Abstract
Glia comprise a conspicuous population of non-neuronal cells in vertebrate and invertebrate nervous systems. Drosophila serves as a favorable model to elucidate basic principles of glial biology in vivo. The Drosophila neuropil glia (NPG), subdivided into astrocyte-like (ALG) and ensheathing glia (EG), extend reticular processes which associate with synapses and sheath-like processes which surround neuropil compartments, respectively. In this paper we characterize the development of NPG throughout fly brain development. We find that differentiated neuropil glia of the larval brain originate as a cluster of precursors derived from embryonic progenitors located in the basal brain. These precursors undergo a characteristic migration to spread over the neuropil surface while specifying/differentiating into primary ALG and EG. Embryonically-derived primary NPG are large cells which are few in number, and occupy relatively stereotyped positions around the larval neuropil surface. During metamorphosis, primary NPG undergo cell death. Neuropil glia of the adult (secondary NPG) are derived from type II lineages during the postembryonic phase of neurogliogenesis. These secondary NPG are much smaller in size but greater in number than primary NPG. Lineage tracing reveals that both NPG subtypes derive from intermediate neural progenitors of multipotent type II lineages. Taken together, this study reveals previously uncharacterized dynamics of NPG development and provides a framework for future studies utilizing Drosophila glia as a model.
Keywords: astrocyte-like glia, ensheathing glia, Repo, brain, Drosophila
INTRODUCTION
Drosophila glial cells have become a genetically-tractable in vivo system to understand fundamental aspects of glial cell biology. Glial cells are divided into three basic classes defined by topology, cell morphology, and function (Awasaki et al., 2008; Ito et al., 1995; Pereanu et al., 2005; Xiong et al., 1994). (1) Surface glia, further subdivided into subperineurial and perineurial glia, have cell bodies which lie on the brain surface. These glia extend flattened processes which encapsulate the entire outer brain surface and together form a structure analogous to the blood-brain-barrier (DeSalvo et al., 2011; Stork et al., 2008). (2) Cortex glia (or cell body-associated glia), of which there is only one subtype, possess cell bodies located within the cellular cortex amongst the somata of differentiated neurons. This class extends processes which encapsulate neuronal cell bodies and neuroblasts, forming the so-called “trophospongium” (Dumstrei et al., 2003; Hoyle, 1986; Hoyle et al., 1986). (3) Neuropil glia have somata at the neuropil-cortex interface and associate with the various neuropil compartments of the fly brain. Two distinct neuropil glia subtypes have been identified (Awasaki et al., 2008; Doherty et al., 2009; Pereanu et al., 2005). One subtype, known as reticular or astrocyte-like glia (ALG), extends processes that extend into the neuropil. These extensively branched processes are in close association with terminal neurites and synapses, situating them in a position to modulate neurotransmission, similarly to the vertebrate astrocyte. The second subtype, known as ensheathing glia (EG), extends sheath-like processes around the neuropil and some of the major axon tracts, but lack processes which penetrate into the neuropil.
A number of recent studies have shown that neuropil glia (ALG in particular) express amino acid transporters important for the reuptake of neurotransmitters, such as glutamate and γ-aminobutyric acid transporter (GABA) (Stacey et al., 2010; Stork et al., 2014). As a result, neuropil glia play a crucial role in controlling the encoding of specific behaviors. The concentration of transmitters, that in turn depends on re-uptake by glial cells, will either strengthen or weaken synaptic transmission and/or neurotransmitter tone (Grosjean et al., 2008; Jackson and Haydon, 2008; Sinakevitch et al., 2010; Stork et al., 2014). In addition to their physiological role in mature brain function, ALG and EG also appear to play multiple roles during neural development. Interestingly, different neuropil glia subtypes phagocytose accumulating neuronal debris in a context dependent manner. EG, which express the engulfment receptor Draper and dCed-6, are important for clearing axonal debris due to injury in adult brains (Doherty et al., 2009), whereas ALG, also expressing Draper, are responsible for the uptake of pruned axons from neurons that are remodeled during metamorphosis (Tasdemir-Yilmaz et al., 2014). Furthermore, neuropil glia also play a part in the construction of neuronal circuitry, by aiding in axonal guidance, terminal branching, and synaptogenesis (Hidalgo et al., 1995; Muthukumar et al., 2014; Spindler et al., 2009).
Studies of the developmental origin, migration patterns and morphogenesis of glia are essential in understanding the role of glia during nervous system development. Such studies will also provide the genetic tools to selectively eliminate groups of glial cells, by, for example, ablating the progenitor type that produces them.
The embryonic origin of glia has been mapped in detail for the embryonic ventral nerve cord (VNC) (Beckervordersandforth et al., 2008; Ito et al., 1995; Schmidt et al., 1997). Here, neuropil glia (also known as longitudinal glia), derive from a single lateral glioblast (LGB). The LGB progeny migrate towards the longitudinal connectives, undergo several rounds of mitotic divisions to produce a cluster of 9 cells per hemineuromere, and subsequently migrate around and encapsulate the neuropil (Beckervordersandforth et al., 2008; Ito et al., 1995; Jacobs et al., 1989). Late during embryogenesis, longitudinal glia are thought to be differentially specified by a largely unknown mechanism to generate ensheathing and astrocyte-like glia. Neuropil glia of the brain originate from one or a small set of neuroblasts at the deutero-tritocerebral boundary, from where they migrate over the brain neuropil surface while likely undergoing several rounds of divisions (Hartenstein et al., 1998).
Studies of postembryonic glial development are restricted to the brain. Pereanu et al. (2005) observed that all glial classes increase in number during late larval stages and attributed this growth primarily to secondary proliferation of neuroblasts, as well as mitotic divisions of differentiated glia and/or persistent glial precursors generated in the embryo. Consistent with this notion, posteriorly-located type II neuroblasts, which generate large lineages via intermediate neural progenitors (INPs), have been shown to generate glia during the early phase of postembryonic proliferation (Bayraktar and Doe, 2013; Izergina et al., 2009; Viktorin et al., 2011; Yu et al., 2013). As they spread out around the neuropil in the pupa, these glial cells continue to proliferate. The molecular mechanisms, such as the FGF, InR/TOR, and Merlin-Hippo signaling pathways, which mediate subtype-specific, post-embryonic gliogenesis are just beginning to be elucidated (Avet-Rochex et al., 2012; Reddy and Irvine, 2011).
In the present study we address several fundamental aspects of neuropil glial origin and development which have remained elusive. Astrocyte-like and ensheathing glia have been described in both the larval and adult brain (Awasaki et al., 2008; Pereanu et al., 2005). At least in the larva, both classes of differentiated neuropil glia comprise a relatively small number of cells (this study). Are these primary glia fixed in number, similar to primary neurons? Is their location, in relation to specific neuropil compartments, invariant? Do both classes contribute equally to the neuropil sheath? In terms of development, a crucial question is how, if at all, are the larval and adult neuropil glia related? Are both derived from the same embryonic progenitor pool? Do larval glial cells proliferate to give rise to adult glia? Does the same class of progenitors produce both ALG and EG? Here, using specific markers for ALG (NP3233-Gal4; alrm-Gal4; anti-GAT) and EG (NP6520-Gal4) (Awasaki et al. 2008; Doherty et al., 2009; Stork et al., 2014) in conjunction with lineage tracing experiments, we have reconstructed the origin and developmental fate of neuropil glia of the Drosophila brain. This analysis demonstrates that a small population of approximately 30 primary ALG and EG originates in the basal brain from where they spread out to occupy stereotyped positions around the neuropil by the early larva. These primary ALG and EG do not proliferate during the larval period, but increase in nuclear/cytoplasmic size and process length to accommodate the increase in neuropil volume. During metamorphosis, primary ALG loose their branched morphology and subsequently undergo programmed cell death. Secondary neuropil glia originate from a different progenitor pool than primary glia, which includes several of the dorsomedial type II lineages. Secondary neuropil glia increase in number while spreading out around the neuropil surface and the emerging compartments of the central complex during the late larval and early pupal period. They extend processes into the neuropil starting around P60–72 (hrs after puparium formation), a developmental time period which corresponds to synaptic maturation.
MATERIALS AND METHODS
Fly Stocks
Flies were grown at 25°C using standard fly media unless otherwise noted. The following fly stocks were used (sources in parentheses): NP3233-Gal4 and NP6520-Gal4 (Awasaki et al., 2008) (Kyoto Drosophila Genetic Resource Center), alrm (astrocytic leucine-rich repeat molecule)-Gal4 (Doherty et al., 2009) (kind gift from M. Freeman, University of Massachusetts Medical School, Worcester, MA), MZ1407 (inscuteable)-Gal4 (kind gift from H. Reichert, Biozentrum, University of Basel, Basel, Switzerland), tub-Gal80ts, UAS-Redstinger6, UAS-mCD8-GFP, UAS-FLPJD1, hs-FLP1, UAS-mCD8-GFP.nls, R9D11 (earmuff)-Gal4 (Bayraktar et al., 2010), acj6-Gal4, sine oculis-Gal4, UAS-EGFRDN, engrailed-Gal4, Act5C>Stop>lacZ (Act5C promoter-FRT-phl[+]-FRT-lacZ.nls), tub-FRT-GAL80-FRT-GAL4 (Zecca and Struhl, 2002) (Bloomington Stock Center).
Clonal Analysis and Lineage Tracing Experiments
Single cell, primary neuropil glia flp-out clones were generated using flies bearing the genotype: hs-FLP1, Wt-/WCD8-GFP/+; UAS-mCD8-GFP/+; tub-FRT-GAL80-FRT-GAL4 (Zecca and Struhl, 2002). Briefly, embryos were heat-shocked at 38°C for 5 minutes and raised in 18°C until wandering third instar larva; third instar larval brains were dissected and processed for immunohistochemistry (see below). Flies bearing the genotype UAS-FLPJD1; tub-Gal80ts/Act5C>Stop>lacZ; alrm-Gal4/+ were used to determine the fate of primary astrocyte-like glia in the pupa. Briefly, embryos were collected for 48 hours in 18°C; freshly hatched larvae were raised in 29°C for 24 hours, and placed back in 18°C until specified extrapolated pupal time points (development in 18°C is roughly twice as long as 25°C). UAS-FLPJD1, Act5C>Stop>lacZ flies were also crossed to appropriate Gal4 lines and stained with anti-GAT antibody at P48 or stained with anti-Repo antibody in the wandering third instar larva, to determine the lineage of secondary astrocyte-like glia or secondary neuropil glia precursors, respectively.
Immunostaining and TUNEL assay
Samples were fixed in 4% methanol-free formaldehyde in phosphate buffered saline (PBS, Fisher-Scientific, pH = 7.4; Cat No. #BP399-4). Tissues were permeabilized in PBT (PBS with 0.1% or 0.3% Triton X-100, pH = 7.4) and immunohistochemistry was performed using standard procedures (Ashburner, 1989). Embryos were dechorionated in 50% bleach, fixed in equal ratio PEMS (.1 M Pipes, 2 mM MgSO4, 1 mM EGTA, pH 7.0) buffered 4% formaldehyde and heptane. Embryos were then devitellinized in methanol, rehydrated in PBT and stained using standard procedures (Ashburner, 1989). The following antibodies were provided by the Developmental Studies Hybridoma Bank (Iowa City, IA): mouse anti-Repo (repo, 1:10), mouse anti-Neurotactin (BP106, 1:10), rat anti-DN-cadherin (DN-EX #8, 1:20), and mouse anti-Bruchpilot (nc82, 1:30). For third instar larval staining of Neurotactin, samples were subjected to a methanol dehydration series (25, 50, and 100% for 20 minutes each) after fixation, prior to staining. We also used mouse or rabbit anti-β-galactosidase (1:100, Promega), rabbit anti- C-terminus γ-aminobutyric acid transporter (GAT, 1:3000) (kind gift from M. Freeman, University of Massachusetts Medical School, Worcester, MA) and rabbit anti-Labial (1:200) (kind gift from H. Reichert, Biozentrum University of Basel, Basel, Switzerland). Secondary antibodies, IgG1 (Jackson ImmunoResearch; Molecular Probes) were used at the following dilutions: Alexa 546-conjugated anti-mouse (1:500), Cy3-conjugated anti-mouse (1:500), Cy5-conjugated anti-rat (1:400), Alexa 568-conjugated anti-mouse (1:500), Alexa 568-conjugated anti-rabbit (1:400), Alexa 488-conjugated anti-mouse (1:400). For terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), we used the In Situ Cell Death Detection Kit, Fluorescein (Roche). Briefly, P24 brains were dissected directly in cold 4% methanol-free formaldehyde and fixed for 45 min, followed by cold PBS washes and an ethanol dehydration series (5, 10, 20, 40, 75, 100% for 5 min each). After an overnight incubation in 100% EtOH at 4°C, samples were rehydrated in 0.3% PBT and stained with antibodies using standard procedures. Immunostained samples were incubated in 1:10 TUNEL reaction mixture for 1 hour and 15 minutes and prepared for imaging.
Confocal Microscopy and Analysis
Staged Drosophila embryonic, larval, pupal, and adult brains labeled with suitable markers were viewed as whole-mounts by confocal microscopy [LSM 700 Imager M2 using Zen 2009 (Carl Zeiss Inc.); lenses: 40× oil (numerical aperture 1.3)]. Complete series of optical sections were taken at 2-µm intervals. Captured images were processed by ImageJ or FIJI (National Institutes of Health, http://rsbweb.nih.gov/ij/ and http://fiji.sc/) and figures assembled in Adobe Photoshop. Cell counts and nuclear size measurements were carried out manually using the “Cell Counter” and “Measure” plugins in ImageJ. All error bars represent ± Stdev. Neuropil glia counts for embryonic brains were carried out for one hemisphere from the deutero-tritocerebral boundary to the supraesophageal commissure. Neuropil glia counts for larval brains were carried out for the supraesophageal ganglion specifically.
Registration and generation of three-dimensional models
Digitized images of confocal sections were processed in ImageJ and imported into FIJI (Schindelin et al., 2012; http://fiji.sc/). Surface renderings of first and third instar larval brains stained with anti-DN-cadherin and anti-Bruchpilot, respectively, were generated as volumes in the 3-dimensional viewer in FIJI. Particularly well oriented samples (for both L1 and L3) in which the peduncles were aligned with the z-axis of the stack, were chosen as the “standard” brains for registration. In FIJI, using the “Name Landmarks and Register” plugin (http://fiji.sc/Name_Landmarks_and_Register), fiducial markers were placed at spatially reproducible landmarks on the standard brain throughout the neuropil surface (ex. entry points for specific lineages). The digitized confocal section images of alrm-Gal4, UAS-mCD8-GFP brains were registered to the standard brains, imported using the TrakEM2 plugin in FIJI software (Cardona et al., 2012), and glial cell bodies were indicated (as spheres) on surface renderings using TrakEM2.
RESULTS
Astrocyte-like and ensheathing glia of the larval and adult brain
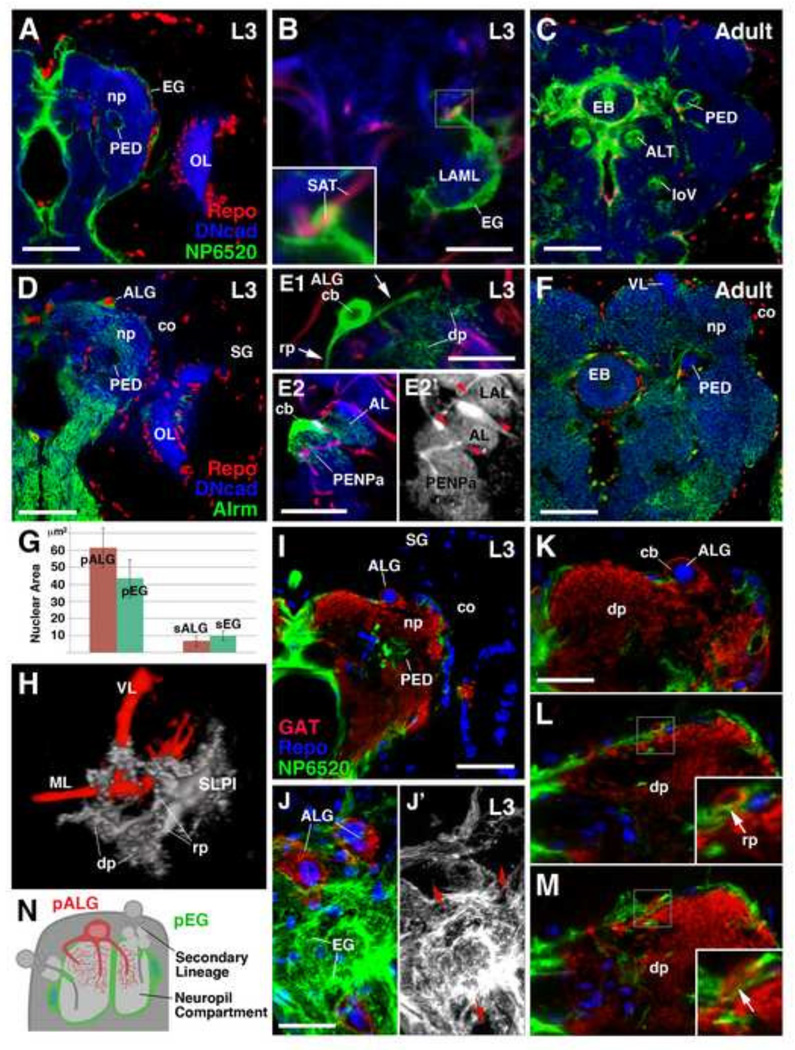
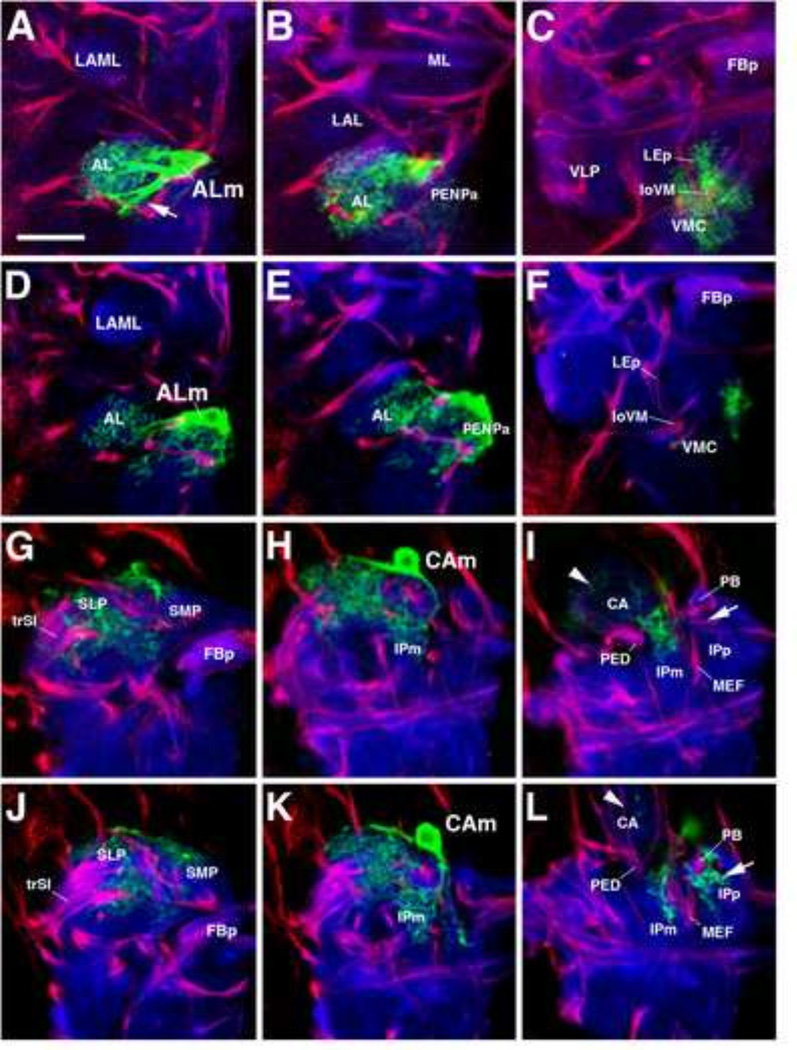
The Drosophila larval and adult brains contain populations of Repo-positive ensheathing and astrocyte-like glia which can be labeled by the neuropil glia subtype-specific Gal4 drivers, NP6520-Gal4 and NP3233 or alrm-Gal4, respectively (Fig. 1A–F). Similarly to neurons, these populations are derived from primary and secondary neuroblast lineages and therefore we classify them as primary (larval) and secondary (adult) neuropil glia (pNPG and sNPG) (see later sections). However, unlike primary neurons, many of which are remodeled and retained into adulthood, pNPG are not retained into adulthood (see later sections). EG extend sheath-like processes around neuropil compartments and axon tracts (Fig. 1A–C; see Table 1 for abbreviations). Single cell clonal analysis in the third instar larva demonstrates that primary EG (pEG) typically have enlarged, flattened cell bodies (Fig. 1B, G). The processes of a single primary ensheathing glial cell can ensheath the border of a neuropil compartment and incoming secondary axon tracts simultaneously (Fig. 1B; inset). Secondary EG (sEG) are prominent around major tracts and central complex of the adult brain (Fig. 1C). ALG extend highly branched processes with dendritic/reticular morphology into neuropil compartments (Fig. 1D–F). Flp-out clonal analysis of a single, primary astrocyte-like glial cell in the third instar larva reveals details of their complex morphology. Primary astrocyte-like glia (pALG) have very large, rounded cell bodies and nuclei in comparison to neurons, and even in comparison to pEG (Fig. 1E, G). They extend a variable number of processes which are relatively thick where they branch off the soma (“root processes”), but branch distally into a dense network of fine fibers (“distal processes”) (Fig. 1E1, H). The processes of a single primary astrocyte-like glial cell typically infiltrates more than one neuropil compartment (e.g., the antennal lobe and anterior peri-esophageal compartment as shown in Fig. 1E2–E2’), and often traverses a large neuropil volume. sALG of the adult brain are smaller in size and far more numerous than the pALG of the larval brain (Fig. 1F–G).
Figure 1.
Cytology of mature Drosophila neuropil glia. (A–C): Ensheathing glia (EG). Frontal confocal sections (dorsal, up; lateral, right) of late third instar larval (A and B) and adult brain hemispheres (C) depicting mature primary ensheathing glia (pEG) (A), a high magnification image of a representative GFP-labeled pEG single cell clone (B), and mature secondary ensheathing glia (sEG) (C). pEG and sEG are labeled in green by NP6520-Gal4 (A and C). (B) pEG single cell clone associated with the neuropil compartment, lateral appendix of the medial lobe (LAML) and is simultaneously associated with a secondary axon tract (SAT) labeled by anti-Neurotactin (red) (boxed inset). (D–F): Astrocyte-like glia (ALG). Equivalent stages depicting mature primary astrocyte-like glia (pALG) in late larval brain (D), representative pALG single cell clones (E1, E2-E2’), and mature secondary astrocyte-like glia (sALG) in adult (F). pALG and sALG are labeled in green by alrm-Gal4 (D and F). (E1) pALG single cell clone demonstrates that pALG have a large nucleus and soma. A single pALG cell extends multiple primary “root processes” (rp, arrows), each with reticular “distal processes” (dp). (E2-E2’) A second pALG single cell clone demonstrating extension of processes into two distinct neuropil compartments labeled by anti-DN-cadherin (E2, blue; E2’, gray) bordered by SATs [anti-Neurotactin (red), red arrowheads]; the antennal lobe and the anterior peri-esophageal neuropil. Compartment definitions have been described previously (Pereanu et al., 2010). The nuclei of all glial cells are labeled by anti-Repo (red) (A, C, D, F). The larval and adult neuropil (np) is labeled with anti-DN-cadherin (blue) (A–F). (G) Nuclear mean area quantifications of primary and secondary neuropil glia (n=39). (H) 3D volume rendering of a spatially identifiable pALG single cell clone, representing the SLPI (gray color; see Fig. 3); the mushroom body is shown for spatial reference (red). (I–M) Frontal (or tangential; J–J’) confocal sections of third instar larval brains in which pEG are labeled by NP6520-GaI4 (green), and pALG are labeled by anti-GAT (red); glial nuclei marked with anti-Repo (blue). (J–J’) Tangential section of the neuropil surface demonstrating that the neuropil surface is predominantly ensheathed by the membranes of pEG; the somata of pALG occupy gaps in the pEG (red arrows) and contribute to the sheath as well. pALG processes predominantly extend directly into the neuropil (K) or stay superficial relative to pEG prior to penetrating through the pEG into neuropil (boxed inset; arrows) (L–M). (N) Schematic depicting the morphology of, and spatial relationship between, astrocyte-like (red) and ensheathing (green) glia relative to other brain structures such as secondary lineages and neuropil compartments. Anatomical abbreviations found in Table 1. Other abbreviations: cb, cell body; co, cortex; SG, surface glia.
Bars: 50µm (A, C, D, F, I); 20µm (B, E); 10µm (J; K–M)
Table 1.
List of abbreviations of neuropil brain compartments (left), fascicles (right), optic lobe-specific terms (right), and other (right).
| Brain Compartments | Abbr. | Brain Fascicles | Abbr. |
|---|---|---|---|
| Antennal lobe | AL | Antennal lobe tract | ALT |
| Anterior optic tubercle | AOTU | Great commissure | GC |
| Anterior periesophageal neuropil | PENPa | Longitudinal superior medial fascicle | loSM |
| Ellipsoid Body | EB | Longitudinal ventral fascicle | loV |
| Fan-shaped body | FB | Medial loV | loVM |
| Fan-shaped body primordium | FBp | Medial equatorial fascicle | MEF |
| Inferior protocerebrum | IP | Posterior lateral ellipsoid fascicle | LEp |
| Medial IP | IPm | Supraellipsoid body commissure | SEC |
| Posterior IP | IPp | Transverse super intermediate fascicle | trSI |
| Lateral accessory lobe | LAL | Optic Lobe | Abbr. |
| Lateral appendix of the medial lobe | LAML | Inner Optic Anlage | IOA |
| Mushroom body | MB | Lobula neuropil | LOnp |
| Calyx of MB | CA | Medula cortex | MEco |
| Medial lobe of MB | ML | Medulla neuropil | MEnp |
| Peduncle of MB | PED | Optic Lobe | OL |
| Vertical lobe of MB | VL | Outer Optic Anlage | OOA |
| Protocerebral bridge | PB | Other | Abbr. |
| Superior lateral protocerebrum | SLP | Abdominal ganglia | ABD |
| Superior lateral protocerebrum/Lateral horn primordium | SLPp/LHp | leg neuropil primordium | Inp |
| Superior medial protocerebrum | SMP | Subesophageal ganglion | SEG |
| Ventro-lateral protocerebrum | VLP | ||
| Ventro-medial cerebrum | VMC | ||
| Postcommissural/Posterior VMC | VMCpo | ||
We utilized double labeling of the two pNPG subpopulations in order to assess the spatial relationship between the pALG and pEG at the neuropil surface (Fig. 1I). ALG stably express the γ-aminobutyric acid transporter (GAT) on their soma as well as on their membrane protrusions; this serves as a specific, reliable marker for ALG (Stork et al., 2014; Supp. Info. Fig.S1). The entire neuropil surface is encapsulated by the membrane contributions of the two NPG subtypes. Most of the neuropil surface is covered with the large, flattened pEG (Fig. 1J–1J’). This EG sheath is disrupted by gaps which are filled by the cell bodies of ALG (Fig. 1J–J’). The processes emanating from pALG somata most frequently extend directly into the neuropil, however, they occasionally grow tangentially along the neuropil surface, passing over pEG sheaths before entering the neuropil through a gap in the pEG (Fig. 1K–M).
Development of neuropil glia around the embryonic brain
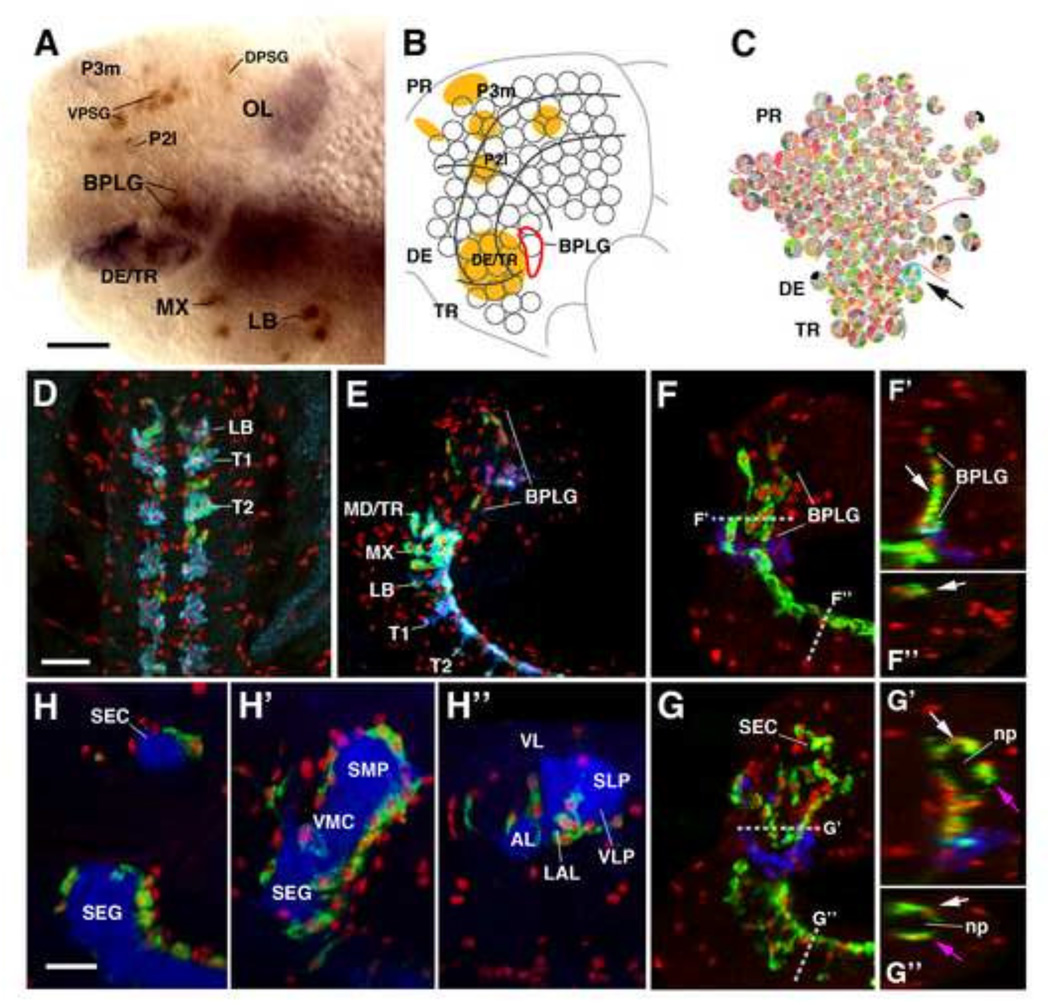
Progenitors of the primary neuropil glia form part of the population of neuroglioblasts/glioblasts that have been described for the embryo. In the thoracic and abdominal neuromeres of the ventral nerve cord, as well as the gnathal neuromeres, neuropil (longitudinal) glia are derived from a single paired cell, the lateral glioblast (Beckervordersandforth et al., 2008; Jacobs et al., 1989). Brain neuropil glial progenitors form a small cluster of Repo-positive cells, called the basal procephalic longitudinal glia (BPLG), located at the boundary between the tritocerebrum and deuterocerebrum (Hartenstein et al., 1998) (Fig. 2A–B). The glial determinant glial cells missing (gcm) is known to be required and sufficient for gliogenesis and is expressed early in glial lineages (Hosoya et al., 1995; Jones et al., 1995; Vincent et al., 1996). The brain neuroblast map by Urbach and Technau, 2003 demonstrated that there is a single gcm-positive neuroblast amongst the tritocerebral neuroblasts which is likely to represent the neuroglioblast (or glioblast) that generates the BPLG (Fig. 2C). It has been shown (Beckervordersandforth et al., 2008; Ito et al., 1995) that neuropil glia of the VNC migrate towards the dorsal surface of the nascent neuropil; by stage 14, they form a continuous double row of Repo-positive cells along the dorsal surface of the VNC (Fig.2D). Likewise, neuropil glia of the brain derived from the BPLG cluster, assemble at the medial surface of the brain neuropil, reaching dorsally to a level where the brain commissure will appear (Fig.2E). 53 ± 9% (n=5) of the Repo-positive neuropil glia expresses alrm/GAT, attesting to the fate of these cells as presumptive pALG. During stages 15 and 16, a subpopulation of neuropil glia migrates around the entire neuropil surface: ventrally in the VNC, laterally and dorsally in the brain (Fig. 2F–F”, 2G–G”). As the neuropil becomes structured into distinct compartments (e.g., antennal lobe, mushroom body, lateral accessory lobe), neuropil glia adopt their final position relative to these compartments (Fig. 2H–H”).
Figure 2.
Embryonic development of primary neuropil glia. (A) Early stage 12 embryo labeled with anti-Repo (brown) and anti-Fasll (purple), depicting specific immature glial clusters. (B) Schematic representation of the brain neuroblast map. (C) Brain neuroblast map of Urbach and Technau (2003). Fasll-positive cells shown in (A), and depicted as orange domains in (B), represent clusters of pioneer neurons that serve as reference markers to follow cell movements in the brain primordium. Gray lines in (B) subdivide the brain neuroblast map into a protocerebral domain (PR), deuterocerebral domain (DE), and tritocerebral domain (TR); the same subdivisions are indicated in (C). Progenitors of brain neuropil glia form the basal procephalic longitudinal glia (BPLG) cluster, outlined by red line in (B). Arrow and cyan circle in (C) points at the only gcm-positive neuro-(glio)blast (Td7) of the neuroblast map. This cell is situated at the location where the BPLG develops, and therefore is most likely the progenitor of the brain neuropil glia. (D–H) Z-projections of confocal sections of stage 15–17 embryonic ventral nerve cords (VNC) and brains labeled with anti-Repo (red) and marker of astrocyte-like glia (alrm-Gal4, green). (D) horizontal section of stage 15 embryonic VNC. Astrocyte-like glia are marked by the expression of GAT (blue), in addition to alrm-Gal4 (green), and appear in cyan color. (F): parasagittal sections of stage 15 brain and VNC. (F’, F”): digital rotations of z-projection shown in (F), showing virtual horizontal section of brain, and frontal section of VNC [see dashed lines in (F) for plane of section of F’ and F”]. Labeling with anti-Labial (blue) was used to delineate the tritocerebral domain, from where the brain neuropil glia originates (see panel (C) above). At the stage shown, cells of the alrm-positive BPLG cluster have spread out dorsally along the emerging brain neuropil; cells are still clustered at the medial surface of the neuropil (arrow in F’). VNC neuropil glia are located at the dorsal surface of the VNC neuropil (arrow in F”). (G–G”): z-projection and digital rotations thereof of parasagittal sections of late stage 16; labeled and oriented like panels (F–F”). The alrm-positive brain neuropil glia have further spread dorsally (G), and also laterally/anteriorly, surrounding the neuropil (np) on all sides (from white to magenta arrow in G’). The spreading of VNC glia around the neuropil also occurs at this stage (from white to magenta arrow in G”). For clarity, non-CNS staining was manually removed (E–G”). (H–H”) Three parasagittal sections of a stage 17 brain, representing level close to the midline (H), level along center of neuropil (H’) and level of lateral neuropil (H”). Neuropil is labeled by anti-DN-cadherin (blue). Alrm-positive glia have reached final position around neuropil compartments. Anatomical abbreviations found in Table 1. Other abbreviations: DPSG, dorsal procephalic subperineurial glia; P2I and P3m, fiber tract founder clusters of the protocerebrum; LB, labial segment; MX, maxillary segment; MD/TR mandibular segment/tritocerebrum; T1, T2, Thoracic segments 1 and 2; VPSG, ventral procephalic subperineurial glia.
Bars: 10µm
Pattern of primary astrocyte-like and ensheathing glia in the larval brain
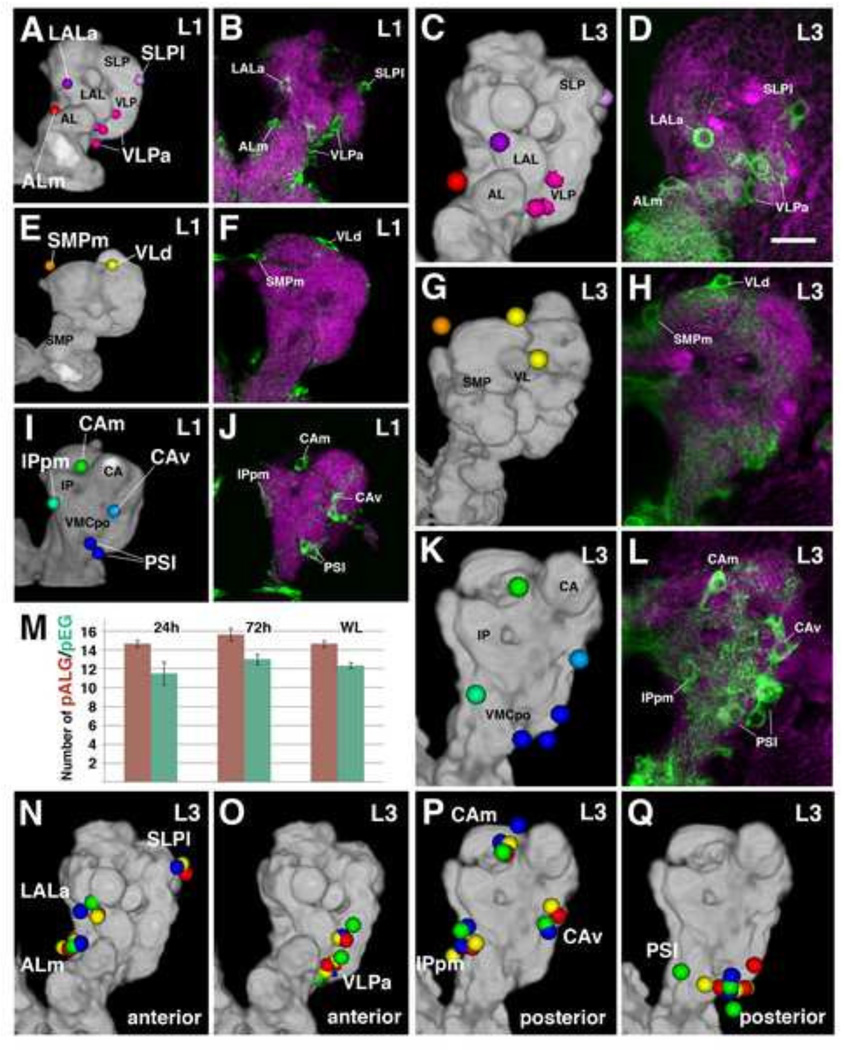
The neuropil of the hatching larva is surrounded by approximately 15 pALG and 12 pEG per brain hemisphere (Fig. 3M). These figures do not increase throughout the larval period (Fig. 3M). Similarly to the pALG of the VNC (Stork et al., 2014), pALG of the brain are distributed in an almost invariant pattern which does not change from early to late larva (Fig. 3). Due to their topological reproducibility, we adopted a nomenclature system for individual or clusters of pALG based on the adjacent neuropil compartment (Table 2). At the anterior neuropil surface we typically find three individual cells flanking the antennal lobe (ALm), the lateral accessory lobe (LALa), and the superior lateral protocerebrum (SLPI; Fig. 3A–D, N), respectively. In addition, a cluster of 3–4 pALG borders the anterior surface of the ventrolateral protocerebrum (VLPa; Fig.3A–D, O). Further posteriorly, one primary astrocyte-like glial cell flanks the crown of the superior medial protocerebrum (SMPm) and 1–2 pALG are grouped around the tip of the vertical lobe of the mushroom body (VLd; Fig. 3E–H). Lastly, the pALG around the posterior neuropil surface include two cells bordering the calyx medially (CAm) and ventrally (CAv), one cell adjacent to the posterior inferior protocerebrum (IPpm), and a group of 2–3 cells covering the ventromedial cerebrum (PSL; Fig. 3l–L, P-Q).
Figure 3.
Primary neuropil glia exhibit a relatively stereotyped topological pattern around the larval neuropil. Anterior pALG clusters (A–D), intermediate clusters (E–H), and posterior clusters (l–L) are shown in the first larval instar (left two columns) and in the third larval instar (right two columns); (dorsal up, lateral right). The first and the third column represent digital 3D models of the first instar (A, E, I) and third instar (C, G, K) larval brain. The neuropil surface is rendered in gray (annotated with black lettering) surrounded by color coded pALG clusters (annotated with white lettering). Panels of the second column (B, F, J) and fourth column (D, H, L) show frontal confocal z-projections of first instar and third instar brains, respectively. The same pALG clusters modeled in (A, E, I) and (C, G, K), respectively, are visualized by alrm-Gal4 in green; the neuropil is labeled by anti-DN-cadherin (magenta). (A–D) Anterior view reveals the anterior clusters of astrocyte-like glial cells, including the ALm (antennal lobe medial; red), LALa (lateral accessory lobe anterior; purple), SLPI (superior lateral protocerebrum lateral; light purple), and VLPa (ventro-lateral protocerebrum anterior; magenta). (E–H) Anterior dorsal view reveals the intermediate clusters; VLd (vertical lobe dorsal; yellow) and SMPm (superior medial protocerebrum medial; orange). (I–L) Posterior view reveals the posterior clusters; CAm (calyx medial; green), CAv (calyx ventral; cyan), PSI (posterior slope lateral; blue), and IPpm (inferior protocerebrum posterior medial; green blue). (M) Numbers of pALG and pEG at early larval stage (24h ah), mid larval stage (72h ah), and at wandering larva stage (WL) (n≥3 for each timepoint). (N–Q) 3D volume renderings of third instar larval brain neuropils (gray) depicting anterior (N–O) and posterior (P–Q) primary astrocyte-like glial clusters. Corresponding pALG from four representative brain hemispheres were registered to a standard brain and are shown in red, blue, yellow, and green. Note the low degree of spatial variability of pALG between samples. Primary astrocyte-like glia nomenclature can be found in Table 2.
Bar: 25µm
Table 2.
Nomenclature and abbreviations for primary astrocyte-like glial clusters.
| Primary ALG Cluster Nomenclature | # of Cells Per Cluster |
Anterior-Posterior Levels |
Color in Fig. 3 |
|---|---|---|---|
| Antennal Lobe medial (ALm) | 1–2 | Anterior | red |
| Lateral Accessory Lobe anterior (LALa) | 1 | Anterior | purple |
| Ventrolateral Protocerebrum anterior (VLPa) | 3–4 | Anterior | magenta |
| Superior Lateral Protocerebrum lateral (SLPI) | 0–1 | Anterior | light purple |
| Vertical Lobe dorsal (VLd) | 1–2 | Intermediate | yellow |
| Superior Medial Protocerebrum medial (SMPm) | 1 | Intermediate | orange |
| Calyx medial (CAm) | 1–2 | Posterior | green |
| Calyx ventral (CAv) | 1 | Posterior | cyan |
| Posterior Slope lateral (PSI) | 2–3 | Posterior | blue |
| Inferior Protocerebrum posterior medial (IPpm) | 1–2 | Posterior | green blue |
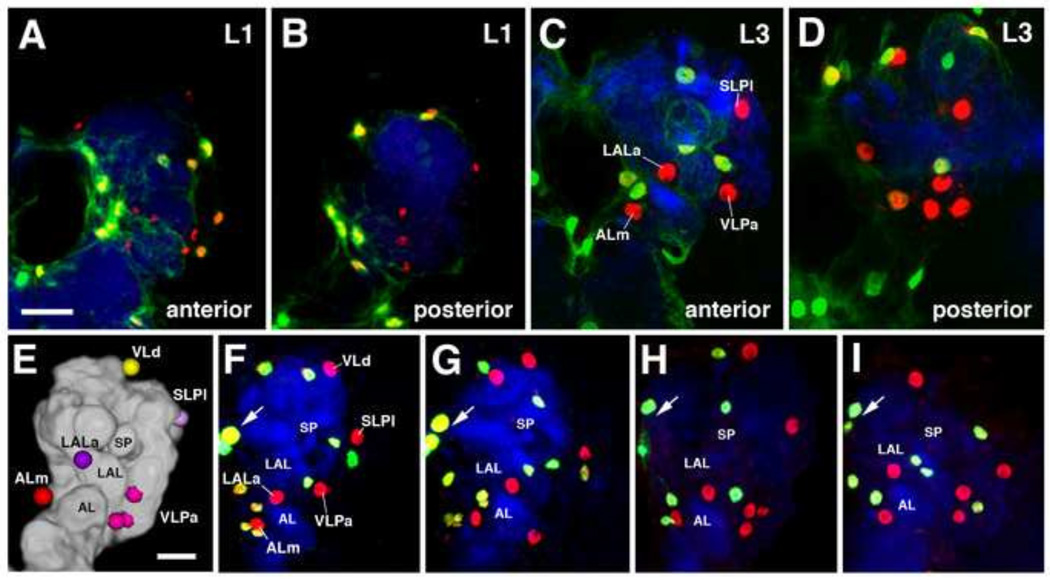
pEG exhibit a higher degree of spatial variability than pALG (Fig. 4). As shown in Fig. 4E–I, even though the overall number of pEG is in the same, low range as pALG, the location of pEG cell bodies and nuclei in relationship to compartments, or nearby pALG, changes between different specimens. We therefore did not establish a specific nomenclature for individual cell clusters in this glial subclass.
Figure 4.
Pattern of primary ensheathing glia (pEG). Frontal confocal z-projections of first instar (A–B) and third instar (C–D) larval brains depicting anterior (A, C) and posterior (B, D) clusters of primary ensheathing glia labeled by NP6520-Gal4 driving nuclear GFP in green relative to the semi-stereotyped primary astrocyte-like glia, astrocyte-like glia labeled by anti-Repo in red, and the neuropil labeled by anti-DN-cadherin in blue. Anatomical abbreviations found in Table 1. (E) 3D digital model of anterior surface of third instar larval brain hemisphere (medial to the left), depicting five pALG clusters. (F–I) Confocal sections of four brain hemispheres, labeled with anti-Repo (red) and NP6520-Gal4 (green). For clarity, non-CNS and non-primary neuropil glial staining was manually removed (A–l). Note invariant pattern of pALG (Repo-positive; NP6520-negative), corresponding to the cells shown in model (E). One cell, ALm, can be always associated with the antennal lobe (AL), another one, LALa, with the lateral accessory lobe (LAL); one, SLPI, with the anterior surface of the SLP compartment; one or two (VLd) with the vertical lobe. By contrast, NP6520-positive pEG, with the exception of the four interhemispheric glia (arrow; two nuclei in focus), show more variability. For example, two pEG are close to the anterior SLP in the hemisphere shown in (F); one in (G); none in (H), and one in (I). Two pEG are close to the vertical lobe in (F), one in (G) and (H), and none in (I). Other abbreviations: SP spur of mushroom body.
Bars: 25µm
The relatively invariant pattern of pALG somata suggests that the central processes of pALG may also associate with neuropil compartments in a similar, relatively invariant fashion. Single cell clonal analysis of equivalent pALG of different brains demonstrates that this is indeed the case. However, the exact trajectory of pALG root processes and the neuropil volume invested by their distal processes, similarly to the topology of somata, can exhibit subtle variability. For example, the cell ALm, shown for two different brains in Fig. 5A–C and Fig. 5D–F, extends processes laterally into the antennal lobe (AL) and medially/posteriorly towards the adjacent neuropil. Distal processes of this cell fill the entire AL. A subtle, but significant difference between the two ALm cells exists in regard to the processes directed towards the neuropil flanking the AL medially. The cell shown in the top row (Fig. 5A–C) branches predominantly in the ventromedial cerebrum (VMC), located posteriorly adjacent to the AL; most extra-antennal processes of the other ALm (Fig. 5D–F) are focused further anteriorly in the peri-esophageal neuropil (PENPa). Note the abundance of ALm processes surrounding the loVM fascicle (penetrating the VMC compartment) in Fig. 5C, and the virtual absence of such processes in Fig. 5F. Shown as a second example in Fig. 5G–l and Fig. 5J–L, respectively, are CAm clones from two different brains. In both cases, CAm sends a major branch laterally into the superior lateral protocerebrum (SLP; Fig. 5G–H, J–K), and another branch postero-medially into the superior medial protocerebrum (SMP) and medial inferior protocerebrum (IPm; Fig. 5H, K). At a posterior level, the branching pattern of both cells differs subtly, in that one projects more prominently into the calyx (CA; Fig. 5I; arrowhead), whereas the other projects into the posterior inferior protocerebrum (IPp; Fig. 5L; arrow).
Figure 5.
Branching pattern of primary astrocyte-like glial cells. (A–L) Z-projections of frontal confocal sections (dorsal, up; lateral, left) of late third instar larval brain hemispheres. GFP-labeled single-cell flp-out pALG clones in green, neuropil labeled by anti-DN-cadherin in blue, and secondary axon tracts labeled by anti-Neurotactin in red. (A–C) and (D–F): Antennal lobe medial (ALm) cell in two different brains. Z-projections represent brain slices of 10–16 µm thickness at three different antero-posterior levels (A, D): anterior part of antennal lobe (AL); (B, E): posterior part of antennal lobe; (C, F): ventromedial cerebrum (VMC), posterior of antennal lobe. Even though both cells are located at the same position, the example shown in (A–C) projects more thick root branches towards the ventrolateral antennal lobe [arrow in (A)] and covers a larger part of the VMC (C) than the second example shown in (D–F). (G–l) and (J–L): Calyx medial (CAm) cell in two different brains. Z-projections taken at three antero-posterior levels [level of fan-shaped body primordium (FBp) (G, J); posterior of FBp, at the level of the medial inferior protocerebrum (IPm) (H, K); calyx (CA) (I, L)]. The projections of both examples of the CAm glia are very similar; note, however, more extensive arborizations of the cell shown in (G–l) in the calyx (CA; arrowheads in I, L) and the cell shown in (J–L) in the posterior inferior protocerebrum (IPp; arrows in I, L). Anatomical abbreviations found in Table 1.
Bar: 20µm
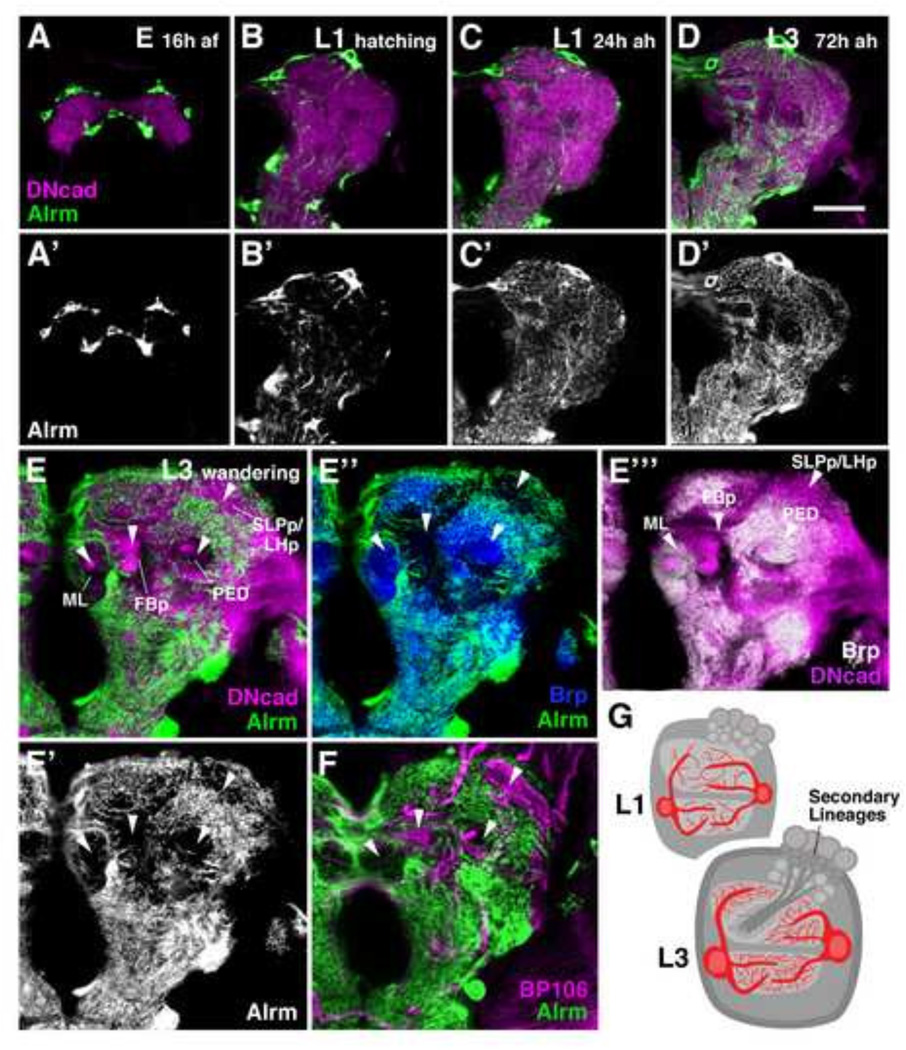
Morphogenesis of primary astrocyte-like glia: progressive densification of processes in the larva, followed by regression and programmed cell death in the pupa
In order to accommodate for the several-fold expansion of the larval neuropil during development, pALG increase in size (Fig. 6), indicating that pALG, as seen for most other differentiated Drosophila cell types, undergo endoreplication (Unhavaithaya and Orr-Weaver, 2012). Concomitant with the increase in cytoplasmic and nuclear volume, there is an increase in density of pALG processes filling the neuropil compartments. In the late embryo, pALG are spindle-shaped cells restricted to the neuropil surface (Fig. 6A–A’). After hatching, pALG project small protrusions which fill the neuropil volume (Fig. 6B–B’). Processes are added continuously throughout the larval period (Fig. 6C–D’). During the third larval instar (Fig. 6E–F), a conspicuous pattern of neuropil domains with high pALG density, separated by domains lacking pALG processes altogether (arrowheads in Fig.6E–E’”), evolves. The domains lacking pALG processes are positive for Neurotactin (BP106; Fig. 6F), which labels secondary neurons, as well as for DN-cadherin (Fig. 6E, E’”), but they are negative for the synaptic marker Bruchpilot (nc82; Fig. 6E”-E’”). We therefore interpret the pALG-negative domains as the massive bundles of secondary axon tracts (SATs), tipped by growth cones and filopodia, that, as described in previous works (Pereanu and Hartenstein, 2006; Pereanu et al., 2010) invade the primary neuropil and generate wide gaps between larval synaptic compartments. Several of the domains filled by SATs form distinctive primordia of adult neuropil compartments, such as the compartment of the fan-shaped body (FBp; Fig. 6E–E’”), or (in the ventral nerve cord) the primordia of the leg neuropils (Supp. Info. Fig.S4A–A’). pALG processes are virtually excluded from these primordia (Fig. 6E–E”). The only compartment that is rich in synapses but contains a low amount of pALG processes are the lobes (ML medial lobe; VL vertical lobe) and peduncle (PED) of the mushroom body (MB; Fig. 6E–E”).
Figure 6.
Morphogenesis of primary astrocyte-like glia. (A–F’) Developmental time course of astrocyte-like glia from late embryo (16 hours after fertilization) to the third instar wandering larva. pALG are labeled by alrm-Gal4 in green (A–F) and in grayscale (A’-D’, E”). The neuropil is labeled by anti-DN-cadherin in magenta (A–E, E’”). pALG processes extend initially around the surface of the late embryonic neuropil (A–A’), followed by the formation of branches that penetrate into the neuropil in the freshly hatched larva (B–B’). Neuropil branches increase in density throughout larval development (C–E’). The third instar larval neuropil contains “gaps” in the density of pALG processes [arrowheads in (E–F)], which correspond to domains where fiber bundles of secondary neurons labeled by Neurotactin (BP106) penetrate into the neuropil (F). These undifferentiated fibers form the primordia of adult neuropil compartments; they lack synapses [see absence of blue and white Brp signal in (E” and E”‘), respectively], but are positive for DN-cadherin [magenta signal in (E, E’”)]. Processes of pALG are restricted to the Brp-positive (synapse-rich) neuropil domains (E”). (G) Schematic depicting the coordinate morphogenesis (increase in cell size and process density) of primary astrocyte-like glia and the increasing neuropil volume from early to late larva. Anatomical abbreviations found in Table 1. Other abbreviations: af, after fertilization; ah, after hatching.
Bar: 25µm
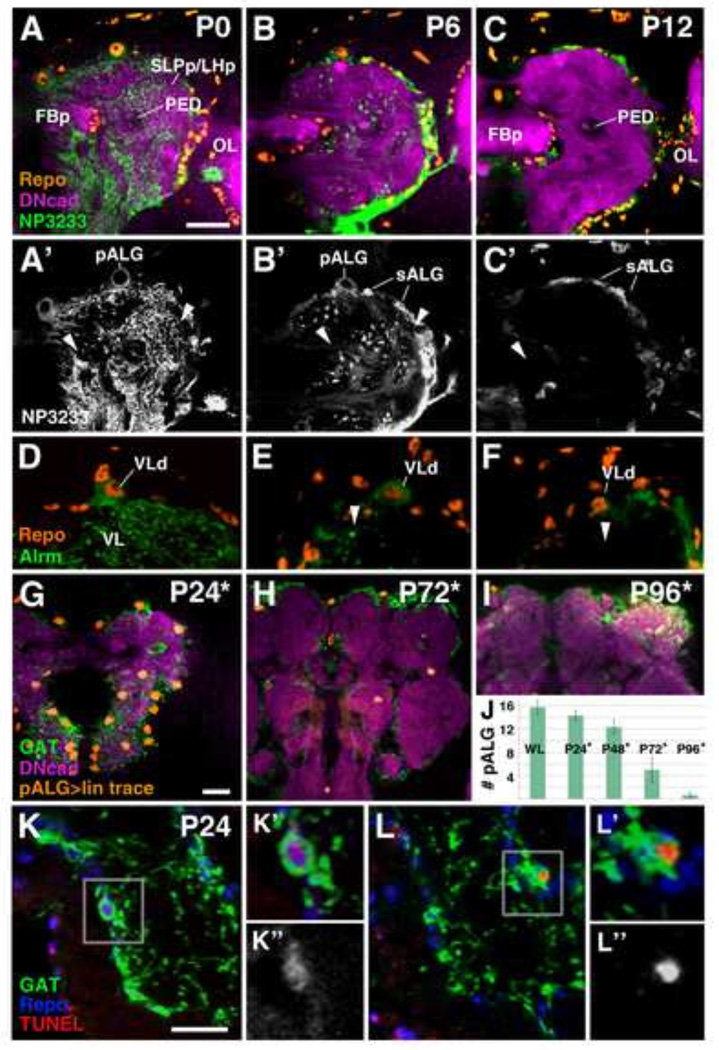
At the onset of metamorphosis, from P0-P6, the distal processes of the pALG begin to fragment (Fig. 7A–F). The network of fine processes disappears, and is replaced by dense spherical bodies (visualized with mcd8-GFP; Fig. 7B’, E). These spherical bodies correspond to the pALG vacuoles that, according to previous studies, are involved in the phagocytosis of neuronal debris (Cantera and Technau, 1996; Hakim et al., 2014; Singh and Singh, 1999; Tasdemir-Yilmaz and Freeman, 2014). By P12, the neuropil is virtually free of any NP3233/alrm-Gal4-driven mcd8-GFP signal (Fig. 7C–C’, F). pALG somata are also difficult to detect after this point, suggesting that they undergo programmed cell death (Fig. 7C–C’, F). This conclusion was confirmed by two experiments, lineage tracing of pALG and TUNEL labeling. We performed lineage tracing experiments, where an Act5C >Stop >lacZ construct was specifically and stably activated in pALG during the early larval period, and not activated in sALG due to the presence of tub-Gal80ts (see Material and Methods). Third instar larvae expressed LacZ only in pALG, confirmed by co-staining with GAT antibody (data not shown). LacZ-positive pALG were still present at relatively normal numbers at P24 (Fig. 7G, J), but became rare around P72 (Fig. 7H, J), and were absent by P96 (Fig. 7I–J). GAT antibody first labels primary ALG and their processes until P24 (Fig. 7G), is absent around P36 (data not shown), and turns back on in secondary ALG by P48 onward (Fig. 7H–I; Muthukumar et al., 2014). Importantly, lineage traced pALG were GAT-positive at P24 (Fig. 7G), but GAT-positive secondary ALG were not LacZ-positive (Fig. 7H–I), demonstrating that pALG undergo cell death and that they do not give rise to sALG. TUNEL staining of pupal brains, double labeled with anti-GAT and anti-Repo, showed that TUNEL positive cells are detectable during early to mid-pupal stages (Fig. 7K–L).
Figure 7.
Primary astrocyte-like glia undergo programmed cell death during metamorphosis. (A–F) Developmental time course of astrocyte-like glia from white prepupa (PO) to P12. ALG are labeled by NP3233-Gal4 in green (A–C) and in grayscale (A’–C). The neuropil is labeled by anti-DN-cadherin in magenta and glial cell nuclei are labeled by anti-Repo in orange in (A–C). (D–F) High magnification images of the dorsal neuropil in context of the vertical lobe (VL, arrowheads); pALG in green labeled by alrm-Gal4 and glial cell nuclei labeled by anti-Repo in orange [looking specifically at the vertical lobe dorsal cell (VLd)] at equivalent stages as (A–C). At PO (A–A’, D) highly branched pALG processes are seen throughout the neuropil. During the first six hours of metamorphosis, pALG processes start to undergo fragmentation and vacuolization (B–B’, E). At P12, pALG processes are almost undetectable in the neuropil using Gal4 drivers (C–C, F). Note also small, NP3233 and Repo-positive sALG precursors emerging at the neuropil-cortex interface (B–C). (G–l) Lineage tracing of alrm-Gal4 expressing pALG. Nuclei of lineage traced cells are labeled in orange, anti-GAT in green, and anti-DN-cadherin in magenta. Asterisk indicates estimated time points based on rate of development at 18°C. (G) Lineage traced pALG are still relatively present at P24. (H–l) By the mid-pupal stage, the number of lineage traced nuclei declines and remaining nuclei appear atrophied; no pALG can be detected in the late pupa or adult. (J) Quantification of lineage traced pALG throughout metamorphosis. pALG numbers steadily decline until mid-metamorphosis, and quickly decline during late metamorphosis (n≥3 for each time point). (K–L) TUNEL labeling of pALG at P24. pALG somata at this stage can still be detected, unlike the Gal4 drivers, using anti-GAT (green). Sparse TUNEL labeling (red) can be observed in pALG (gray in K” and L”) at this stage. Anti-Repo is shown in blue. Anatomical abbreviations found in Table 1.
Bar: 25µm
Pattern and morphogenesis of secondary neuropil glia throughout metamorphosis
Until mid-larval development (48 hrs after hatching (ah), late first/early second instar), most, if not all Repo-positive neuropil glia express markers for differentiated pALG or pEG (GAT or NP6520-positive) (Supp. Info. Fig.S2A). During the third larval instar, an increasing number of small, Repo-positive cells, clearly distinguishable from the polyploid primary NPG, appear on the surface of neuropil compartments (Fig. 7A; Supp. Info. Fig.S2B). These cells, which are initially negative for alrm/NP3233/GAT and NP6520, constitute the precursors of secondary ALG and EG. In the late larva/early pupa, they are clustered at the posterior and lateral surface of the neuropil and, inside the neuropil, around the primordium of the central complex (Fig. 7A; Supp. Info. Fig.S2B). During early metamorphosis, a subset of these secondary neuropil glia precursors become NP3233-positive and, somewhat later, alrm-positive, demonstrating that they are secondary astrocyte-like glia (sALG) precursors (Fig. 7B–C’; Supp. Info. Fig.S3). NP3233-positive sALG precursors are small, spindle-shaped cells which are much more numerous than pALG (Fig. 7B–C’) and can also be observed in the VNC (Supp. Info. Fig.S4).
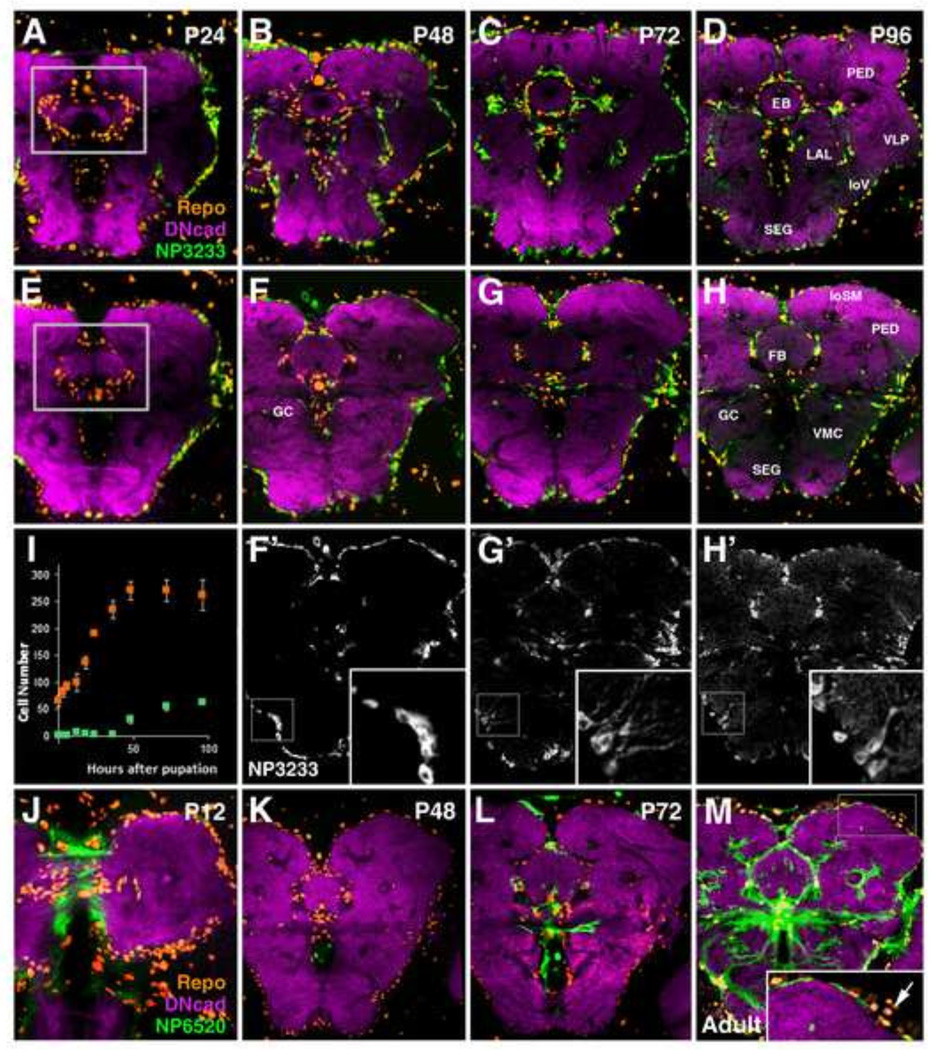
As metamorphosis proceeds, between P12 and P48, secondary NPG precursors increase in number and become evenly distributed over the outer neuropil surface (Fig. 8A–H). During later pupal stages, sNPG also spread within the neuropil, migrating along major axon bundles, including the great commissure, antennal lobe tract, peduncle, longitudinal ventral fascicle, and longitudinal superior medial fascicle (Fig. 8F–H). Counts of sNPG precursors around the enlarging compartments of the central complex (fan-shaped body, ellipsoid body, noduli) demonstrate that, although proliferation of neuroblasts has terminated (Ito and Hotta, 1992), a nearly 3-fold increase in sNPG precursors occurs during this time (99 to 270 cells) (Fig. 8I). This implies that following Repo expression, neuropil glia precursors proliferate, confirming the descriptions of Viktorin et al. (2011).
Figure 8.
Pattern of secondary neuropil glia throughout metamorphosis. (A–H) Z-projections of frontal confocal sections of pupal brains in which sALG are labeled by NP3233-Gal4 (green); the neuropil is visualized by anti-DN-cadherin (magenta), and glial cell nuclei are labeled by anti-Repo (orange). sALG from (F–H) are shown in grayscale with high magnification insets of boxed regions in (F’-H’). Panels of first row (A–D) show z-projections at level of ellipsoid body (EB); panels of second and third row (E–H, F’-H’) show slightly more posterior level at fan-shaped body (FB). Boxed regions in (A, E) depict the domains in which cell counts were conducted (I); (I) total Repo-positive cells associated with the central complex (i.e. EB, FB, noduli) are shown in orange, fraction of Repo-positive cells which co-express NP3233-Gal4 is shown in light green (n>3 for each time point). At early and mid-pupal stages, sALG spread out around neuropil surface and central complex primordium while at the same time increasing in number (A, E, B, F–F’). sALG processes do not yet enter the neuropil at significant numbers (see inset in F’). During the second half of the pupal stage (C, G–G’), radial processes emanate from the sALG into the neuropil (see inset in G’). Glial processes form a rich network of branches during the last day of metamorphosis (D, H–H’; see inset in H’). (J–M) Developmental time course of primary and secondary ensheathing glia (sEG) from P12 to adult at the level of the fan-shaped body (FB) and great commissure (GC). EG are labeled by NP6520-Gal4 in green, neuropil is labeled by anti-DN-cadherin in magenta, and glial cell nuclei are labeled by anti-Repo in orange. At P12 (J) primary EG of the larva are still present. During mid-metamorphosis (K), NP6520-positive pEG are no longer seen. NP6520 expression in secondary EG appears first around the central complex (L), followed by expression at the neuropil surface (M). Clusters of NP6520-Gal4 negative neuropil glia, interpreted as sALG, occupy regions of low sEG density at the adult neuropil surface (M, inset; arrow). Anatomical abbreviations found in Table 1.
Bar: 50µm
Morphogenesis of sALG proceeds similar to the pattern described for pALG in the larva. As reported previously (Muthukumar et al., 2014), up until around P48, sALG are flat or spindle-shaped cells (Fig. 8F’). From P48-P72, sALG elaborate a few primary processes that radiate into the neuropil (Fig. 8G’); during the last day of the pupal period, these processes form branches of increasing density in the neuropil (Fig. 8H’). High magnification images of the developing antennal lobe clearly illustrate the dynamics of sALG morphogenesis (Supp. Info. Fig.S5). Oland et al. (2008) demonstrated that primary processes of sALG form from P60-P72 around the glomeruli, which had appeared much earlier (Supp. Info. Fig.S5B–C’). The interior of glomeruli becomes penetrated by the fine branches of sALG between P72 and P96 (Supp. Info. Fig.S5D–D’).
Expression of the EG marker NP6520 begins later and shows a different dynamic than that of NP3233. NP6520 expression decreases steadily from late larva to around P12 (Fig. 8J). No expression is visible during mid-metamorphosis (Fig. 8K). Around P60-P72, NP6520 appears in centrally located neuropil glia surrounding the central complex, and only later, from P96 onward, in glia at the neuropil surface (Fig. 8M). At this stage, most of the external neuropil surface, in addition to the compartments of the central complex, mushroom body and adjoining major tracts, are covered by sEG. Only at the dorsal neuropil surface the sEG sheath remains thin, and small gaps appear at locations where there is a high concentration of (NP6520-negative) sALG (arrow in inset of Fig. 8M).
Origin and fate of secondary neuropil glia from gliogenic type II lineages
Primary neuropil glia do not divide, and undergo cell death during the pupal period (see above). This prompts the question for the origin of secondary NPG. Previous studies had suggested that these cells are formed as part of secondary neuro-(glio)blast lineages (Pereanu et al., 2005); the formation of putative central complex-associated sEG by a subset of type II lineages, utilizing lineage tracing and MARCM analysis, had been followed in detail (Izergina et al., 2009; Viktorin et al., 2011). To systematically address the origin and migration of sNPG, in particular sALG, we followed the association of Repo-positive sNPG with secondary lineages at different larval stages, and carried out lineage tracing experiments to confirm the origin of these cells.
As expected, using the secondary lineage-specific driver, insc-Gal4, we could identify Repo-positive nuclei of sNPG precursors as part of the type II lineages DPMm1 (DM1), DPMpm1 (DM2), DPMpm2 (DM3), CM1 (DM5), and CP2/3 (DL1/2), from about 60hr after hatching onward (Fig. 8A–C). This confirms the findings of Izergina et al., (2009) and Viktorin et al., (2011), and in part, the analysis of secondary lineage clones in the adult brain by Yu et al., (2013). Sparse Repo-positive nuclei were also occasionally observed in lineages CM4 (DM4) and CM3 (DM6). None of the type I lineages showed Repo labeling, corroborating the findings of Izergina et al., (2009). Repo-positive sNPG precursors are located distally within their respective lineages, indicating that they are among the first born cells (Fig. 9L). As development proceeds, lineages add new cells at the periphery (i.e., close to the neuroblast and intermediate progenitors). These newly born cells turn on the insc marker, whereas earlier born cells, including sNPG precursors, lose insc expression (Fig. 9L). Among the later born cells, none expressed Repo, excepting in one lineage, CM1 (DM5), which continued to produce new Repo-positive sNPG until the late larval stage (Fig. 9J).
Figure 9.
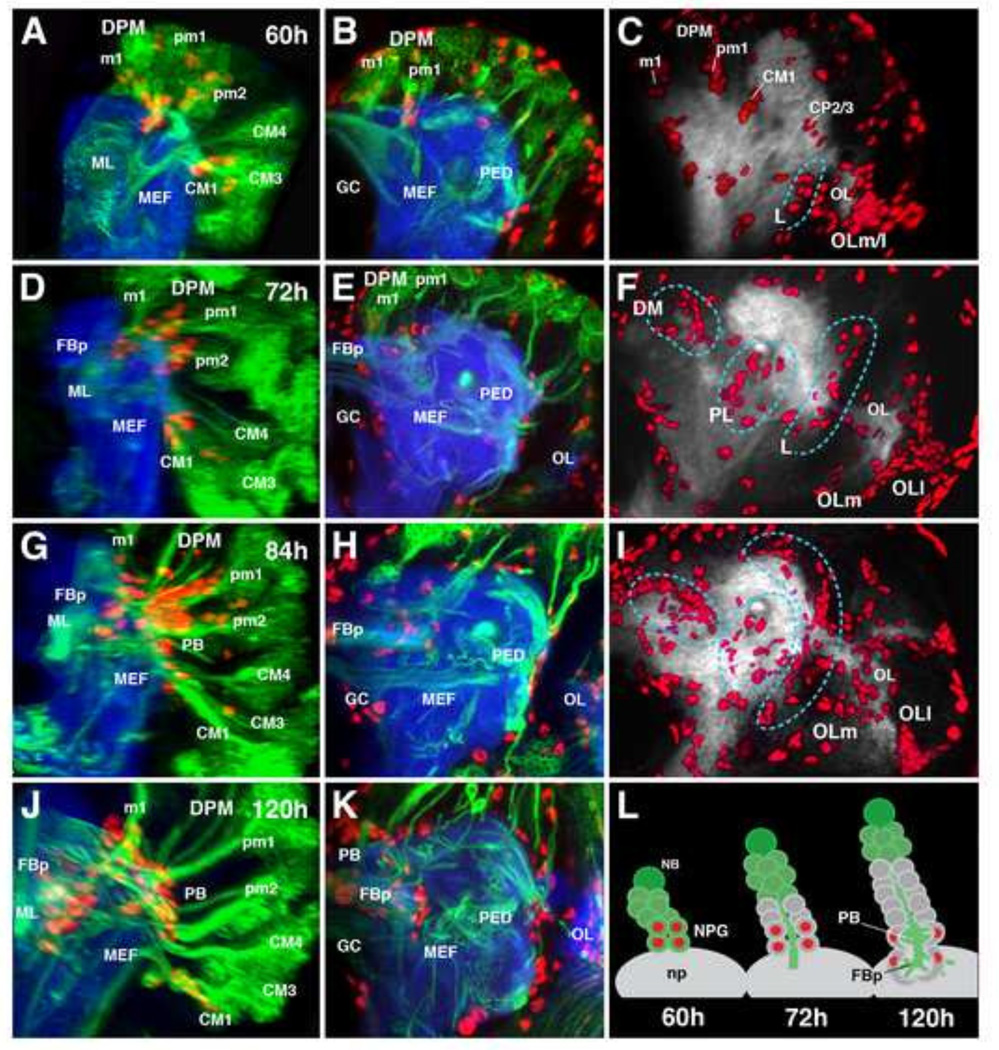
Origin and migration of secondary neuropil glial precursors throughout larval development. (A–K) Confocal z-projections depicting parasagittal views (first column) frontal view at the level of the fan-shaped body primordium (second column) and frontal view of anti-Repo volume renderings (third column) of larval development from 60h ah (A–C) to 120h ah (J–K). Neuro-(glio)blast lineages are labeled by insc-Gal4 in green, glial nuclei by anti-Repo in red, and the neuropil by anti-DN-cadherin in blue (A–B, D–E, G–H, J–K). (C, F, I) Neuropil is labeled by anti-DN-cadherin in gray and volume renderings of glial nuclei labeled by anti-Repo in red. For clarity, in parasagittal views (A, D, G, J), type I lineages were manually erased, along with cortex and surface glia. Secondary neuropil glia emerge within identifiable type II lineage clusters (A–C, L). Secondary neuropil glia remain associated with the axon tracts of the lineages from which they emerge and reach the neuropil surface (D–E, L), followed by invasion into the brain to associate with emerging neuropil compartments such as the fan-shaped body primordium (G–H, J–K, L), and spread out onto the posterior lateral surface of the neuropil (l–L). Neuropil glia can initially be identified as constituents of specific lineages (C), and subsequently distribute into rough categories (demarcated by cyan hashed lines): dorsomedial (DM), postero-lateral (PL), and lateral (L) clusters (F). Anatomical abbreviations found in Table 1. Other abbreviations: np, neuropil; OLm/l, medial and lateral optic lobe glia.
Throughout larval development, sNPG precursors remain close to the neuropil surface, clustered around the entry points of lineage tracts into the neuropil (Fig. 9A, D, L). Starting around 84h ah, sNPG precursors invade the neuropil and spread out around the posterolateral neuropil surface and emerging neuropil compartments (e.g., primordia of the protocerebral bridge and fan-shaped body; Fig. 9G–I, J–L). The contribution of each lineage to the total sNPG population becomes difficult to assess due to the subsequent distribution of glial cells over the neuropil surface (Fig. 9F, I). However, one can, at least initially, roughly categorize sNPG precursors into dorsomedial (DM), posterolateral (PL), and lateral (L) groups (Fig. 9F). The DPM lineages appear to make significant contributions to the DM group which populate the central complex primordium, the CM lineages to the PL group which splay across the posterior lateral protocerebrum, and the CP lineages to the L group which cluster around the lateral regions of the brain neuropil (Fig. 9F, I).
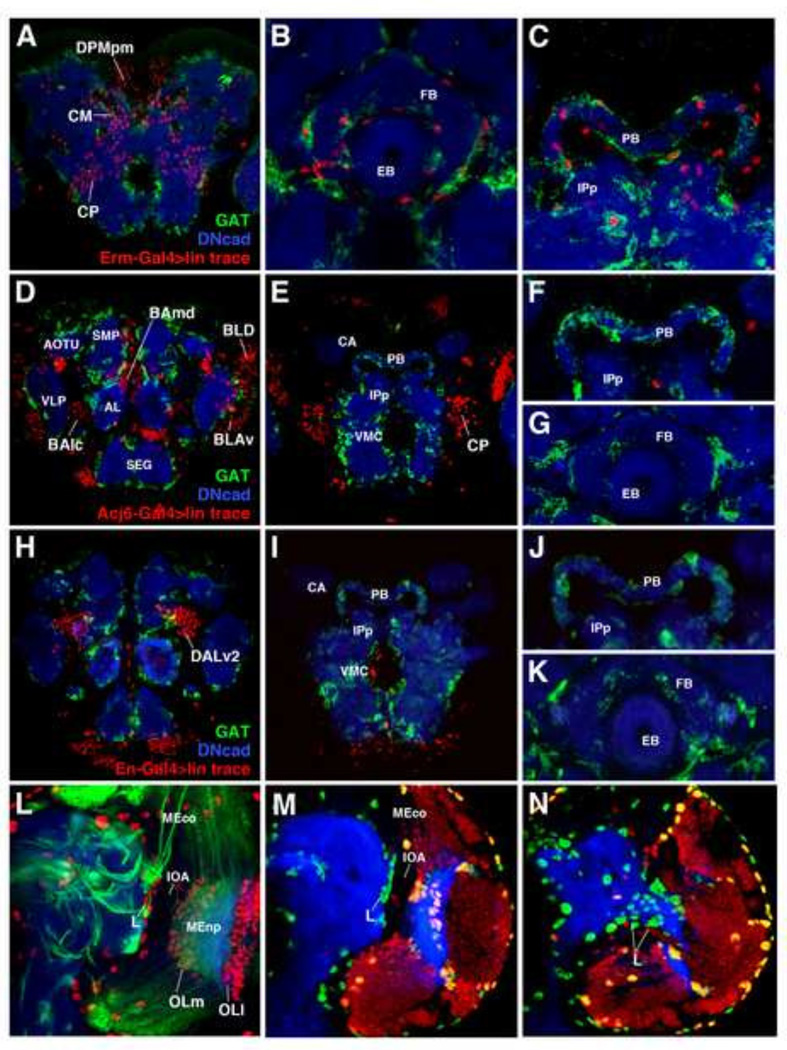
To critically address the question whether both types of sNPG, sALG and sEG, are derived from type II lineages, we traced intermediate precursors (INPs) of type II lineages, using the INP-specific driver 9D11(erm)-Gal4, and determined their fate using the ALG-specific marker, GAT. We find that, as expected, many neurons located in the posterior cortex are LacZ positive (Fig. 10A). We also find many GAT-positive and GAT-negative nuclei surrounding the central complex, suggesting that type II lineages can indeed generate both sNPG subtypes (Fig. 10B–C). To further support the notion that type I lineages do not give rise to sNPG, we used a number of drivers expressed in sets of type I lineages, among them acj6-Gal4 [expressed in secondary lineages BAmv3, BLD5, BLD6, BLP3, BLP5, BLVp1, BLVp2 (Supp. Info. Fig. 6)] and engrailed-Gal4 [expressed in secondary lineages BAla3, DALv2, DALv3, DPLam (Kumar et al., 2009a; 2009b)]. Lineage tracing using both lines revealed predictably distributed neuronal progeny (Fig. 10D–E, H–I). However, neither line produced any sALG or sEG (Fig. 10F–G, J–K).
Figure 10.
Both neuropil glia subtypes of the central complex derive from intermediate progenitors of multipotent type II neuroglioblasts, but do not derive from the optic lobe (A–K) Frontal confocal z-projections of P48 brains. The neuropil is labeled by anti-DN-cadherin in blue, secondary astrocyte-like glia by GAT in green, and lineage traced cells from specific progenitor populations by anti-β-gal in red. Progeny of type II lineage intermediate progenitors (INPs) (A–C), and the type I neuroblast lineages BLD, BAmd, BALc, BLAv (D–G), and DALv2 (H–K), were labeled by erm-Gal4, acj6-Gal4, and engrailed-Gal4, respectively. Neuronal progeny of INPs can be observed by a z-projection of posterior brain (A), and neuropil glial progeny can be observed around compartments of the central complex [(ellipsoid body and fan-shaped body, B; protocerebral bridge, C)]. Neuronal progeny of acj6- and engrailed-positive lineages can be observed in anterior (D, H) and posterior (E, I) z-projections of the brain, but no glial progeny are associated with the compartments of the central complex (F–G, J–K). (L) Frontal confocal z-projection of a third instar larval brain at the level of the inner optic anlage (IOA) depicting medial (OLm) and lateral (OLI) populations of optic lobe glia. Neuro-(glio)blast lineages are labeled by insc-Gal4 in green, glial nuclei by anti-Repo in red, and the neuropil by anti-DN-cadherin in blue. Lateral population of secondary neuropil glia precursors (L) is indicated and, posteriorly (not shown), exhibits a continuity with medial glial cells of the optic lobe (OLm). Confocal z-projections of the intermediate (M) and posterior (N) neuropil of a third instar larval brain. Glial nuclei labeled by anti-Repo in green, the neuropil by anti-DN-cadherin in blue, and neuronal/glial progeny of sine oculis-positive progenitors (optic lobe placode) labeled in red. Optic lobe placode-derived neurons (red) and glia (orange) revealed by sine oculis lineage tracing reveals that the lateral population (L) of secondary neuropil glia are not optic lobe-derived. Anatomical abbreviations found in Table 1.
The optic lobe primordium does not contribute to the neuropil glia of the central brain
The optic lobe generates numerous specialized populations of glia (Edwards et al., 2010; Hartenstein, 2011). Distinguished in Fig. 10L are the conspicuous bands of glial precursors produced at an early stage by the inner and outer optic anlage [subsumed as lateral optic lobe glial precursors (OLl)], and the medial glial precursors (OLm), born later over a long period of time from the large number of lineages that form the medulla. An increasing population of lateral Repo-positive sNPG of the central brain neuropil (“L” in Fig. 9C, F, I) exhibits a continuity with the Repo-positive cells within the optic lobe, specifically, with the cells of the Olm. This observation is consistent with Viktorin et al., (2013) which demonstrated that the type II lineage CP2/3, specifically DL1, gives rise to Repo-positive cells which migrate from the central brain into the optic lobe to form a subpopulation of optic lobe glia. It is also possible that a subpopulation of the large numbers of glial cells born in the optic lobe migrate into the opposite direction, contributing to the glial population of the central brain. To address this possibility, we lineage-traced using sine oculis-Gal4 to identify all cells derived from the optic lobe placode of the embryo (Chang et al., 2003). Although a large number of neurons and glia (OLl and OLm) were derived from sine oculis-Gal4 expressing progenitors, lateral sNPG were not (Fig. 10M–N). This finding indicates that glia born in the optic lobe primordium remains confined to this domain and does not contribute to central brain neuropil glia.
DISCUSSION
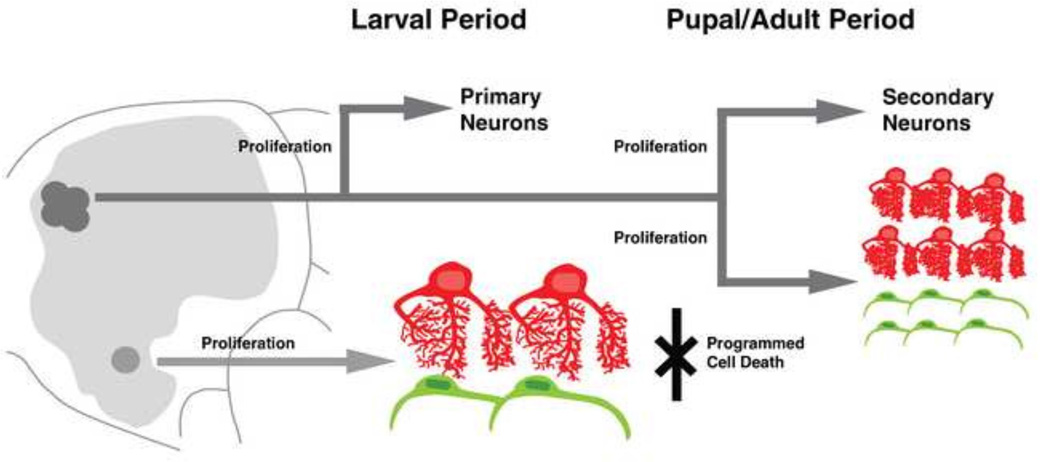
Glia are intimately associated with neurons and represent a major cellular constituent of both vertebrate and invertebrate nervous systems. Invertebrate model organisms such as Drosophila, with their wealth of genetic tools, are gaining momentum to elucidate fundamental aspects of glial biology. A number of recent studies (Avet-Rochex et al., 2012; Awasaki et al., 2008; Beckervordersandforth et al., 2008; Doherty et al., 2009; Muthukumar et al., 2014; Stork et al. 2008; Stork et al., 2014; Tasdemir-Yilmaz and Freeman, 2014), emphasize the ample heterogeneity of Drosophila glia, and initiated questions regarding their origin and developmentally changing patterns. In the present study, we establish a developmental/morphological framework for neuropil glia of the brain, the glial class most intimately associated with neurites. We demonstrate that the larval brain is associated with large, primary neuropil glia which are few in number and relatively stereotyped in position. These cells undergo programmed cell death and do not contribute to neuropil glia of the adult. The adult brain is invested with a much larger population of secondary neuropil glia which is generated during larval stages from a relatively small set of neuroglioblasts, the type II lineages, different from those that gave rise to primary neuropil glia.
Origin and morphogenesis of primary neuropil glia
Primary NPG has been reconstructed for the embryonic ventral nerve cord in a number of previous studies. Jacobs and Goodman (1989) originally classified glia associated with the neuropil of the embryonic VNC as longitudinal, midline, and nerve root glia (Jacobs and Goodman, 1989). Longitudinal glia, also called “interface glia”, envelop the connectives, midline glia the commissures, and nerve root glia the points of exit of peripheral nerves (Beckervordersandforth et al., 2008; Ito et al., 1995; Jacobs and Goodman, 1989). It was shown that the longitudinal glia consist of 9 cells per hemineuromere and are produced by a dedicated glioblast termed, the “lateral glioblast” (LGB) (Beckervordersandforth et al., 2008; Jacobs et al., 1989). Progeny of the LGB migrate from their lateral point of origin to the location where the longitudinal axons pioneering the connectives appear, and subsequently undergo multiple rounds of equal mitoses to produce the longitudinal glia (Griffiths and Hidalgo, 2004). Longitudinal glia likely comprise an equivalence group which differentially specifies into primary astrocyte-like and ensheathing glia (Stork et al., 2014; this study). Although a more descriptive term such as the previously suggested “reticular glia” for ALG may be more suitable considering they may or may not be homologous to vertebrate astrocytes by definition, we adopt this terminology to maintain consistency in the field (Hartenstein, 2011; Hartline, 2011). Midline glia (the only type of glia in Drosophila that does not express Gcm or Repo) comprises 3–4 cells in each hemineuromere and has its origin from among the so-called midline progenitors, peculiar cells which divide only once before differentiating (Bossing and Technau, 1994). Nerve root glia originates from one of the lateral neuroglioblasts, Nb1–3 (Beckervordersandforth et al., 2008). The origin and fate of embryonic midline and nerve root glia are not addressed in this study, although they were defined as members of the embryonic “neuropil-associated glia” class. A dedicated (neuro)-glioblast which generates neuropil glia precursors of the brain, the BPLG, has yet to be identified. However, a strong candidate includes Td7 based on its expression of glial-specific genes and proximity to the emerging BPLG cluster (Hartenstein et al., 1998; Urbach and Technau, 2003).
Previous counts of Repo-positive neuropil glia throughout larval development demonstrates that the number of NPG remains relatively constant (<30) until the early third larval instar, at which point the number of NPG nearly triples (Pereanu et al., 2005). Our analysis using markers of differentiated NPG subtypes demonstrates that the number of differentiated neuropil glia does not change throughout larval development in the VNC (data not shown) or brain. In addition, double labeling experiments demonstrate that all NPG of the second instar larval brain (prior to this massive increase in number) are pEG and pALG. Therefore, the newly generated NPG in the third larval instar represent recently born secondary neuropil glia precursors (see later section). We propose that a more or less invariant number of pNPG in the CNS which undergo a late embryonic migration to stereotyped positions around the neuropil are generated by dedicated primary glioblasts.
Although the number of pNPG does not change throughout development, they exhibit significant morphogenetic changes. Both neuropil glial subtypes exhibit an increase in nuclear size and progressive elaboration of their membranous processes. These changes are likely carried out to accommodate the many-fold expansion of the larval neuropil volume. Previous studies examining incorporation of BrdU during larval development suggested that neuropil glia of the third instar VNC, confirmed to represent mature longitudinal glia, replicate DNA content without cellular division (Prokop and Technau, 1994). Subperineurial glia (SPG), which, similarly to pNPG do not execute cytokinesis, also undergo polyploidization (Unhavaithaya and Orr-Weaver, 2012). In this case, polyploidization is required to sustain the massive increase in cell size necessary to maintain the blood-brain barrier around a progressively expanding larval brain. Interestingly, SPG evidently increase ploidy in response to an increase in neuronal mass, suggesting a feedback mechanism to sustain coordinate growth during organogenesis (Unhavaithaya and Orr-Weaver, 2012). It will be interesting to address the possibility that endoreplication in pNPG is required in a similar way for the expanding larval neuropil.
At the onset of metamorphosis, pALG drastically change morphology. Tasdemir-Yilmaz and Freeman (2014) demonstrated that the processes of primary ALG (called “larval astrocytes” in their study) vacuolate in an ecdysone-dependent manner at the onset of metamorphosis and become sparser as metamorphosis proceeds, consistent with our findings. However, it was suggested that the same population of cells re-extends their processes into the neuropil during late metamorphosis, prior to adulthood. Our analysis suggests that pALG do not re-infiltrate the neuropil, and in fact they are lost by programmed cell death. This represents, to our knowledge, the first description of cell death of an entire cellular class in the Drosophila brain. A similar developmental scenario is likely to be true of ensheathing glia based on expression of the NP6520-Gal4 driver. Similarly to alrm-Gal4, NP6520-Gal4 is expressed throughout larval development in a small number of primary EG with progressively enlarging nuclei and turns off during mid-metamorphosis (P24-P48). However, further lineage tracing experiments of pEG must be conducted to confirm this hypothesis.
Origin and morphogenesis of secondary neuropil glia
Previous studies in the developing larval brain suggest that all three glial classes increase in number, particularly during the third instar (Pereanu et al., 2005). The observed increase in Repo-positive neuropil glia can be due to either (or a combination of) 1) mitotic divisions of differentiated primary glia, 2) proliferation of secondary multipotent neural/glial progenitors (neuroglioblasts), 3) mitotic divisions of undifferentiated, secondary NPG precursors. Based on our comprehensive analysis of various neuropil glia-subtype specific markers throughout development in combination with lineage tracing, we suggest that the increase in neuropil glia number is primarily due to proliferation of neuroglioblast lineages rather than division of differentiated pNPG. In addition, mitotic divisions of sNPG precursors are also thought to contribute to the final population (Pereanu et al., 2005; Viktorin et al., 2011). Thus, after the Repopositive sNPG precursors have separated from their lineages of origin and started to spread around the neuropil, there is a further, substantial increase in their number during the early pupal period, (Fig. 8I), even though proliferation of neuroblasts and neuroglioblasts has ceased (Ito and Hotta, 1992). Therefore, clonal expansion of sNPG precursors likely contributes greatly (up to 75%) of the mature population (Viktorin et al., 2011; this study). The molecular mechanisms which dictate proliferation, although elucidated for other glial subtypes, require further investigation for sALG and sEG (Avet-Rochex et al., 2012). Therefore, we are currently pursuing the role of FGF signaling in mediating proliferation of post-embryonic sALG precursors (Gibson et al., 2012; unpublished data).
As previously reported (Izergina et al., 2009, Viktorin et al., 2011) and confirmed in this study, a subset of the posteriorly located type II lineages constitute the source of central complex-associated sNPG. Our data suggests that type I lineages do not give rise to any sNPG. Based on cell morphology at P20, type II lineages were suggested to exclusively give rise to ensheathing glia (Viktorin et al., 2011). However, we have demonstrated, using NPG subtype-specific markers, that type II lineages can give rise to both sNPG subtypes. We surmise that this discrepancy is based on the fact that one cannot fully distinguish sALG and sEG at this stage in metamorphosis, given that sALG have not fully elaborated their processes. It remains to determined, however, if sALG and sEG are derived from specific, distinct type II neuroblasts or potentially from distinct INPs from individual type II neuroblasts.
MARCM analysis suggested that precursors of sNPG (postembryonic neuropil glia) proliferate in stereotyped regions in the larval brain (posteromedial, lateral, and dorsal interface), followed by migration during metamorphosis (Awasaki et al., 2008). We documented this migration with preparations representing multiple discrete time windows throughout metamorphosis. Our results suggest that sNPG, albeit more complex in nature in comparison to pNPG, also exhibit a stereotyped migratory pattern emanating from the previously described posteromedial, lateral, and dorsal loci. This pattern is characterized by Repo-positive sNPG gradually distributing equally over the neuropil surface and invading crevices between neuropil compartments. Interestingly, sALG retain a spindle-shaped morphology until late metamorphosis (P48-P72), at which point they infiltrate the neuropil and remain for the rest of adulthood. Recent work has demonstrated that primary ALG require fibroblast growth factor receptor signaling in order to extend processes into the late embryonic neuropil (Stork et al., 2014). However, the molecular mechanisms responsible for prompting sALG processes to infiltrate the neuropil during metamorphosis have yet to be determined. Notably, the developmental time windows when synaptogenesis and elaboration of sALG processes occurs are coincident (Muthukumar et al., 2014; Singh and Singh, 1999). Although specific sALG-derived synaptogenic molecules remain to be determined, recent work has demonstrated that sALG influence synaptogenesis of adult-specific neurons during metamorphosis, akin to what has been described for vertebrate astrocytes (Clarke and Barres, 2013; Muthukumar et al., 2014).
Relationship between astrocyte-like and ensheathing glia
A morphological distinction among the neuropil glia between ensheathing glia, whose processes largely remain confined to the neuropil surface, and astrocyte-like glia with neuron-like, highly branched morphology was prompted by clonal labeling of, and specific markers for these cells (Awasaki et al., 2008; Pereanu et al., 2005). It appears likely that for both brain and VNC, both cell types are derived from the same progenitors. Thus, as shown in the present study, a small number of type II lineages in the posterior brain generate the large majority of sEG and sALG. In the embryonic VNC, the lateral glioblast generates both pEG and pALG.
What is the significance of two different NPG cell types, and how is the differentiation between EG and ALG specified? In the embryonic VNC, it appears that initially, all descendants of the lateral glioblast express the ALG marker alrm-Gal4 (Stork et al., 2014; this study), and that the separation between the two NPG subtypes, manifested by the restriction of alrm to just anterior longitudinal glia, occurs secondarily. Presumptive primary ALG can be distinguished from EG by the expression of Prospero (Pros) (Griffiths and Hidalgo, 2004). Notch signaling appears likely to be involved in this separation, since Notch activity is required independently for the expression of Pros and the glutamate transporter, excitatory amino acid transporter 1 (Eaat1), in the anterior longitudinal glia (presumptive pALG) (Stacey et al., 2010; Thomas and Van Meyel, 2007). Therefore, longitudinal glia are initially equivalent cells which require environmental cues such as fringe-mediated serrate/delta-notch signaling to promote pNPG subtype-specific gene expression. It is currently unknown whether similar mechanisms dictate the differential specification of brain primary neuropil glia. In addition, nothing is known about the corresponding specification of type II-derived secondary ALG and EG. The concept of the longitudinal glia as an equivalence group prompts the question of the true identity of ensheathing glia. For example, are EG and ALG actually different cells, or is EG the “default” fate? Is it possible that EG are simply a reserve population which are competent to elaborate processes into the neuropil, but do not because they lack appropriate environmental cues during a specific phenocritical period? Further studies ablating ALG populations need to be conducted to answer this question.
Neuropil glia in Drosophila and other insects
In this study, we provide a developmental framework for the study of neuropil glia in Drosophila. Neuropil glia are defined by their somata location (located at neuropil boundaries) and morphology (dendritic and invested in the neuropil or sheath-like and surrounding neuropil boundaries for ALG and EG, respectively). We describe two disparate glial strategies for “supporting” the neuropil of the insect nervous system. The larval brain is associated with few, large pNPG which likely undergo endoreplication to accommodate the growing CNS rather than proliferate. The adult brain is associated with many more, small sNPG which derive from secondary lineages and clonally expand to generate the final population. Do other insects have analogous neuropil glia subtypes? If so, what is their cellular origin? What “strategy” do various insects use, especially considering many are hemimetabolous and do not undergo a metamorphosis?
Studies in moth (Manduca sexta) larvae have described “glia of the neuropil cover” which consisted of multiple subtypes roughly corresponding to Drosophila primary NPG (Cantera, 1992). Although primary NPG in Manduca have not been followed throughout larval development, a recent study demonstrated GAT (MasGAT in Manduca) immunoreactivity exclusively in the “complex” glial cells (analogous to pALG) and absent in the “simple” glial cells (analogous to pEG) of the late larval and early metamorphosing antennal lobe (Oland et al., 2010). Although not comprehensively investigated, the striking phenomena of glial process vacuolization (likely corresponding to clearance of neuronal debris) and clearance has been documented (Fig. 4 in Oland et al., 2010). In addition, the presence of what are most likely secondary neuropil glia analogous in morphology to the subtypes we observe in Drosophila have been described in the developing adult Manduca antennal lobe (Oland et al., 1999). This demonstrates the striking similarity between glial morphology and gene expression between holometabolous insects.
Studies of neuropil glia in the hemimetabolan grasshopper Schistocerca americana have shown the presence of “primitive” glia (“glia of the connective”) associated with the early embryonic longitudinal connectives. These glia, similarly to Drosophila, are thought to be required for establishing reciprocal cellular interactions with neurons imperative for normal development of the longitudinal connectives (Bastiani and Goodman, 1986; Hidalgo and Booth, 2000). However, the fate of the grasshopper “glia of the connective” or their unknown brain counterparts, which likely correspond to Drosophila primary NPG, is unknown. Large glial cells reminiscent of pNPG have thus far, not been reported. Unlike Drosophila, the grasshopper Schistocerca gregaria, like other hemimetabolan insects, generates brain structures required for complex adult motor control such as the central complex in the embryo. This structure has been shown to be associated with neuropil glia strikingly similar to the secondary neuropil glia which we have described during metamorphosis (Boyan et al., 2013). In fact, like Drosophila sALG, these glia first migrate to the central complex from other brain regions and only elaborate their processes into the central complex neuropil after the columnar neuroarchitecture has formed (Boyan et al., 2011). In addition, Repo-positive cells have been shown to intermingle with neurons within developing central complex lineages and proliferate, thus demonstrating the presence of multipotent neuroglioblasts in the grasshopper (Liu and Boyan, 2013). Thus, we hypothesize that grasshoppers and other hemimetabolans likely generate (primary) NPG at an early stage for the purpose of establishing early neuroarchitecture, but in contrast to holometabolans, do not maintain and enlarge these cells for functional use in a larval stage that is lacking in these insects.
Supplementary Material
Figure 11.
Working model for the lineage and development of neuropil glia throughout Drosophila brain development. Primary neuropil glia of the brain [pALG (red) and pEG (green)] are derived from dedicated glioblasts located in the basal brain located at the tritocerebrum and deuterocerebrum boundary. Primary neuropil glia are large cells which are few in number; they increase cellular size likely via endoreplication to account for the expanding neuropil. Primary neuropil glia undergo programmed cell death and do not contribute to neuropil glia of the adult. Type II lineages, which likely give rise solely to neurons during the embryonic (primary) phase of progenitor proliferation, undergo a secondary phase of multipotent proliferation in which they give rise to secondary neurons and secondary neuropil glia precursors. Secondary neuropil glia precursors are small, spindle-shaped cells which greatly expand in number around the developing neuropil compartments and ultimately differentiate into secondary astrocyte-like and ensheathing glia [sALG (red) and sEG (green)] of the adult brain.
Highlights.
The Drosophila larval and adult brain contain neuropil glia which are derived from distinct embryonic and postembryonic progenitors.
Larval (primary) neuropil glia are large, few in number, and undergo cell death during metamorphosis.
Adult (secondary) neuropil glia are type II lineage-derived cells which are small in size but large in number.
ACKNOWLEDGEMENTS
We thank the members of the Hartenstein laboratory and Philipp Kuert for critical discussions during the preparation of this manuscript. We are grateful to the Bloomington Stock Center and the Developmental Studies Hybridoma Bank for fly strains and antibodies. We thank Dr. Heinrich Reichert for anti-Labial antibody. We thank Diana Rigueur and Karen Lyons for assistance with the TUNEL assay. We also thank Dr. Marc Freeman for critical fly strains (alrm-Gal4) and antibodies (rabbit anti-GAT). This work was supported by NIH grant (R01 NS29357-15). J.J.O. is supported by the Ruth L. Kirschstein National Research Service Award (No. GM007185).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ashburner M. A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Drosophila; pp. 214–217. [Google Scholar]
- Avet-Rochex A, Kaul AK, Gatt AP, McNeill H, Bateman JM. Concerted control of gliogenesis by InR/TOR and FGF signalling in the Drosophila post-embryonic brain. Development. 2012;139:2763–2772. doi: 10.1242/dev.074179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T, Lai SL, Ito K, Lee T. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J Neurosci. 2008;28:13742–13753. doi: 10.1523/JNEUROSCI.4844-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiani MJ, Goodman CS. Guidance of neuronal growth cones in the grasshopper embryo. III. Recognition of specific glial pathways. J Neurosci. 1986;6:3542–3551. doi: 10.1523/JNEUROSCI.06-12-03542.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar OA, Boone JQ, Drummond ML, Doe CQ. Drosophila type II neuroblast lineages keep Prospero levels low to generate large clones that contribute to the adult brain central complex. Neural Dev. 2010;5:26. doi: 10.1186/1749-8104-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar OA, Doe CQ. Combinatorial temporal patterning in progenitors expands neural diversity. Nature. 2013;498:449–455. doi: 10.1038/nature12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth RM, Rickert C, Altenhein B, Technau GM. Subtypes of glial cells in the Drosophila embryonic ventral nerve cord as related to lineage and gene expression. Mech Dev. 2008;125:542–557. doi: 10.1016/j.mod.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Bossing T, Technau GM. The fate of the CNS midline progenitors in Drosophila as revealed by a new method for single cell labelling. Development. 1994;120:1895–1906. doi: 10.1242/dev.120.7.1895. [DOI] [PubMed] [Google Scholar]
- Boyan G, Loser M, Williams L, Liu Y. Astrocyte-like glia associated with the embryonic development of the central complex in the grasshopper Schistocerca gregaria. Dev Genes Evol. 2011;221:141–155. doi: 10.1007/s00427-011-0366-4. [DOI] [PubMed] [Google Scholar]
- Boyan G, Williams L, Götz S. Postembryonic development of astrocyte-like glia of the central complex in the grasshopper Schistocerca gregaria. Cell Tissue Res. 2013;351:361–372. doi: 10.1007/s00441-012-1535-0. [DOI] [PubMed] [Google Scholar]
- Cantera R. Glial cells in adult and developing prothoracic ganglion of the hawk moth Manduca sexta. Cell Tissue Res. 1992;272:93–108. [Google Scholar]
- Cantera R, Technau GM. Glial cells phagocytose neuronal debris during the metamorphosis of the central nervous system in Drosophila melanogaster. Dev Genes Evol. 1996;206:277–280. doi: 10.1007/s004270050052. [DOI] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M, Tomancak P, Hartenstein V, Douglas RJ. TrakEM2 software for neural circuit reconstruction. PLoS ONE. 2012;7(6):e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T, Shy D, Hartenstein V. Antagonistic relationship between Dpp and EGFR signaling in Drosophila head patterning. Dev Biol. 2003;263:103–113. doi: 10.1016/s0012-1606(03)00448-2. [DOI] [PubMed] [Google Scholar]
- Chen W, Hing H. The L1-CAM, Neuroglian functions in glial cells for Drosophila antennal lobe development. Devel Neurobio. 2008;68:1029–1045. doi: 10.1002/dneu.20644. [DOI] [PubMed] [Google Scholar]
- Clarke LE, Barres BA. Emerging roles of astrocytes in neural circuit development. Nat Rev Neurosci. 2013;14:311–321. doi: 10.1038/nrn3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSalvo MK, Mayer N, Mayer F, Bainton RJ. Physiologic and anatomic characterization of the brain surface glia barrier in Drosophila. Glia. 2011;59:1322–1340. doi: 10.1002/glia.21147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty J, Logan MA, Taşdemir OE, Freeman MR. Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci. 2009;29:4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Hartenstein V. Role of DE-cadherin in neuroblast proliferation, neural morphogenesis, and axon tract formation in Drosophila larval brain development. J Neurosci. 2003;23:3325–3335. doi: 10.1523/JNEUROSCI.23-08-03325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TN, Meinertzhagen IA. The functional organisation of glia in the adult brain of Drosophila and other insects. Prog Neurobiol. 2010;90:471–497. doi: 10.1016/j.pneurobio.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson NJ, Tolbert LP, Oland LA. Activation of glial FGFRs is essential in glial migration, proliferation, and survival and in glia-neuron signaling during olfactory system development. PLoS ONE. 2012;7(4):e33828. doi: 10.1371/journal.pone.0033828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths RL, Hidalgo A. Prospero maintains the mitotic potential of glial precursors enabling them to respond to neurons. EMBO J. 2004;23:2440–2450. doi: 10.1038/sj.emboj.7600258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean Y, Grillet M, Augustin H, Ferveur JF, Featherstone DE. A glial amino-acid transporter controls synapse strength and courtship in Drosophila. Nat Neurosci. 2008;11:54–61. doi: 10.1038/nn2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim Y, Yaniv SP, Schuldiner O. Astrocytes play a key role in Drosophila mushroom body axon pruning. PLoS ONE. 2014;9(1):e86178. doi: 10.1371/journal.pone.0086178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V. Morphological diversity and development of glia in Drosophila. Glia. 2011;59:1237–1252. doi: 10.1002/glia.21162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Nassif C, Lekven A. Embryonic development of the Drosophila brain. II. Pattern of glial cells. J Comp Neurol. 1998;402:32–47. [PubMed] [Google Scholar]
- Hartenstein V, Spindler S, Pereanu W, Fung S. The development of the Drosophila larval brain. Adv Exp Med Biol. 2008;628:1–31. doi: 10.1007/978-0-387-78261-4_1. [DOI] [PubMed] [Google Scholar]
- Hartline DK. The evolutionary origins of glia. Glia. 2011;59:1215–1236. doi: 10.1002/glia.21149. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Booth GE. Glia dictate pioneer axon trajectories in the Drosophila embryonic CNS. Development. 2000;127:393–402. doi: 10.1242/dev.127.2.393. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Urban J, Brand AH. Targeted ablation of glia disrupts axon tract formation in the Drosophila CNS. Development. 1995;121:3703–3712. doi: 10.1242/dev.121.11.3703. [DOI] [PubMed] [Google Scholar]
- Hosoya T, Takizawa K, Nitta K, Hotta Y. Glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell. 1995;82:1025–1036. doi: 10.1016/0092-8674(95)90281-3. [DOI] [PubMed] [Google Scholar]
- Hoyle G. Glial cells of an insect ganglion. J Comp Neurol. 1986;246:85–103. doi: 10.1002/cne.902460106. [DOI] [PubMed] [Google Scholar]
- Hoyle G, Williams M, Phillips C. Functional morphology of insect neuronal cell-surface/glial contacts: the trophospongium. J Comp Neurol. 1986;246:113–128. doi: 10.1002/cne.902460108. [DOI] [PubMed] [Google Scholar]
- Ito K, Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- Ito K, Urban J, Technau GM. Distribution, classification, and development of Drosophila glial cells in the late embryonic and early larval ventral nerve cord. Roux’s Arch Dev Biol. 1995;204:284–307. doi: 10.1007/BF02179499. [DOI] [PubMed] [Google Scholar]
- Izergina N, Balmer J, Bello B, Reichert H. Postembryonic development of transit amplifying neuroblast lineages in the Drosophila brain. Neural Dev. 2009;4:44. doi: 10.1186/1749-8104-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson FR, Haydon PG. Glial cell regulation of neurotransmission and behavior in Drosophila. Neuron Glia Biol. 2008;4:11–17. doi: 10.1017/S1740925X09000027. [DOI] [PubMed] [Google Scholar]
- Jacobs JR, Goodman CS. Embryonic development of axon pathways in the Drosophila CNS. I. A glial scaffold appears before the first growth cones. J Neurosci. 1989;9:2402–2411. doi: 10.1523/JNEUROSCI.09-07-02402.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JR, Hiromi Y, Patel NH, Goodman CS. Lineage, migration, and morphogenesis of longitudinal glia in the Drosophila CNS as revealed by a molecular lineage marker. Neuron. 1989;2:1625–1631. doi: 10.1016/0896-6273(89)90051-2. [DOI] [PubMed] [Google Scholar]
- Jones BW, Fetter RD, Tear G, Goodman CS. Glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell. 1995;82:1013–1023. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- Kumar A, Bello B, Reichert H. Lineage-specific cell death in postembryonic brain development of Drosophila. Development. 2009a;136:3433–3442. doi: 10.1242/dev.037226. [DOI] [PubMed] [Google Scholar]
- Kumar A, Fung S, Lichtneckert R, Reichert H, Hartenstein V. Arborization pattern of engrailed-positive neural lineages reveal neuromere boundaries in the Drosophila brain neuropil. J Comp Neurol. 2009b;517:87–104. doi: 10.1002/cne.22112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Boyan G. Glia associated with central complex lineages in the embryonic brain of the grasshopper Schistocerca gregaria. Dev Genes Evol. 2013;223:213–223. doi: 10.1007/s00427-013-0439-7. [DOI] [PubMed] [Google Scholar]
- Muthukumar AK, Stork T, Freeman MR. Activity-dependent regulation of astrocyte GAT levels during synaptogenesis. Nat Neurosci. 2014 doi: 10.1038/nn.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oland LA, Biebelhausen JP, Tolbert LP. Glial investment of the adult and developing antennal lobe of Drosophila. J Comp Neurol. 2008;509:526–550. doi: 10.1002/cne.21762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oland LA, Gibson NJ, Tolbert LP. Localization of a GABA transporter to glial cells in the developing and adult olfactory pathway of the moth Manduca sexta. J Comp Neurol. 2010;518:815–838. doi: 10.1002/cne.22244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oland LA, Marrero HG, Burger I. Glial cells in the developing and adult olfactory lobe of the moth Manduca sexta. Cell Tissue Res. 1999;297:527–545. doi: 10.1007/s004410051379. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J Neurosci. 2006;26:5534–5553. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Kumar A, Jennett A, Reichert H, Hartenstein V. Development-based compartmentalization of the Drosophila central brain. J Comp Neurol. 2010;518:2996–3023. doi: 10.1002/cne.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Shy D, Hartenstein V. Morphogenesis and proliferation of the larval brain glia in Drosophila. Dev Biol. 2005;283:191–203. doi: 10.1016/j.ydbio.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Prokop A, Technau GM. BrdU incorporation reveals DNA replication in non dividing glial cells in the larval abdominal CNS of Drosophila. Roux’s Arch Dev Biol. 1994;204:54–61. doi: 10.1007/BF00189068. [DOI] [PubMed] [Google Scholar]
- Reddy BV, Irvine KD. Regulation of Drosophila glial cell proliferation by Merlin-Hippo signaling. Development. 2011;138:5201–5212. doi: 10.1242/dev.069385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Sinakevitch I, Grau Y, Strausfeld NJ, Birman S. Dynamics of glutamatergic signaling in the mushroom body of young adult Drosophila. Neural Dev. 2010;5:10. doi: 10.1186/1749-8104-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Singh RN. Metamorphosis of the central nervous system of Drosophila melanogaster Meigen (Diptera: Drosophilidae) during pupation. J Biosci. 1999;24:345–360. [Google Scholar]
- Soustelle L, Giangrande A. Novel gcm-dependent lineages in the postembryonic nervous system of Drosophila melanogaster. Dev Dyn. 2007;236:2101–2108. doi: 10.1002/dvdy.21232. [DOI] [PubMed] [Google Scholar]
- Spindler SR, Ortiz I, Fung S, Takashima S, Hartenstein V. Drosophila cortex and neuropile glia influence secondary axon tract growth, pathfinding, and fasciculation in the developing larval brain. Dev Biol. 2009;334:335–368. doi: 10.1016/j.ydbio.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey SM, Muraro Nl, Peco E, Labbé A, Thomas GB, Baines RA, van Meyel DJ. Drosophila glial glutamate transporter Eaat1 is regulated by fringe-mediated notch signaling and is essential for larval locomotion. J Neurosci. 2010;30:14446–14457. doi: 10.1523/JNEUROSCI.1021-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork T, Engelen D, Krudewig A, Silies M, Bainton RJ, Klämbt C. Organization and function of the blood-brain barrier in Drosophila. J Neurosci. 2008;28:587–597. doi: 10.1523/JNEUROSCI.4367-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork T, Sheehan A, Tasdemir-Yilmaz OE, Freeman MR. Neuron-glia interactions through the Heartless FGF receptor signaling pathway mediate morphogenesis of Drosophila astrocytes. Neuron. 2014;83:388–403. doi: 10.1016/j.neuron.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir-Yilmaz OE, Freeman MR. Astrocytes engage unique molecular programs to engulf pruned neuronal debris from distinct subsets of neurons. Genes Dev. 2014;28:20–33. doi: 10.1101/gad.229518.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GB, van Meyel DJ. The glycosyltransferase Fringe promotes Delta-Notch signaling between neurons and glia, and is required for subtype-specific glial gene expression. Development. 2007;134:591–600. doi: 10.1242/dev.02754. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Orr-Weaver TL. Polyploidization of glia in neural development links tissue growth to blood-brain barrier integrity. Genes Dev. 2012;26:31–36. doi: 10.1101/gad.177436.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development. 2003;130:3621–3637. doi: 10.1242/dev.00533. [DOI] [PubMed] [Google Scholar]
- Viktorin G, Riebli N, Popkova A, Giangrande A, Reichert H. Multipotent neural stem cells generate glial cells of the central complex through transit amplifying intermediate progenitors in Drosophila brain development. Dev Biol. 2011;356:553–565. doi: 10.1016/j.ydbio.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Viktorin G, Riebli N, Reichert H. A multipotent transit-amplifying neuroblast lineage in the central brain gives rise to optic lobe glial cells in Drosophila. Dev Biol. 2013;379:182–194. doi: 10.1016/j.ydbio.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Vincent S, Vonesch JL, Giangrande A. Glide directs glial fate commitment and cell fate switch between neurones and glia. Development. 1996;122:131–139. doi: 10.1242/dev.122.1.131. [DOI] [PubMed] [Google Scholar]
- Xiong WC, Okano H, Patel NH, Blendy JA, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
- Yu HH, Awasaki T, Schroeder MD, Long F, Yang JS, He Y, Ding P, Kao JC, Wu GY, Peng H, Myers G, Lee T. Clonal development and organization of the adult Drosophila central brain. Curr Biol. 2013;23:633–643. doi: 10.1016/j.cub.2013.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M, Struhl G. Subdivision of the Drosophila wing imaginal disc by EGFR-mediated signaling. Development. 2002;129:1357–1368. doi: 10.1242/dev.129.6.1357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.