMSO-d6, 500 MHz) and 13C NMR (DMSO-d6, 100 MHz), see Table 1; (+)-HRESIMS m/z 550.1381 [M + H]+ (calcd for C28H25ClN3O7, 550.1376); (−)-HRESIMS m/z 548.1222 [M – H] − (calcd for C28H23ClN3O7, 548.1230).
AT2433-A5 (3): yellow solid; Rf 0.58 (silica gel, CH2Cl2); UV/vis (MeOH) λmax (log ε) 236 (3.79), 285 (3.65), 316 (3.74), 400 (2.73) nm; 1H NMR (DMSO-d6, 500 MHz) and 13C NMR (DMSO-d6, 100 MHz), see Table 1; (−)-HRESIMS m/z 372.0536 [M – H] − (calcd for C21H11ClN3O2, 372.0545).
AT2433-B3 (4): yellow solid; UV/vis (MeOH) λmax (log ε) 237 (3.56), 286 (3.38), 319 (3.53), 406 (2.57) nm; 1H NMR (CD3OD, 500 MHz) and 13C NMR (CD3OD, 100 MHz), see Table 1; (+)-APCI-MS m/z 516 [M + H]+; (−)-APCI-MS m/z 514 [M – H] −; (+)-HRESIMS m/z 516.1769 [M + H]+ (calcd for C28H26N3O7, 516.1769); (−)-HRESIMS m/z 514.1611 [M – H] − (calcd for C28H24N3O7, 514.1620).
Cancer Cell Line Viability Assays
Cytotoxicity screening was accomplished using previously reported methods for both the single-dose (10 μM) screen and full IC50 determinations (0.03 to 100 000 nM) in triplicate.88
Antimicrobial Assay
Antimicrobial assays were accomplished in triplicate following previously reported methods.88 Antibacterial MIC values were obtained after 16–48 h of incubation, while antifungal MIC values were obtained after 24 h of incubation.
Supplementary Material
Acknowledgments
This work was supported in part by the University of Kentucky College of Pharmacy, the University of Kentucky Markey Cancer Center, and the National Center for Advancing Translational Sciences (UL1TR000117). We also thank Prof. J. Rohr and Prof. S. G. Van Lanen (University of Kentucky, College of Pharmacy) for access to routine HPLC-MS and Dr. J. P. Goodman (University of Kentucky Mass Spectrometry Facility) for the HRESIMS.
Footnotes
Notes
The authors declare the following competing financial interest(s): The authors report competing interests. J.S.T. is a co-founder of Centrose (Madison, WI, USA).
ASSOCIATED CONTENT
UV, HPLC, HPLC-MS, HRESIMS, and NMR (1D and 2D) spectra of compounds 1–7 and harman. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jnatprod.5b00429.
References
- 1.Nakano H, Omura S. J Antibiot. 2009;62:17–26. doi: 10.1038/ja.2008.4. [DOI] [PubMed] [Google Scholar]
- 2.Speck K, Magauer T. Beilstein J Org Chem. 2013;9:2048–2078. doi: 10.3762/bjoc.9.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sánchez C, Méndez C, Salas JA. Nat Prod Rep. 2006;23:1007–1045. doi: 10.1039/b601930g. [DOI] [PubMed] [Google Scholar]
- 4.Ryan KS, Drennan CL. Chem Biol. 2009;16:351–364. doi: 10.1016/j.chembiol.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer T, Stone RM, Deangelo DJ, Galinsky I, Estey E, Lanza C, Fox E, Ehninger G, Feldman EJ, Schiller GJ, Klimek VM, Nimer SD, Gilliland DG, Dutreix C, Huntsman-Labed A, Virkus J, Giles FJ. J Clin Oncol. 2010;28:4339–4345. doi: 10.1200/JCO.2010.28.9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shabbir M, Stuart R. Expert Opin Investig Drugs. 2010;19:427–436. doi: 10.1517/13543781003598862. [DOI] [PubMed] [Google Scholar]
- 7.Chisholm JD, Golik J, Krishnan B, Matson JA, Van Vranken DL. J Am Chem Soc. 1999;121:3801–3802. [Google Scholar]
- 8.Zhang C, Albermann C, Fu X, Peters NR, Chisholm JD, Zhang G, Gilbert EJ, Wang PG, Van Vranken DL, Thorson JS. ChemBioChem. 2006;7:795–804. doi: 10.1002/cbic.200500504. [DOI] [PubMed] [Google Scholar]
- 9.Facompre M, Carrasco C, Vezin H, Chisholm JD, Yoburn JC, Van Vranken DL, Bailly C. ChemBioChem. 2003;4:386–395. doi: 10.1002/cbic.200200478. [DOI] [PubMed] [Google Scholar]
- 10.Anizon F, Belin L, Moreau P, Sancelme M, Voldoire A, Prudhomme M, Ollier M, Sevére D, Riou JF, Bailly C, Fabbro D, Meyer T. J Med Chem. 1997;40:3456–3465. doi: 10.1021/jm9702084. [DOI] [PubMed] [Google Scholar]
- 11.Pereira ER, Belin L, Sancelme M, Prudhomme M, Ollier M, Rapp M, Severe D, Riou JF, Fabbro D, Meyer T. J Med Chem. 1996;39:4471–4477. doi: 10.1021/jm9603779. [DOI] [PubMed] [Google Scholar]
- 12.Wood JL, Stoltz BM, Dietrich HJ, Pflum DA, Petsch DT. J Am Chem Soc. 1997;119:9641–9651. [Google Scholar]
- 13.Wood JL, Stoltz BM, Goodman SN, Onwneme K. J Am Chem Soc. 1997;119:9652–9661. [Google Scholar]
- 14.Laatsch H. AntiBase 2014. Wiley-VCH; Weinheim: 2014. The natural compound identifier. [Google Scholar]
- 15.Elshahawi SI, Shaaban KA, Kharel MK, Thorson JS. Chem Soc Rev. 2015 doi: 10.1039/C4CS00426D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golik J, Doyle TW, Krishnan B, Dubay G, Matson JA. J Antibiot. 1989;42:1784–1789. doi: 10.7164/antibiotics.42.1784. [DOI] [PubMed] [Google Scholar]
- 17.Matson JA, Claridge C, Bush JA, Titus J, Bradner WT, Doyle TW, Horan AC, Patel M. J Antibiot. 1989;42:1547–1555. doi: 10.7164/antibiotics.42.1547. [DOI] [PubMed] [Google Scholar]
- 18.Qu X, Chaires JB, Ohkubo M, Yoshinari T, Nishimura S, Bailly C. Anti-Cancer Drug Des. 1999;14:433–442. [PubMed] [Google Scholar]
- 19.Bailly C, Qu X, Anizon F, Prudhomme M, Riou JF, Chaires JB. Mol Pharmacol. 1999;55:377–385. doi: 10.1124/mol.55.2.377. [DOI] [PubMed] [Google Scholar]
- 20.Facompre M, Carrasco C, Colson P, Houssier C, Chisholm JD, Van Vranken DL, Bailly C. Mol Pharmacol. 2002;62:1215–1227. doi: 10.1124/mol.62.5.1215. [DOI] [PubMed] [Google Scholar]
- 21.Pearce CJ, Doyle TW, Forenza S, Lam KS, Schroeder DR. J Nat Prod. 1988;51:937–940. doi: 10.1021/np50059a020. [DOI] [PubMed] [Google Scholar]
- 22.Lam KS, Forenza S, Doyle TW, Pearce CJ. J Ind Microbiol. 1990;6:291–294. doi: 10.1007/BF01575876. [DOI] [PubMed] [Google Scholar]
- 23.Hyun CG, Bililign T, Liao J, Thorson JS. ChemBioChem. 2003;4:114–117. doi: 10.1002/cbic.200390004. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez C, Zhu LL, Brana AF, Salas AP, Rohr J, Mendez C, Salas JA. Proc Natl Acad Sci USA. 2005;102:461–466. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howard-Jones AR, Walsh CT. Biochemistry. 2005;44:15652–15663. doi: 10.1021/bi051706e. [DOI] [PubMed] [Google Scholar]
- 26.Salas AP, Zhu L, Sanchez C, Brana AF, Rohr J, Mendez C, Salas JA. Mol Microbiol. 2005;58:17–27. doi: 10.1111/j.1365-2958.2005.04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Q, Zhang C, Blanchard S, Thorson JS. Chem Biol. 2006;13:733–743. doi: 10.1016/j.chembiol.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Nishizawa T, Gruschow S, Jayamaha DH, Nishizawa-Harada C, Sherman DH. J Am Chem Soc. 2006;128:724–725. doi: 10.1021/ja056749x. [DOI] [PubMed] [Google Scholar]
- 29.Zhang C, Weller RL, Thorson JS, Rajski SR. J Am Chem Soc. 2006;128:2760–2761. doi: 10.1021/ja056231t. [DOI] [PubMed] [Google Scholar]
- 30.Kim SY, Park JS, Chae CS, Hyun CG, Choi BW, Shin J, Oh KB. Appl Microbiol Biotechnol. 2007;75:1119–1126. doi: 10.1007/s00253-007-0924-x. [DOI] [PubMed] [Google Scholar]
- 31.Bitto E, Huang Y, Bingman CA, Singh S, Thorson JS, Phillips GN., Jr Proteins. 2008;70:289–293. doi: 10.1002/prot.21627. [DOI] [PubMed] [Google Scholar]
- 32.Singh S, McCoy JG, Zhang C, Bingman CA, Phillips GN, Jr, Thorson JS. J Biol Chem. 2008;283:22628–22636. doi: 10.1074/jbc.M800503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onaka H. Biosci Biotechnol Biochem. 2009;73:2149–2155. doi: 10.1271/bbb.90263. [DOI] [PubMed] [Google Scholar]
- 34.Goldman PJ, Ryan KS, Hamill MJ, Howard-Jones AR, Walsh CT, Elliott SJ, Drennan CL. Chem Biol. 2012;19:855–865. doi: 10.1016/j.chembiol.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh S, Zhang J, Huber TD, Sunkara M, Hurley K, Goff RD, Wang G, Zhang W, Liu C, Rohr J, Van Lanen SG, Morris AJ, Thorson JS. Angew Chem, Int Ed. 2014;53:3965–3969. doi: 10.1002/anie.201308272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shepherd SA, Karthikeyan C, Latham J, Struck AW, Thompson ML, Menon BRK, Styles MQ, Levy C, Leys D, Micklefield J. Chem Sci. 2015;6:3454–3460. doi: 10.1039/c5sc00913h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lam KS, McDonald LA, Mattei J, Forenza S, Matson JA. Antitumor antibiotic BMY-41219 and its manufacture with tryptophan-supplemented Saccharothrix aerocolonigenes cultures. 5326754 A. US. 1994
- 38.Aassila H, Bourguet-Kondracki ML, Rifai S, Fassouane A, Guyot M. Mar Biotechnol. 2003;5:163–166. doi: 10.1007/s10126-002-0060-7. [DOI] [PubMed] [Google Scholar]
- 39.Seki H, Hashimoto A, Hino T. Chem Pharm Bull. 1993;41:1169–1172. [Google Scholar]
- 40.Welti DH. Magn Reson Chem. 1985;23:872–874. [Google Scholar]
- 41.Rocca P, Marsais F, Godard A, Queguiner G. Tetrahedron Lett. 1994;35:2003–2004. [Google Scholar]
- 42.Cao R, Peng W, Wang Z, Xu A. Curr Med Chem. 2007;14:479–500. doi: 10.2174/092986707779940998. [DOI] [PubMed] [Google Scholar]
- 43.Bohlendorf B, Forche E, Bedorf N, Gerth K, Irschik H, Jansen R, Kunze B, TrowitzschKienast W, Reichenbach H, Hofle G. Liebigs Ann. 1996:49–53. [Google Scholar]
- 44.Shaaban KA. PhD Thesis. University of Göttingen; 2009. [Google Scholar]
- 45.Robins MJ, Hansske F, Wnuk SF, Kanai T. Can J Chem. 1991;69:1468–1474. [Google Scholar]
- 46.Stroch K, Zeeck A, Antal N, Fiedler HP. J Antibiot. 2005;58:103–110. doi: 10.1038/ja.2005.13. [DOI] [PubMed] [Google Scholar]
- 47.Moreau P, Anizon F, Sancelme M, Prudhomme M, Severe D, Riou JF, Goossens JF, Henichart JP, Bailly C, Labourier E, Tazzi J, Fabbro D, Meyer T, Aubertin AM. J Med Chem. 1999;42:1816–1822. doi: 10.1021/jm980702n. [DOI] [PubMed] [Google Scholar]
- 48.Moreau P, Anizon F, Sancelme M, Prudhomme M, Bailly C, Carrasco C, Ollier M, Severe D, Riou JF, Fabbro D, Meyer T, Aubertin AM. J Med Chem. 1998;41:1631–1640. doi: 10.1021/jm970843+. [DOI] [PubMed] [Google Scholar]
- 49.Messaoudi S, Anizon F, Peixoto P, David-Cordonnier MH, Golsteyn RM, Leonce S, Pfeiffer B, Prudhomme M. Bioorg Med Chem. 2006;14:7551–7562. doi: 10.1016/j.bmc.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 50.Messaoudi S, Anizon F, Leonce S, Pierre A, Pfeiffer B, Prudhomme M. Eur J Med Chem. 2005;40:961–971. doi: 10.1016/j.ejmech.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 51.Akinaga S, Sugiyama K, Akiyama T. Drug Des. 2000;15:43–52. [PubMed] [Google Scholar]
- 52.Takahashi I, Asano K, Kawamoto I, Tamaoki T, Nakano H. J Antibiot. 1989;42:564–570. doi: 10.7164/antibiotics.42.564. [DOI] [PubMed] [Google Scholar]
- 53.Omura S, Sasaki Y, Iwai Y, Takeshima H. J Antibiot. 1995;48:535–548. doi: 10.7164/antibiotics.48.535. [DOI] [PubMed] [Google Scholar]
- 54.Cai Y, Fredenhagen A, Hug P, Meyer T, Peter HH. J Antibiot. 1996;49:519–526. doi: 10.7164/antibiotics.49.519. [DOI] [PubMed] [Google Scholar]
- 55.Funato N, Takayanagi H, Konda Y, Toda Y, Harigaya Y, Iwai Y, Ōmura S. Tetrahedron Lett. 1994;35:1251–1254. [Google Scholar]
- 56.Kinnel RB, Scheuer PJ. J Org Chem. 1992;57:6327–6329. [Google Scholar]
- 57.Cai Y, Fredenhagen A, Hug P, Peter HH. J Antibiot. 1996;49:1060–1062. doi: 10.7164/antibiotics.49.1060. [DOI] [PubMed] [Google Scholar]
- 58.Hoehn P, Ghisalba O, Moerker T, Peter HH. J Antibiot. 1995;48:300–305. doi: 10.7164/antibiotics.48.300. [DOI] [PubMed] [Google Scholar]
- 59.Omura S, Iwai Y, Hirano A, Nakagawa A, Awaya J, Tsuchya H, Takahashi Y, Masuma R. J Antibiot. 1977;30:275–282. doi: 10.7164/antibiotics.30.275. [DOI] [PubMed] [Google Scholar]
- 60.Hernandez LM, Blanco JA, Baz JP, Puentes JL, Millan FR, Vazquez FE, Fernandez-Chimeno RI, Gravalos DG. J Antibiot. 2000;53:895–902. doi: 10.7164/antibiotics.53.895. [DOI] [PubMed] [Google Scholar]
- 61.Nakanishi S, Matsuda Y, Iwahashi K, Kase H. J Antibiot. 1986;39:1066–1071. doi: 10.7164/antibiotics.39.1066. [DOI] [PubMed] [Google Scholar]
- 62.Yasuzawa T, Iida T, Yoshida M, Hirayama N, Takahashi M, Shirahata K, Sano H. J Antibiot. 1986;39:1072–1078. doi: 10.7164/antibiotics.39.1072. [DOI] [PubMed] [Google Scholar]
- 63.Cai Y, Fredenhagen A, Hug P, Peter HH. J Antibiot. 1995;48:143–148. doi: 10.7164/antibiotics.48.143. [DOI] [PubMed] [Google Scholar]
- 64.Bush JA, Long BH, Catino JJ, Bradner WT, Tomita K. J Antibiot. 1987;40:668–678. doi: 10.7164/antibiotics.40.668. [DOI] [PubMed] [Google Scholar]
- 65.Lam KS, Schroeder DR, Veitch JM, Matson JA, Forenza S. J Antibiot. 1991;44:934–939. doi: 10.7164/antibiotics.44.934. [DOI] [PubMed] [Google Scholar]
- 66.Koshino H, Osada H, Amano S, Onose R, Isono K. J Antibiot. 1992;45:1428–1432. doi: 10.7164/antibiotics.45.1428. [DOI] [PubMed] [Google Scholar]
- 67.Osada H, Satake M, Koshino H, Onose R, Isono K. J Antibiot. 1992;45:278–279. doi: 10.7164/antibiotics.45.278. [DOI] [PubMed] [Google Scholar]
- 68.Takahashi H, Osada H, Uramoto M, Isono K. J Antibiot. 1990;43:168–173. doi: 10.7164/antibiotics.43.168. [DOI] [PubMed] [Google Scholar]
- 69.Bonjouklian R, Smitka TA, Doolin LE, Molloy RM, Debono M, Shaffer SA, Moore RE, Stewart JB, Patterson GML. Tetrahedron. 1991;47:7739–7750. [Google Scholar]
- 70.Tanida S, Takizawa M, Takahashi T, Tsubotani S, Harada S. J Antibiot. 1989;42:1619–1630. doi: 10.7164/antibiotics.42.1619. [DOI] [PubMed] [Google Scholar]
- 71.Fu P, Zhuang Y, Wang Y, Liu P, Qi X, Gu K, Zhang D, Zhu W. Org Lett. 2012;14:6194–6197. doi: 10.1021/ol302940y. [DOI] [PubMed] [Google Scholar]
- 72.Feng Y, Matsuura N, Ubukata M. J Antibiot. 2004;57:627–633. doi: 10.7164/antibiotics.57.627. [DOI] [PubMed] [Google Scholar]
- 73.Matsuura N, Tamehiro N, Andoh T, Kawashima A, Ubukata M. J Antibiot. 2002;55:355–362. doi: 10.7164/antibiotics.55.355. [DOI] [PubMed] [Google Scholar]
- 74.Tamehiro N, Matsuura N, Feng Y, Nakajima N, Ubukata M. J Antibiot. 2002;55:363–370. doi: 10.7164/antibiotics.55.363. [DOI] [PubMed] [Google Scholar]
- 75.Ubukata M, Tamehiro N, Matsuura N, Nakajima N. J Antibiot. 1999;52:921–924. doi: 10.7164/antibiotics.52.921. [DOI] [PubMed] [Google Scholar]
- 76.Han XX, Cui CB, Gu QQ, Zhu WM, Liu HB, Gu JY, Osada H. Tetrahedron Lett. 2005;46:6137–6140. [Google Scholar]
- 77.Lam KS, Schroeder DR, Veitch JM, Colson KL, Matson JA, Rose WC, Doyle TW. J Antibiot. 2001;54:1–9. doi: 10.7164/antibiotics.54.1. [DOI] [PubMed] [Google Scholar]
- 78.Williams DE, Bernan VS, Ritacco FV, Maiese WM, Greenstein M, Andersen RJ. Tetrahedron Lett. 1999;40:7171–7174. [Google Scholar]
- 79.McAlpine JB, Karwowski JP, Jackson M, Mullally MM, Hochlowski JE, Premachandran U, Burres NS. J Antibiot. 1994;47:281–288. doi: 10.7164/antibiotics.47.281. [DOI] [PubMed] [Google Scholar]
- 80.Kojiri K, Kondo H, Yoshinari T, Arakawa H, Nakajima S, Satoh F, Kawamura K, Okura A, Suda H, Okanishi M. J Antibiot. 1991;44:723–728. doi: 10.7164/antibiotics.44.723. [DOI] [PubMed] [Google Scholar]
- 81.Tanaka S, Ohkubo M, Kojiri K, Suda H, Yamada A, Uemura D. J Antibiot. 1992;45:1797–1798. doi: 10.7164/antibiotics.45.1797. [DOI] [PubMed] [Google Scholar]
- 82.O’Brien PJ. Basic Clin Pharmacol Toxicol. 2014;115:4–17. doi: 10.1111/bcpt.12227. [DOI] [PubMed] [Google Scholar]
- 83.Schwandt A, Mekhail T, Halmos B, O’Brien T, Ma PC, Fu P, Ivy P, Dowlati A. J Thorac Oncol. 2012;7:751–754. doi: 10.1097/JTO.0b013e31824abca2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dowlati A, Posey J, Ramanathan RK, Rath L, Fu P, Chak A, Krishnamurthi S, Brell J, Ingalls S, Hoppel CL, Ivy P, Remick SC. Pharmacology. 2009;65:73–78. doi: 10.1007/s00280-009-1005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saif MW, Diasio RB. Clin Colorectal Cancer. 2005;5:27–36. doi: 10.3816/ccc.2005.n.014. [DOI] [PubMed] [Google Scholar]
- 86.Arakawa H, Iguchi T, Morita M, Yoshinari T, Kojiri K, Suda H, Okura A, Nishimura S. Cancer Res. 1995;55:1316–1320. [PubMed] [Google Scholar]
- 87.Prudhomme M. Eur J Med Chem. 2003;38:123–140. doi: 10.1016/s0223-5234(03)00011-4. [DOI] [PubMed] [Google Scholar]
- 88.Wang X, Reynolds AR, Elshahawi SI, Shaaban KA, Ponomareva LV, Saunders MA, Elgumati IS, Zhang Y, Copley GC, Hower JC, Sunkara M, Morris AJ, Kharel MK, Van Lanen SG, Prendergast MA, Thorson JS. Org Lett. 2015;17:2796–2799. doi: 10.1021/acs.orglett.5b01203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.