Abstract
Background
Alcohol dependence (AD) is a complex psychiatric disorder and a significant public health problem. Twin and family-based studies have consistently estimated its heritability to be approximately 50%, and many studies have sought to identify specific genetic variants associated with susceptibility to AD. These studies have been primarily linkage or candidate gene-based, and have been mostly unsuccessful in identifying replicable risk loci. Genome-wide association studies (GWAS) have improved the detection of specific loci associated with complex traits, including AD. However, findings from GWAS explain only a small proportion of phenotypic variance and alternative methods have been proposed to investigate the associations that do not meet strict genome-wide significance criteria.
Methods
This review summarizes all published AD GWAS and post-GWAS analyses that have sought to exploit GWAS data to identify AD-associated loci.
Results
Findings from AD GWAS have been largely inconsistent, with the exception of variants encoding the alcohol metabolizing enzymes. Analyses of GWAS data that go beyond single SNP association testing have demonstrated the polygenic nature of AD and the large contribution of common variants to risk, nominating novel genes and pathways for AD susceptibility.
Conclusions
Findings from AD GWAS and post-GWAS analyses have greatly increased our understanding of the genetic etiology of AD. However, it is clear that larger samples will be necessary to detect loci in addition to those that encode alcohol metabolizing enzymes, which may only be possible through consortium-based efforts. Post-GWAS approaches to studying the genetic influences on AD are increasingly common and could greatly increase our knowledge of both the genetic architecture of AD and the specific genes and pathways that influence risk.
Keywords: Alcohol dependence, GWAS, SNP heritability, risk profile scoring, pathway analysis
Introduction
Alcohol dependence (AD) is a common, complex psychiatric disorder characterized by the excessive and compulsive use of alcohol, which often results in physical and social consequences. The National Epidemiologic Survey on Alcohol and Related Conditions estimated the lifetime prevalence of AD in the general American population to be 12.5% and the 12-month prevalence to be 3.8% (Hasin et al., 2007). Thus, AD represents a significant public health problem.
Early evidence that AD has a genetic component came from studies showing significant familial aggregation of the disorder (Cotton, 1979). Although environmental factors play a significant role in AD risk, twin and family-based studies have consistently demonstrated a heritability of approximately 50% (Heath et al., 1997; Kendler et al., 1992; McGue et al., 1992; Prescott and Kendler, 1999; Reed et al., 1996; True et al., 1999; Verhulst et al., 2014). Identifying genes that influence risk for AD will inform our knowledge of the mechanisms underlying the disorder, potentially improving its diagnosis, treatment and prevention.
Significant advances in the identification of risk loci for AD in the past decade can be largely attributed to the advent of genome-wide association studies (GWAS). However, the individual loci that have been identified explain just a fraction of the variance in AD risk. This has led to efforts to exploit GWAS data using different analytic approaches, including studies of the heritability, risk prediction, and pathways involved in AD.
Here we summarize findings from all identified published classical GWAS of AD, discuss the knowledge that has been gained and the lessons learned from these studies, and provide an overview of the emerging post-GWAS approaches being used to study the genetics of AD. To identify studies, we used the PubMed and Google Scholar databases and searched for all English-language articles using combinations of the following search terms: genome-wide association study (GWAS), alcohol, alcohol dependence (AD), GCTA, profile score, gene set, and pathway analysis.
Genome-wide association studies
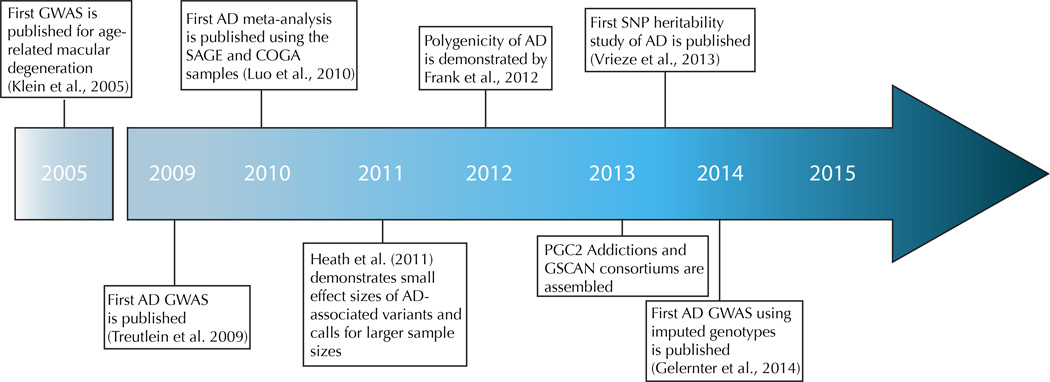
Genome-wide linkage and candidate gene association studies (reviewed by Agrawal and Bierut, 2012; Enoch, 2013; Rietschel and Treutlein, 2013) have been largely unsuccessful in identifying replicable risk loci for AD, with the exception of studies of the alcohol metabolizing enzyme genes, which showed strong association in populations of Asian, European, and African ancestry (Thomasson et al., 1991; Chen et al., 1999; Edenberg, 2007, Bierut et al. 2012). We will not review those studies in detail here, however, as the focus is on genome-wide studies. Both linkage and candidate gene methods have major limitations. Linkage studies necessitate the recruitment of family-based samples with several affected individuals and typically identify regions containing multiple genes. Candidate gene studies are biased in that a specific gene of interest must be specified a priori, limiting the conclusions that can be drawn from the findings. The genome-wide association study (GWAS) approach was proposed to identify loci associated with complex traits without a dependence on prior hypotheses, and became feasible due to the completion of the Human Genome Project in 2001 and subsequent rapid advancements in genotyping technology. In contrast to linkage studies, GWAS can be performed with unrelated subjects, who can be recruited more readily than families, and identifies specific risk alleles. The foundation of GWAS lies in the “common disease, common variant” hypothesis, i.e., that relatively common variants in the population with low penetrance contribute to genetic susceptibility to common diseases (Chakravarti, 1999; Lander, 1996; Pritchard and Cox, 2002; Reich and Lander, 2001). Early GWAS operated under the expectation that the variants underlying complex traits would have moderate-to-large effect sizes and would be detectable in relatively small samples. Although this was true for some phenotypes, such as age-related macular degeneration and exfoliation glaucoma (Haines et al., 2005; Thorleifsson et al., 2007), it has become clear that, for the majority of complex traits, much larger samples are needed to achieve the statistical power necessary to detect the small effects of risk loci. It is now recognized that, for the most part, the risk loci identified could explain only a small portion of the variance in the traits (i.e., the “missing heritability problem”) (Manolio et al., 2009). Thus, the focus has been broadened from use of a standard, Bonferroni-corrected genome-wide significance threshold (generally 5 × 10−8 for GWAS) to the use of other indicators of association (Yang et al., 2011). Additionally, results from early GWAS led to the development of large meta- and mega-analysis-based collaborative efforts, which have identified replicable risk loci for a variety of complex traits. Following an overview of the 12 AD GWAS that have been published using a classical approach, we will consider these approaches in detail in relation to the lessons learned in the study of AD genetics. Figure 1 describes the seminal events in AD GWAS discovery.
Figure 1.
Findings from classical AD GWAS
Table 1 provides an overview of the 12 published AD GWAS. The first GWAS of AD was published in 2009 in a sample of German men, including 476 cases and 1,358 controls (Treutlein et al., 2009). The discovery phase involved over 500,000 SNPs and was followed by a replication analysis of 139 SNPs in 1,024 AD cases and 996 controls. Although no associations were genome-wide significant (GWS) in the discovery sample, two SNPs that mapped to the PECR gene were GWS in a meta-analysis of the discovery and replication samples. Overall, 15 top-ranked SNPs located in or near several genes (CAST, ERAP1, PPP2R2B, ESR1, CCD41, ADH1C, GATA4, and CDH13) were GWS in a meta-analysis of the discovery and replication samples.
Table 1.
Overview of AD genome-wide association studies (GWAS).
| Reference | Phenotype | Study | Discovery sample size | GWS findings (at P<5×10−8) | Replicated? |
|---|---|---|---|---|---|
| Treutlein et al., 2009 | AD case-control status | German GWAS | 1,024 German ancestry cases, 996 German ancestry controls (male) | rs7590720, rs1344694 (PECR) | Not attempted |
| Bierut et al., 2010 | AD case-control status | SAGE | 1,235 EA cases, 1,433 EA controls and 662 AA cases, 449 AA controls | None | n/a |
| Edenberg et al., 2010 | AD case-control status | COGA | 847 EA cases, 552 EA controls | None | n/a |
| Kendler et al., 2011 | AD criteria factor score | MGS2 controls | 2,357 EAs; 812 AAs | None | n/a |
| Heath et al., 2011 | AD factor score | OZALC | 2,062 Australian cases, 3,393 Australian controls | None | n/a |
| Frank et al, 2012 | AD case-control status | German GWAS | 1,333 German ancestry cases, 2,168 German ancestry controls (male) | rs1789891 (ADH1C) | Yes, in COGA |
| Zuo et al., 2012 | AD case-control status | SAGE and COGA meta-analysis | 1,409 EA cases, 1,518 EA controls | None | n/a |
| Wang et al., 2013 | DSM-IV AD criteria count | COGA | 2,322 EAs | None | n/a |
| McGue et al., 2013 | AD factor score | MCTFR | 7,188 EAs | None | n/a |
| Park et al., 2013 | AD case-control status | Korean GWAS | 117 Korean cases, 279 Korean controls | rs1442492 and rs10516441 (ADH7), rs10516441 (ALDH2) | Yes, in an independent Korean sample |
| Quillen et al., 2014 | AD case-control status | Chinese GWAS | 102 Chinese cases, 212 Chinese controls (male) | rs3782886 and rs671 (ALDH2) | Not attempted |
| Gelernter et al., 2014 | DSM-IV AD criteria count; AD case-control status | Yale-Penn and SAGE | 2,379 EAs, 3,318 AAs (Yale-Penn); 2,752 EAs, 1,311 AAs (SAGE) | 14 SNPs in chromosome 4 in AAs including rs28542574 (LOC100507053) and rs2066702 (ADH1B) and rs1229984 (ADH1B) in EAs; rs1437396 on chromosome 2 (intergenic) | Yes, in both an independent, identically ascertained sample and in the German GWAS sample (case-control only) |
Abbreviations: AA, African-American; EA, European-American; AD, alcohol dependence; GWS, genome-wide significant
Bierut et al. (2010) performed a GWAS of AD in the Study of Addiction: Genetics and Environment (SAGE) sample, tested over 900,000 SNPs in 1,235 European-American (EA) and 662 African-American (AA) cases and 1,433 EA and 499 AA alcohol-exposed controls. Although, in the combined EA/AA sample, 15 SNPs were identified with P < 10−5, the findings were not replicated in two independent datasets. There was modest evidence to replicate a SNP identified by Treutlein et al. (rs13160562 in ERAP1, P=0.03), but no other findings from that GWAS were replicated.
Edenberg et al. (2010) performed a GWAS using over 800,000 SNPs in a family-based sample of 847 AD EA cases and 552 EA controls and 345 AA cases and 140 AA controls from the Collaborative Study on the Genetics of Alcoholism (COGA) sample, which included 612 EA AD cases and 413 EA controls previously analyzed in Bierut et al. (2010). Their primary analysis was a case-control study in the EA sample. They also repeated the analyses restricting the cases to those meeting early onset-AD and extended the analysis to the AA sample. They found no GWS associations in any of the analyses, and although the EA sample overlapped substantially with that of Bierut et al. (2010), results from the two studies differed substantially.
Kendler et al. (2011) performed an AD GWAS in a population-based sample of 2,357 EAs and 812 AAs from the Molecular Genetics of Schizophrenia (MGS2) control sample. Rather than examining association with case-control status, this study examined a quantitative AD criterion factor score. Although no GWS SNPs were identified in either sample, three genes (PITRM1, PIGG, and AKAP9) were significant in the gene-based analysis in EAs.
In one of the largest AD GWAS to date, Heath et al. (2011) included 8,754 European ancestry family members (2,062 AD cases) and 3,393 Australian population-based controls to test the association of 300,000 SNPs with three phenotypes: AD case-control status, a quantitative AD factor score, and a quantitative “heaviness of drinking” factor score. No GWS loci were identified for any of the phenotypes, and the authors were unable to replicate any of the SNPs identified in previous GWAS (Treutlein et al., 2009; Bierut et al., 2010; Edenberg et al., 2010). This study showed that effects, as expected, are very small (0.15–0.25% of the variance in AD risk), and that larger samples are needed to detect them.
Frank et al. (2012) performed a GWAS with over 400,000 SNPs in a German sample of 1,333 men with severe AD and 2,168 controls, including 487 cases and 1,358 controls from Treuitlen et al. (2009). Testing for an association of AD case-control status, they identified one SNP that was GWS (rs1789891, P=1.27 × 10−8). Interestingly, the SNP is located in the region between ADH1B and ADH1C, two alcohol metabolizing enzyme genes and in complete linkage disequilibrium (LD) with a missense polymorphism in ADH1C (rs1693482, D’=1.0, r2=0.27). Although this study replicated a finding in the COGA sample (P=0.015), it did not replicate the top (non-GWS) findings in either the COGA (Edenberg et al., 2010) or SAGE (Bierut et al., 2010) datasets. This was the first AD GWAS to identify a GWS association in the discovery sample that was replicated in an independent sample and the first to identify significant associations in alcohol metabolizing enzyme genes, importantly replicating the findings from candidate gene studies (Thomasson et al., 1991; Chen et al., 1999; Edenberg, 2007, Bierut et al. 2012).
Zuo et al. (2012) performed an AD meta-analysis in the SAGE and COGA datasets (1,409 EA AD cases, 1,518 EA controls, 681 AA AD cases and 608 AA controls), the primary results from which are described above, in which the EA sample served as the discovery sample and the AA sample as the replication sample. At a significance threshold of 5 × 10−7, which takes into account the non-independence of SNP markers, this study identified eight SNPs that they considered GWS (although none reached a conventional threshold): two in KIAA0040, five in SERINC2, and one in HTR1A. Although no association was replicated in AAs, signals at the KIAA0040 locus were enriched in AAs, and several of the KIAA0040 SNPs were demonstrated to be cis eQTLs in HapMap cell lines, which the authors concluded suggested a functional link with AD.
Wang et al. (2013) performed an AD GWAS with approximately 600,000 SNPs in an expanded COGA sample of 2,322 EAs from 118 families, including 275 subjects from Edenberg et al. (2010). Testing for association with a quantitative DSM-IV criterion count phenotype, they found no GWS associations.
McGue et al. (2013) performed an AD GWAS of “behavioral disinhibition” traits including AD and alcohol consumption in a twin and adoptive family sample of 7,188 EA subjects clustered in 2,300 nuclear families from the Minnesota Center for Twin and Family Research (MCTFR). The authors tested for association between an AD factor score and over 500,000 SNPs, but found no GWS associations with AD.
Park et al. (2013) conducted an AD GWAS in 117 Korean AD cases and 279 Korean controls, testing for association of AD case-control status with over 400,000 SNPs. Despite the small sample, this study identified three GWS associations: two SNPs in the intergenic region flanking the 3’ end of the alcohol metabolizing enzyme gene ADH7 (rs1442492, P = 6.28 × 10−8 and rs10516441, P = 6.46 × 10−8) and a missense SNP in another alcohol metabolizing enzyme gene, ALDH2 (rs671, P=8.42 × 10−8). Similar to Frank et al. (2012), this study suggested that variants in alcohol metabolizing enzyme genes are among the strongest AD risk loci.
Quillen et al. (2014) performed an AD GWAS in 102 Chinese male AD cases and 212 Chinese male controls from extended pedigrees enriched for AD. Testing for association of AD case-control status with 300,000 SNPs, they identified two GWS loci in the alcohol metabolizing enzyme gene ALDH2, rs3782886 (P=4.74 × 10−8) and rs671 (P=4.73 × 10−8). They also found these SNPs to be strongly associated with the flushing response and maximum drinks consumed in a 24-hour period. The associations with AD are consistent with those from the study by Park et al. (2013) in a Korean sample.
The largest AD GWAS to date was performed by Gelernter et al. (2014a). The discovery sample consisted of 2,379 EA and 3,318 AA subjects (“Yale-Penn”) and 2,752 EA and 1,311 AA subjects from the SAGE sample (Bierut et al., 2010). The replication sample consisted of 1,746 EA and 803 AA subjects (Yale-Penn replication) and 1,806 EA AD cases and 1,978 EA controls from a previously analyzed German sample (Frank et al., 2012), which were used to replicate the results for the case-control phenotype. The Yale-Penn samples were not previously included in any other GWAS. In addition to being the largest AD GWAS to date, this was the first to impute the SNPs tested to include over 9 million in AAs and over 6 million in EAs. Similar to their previous analyses for cocaine and opioid dependence (Gelernter et al., 2014b; Gelernter et al., 2014c) and the analysis by Wang et al. (2013), the authors tested for association with a quantitative DSM-IV AD criterion count phenotype. They controlled for cocaine, opioid, and nicotine dependence criteria in the sample and meta-analyzed the results with those from the SAGE sample (Bierut et al., 2010) and ultimately with their discovery and replication samples. In AAs, they identified several GWS findings at the ADH locus on chromosome 4, with the strongest finding in ADH1B (rs2066702, P=1.50 × 10−23). In EAs, the strongest finding was also in ADH1B (rs1229984, P=1.52 × 10−22). Both ADH1B SNPs are missense polymorphisms and both associations replicated in an independent sample. Thus, as in other AD GWAS (Frank et al., 2012, Park et al., 2013), the ADH gene cluster appears to have the greatest effect on AD risk.
Lessons learned from classical AD GWAS
The only consistent findings from classical GWAS of AD are those implicating the alcohol metabolizing enzyme genes, which have shown association in GWAS for AD and a proposed intermediate phenotype for AD, alcohol consumption (Baik et al., 2011; Frank et al., 2012; Gelernter et al., 2014a; Kapoor et al., 2013; Park et al., 2013; Quillen et al., 2014; Schumann et al., 2011; Takeuchi et al., 2011), replicating findings from candidate gene studies of AD. The lack of consistent findings overall may reflect low power to detect variants of small effect and genetic and phenotypic heterogeneity among studies. Thus, the SNPs in the genes encoding metabolizing enzymes are among the common variants with the largest effects on AD risk. Interestingly, associations with other candidate genes for AD such as DRD2, OPRM1, and COMT have not been replicated in GWAS (Olfson and Bierut, 2012), suggesting that their presumed effect sizes have been greatly overestimated in candidate gene studies, where small sample sizes have been used to detect significant effects (P<0.05), or they are false positives.
A major lesson from GWAS is that the initial studies of almost all complex traits were underpowered to detect significant associations given the small effects of individual risk loci and the stringent genome-wide significance threshold. Collaborative efforts will be needed to assemble the large samples necessary for GWAS to identify replicable loci and pathways associated with AD risk. Meta- and mega-analyses have elucidated the genetic architecture and identified variants associated with complex traits like type 2 diabetes (Morris et al., 2012), height (Lango Allen et al., 2010; Wood et al., 2014), schizophrenia (Ripke et al., 2011; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), and bipolar disorder (Sklar et al., 2011). Although no large-scale meta-analyses of AD have yet been performed, the Psychiatric Genomics Consortium (PGC) has assembled a working group to conduct mega-analyses of alcohol and illicit drug phenotypes. Another collaborative group, the GWAS and Sequencing Consortium of Alcohol and Nicotine use (GSCAN), has also formed to conduct GWAS and exome meta-analyses and whole genome sequencing studies of alcohol and nicotine use.
Although the inclusion of multiple populations in GWAS is important to gain a comprehensive understanding of the genetic influences on disease risk, there are few studies of AD and other phenotypes in non-European ancestry populations, the most studied of which are African-ancestry populations, which account for only 3% of GWAS reported in the PubMed database (Peprah et al., 2015). To date, only one AD GWAS has identified GWS loci in AAs (Gelernter et al., 2014a), probably because the size of available African ancestry samples is generally much smaller than for European ancestry. We urge the newly formed collaborative groups to perform analyses in subjects of non-European ancestry, if feasible, as the identification of loci contributing to AD risk in a range of populations will yield a fuller understanding of the disorder.
Post-GWAS approaches to studying the genetics of AD risk
Classical GWAS approaches have identified loci associated with AD risk, but these explain only a small proportion of the risk for AD and related phenotypes (Agrawal et al., 2012; Bühler et al., 2015). These studies suggest that, as with other complex traits, a large number of causal risk variants contribute to the development of AD and that variants individually have comparatively small effects on risk (Manolio et al., 2009). Because of this and the stringent correction for the large number of SNPs tested in GWAS, large samples are necessary to detect effects reliably. Alternatives to single SNP analysis of GWAS data have examined individually non-significant loci en masse, which are largely disregarded in traditional GWAS. Here, we discuss two categories of post-GWAS approaches: 1) studies that focus on so-called polygenic methods, heritability analyses and genetic risk prediction, and 2) studies that focus on identifying sets of genes and/or pathways influencing risk. Table 2 provides an overview of these studies.
Table 2.
Post-genome-wide association study (GWAS) analyses of AD GWAS datasets.
| Post-GWAS method | Reference | Methods and findings |
|---|---|---|
| SNP heritability | Vrieze et al., 2013 | Using Genome-wide Complex Trait Analysis (GCTA), estimated a SNP heritability of 16% for AD and 18% for alcohol consumption in the MCTFR family-based sample (unrelated parents only); the estimate for AD was not significant. Estimates in the full family-based sample were high and likely reflect shared environmental effects. |
| Yang et al., 2014 | Using GCTA, estimated a SNP heritability of 22.1% for AD in the unrelated AA subset of the Yale-UPenn sample (n=1,838). | |
| Risk profile scoring | Frank et al., 2012 | Split the German GWAS sample into a discovery sample and a target sample; constructed a polygenic risk score in the discovery sample that significantly predicted AD case-control status in the target sample; used the full German GWAS sample as a discovery sample and significantly predicted AD case-control status in SAGE and COGA samples. |
| Kos et al., 2013 | Significantly predicted AD case-control status using EA and AA discovery samples from COGA and EA and AA target samples from SAGE. Although significant, the variance explained was small (0.73% in EAs and 2.14% in AAs). | |
| Vrieze et al., 2013 | Used k-fold cross-validation within the Minnesota Center for Twin and Family Research (MCTFR) sample and observed polygenicity; as the P-value threshold increased, the percent of variation increased. | |
| Yan et al., 2014 | Used the COGA and SAGE samples to conduct risk profile scoring for candidate gene SNPs as well as with SNPs from GWAS. Did not observe significant risk prediction from a set of 21 candidate gene SNPs. When combined COGA and SAGE datasets and split this sample in half, able to significantly predict AD case-control status in the target sample using results from the discovery sample. | |
| Levey et al., 2014 | Used a Convergent Functional Genomic approach to integrate multiple sources of information and generated a list of 713 nominally significant SNPs in 135 candidate genes. Were unable to predict case-control status in the German GWAS target sample, but when this list was prioritized to a set of 11 genes using data from a DBP stress-reactive knockout mouse model for alcoholism, significantly predicted risk in German sample and two additional samples. | |
| Gene set analysis | McGue et al., 2013 | Used the versatile gene-based association study gene set method in the MCTFR sample but did not identify any significant genes after Bonferroni correction. Did not identify enrichment of candidate genes for substance abuse or related phenotypes. |
| Han et al., 2013 | Used a network-based gene set analysis approach to identify human protein interaction networks (HPIN) enriched for AD-associated genes in the SAGE and COGA EA and AA datasets. Identified seven HPIN that were enriched for AD-associated genes within EAs and AAs. Found that this subnetwork was associated AD case-control status in EAs and AAs, and replicated this finding in two additional samples. | |
| Pathway analysis | Kendler et al., 2011 | Conducted pathway analysis in MGS2 controls GWAS sample using ALIGATOR and found significant enrichment of Gene Ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in EAs and AAs, including those related to lipid and cholesterol metabolism and cell adhesion. |
| Biernacka et al., 2013 | Used the EA SAGE dataset and grouped genes into sets based on KEGG pathway annotations, but did not observe significant association with AD case-control status. Several gene sets showed suggestive association including “synthesis and degradation of ketone bodies” and “neuroactivated ligand-receptor interaction”. | |
| Kos et al., 2013 | Evaluated enrichment of pathways/ontologies enriched among SNPs identified in risk profile scoring analysis using the COGA discovery sample and SAGE target sample. Identified four ontologies relevant to brain development and inhibitory neurotransmission with significant enrichment among the gene sets from EAs and AAs. | |
| Juraeva et al., 2015 | Conducted pathway analysis in AD cases and controls from the German GWAS sample using annotations from KEGG, Reactome, Gene Ontology, Biocarta, microRNA targets, transcription factor targets, and positional information. Found that 19 gene sets contained the gene XRCC5, and followed up on this gene with functional studies. Found that Drosophila knockdown model of the XRCC5 ortholog Ku80 had lower sensitivity to alcohol than controls. In a human laboratory-based self-admnistration study, found significant association between maximum blood alcohol concentration and the top XRCC5 SNP from the GWAS, rs828701. |
Abbreviations: GWAS, genome-wide association study; AD, alcohol dependence
SNP heritability and genomic risk profile scoring methods
Two complementary methods have been proposed to investigate the genetic architecture of complex traits by considering large sets of SNPs simultaneously. The first, the SNP (or sometimes chip) heritability method, estimates the proportion of additive genetic variance attributable to all measured SNPs in a GWAS. This analysis is most often performed using a Genome-wide Complex Trait Analysis (GCTA; Yang et al., 2011). GCTA uses a mixed linear model to fit the effects of all SNPs as random effects and estimates the variance attributable to the aggregate random effect. A genetic relationship matrix (GRM) is estimated from the SNP data of all pairs of individuals. GCTA is typically performed on a set of unrelated subjects to estimate the variance attributable to common SNPs in the absence of effects due to shared environment or non-common SNPs (e.g., rare variants). It can be extended to compute the genetic correlation between traits measured in the same study or different studies.
Two SNP heritability studies have been performed for AD. McGue et al. (2013) used data from a GWAS that yielded no GWS associations to estimate SNP heritability with GCTA. Restricting their analysis to subjects with a genetic relatedness < 0.025, they found an estimate of 8% for AD. In the same sample, Vrieze et al. (2013) investigated genetic influences on behavioral disinhibition, AD, and alcohol consumption factor scores using both SNP heritability and profile scoring methods (discussed below). To address confounding by shared environment and non-SNP genetic influences, these investigators performed four separate GCTA analyses. Analysis in the full sample (n=7,188) yielded estimates very similar to those obtained using biometric methods in the subsample of twins (n= 2,877), likely reflecting shared environmental and non-additive, non-SNP genetic influences. The estimates in the unrelated parent sample (n=3,542) were 16% for AD and 18% for alcohol consumption; only the latter estimate was statistically significant. In the unrelated offspring sample (n=1,784), the estimates were unstable and non-significant, most likely due to the small sample. Estimates in the full offspring sample (n=3,336) were the highest of all, potentially reflecting greater shared environmental influences than obtained in the full sample including parents.
Yang et al. (2014) explored the genetic architecture of AD in 1,719 AA AD cases and 1,156 AA controls that were included in a previous GWAS on AD (Gelernter et al., 2014a). Here, they used the GCTA method and estimated that, in the unrelated subjects (n=1,838), 22.1% (s.e. 17.7%) of the risk for AD was attributable to common SNPs. A related analysis using a common variance component to account for shared environment (Do et al. 2012), yielded a very similar estimate (i.e., 23.9%; s.e. 9.3%). The higher estimates of SNP heritability in this study than those in Vrieze et al. (2013) potentially reflect the samples’ different population composition, affection rate, and statistical power. Estimating the variance explained by top-ranking SNPs for nine different P-value thresholds, Yang et al. (2014) found that, at the least stringent threshold (P≤0.001), the top SNPs explained only 0.68% of the phenotypic variance, and that the variance explained plateaued at P≤0.01 (17.6%), suggesting that the SNP heritability signal is primarily coming from SNPs with P≤0.01. When they partitioned the genome by chromosome and by genic/intergenic regions to estimate the variance explained by each chromosome/region, they found a relationship between the length of the chromosome and the amount of phenotypic variance explained. However, they found no relationship between the number of known genes on a chromosome and the explained phenotypic variance.
A second method developed to investigate the polygenic nature of complex traits is the risk profile scoring method. Similar to the SNP heritability method, risk profile scoring utilizes associations that do not necessarily meet the strict genome-wide significance threshold, with the hypothesis that this set of nominally associated SNPs may be enriched for true positives. Here, these associations are used to predict risk for a disease trait in a “target” sample given results from a “discovery” sample. For this analysis, a GWAS is conducted in the discovery sample, and using a P-value threshold, the results are used to generate a risk profile score for each subject in the target sample. This score is equal to the sum of the count of risk alleles for that subject weighted by their effect sizes in the discovery sample. The score is then typically used in a regression analysis to evaluate its association with the phenotype of interest in the target sample, as well as the proportion of variance explained (e.g., R2). This approach has been used to investigate the polygenicity of complex traits such as schizophrenia and bipolar disorder (International Schizophrenia Consortium et al., 2009), multiple sclerosis (International Multiple Sclerosis Genetics Consortium et al., 2010), height (Lango Allen et al., 2010), body mass index (Speliotes et al., 2010), and rheumatoid arthritis (Stahl et al., 2012). Frank et al. (2012) first applied this method to AD. They randomly divided their German GWAS sample into a discovery sample (667 cases and 1084 controls) and a target sample (666 cases and 1084 controls). The polygenic risk score was computed in the target sample using SNPs with P<0.5. The score significantly predicted case-control status in the target sample (P=9.66 × 10−9). In a similar analysis using the full German sample as the discovery sample and the COGA and SAGE samples as the target samples, profile scores significantly predicted case-control status in both samples (P=3.9 × 10−2 and P=1.1 × 10−4, respectively).
Kos et al. (2013) were the first to examine the polygenic architecture of AD in both EA and AA populations. In their risk profile scoring analysis, the discovery sample consisted of 1,274 EA and 285 AA subjects from COGA, and the target sample consisted of 1,573 EA and 841 AA subjects from SAGE. GWAS was performed in the discovery sample and the results were LD pruned and binned according to P-value and minor allele frequency (MAF). Risk prediction scores derived from GWAS results in the discovery sample significantly predicted case-control status in both the EA and AA target samples, although the maximum variance explained in EAs (at a P-value threshold of 0.05) was only 0.73% (P=1.64 × 10−3) and in AAs (at a threshold of < 0.3) it was 2.14% (P=2.08 × 10−4). When binned by MAF, SNPs with higher allele frequencies explained more variance, with a peak of 0.3–0.4% in both populations (EA: 0.57%, P=0.0047; AA: 2.13%, P=1.3 × 10−4).
Vrieze et al. (2013) conducted risk profile scoring in the MCTFR sample using a subsampling approach termed k-fold cross-validation (Breiman and Spector, 1992; Hastie et al., 2009). This approach split the sample into 10 equal subsamples, keeping family members within a subsample to control for correlated genotypes and phenotypes. Nine subsamples were combined to generate a set of associations and effect size weights, and the tenth sample was used as the target sample. The procedure was repeated for every combination of the 10 samples with each subsample serving as the discovery sample nine times and as the target sample once. SNPs were filtered based on LD and genetic risk scores were computed in the target set at multiple P-value thresholds. Similar to the results from the other polygenic score analyses discussed here, the authors observed a polygenic effect for AD, such that as the P-value threshold increased, the percent of variance explained also increased; this effect was also seen for the alcohol consumption phenotype.
Yan et al. (2014) used the SAGE EA and COGA EA datasets to conduct risk profile scoring analyses for both a set of candidate gene SNPs and SNPs from the GWAS using various P-value thresholds. In the first analysis, using a set of 21 SNPs associated with AD from candidate gene studies in the family-based COGA dataset to predict AD, they found no association with AD in either the COGA or SAGE samples. In a second analysis, they combined the SAGE and COGA datasets and randomly split it equally into case and control groups. The GWAS datasets were pruned for LD and genetic risk scores were computed in the target sample at several P-value thresholds. Here, in contrast to their results from candidate gene SNPs, they observed significant prediction of AD in the target sample for all thresholds with P ≥ 0.01, supporting a polygenic model of AD risk.
Levey et al. (2014) used a Convergent Functional Genomics (CFG) approach to conduct a risk profile scoring analysis for AD. Here, they used the German GWAS dataset (Treutlein et al., 2009) and information from other association and linkage studies of AD, gene expression (including post-mortem brain expression and peripheral tissue expression), and genetic studies in animal models to generate a list of 135 candidate genes, and assembled risk profile scores from 713 nominally significant SNPs (P≤0.05) within these genes. They then tested for the ability of this score to predict case-control status in an independent German target sample (Frank et al., 2012). Overall, they found that they were unable to predict case-control status significantly with the risk prediction score, similar to the findings from the candidate gene study-derived risk prediction scores in Yan et al. (2014). However, when they prioritized the genes in the risk prediction score to a set of 11 cross-validated findings from the DBP stress-reactive knockout mouse model for alcoholism, they found significant prediction in the German target sample (P=0.041) and alcohol abuse and dependence samples from the United States (Gelernter et al., 2014a).
SNP heritability studies of AD have demonstrated its highly polygenic nature. Risk profile scoring analyses support the polygenicity of AD by demonstrating that scores generated using nominally significant SNPs from GWAS significantly predict risk in independent samples. Thus far, SNP heritability studies have estimated the heritability from common SNPs to be up to 23%, which with better-powered samples could increase. Thus, SNP heritability accounts for roughly half of the heritability of AD estimated in family studies, which is in line with estimates for phenotypes such as height, where the SNP heritability was estimated at 45%, about half of the overall heritability estimated in twin studies of 80% (Yang et al., 2010). To date, all such studies of AD have used comparatively small samples. Studies in larger samples, potentially available through collaborations such as the PGC Addictions group and GSCAN, should provide more accurate estimates of the SNP heritability.
Gene-set and pathway based methods
Gene-set and pathway analyses have also been used to investigate genetic influences on complex traits by utilizing associations that are not GWS, based on the principle that genes belonging to the same molecular and cellular pathways work together to influence disease susceptibility. Although the effects of single genes may be too small to detect individually, they may be detectable when their effects are aggregated (Wang et al., 2011). Gene-set analysis identifies genes that contain an excess of SNPs with low P-values, in contrast to traditional GWAS analysis, which tests for association with individual SNPs. This reduces the multiple testing burden from ∼1 million tests to ∼20,000 tests. Pathway analysis, an extension of gene-set analysis, models the relationships between genes in a set to gain insights into the underlying biology of a phenotype. Although the terms “gene-set” and “pathway” analysis are often used synonymously, we limit the term “gene-set analysis” to those that do not incorporate pathway-level information.
McGue et al. (2013) applied the versatile gene-based association study gene set method (VEGAS; Liu et al., 2010) in their AD/alcohol consumption GWAS in the MCTFR sample. VEGAS uses P-values for individual SNP associations to compute a per-gene association P-value and can be applied to all SNPs in a gene or restricted to SNPs associated with a phenotype at a certain threshold. After correcting for 17,567 genes tested, no genes reached statistical significance and there was no evidence that candidate genes for substance abuse or related phenotypes were enriched in their sample.
Han et al. (2013) used a network-based approach to identify potential pathways underlying AD risk, which they argued is more flexible in gene set definition, less biased, and better at detecting genes that work across multiple pathways than standard pathway analysis. They focused on human protein interaction networks (HPIN) to identify an HPIN network enriched for AD-associated genes using the 2,332 unrelated EA and 1,088 unrelated AA subjects from the SAGE and COGA datasets. Using the R package dmGWAS (Jia et al., 2011), they identified 429 HPIN modules significantly enriched for AD-associated genes in EAs, seven of which were also significantly enriched for AD-associated genes in AAs. Combining these seven modules yielded a set of 39 genes, which showed significant association with AD in EAs (P<0.0001) and AAs (P=0.0008), findings that were replicated in the European OZALC GWAS sample (P=0.006) and the EA and AA Yale-Penn samples (P=0.001 and 0.007, respectively). The subnetwork was not associated with other complex disorders, including bipolar disorder, major depressive disorder, and type 2 diabetes, supporting its specificity to AD.
Kendler et al. (2011) conducted a pathway enrichment analysis using ALIGATOR (Holmans et al., 2009) to test whether specific gene-sets defined by Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG), corresponding to specific categories and pathways, were enriched for association with AD in their GWAS sample. They found a significant enrichment in both AAs and EAs, with 62 and 96 significantly enriched categories/pathways at P≤0.01, respectively, including those involving lipid and cholesterol metabolism and cell adhesion, the latter having been previously associated with several addiction-related phenotypes (Gelernter et al., 2006; Gelernter et al., 2014b; Hart et al., 2012; Johnson et al., 2006; Yang et al., 2007; Yang et al., 2008).
Biernacka et al. (2013) used GWAS data from the EA subset of the SAGE sample (n=1,165 AD cases, 1,379 controls) to investigate genetic influences on AD risk. They grouped genes into sets based on all pathway annotations from KEGG, which generated 200 gene sets. They used a “two-step” association approach, wherein SNPs within a gene were first tested for association of that gene with AD, and then gene-level tests conducted to evaluate the association of a gene set with AD. They also used a “one-step” approach, in which all SNPs in a gene set were included in the analysis irrespective of individual gene-level associations with AD. Although neither method showed significant associations after correction for the number of pathways tested, several gene sets yielded suggestive evidence for association with AD. For the two-step analysis, the “synthesis and degradation of ketone bodies” gene set was the most significant (P=0.0009), while for the one-step analysis, the most significant gene set was the “neuroactivated ligand-receptor interaction” pathway (P=0.008). Interestingly, many of the strongly associated SNPs in this gene set were in glutamate receptor genes, including GRIK1, which has been associated with AD (Kranzler et al., 2009) and the response to topiramate treatment for heavy drinkers (Kranzler, 2014).
Kos et al. (2013), using pathway analysis, aimed to identify specific ontologies and/or pathways (defined by ResNet Mammalian, Ariadne Genomics) that were enriched among the groups of SNPs identified in their risk profile scoring analyses in the COGA discovery and SAGE target samples. For each bin (n=20, defined by GWAS P-value threshold), they retained SNPs with consistent directions of effect in the discovery and target samples, assigned them to their corresponding genes, and evaluated the significance of enrichment via permutation. Although the majority of ontologies and pathways showed no enrichment, four ontologies displayed significant enrichment in the gene sets from both EAs and AAs. The pathways identified as enriched among the GWAS associations [Maf transcription factors, homeotic (Hox) AbdB genes, chloride transport, and glycine and serine metabolism] relate to brain development and inhibitory neurotransmission and may represent future targets for AD research.
Juraeva et al. (2015) combined pathway analysis with functional follow-up studies in a Drosophila model and a human sample to identify AD risk genes. They conducted gene set analysis on 1,333 AD cases and 2,168 controls from the German GWAS sample (Frank et al., 2012) using the R package globaltest (Goeman et al., 2004). Gene sets were constructed using multiple sources such as KEGG, Reactome, Gene Ontology, Biocarta, microRNA targets, transcription factor targets, and positional information. The analysis identified 19 significant gene sets (FDR ≤ 0.05), with the gene XRCC5 present in six of these. In a Drosophila knockdown model of the XRCC5 ortholog, Ku80, the mutants had lower sensitivity to ethanol than controls. An alcohol intravenous self-administration study in 85 healthy humans showed a significant association between maximum blood alcohol concentration and XRCC5 genotype for the top SNP from their GWAS, rs828701 (P=0.03), indicating that XRCC5 may be an AD candidate gene.
Gene set and pathway analyses of GWAS data are potentially powerful tools that have made several contributions to our understanding of the genetics of AD. The analyses summarized here have nominated novel candidate genes for AD and have highlighted pathways of potential importance. Although gene set and pathway analyses are leverage GWAS data to gain mechanistic insight into the biology of complex traits, they are analytically limited, failing to capture alternative transcripts of genes, providing incomplete and inaccurate annotations, and lacking information on the role of pathways in specific cell types and under specific conditions (Khatri et al., 2012). The recently completed ENCODE project (ENCODE Project Consortium, 2012), a major source of new information that may greatly increase our knowledge and understanding of biological pathways, will likely contribute to the improvement of currently available methods. Thus, pathway and related analyses are likely to become increasingly important approaches to gene finding for AD.
Conclusions and future considerations
GWAS has greatly increased our understanding of the genetic etiology of AD. The most robust associations for AD have been with the alcohol metabolizing enzyme genes, especially ALDH2 in East Asian populations (Park et al., 2013; Quillen et al., 2014), and ADH1B in European-American and African-American populations (Frank et al., 2012; Gelernter et al., 2014a). Samples in the tens of thousands will likely be necessary to detect additional loci beyond those identified by the best-powered AD GWAS to date (Gelernter et al. 2014a).
Studies of sufficiently large samples are currently being organized by the PGC Addictions group and GSCAN for large meta- and mega-analyses combining multiple GWAS datasets. Although these efforts will substantially increase the size of the study sample, they also introduce between-study heterogeneity, which could be mitigated by homogeneous subtype analysis. Latent class analysis was recently applied to an AD GWAS using the COGA sample (Wetherill et al., 2013), which identified a GWS association between “high risk” AD and a SNP in the NALCN gene. We have used cluster analysis to identify heritable, homogeneous subtypes of cocaine and opioid dependence (Bi et al., 2014; Sun et al., 2012; 2014). These methods may be useful in increasing statistical power in consortium-based analyses, including those focusing on AD.
A related impediment to AD gene finding is the phenotypic complexity of the disorder, especially when using a diagnosis-based phenotype. An alternative to DSM diagnosis-based phenotypes are intermediate phenotypes, which have been proposed to be more closely related to the specific genetic mechanisms and biological pathways underlying disease risk, thus having the potential to enhance gene identification (Goldman and Ducci, 2007). Several GWAS have already been performed for AD intermediate phenotypes such as alcohol consumption, briefly mentioned above (Baik et al., 2011; Frank et al., 2012; Gelernter et al., 2014a; Kapoor et al., 2013; Park et al., 2013; Quillen et al., 2014; Schumann et al., 2011; Takeuchi et al., 2011), and are likely to become increasingly popular in light of initiatives such as the NIMH Research Domain Criteria (RDoC) Project (Insel et al., 2010).
Analyses of GWAS data beyond traditional SNP association testing have increased our understanding of the genetic architecture of complex traits, including AD. As described above, results from polygenic analyses of GWAS data are consistent with the polygenic nature of AD and have shown that the many individual loci that influence AD risk exert small effects. Nonetheless, overall, estimates of the variance in AD risk attributable to common variants have been as high as 23% (Yang et al., 2014). Polygenic methods can be extended to answer questions such as the degree of genetic sharing between AD and other traits, similar to what has been done for major psychiatric disorders in the PGC dataset using the bivariate GCTA method (Lee et al., 2013) and risk profile scoring (Smoller et al., 2013). Common genetic variation may regulate or encode proteins found within important biological pathways for addiction. Motivated by this hypothesis, pathway analysis of GWAS data has led to new discoveries in the genetics of AD, and will likely become more important as pathway annotations and other resources continue to improve. The integration of data from GWAS, gene expression and whole-genome sequencing, which is becoming increasingly more available, will present new opportunities and challenges in the discovery of novel loci and pathways associated with AD.
Acknowledgements
This work was supported by NIH grants T32 DA028874 and R01 AA021164 and the VISN 4 MIRECC of the Philadelphia VAMC. We thank Dr. Joel Gelernter for reading and commenting on a draft of this review. Dr. Kranzler has been a consultant or advisory board member for Alkermes, Lilly, Lundbeck, Otsuka, and Pfizer. He is a member of the American Society of Clinical Psychopharmacology's Alcohol Clinical Trials Initiative, which is supported by AbbVie, Ethypharm, Lilly, Lundbeck, and Pfizer.
References
- Agrawal A, Bierut LJ. Identifying genetic variation for alcohol dependence. Alcohol Res. 2012;34:274–281. [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Verweij KJ, Gillespie NA, Heath AC, Lessov-Schlaggar CN, Martin NG, Nelson EC, Slutske WS, Whitfield JB, Lynskey MT. The genetics of addiction-a translational perspective. Transl Psychiatry. 2012;2:e140. doi: 10.1038/tp.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baik I, Cho NH, Kim SH, Han B-G, Shin C. Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. Am J Clin Nutr. 2011;93:809–816. doi: 10.3945/ajcn.110.001776. [DOI] [PubMed] [Google Scholar]
- Bi J, Gelernter J, Sun J, Kranzler HR. Comparing the utility of homogeneous subtypes of cocaine use and related behaviors with DSM-IV cocaine dependence as traits for genetic association analysis. Am J Med Genet B Neuropsychiatr Genet. 2014;165:148–156. doi: 10.1002/ajmg.b.32216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernacka JM, Geske J, Jenkins GD, Colby C, Rider DN, Karpyak VM, Choi DS, Fridley BL. Genome-wide gene-set analysis for identification of pathways associated with alcohol dependence. Int J Neuropsychopharmacol. 2013;16:271–278. doi: 10.1017/S1461145712000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nöthen MM, Nurnberger JI, Jr, Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP. Gene, Environment Association Studies Consortium A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L, Spector P. Submodel Selection and Evaluation in Regression - the X-Random Case. International Statistical Review. 1992;60:291–319. [Google Scholar]
- Bühler KM, Giné E, Echeverry-Alzate V, Calleja-Conde J, de Fonseca FR, López-Moreno Common single nucleotide variants underlying drug addiction: more than a decade of research. Addict Biol. 2015 doi: 10.1111/adb.12204. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- JA Chakravarti A. Population genetics—making sense out of sequence. Nature Genetics. 1999;21:56–60. doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- Chen CC, Lu RB, Chen YC, Wang MF, Chang YC, Li TK, Yin SJ. Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. Am J Hum Genet. 1999;65:795–807. doi: 10.1086/302540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton NS. The familial incidence of alcoholism: a review. J Stud Alcohol. 1979;40:89–116. doi: 10.15288/jsa.1979.40.89. [DOI] [PubMed] [Google Scholar]
- Do CB, Hinds DA, Francke U, Eriksson N. Comparison of family history and SNPs for predicting risk of complex disease. PLoS Genet. 2012;8:e1002973. doi: 10.1371/journal.pgen.1002973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Research & Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI, Jr, Rice JP, Schuckit MA, Taylor R, Todd Webb B, Tischfield JA, Porjesz B, Foroud T. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch M-A. Genetic influences on the development of alcoholism. Curr Psychiatry Rep. 2013;15:412. doi: 10.1007/s11920-013-0412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Cichon S, Treutlein J, Ridinger M, Mattheisen M, Hoffmann P, Herms S, Wodarz N, Soyka M, Zill P, Maier W, Mössner R, Gaebel W, Dahmen N, Scherbaum N, Schmäl C, Steffens M, Lucae S, Ising M, Müller-Myhsok B, Nöthen MM, Mann K, Kiefer F, Rietschel M. Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addiction Biology. 2012;17:171–180. doi: 10.1111/j.1369-1600.2011.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang BZ, Kranzler HR, Farrer L. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Hum Mol Genet. 2006;15:3498–3507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, Wodarz N, Zill P, Zhao H, Farrer LA. Genome-wide association study of alcohol dependence: significant findings in African-and European-Americans including novel risk loci. Molecular Psychiatry. 2014a;19:41–49. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR, Farrer L. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Molecular Psychiatry. 2014b;19:717–723. doi: 10.1038/mp.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H, Farrer LA. Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biological Psychiatry. 2014c;76:66–74. doi: 10.1016/j.biopsych.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- Goldman D, Ducci F. Deconstruction of vulnerability to complex diseases: enhanced effect sizes and power of intermediate phenotypes. ScientificWorldJournal. 2007;7:124–130. doi: 10.1100/tsw.2007.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- Han S, Yang BZ, Kranzler HR, Liu X, Zhao H, Farrer LA, Boerwinkle E, Potash JB, Gelernter J. Integrating GWASs and human protein interaction networks identifies a gene subnetwork underlying alcohol dependence. American Journal of Human Genetics. 2013;93:1027–1034. doi: 10.1016/j.ajhg.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AB, Engelhardt BE, Wardle MC, Sokoloff G, Stephens M, de Wit H, Palmer AA. Genome-wide association study of d-amphetamine response in healthy volunteers identifies putative associations, including cadherin 13 (CDH13) PLoS ONE. 2012;7:e42646. doi: 10.1371/journal.pone.0042646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin DS, Stinson FS, Ogburn E, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch Gen Psychiatry. 2007;64:830–842. doi: 10.1001/archpsyc.64.7.830. [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd edition. New York: Springer; 2009. Model assessment and selection; pp. 219–260. [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PA, Montgomery GW. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol Psychiatry. 2011;70:513–518. doi: 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P Wellcome Trust Case-Control Consortium. Owen MJ, O'Donovan MC, Craddock N. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. The American Journal of Human Genetics. 2009;85:13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, Sanislow C, Wang P. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium (IMSGC) Bush WS, Sawcer SJ, de Jager PL, Oksenberg JR, McCauley JL, Pericak-Vance MA, Haines JL. Evidence for polygenic susceptibility to multiple sclerosis--the shape of things to come. American Journal of Human Genetics. 2010;86:621–625. doi: 10.1016/j.ajhg.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Zheng S, Long J, Zheng W, Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;27:95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Liu Q-R, Walther D, Edenberg H, Rice J, Foroud T, Uhl GR. Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:844–853. doi: 10.1002/ajmg.b.30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juraeva D, Treutlein J, Scholz H, Frank J, Degenhardt F, Cichon S, Ridinger M, Mattheisen M, Witt SH, Lang M, Sommer WH, Hoffmann P, Herms S, Wodarz N, Soyka M, Zill P, Maier W, Jünger E, Gaebel W, Dahmen N, Scherbaum N, Schmäl C, Steffens M, Lucae S, Ising M, Smolka MN, Zimmermann US, Müller-Myhsok B, Nöthen MM, Mann K, Kiefer F, Spanagel R, Brors B, Rietschel M. XRCC5 as a risk gene for alcohol dependence: evidence from a genome-wide gene-set-based analysis and follow-up studies in Drosophila and humans. Neuropsychopharmacology. 2015;40:361–371. doi: 10.1038/npp.2014.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Wang JC, Wetherill L, Le N, Bertelsen S, Hinrichs AL, Budde J, Agrawal A, Bucholz K, Dick D, Harari O, Hesselbrock V, Kramer J, Nurnberger JI, Jr, Rice J, Saccone N, Schuckit M, Tischfield J, Porjesz B, Edenberg HJ, Bierut L, Foroud T, Goate A. A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum Genet. 2013;132:1141–1151. doi: 10.1007/s00439-013-1318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Heath AC, Neale MC, Kessler RC, Eaves LJ. A population-based twin study of alcoholism in women. Jama. 1992;268:1877–1882. [PubMed] [Google Scholar]
- Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, Dick DM, Aliev F, Shi J, Levinson DF, Gejman PV. Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. Alcohol Clin Exp Res. 2011;35:963–975. doi: 10.1111/j.1530-0277.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 2012;8:e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos MZ, Yan J, Dick DM, Agrawal A, Bucholz KK, Rice JP, Johnson EO, Schuckit M, Kuperman S, Kramer J, Goate AM, Tischfield JA, Foroud T, Nurnberger J, Jr, Hesselbrock V, Porjesz B, Bierut LJ, Edenberg HJ, Almasy L. Common biological networks underlie genetic risk for alcoholism in African–and European–American populations. Genes Brain Behav. 2013;12:532–542. doi: 10.1111/gbb.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR. Topiramate treatment for heavy drinkers: moderation by a GRIK1 polymorphism. Am J Psychiatry. 2014;171:585–585. doi: 10.1176/appi.ajp.2013.13081014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR, Gelernter J, Anton RF, Arias AJ, Herman A, Zhao H, Burian L, Covault J. Association of markers in the 3' region of the GluR5 kainate receptor subunit gene to alcohol dependence. Alcohol Clin Exp Res. 2009;33:925–930. doi: 10.1111/j.1530-0277.2009.00913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES. The new genomics: global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, Ferreira T, Wood AR, Weyant RJ, Segrè AV, Speliotes EK, Wheeler E, Soranzo N, Park JH, Yang J, Gudbjartsson D, Heard-Costa NL, Randall JC, Qi L, Vernon Smith A, Mägi R, Pastinen T, Liang L, Heid IM, Luan J, Thorleifsson G, Winkler TW, Goddard ME, Sin Lo K, Palmer C, Workalemahu T, Aulchenko YS, Johansson A, Zillikens MC, Feitosa MF, Esko T, Johnson T, Ketkar S, Kraft P, Mangino M, Prokopenko I, Absher D, Albrecht E, Ernst F, Glazer NL, Hayward C, Hottenga JJ, Jacobs KB, Knowles JW, Kutalik Z, Monda KL, Polasek O, Preuss M, Rayner NW, Robertson NR, Steinthorsdottir V, Tyrer JP, Voight BF, Wiklund F, Xu J, Zhao JH, Nyholt DR, Pellikka N, Perola M, Perry JR, Surakka I, Tammesoo ML, Altmaier EL, Amin N, Aspelund T, Bhangale T, Boucher G, Chasman DI, Chen C, Coin L, Cooper MN, Dixon AL, Gibson Q, Grundberg E, Hao K, Juhani Junttila M, Kaplan LM, Kettunen J, König IR, Kwan T, Lawrence RW, Levinson DF, Lorentzon M, McKnight B, Morris AP, Müller M, Suh Ngwa J, Purcell S, Rafelt S, Salem RM, Salvi E, Sanna S, Shi J, Sovio U, Thompson JR, Turchin MC, Vandenput L, Verlaan DJ, Vitart V, White CC, Ziegler A, Almgren P, Balmforth AJ, Campbell H, Citterio L, De Grandi A, Dominiczak A, Duan J, Elliott P, Elosua R, Eriksson JG, Freimer NB, Geus EJ, Glorioso N, Haiqing S, Hartikainen AL, Havulinna AS, Hicks AA, Hui J, Igl W, Illig T, Jula A, Kajantie E, Kilpeläinen TO, Koiranen M, Kolcic I, Koskinen S, Kovacs P, Laitinen J, Liu J, Lokki ML, Marusic A, Maschio A, Meitinger T, Mulas A, Paré G, Parker AN, Peden JF, Petersmann A, Pichler I, Pietiläinen KH, Pouta A, Ridderstråle M, Rotter JI, Sambrook JG, Sanders AR, Schmidt CO, Sinisalo J, Smit JH, Stringham HM, Bragi Walters G, Widen E, Wild SH, Willemsen G, Zagato L, Zgaga L, Zitting P, Alavere H, Farrall M, McArdle WL, Nelis M, Peters MJ, Ripatti S, van Meurs JB, Aben KK, Ardlie KG, Beckmann JS, Beilby JP, Bergman RN, Bergmann S, Collins FS, Cusi D, den Heijer M, Eiriksdottir G, Gejman PV, Hall AS, Hamsten A, Huikuri HV, Iribarren C, Kähönen M, Kaprio J, Kathiresan S, Kiemeney L, Kocher T, Launer LJ, Lehtimäki T, Melander O, Mosley TH, Jr, Musk AW, Nieminen MS, O'Donnell CJ, Ohlsson C, Oostra B, Palmer LJ, Raitakari O, Ridker PM, Rioux JD, Rissanen A, Rivolta C, Schunkert H, Shuldiner AR, Siscovick DS, Stumvoll M, Tönjes A, Tuomilehto J, van Ommen GJ, Viikari J, Heath AC, Martin NG, Montgomery GW, Province MA, Kayser M, Arnold AM, Atwood LD, Boerwinkle E, Chanock SJ, Deloukas P, Gieger C, Grönberg H, Hall P, Hattersley AT, Hengstenberg C, Hoffman W, Lathrop GM, Salomaa V, Schreiber S, Uda M, Waterworth D, Wright AF, Assimes TL, Barroso I, Hofman A, Mohlke KL, Boomsma DI, Caulfield MJ, Cupples LA, Erdmann J, Fox CS, Gudnason V, Gyllensten U, Harris TB, Hayes RB, Jarvelin MR, Mooser V, Munroe PB, Ouwehand WH, Penninx BW, Pramstaller PP, Quertermous T, Rudan I, Samani NJ, Spector TD, Völzke H, Watkins H, Wilson JF, Groop LC, Haritunians T, Hu FB, Kaplan RC, Metspalu A, North KE, Schlessinger D, Wareham NJ, Hunter DJ, O'Connell JR, Strachan DP, Wichmann HE, Borecki IB, van Duijn CM, Schadt EE, Thorsteinsdottir U, Peltonen L, Uitterlinden AG, Visscher PM, Chatterjee N, Loos RJ, Boehnke M, McCarthy MI, Ingelsson E, Lindgren CM, Abecasis GR, Stefansson K, Frayling TM, Hirschhorn JN. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, Mowry BJ, Thapar A, Goddard ME, Witte JS, Absher D, Agartz I, Akil H, Amin F, Andreassen OA, Anjorin A, Anney R, Anttila V, Arking DE, Asherson P, Azevedo MH, Backlund L, Badner JA, Bailey AJ, Banaschewski T, Barchas JD, Barnes MR, Barrett TB, Bass N, Battaglia A, Bauer M, Bayés M, Bellivier F, Bergen SE, Berrettini W, Betancur C, Bettecken T, Biederman J, Binder EB, Black DW, Blackwood DH, Bloss CS, Boehnke M, Boomsma DI, Breen G, Breuer R, Bruggeman R, Cormican P, Buccola NG, Buitelaar JK, Bunney WE, Buxbaum JD, Byerley WF, Byrne EM, Caesar S, Cahn W, Cantor RM, Casas M, Chakravarti A, Chambert K, Choudhury K, Cichon S, Cloninger CR, Collier DA, Cook EH, Coon H, Cormand B, Corvin A, Coryell WH, Craig DW, Craig IW, Crosbie J, Cuccaro ML, Curtis D, Czamara D, Datta S, Dawson G, Day R, De Geus EJ, Degenhardt F, Djurovic S, Donohoe GJ, Doyle AE, Duan J, Dudbridge F, Duketis E, Ebstein RP, Edenberg HJ, Elia J, Ennis S, Etain B, Fanous A, Farmer AE, Ferrier IN, Flickinger M, Fombonne E, Foroud T, Frank J, Franke B, Fraser C, Freedman R, Freimer NB, Freitag CM, Friedl M, Frisén L, Gallagher L, Gejman PV, Georgieva L, Gershon ES, Geschwind DH, Giegling I, Gill M, Gordon SD, Gordon-Smith K, Green EK, Greenwood TA, Grice DE, Gross M, Grozeva D, Guan W, Gurling H, De Haan L, Haines JL, Hakonarson H, Hallmayer J, Hamilton SP, Hamshere ML, Hansen TF, Hartmann AM, Hautzinger M, Heath AC, Henders AK, Herms S, Hickie IB, Hipolito M, Hoefels S, Holmans PA, Holsboer F, Hoogendijk WJ, Hottenga JJ, Hultman CM, Hus V, Ingason A, Ising M, Jamain S, Jones EG, Jones I, Jones L, Tzeng JY, Kähler AK, Kahn RS, Kandaswamy R, Keller MC, Kennedy JL, Kenny E, Kent L, Kim Y, Kirov GK, Klauck SM, Klei L, Knowles JA, Kohli MA, Koller DL, Konte B, Korszun A, Krabbendam L, Krasucki R, Kuntsi J, Kwan P, Landén M, Långström N, Lathrop M, Lawrence J, Lawson WB, Leboyer M, Ledbetter DH, Lee PH, Lencz T, Lesch KP, Levinson DF, Lewis CM, Li J, Lichtenstein P, Lieberman JA, Lin DY, Linszen DH, Liu C, Lohoff FW, Loo SK, Lord C, Lowe JK, Lucae S, MacIntyre DJ, Madden PA, Maestrini E, Magnusson PK, Mahon PB, Maier W, Malhotra AK, Mane SM, Martin CL, Martin NG, Mattheisen M, Matthews K, Mattingsdal M, McCarroll SA, McGhee KA, McGough JJ, McGrath PJ, McGuffin P, McInnis MG, McIntosh A, McKinney R, McLean AW, McMahon FJ, McMahon WM, McQuillin A, Medeiros H, Medland SE, Meier S, Melle I, Meng F, Meyer J, Middeldorp CM, Middleton L, Milanova V, Miranda A, Monaco AP, Montgomery GW, Moran JL, Moreno-De-Luca D, Morken G, Morris DW, Morrow EM, Moskvina V, Muglia P, Mühleisen TW, Muir WJ, Müller-Myhsok B, Murtha M, Myers RM, Myin-Germeys I, Neale MC, Nelson SF, Nievergelt CM, Nikolov I, Nimgaonkar V, Nolen WA, Nöthen MM, Nurnberger JI, Nwulia EA, Nyholt DR, O'Dushlaine C, Oades RD, Olincy A, Oliveira G, Olsen L, Ophoff RA, Osby U, Owen MJ, Palotie A, Parr JR, Paterson AD, Pato CN, Pato MT, Penninx BW, Pergadia ML, Pericak-Vance MA, Pickard BS, Pimm J, Piven J, Posthuma D, Potash JB, Poustka F, Propping P, Puri V, Quested DJ, Quinn EM, Ramos-Quiroga JA, Rasmussen HB, Raychaudhuri S, Rehnström K, Reif A, Ribasés M, Rice JP, Rietschel M, Roeder K, Roeyers H, Rossin L, Rothenberger A, Rouleau G, Ruderfer D, Rujescu D, Sanders AR, Sanders SJ, Santangelo SL, Sergeant JA, Schachar R, Schalling M, Schatzberg AF, Scheftner WA, Schellenberg GD, Scherer SW, Schork NJ, Schulze TG, Schumacher J, Schwarz M, Scolnick E, Scott LJ, Shi J, Shilling PD, Shyn SI, Silverman JM, Slager SL, Smalley SL, Smit JH, Smith EN, Sonuga-Barke EJ, St Clai D, State M, Steffens M, Steinhausen HC, Strauss JS, Strohmaier J, Stroup TS, Sutcliffe JS, Szatmari P, Szelinger S, Thirumalai S, Thompson RC, Todorov AA, Tozzi F, Treutlein J, Uhr M, van den Oord EJ, Van Grootheest G, Van Os J, Vicente AM, Vieland VJ, Vincent JB, Visscher PM, Walsh CA, Wassink TH, Watson SJ, Weissman MM, Werge T, Wienker TF, Wijsman EM, Willemsen G, Williams N, Willsey AJ, Witt SH, Xu W, Young AH, Yu TW, Zammit S, Zandi PP, Zhang P, Zitman FG, Zöllner S International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) Devlin B, Kelsoe JR, Sklar P, Daly MJ, O’Donovan MC, Craddock N, Sullivan PF, Smoller JW, Kendler KS, Wray NR. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nature Publishing Group. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey DF, Le-Niculescu H, Frank J, Ayalew M, Jain N, Kirlin B, Learman R, Winiger E, Rodd Z, Shekhar A, Schork N, Kiefer F, Wodarz N, Müller-Myhsok B, Dahmen N GESGA Consortium. Nöthen M, Sherva R, Farrer L, Smith AH, Kranzler HR, Rietschel M, Gelernter J, Niculescu AB. Genetic risk prediction and neurobiological understanding of alcoholism. Transl Psychiatry. 2014;4:e391. doi: 10.1038/tp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, AMFS Investigators. Hayward NK, Montgomery GW, Visscher PM, Martin NG, MacGregor S. A versatile gene-based test for genome-wide association studies. American Journal of Human Genetics. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Pickens RW, Svikis DS. Sex and age effects on the inheritance of alcohol problems: a twin study. J Abnorm Psychol. 1992;101:3–17. doi: 10.1037//0021-843x.101.1.3. [DOI] [PubMed] [Google Scholar]
- McGue M, Zhang Y, Miller MB, Basu S, Vrieze S, Hicks B, Malone S, Oetting WS, Iacono WG. A genome-wide association study of behavioral disinhibition. Behav Genet. 2013;43:363–373. doi: 10.1007/s10519-013-9606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Kang HM, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan J, Lindgren CM, Müller-Nurasyid M, Pechlivanis S, Rayner NW, Scott LJ, Wiltshire S, Yengo L, Kinnunen L, Rossin EJ, Raychaudhuri S, Johnson AD, Dimas AS, Loos RJ, Vedantam S, Chen H, Florez JC, Fox C, Liu CT, Rybin D, Couper DJ, Kao WH, Li M, Cornelis MC, Kraft P, Sun Q, van Dam RM, Stringham HM, Chines PS, Fischer K, Fontanillas P, Holmen OL, Hunt SE, Jackson AU, Kong A, Lawrence R, Meyer J, Perry JR, Platou CG, Potter S, Rehnberg E, Robertson N, Sivapalaratnam S, Stančáková A, Stirrups K, Thorleifsson G, Tikkanen E, Wood AR, Almgren P, Atalay M, Benediktsson R, Bonnycastle LL, Burtt N, Carey J, Charpentier G, Crenshaw AT, Doney AS, Dorkhan M, Edkins S, Emilsson V, Eury E, Forsen T, Gertow K, Gigante B, Grant GB, Groves CJ, Guiducci C, Herder C, Hreidarsson AB, Hui J, James A, Jonsson A, Rathmann W, Klopp N, Kravic J, Krjutškov K, Langford C, Leander K, Lindholm E, Lobbens S, Männistö S, Mirza G, Mühleisen TW, Musk B, Parkin M, Rallidis L, Saramies J, Sennblad B, Shah S, Sigurðsson G, Silveira A, Steinbach G, Thorand B, Trakalo J, Veglia F, Wennauer R, Winckler W, Zabaneh D, Campbell H, van Duijn C, Uitterlinden AG, Hofman A, Sijbrands E, Abecasis GR, Owen KR, Zeggini E, Trip MD, Forouhi NG, Syvänen AC, Eriksson JG, Peltonen L, Nöthen MM, Balkau B, Palmer CN, Lyssenko V, Tuomi T, Isomaa B, Hunter DJ, Qi L, et al. Wellcome Trust Case Control Consortium; Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium. Shuldiner AR, Roden M, Barroso I, Wilsgaard T, Beilby J, Hovingh K, Price JF, Wilson JF, Rauramaa R, Lakka TA, Lind L, Dedoussis G, Njølstad I, Pedersen NL, Khaw KT, Wareham NJ, Keinanen-Kiukaanniemi SM, Saaristo TE, Korpi-Hyövälti E, Saltevo J, Laakso M, Kuusisto J, Metspalu A, Collins FS, Mohlke KL, Bergman RN, Tuomilehto J, Boehm BO, Gieger C, Hveem K, Cauchi S, Froguel P, Baldassarre D, Tremoli E, Humphries SE, Saleheen D, Danesh J, Ingelsson E, Ripatti S, Salomaa V, Erbel R, Jöckel KH, Moebus S, Peters A, Illig T, de Faire U, Hamsten A, Morris AD, Donnelly PJ, Frayling TM, Hattersley AT, Boerwinkle E, Melander O, Kathiresan S, Nilsson PM, Deloukas P, Thorsteinsdottir U, Groop LC, Stefansson K, Hu F, Pankow JS, Dupuis J, Meigs JB, Altshuler D, Boehnke M, McCarthy MI. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Nature Genetics. 2012;44:981–990. [Google Scholar]
- Olfson E, Bierut LJ. Convergence of genome-wide association and candidate gene studies for alcoholism. Alcohol Clin Exp Res. 2012;36:2086–2094. doi: 10.1111/j.1530-0277.2012.01843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BL, Kim JW, Cheong HS, Kim LH, Lee BC, Seo CH, Kang TC, Nam YW, Kim GB, Shin HD, Choi IG. Extended genetic effects of ADH cluster genes on the risk of alcohol dependence: from GWAS to replication. Hum Genet. 2013;132:657–668. doi: 10.1007/s00439-013-1281-8. [DOI] [PubMed] [Google Scholar]
- Peprah E, Xu H, Tekola-Ayele F, Royal CD. Genome-wide association studies in Africans and African Americans: expanding the framework of the genomics of human traits and disease. Public Health Genomics. 2015;18:40–51. doi: 10.1159/000367962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry. 1999;156:34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant…or not? Hum Mol Genet. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillen EE, Chen XD, Almasy L, Yang F, He H, Li X, Wang XY, Liu TQ, Hao W, Deng HW, Kranzler HR, Gelernter J. ALDH2 is associated to alcohol dependence and is the major genetic determinant of “daily maximum drinks” in a GWAS study of an isolated rural Chinese sample. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:103–110. doi: 10.1002/ajmg.b.32213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed T, Page WF, Viken RJ, Christian JC. Genetic predisposition to organ-specific endpoints of alcoholism. Alcohol Clin Exp Res. 1996;20:1528–1533. doi: 10.1111/j.1530-0277.1996.tb01695.x. [DOI] [PubMed] [Google Scholar]
- Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- Rietschel M, Treutlein J. The genetics of alcohol dependence. Ann N Y Acad Sci. 2013;1282:39–70. doi: 10.1111/j.1749-6632.2012.06794.x. [DOI] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, Lin DY, Duan J, Ophoff RA, Andreassen OA, Scolnick E, Cichon S, St Clair D, Corvin A, Gurling H, Werge T, Rujescu D, Blackwood DH, Pato CN, Malhotra AK, Purcell S, Dudbridge F, Neale BM, Rossin L, Visscher PM, Posthuma D, Ruderfer DM, Fanous A, Stefansson H, Steinberg S, Mowry BJ, Golimbet V, De Hert M, Jönsson EG, Bitter I, Pietiläinen OP, Collier DA, Tosato S, Agartz I, Albus M, Alexander M, Amdur RL, Amin F, Bass N, Bergen SE, Black DW, Børglum AD, Brown MA, Bruggeman R, Buccola NG, Byerley WF, Cahn W, Cantor RM, Carr VJ, Catts SV, Choudhury K, Cloninger CR, Cormican P, Craddock N, Danoy PA, Datta S, de Hann L, Demontis D, Dikeos D, Djurovic S, Donnelly P, Donohoe G, Duong L, Dwyer S, Fink-Jensen A, Freedman R, Freimer NB, Friedl M, Georgieva L, Giegling I, Gill M, Glenthøj B, Godard S, Hamshere M, Hansen M, Hansen T, Hartmann AM, Henskens FA, Hougaard DM, Hultman CM, Ingason A, Jablensky AV, Jakobsen KD, Jay M, Jürgens G, Kahn RS, Keller MC, Kenis G, Kenny E, Kim Y, Kirov GK, Konnerth H, Konte B, Krabbendam L, Krausucki R, Lasseter VK, Laurent C, Lawrence J, Lencz T, Lerer FB, Liang KY, Lichtenstein P, Lieberman JA, Linszen DH, Lönnqvist J, Loughland CM, Maclean AW, Maher BS, Maier W, Mallet J, Malloy P, Mattheisen M, Mattinsgsdal M, McGhee KA, McGrath JJ, McIntosh A, McLean DE, McQuillin A, Melle I, Michie PT, Milanova V, Morris DW, Mors O, Mortensen PB, Moskvina V, Muglia P, Myin-Germeys I, Nertney DA, Nestadt G, Nielsen J, Nikolov I, Nordentroft M, Norton N, Nöthen MM, O’Dushlaine CT, Olincy A, Olsen L, O’Neill FA, Ørntoft T, Owen MJ, Pantelis C, Papadimitriou G, Pato MT, Peltonen L, Petursson H, Pickard B, Pimm J, Pulver AE, Puri V, Quested D, Quinn EM, Rasmussen HB, Réthelyi JM, Ribble R, Rietschel M, Riley BP, Ruggeri M, Schall U, Schulze TG, Schwab SG, Scott RJ, Shi J, Sigurdsson E, Silverman JM, Spencer CC, Stefansson K, Strange A, Strengman E, Stroup TS, Suvisaari J, Tereniuis L, Thirumalai S, Thygesen JH, Timm S, Toncheva D, van den Oord E, van Os J, van Winkel R, Veldink J, Walsh D, Wang AG, Wiersma D, Wildenauer DB, Williams HJ, Williams NM, Wormley B, Zammit S, Sullivan PF, O’Donovan MC, Daly MJ, Gejman PV. Genome-wide association study identifies five new schizophrenia loci. Nature Genetics. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivières S, Aliev FA, Khan AA, Amin N, Aulchenko YS, Bakalkin G, Bakker SJ, Balkau B, Beulens JW, Bilbao A, de Boer RA, Beury D, Bots ML, Breetvelt EJ, Cauchi S, Cavalcanti-Proença C, Chambers JC, Clarke TK, Dahmen N, de Geus EJ, Dick D, Ducci F, Easton A, Edenberg HJ, Esko T, Fernández-Medarde A, Foroud T, Freimer NB, Girault JA, Grobbee DE, Guarrera S, Gudbjartsson DF, Hartikainen AL, Heath AC, Hesselbrock V, Hofman A, Hottenga JJ, Isohanni MK, Kaprio J, Khaw KT, Kuehnel B, Laitinen J, Lobbens S, Luan J, Mangino M, Maroteaux M, Matullo G, McCarthy MI, Mueller C, Navis G, Numans ME, Núñez A, Nyholt DR, Onland-Moret CN, Oostra BA, O’Reilly PF, Palkovits M, Penninx BW, Polidoro S, Pouta A, Prokopenko I, Ricceri F, Santos E, Smit JH, Soranzo N, Song K, Sovio U, Stumvoll M, Surakk I, Thorgeirsson TE, Thorsteinsdottir U, Troakes C, Tyrfingsson T, Tönjes A, Uiterwaal CS, Uitterlinden AG, van der Harst P, van der Schouw YT, Staehlin O, Vogelzangs N, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Whitfield JB, Wichmann EH, Willemsen G, Witteman JC, Yuan X, Zhai G, Zhao JH, Zhang W, Martin NG, Metspalu A, Doering A, Scott J, Spector TD, Loos RJ, Boomsma DI, Mooser V, Peltonen L, Stefansson K, van Duijn CM, Vineis P, Sommer WH, Kooner JS, Spanagel R, Heberlein UA, Jarvelin MR, Elliott P. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci. 2011;108:7119–7124. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, Edenberg HJ, Jr, Nurnberger JI, Rietschel M, Blackwood D, Corvin A, Flickinger M, Guan W, Mattingsdal M, McQuillen A, Kwan P, Wienker TF, Daly M, Dudbridge F, Holmans PA, Lin D, Burmeister M, Greenwood TA, Hamshere ML, Muglia P, Smith EN, Zandi PP, Nievergelt CM, McKinney R, Shilling PD, Schork NJ, Bloss CS, Foroud T, Koller DL, Gershon ES, Liu C, Badner JA, Scheftner WA, Lawson WB, Nwulia EA, Hipolito M, Coryell W, Rice J, Byerley W, McMahon FJ, Schulze TG, Berrettini W, Lohoff FW, Potash JB, Mahon PB, McInnis MG, Zöllner S, Zhang P, Craig DW, Szelinger S, Barrett TB, Breuer R, Meier S, Strohmaier J, Witt SH, Tozzi F, Farmer A, McGuffin P, Strauss J, Xu W, Kennedy JL, Vincent JB, Matthews K, Day R, Ferreira MA, O’Dushlaine C, Perlis R, Raychaudhuri S, Ruderfer D, Lee PH, Smoller JW, Li J, Absher D, Bunny WE, Barchas JD, Schatzberg AF, Jones EG, Meng F, Thompson RC, Watson SJ, Myers RM, Akil H, Boehnke M, Chambert K, Moran J, Scolnick E, Djurovic S, Melle I, Morken G, Gill M, Morris D, Quinn E, Mühleisen TW, Degenhardt FA, Mattheisen M, Schumacher J, Maier W, Steffans M, Propping P, Nöthen MM, Anjorin A, Bass N, Gurling H, Kandaswamy R, Lawrence J, McGhee K, McIntosh A, McLean AW, Muir WJ, Pickard BS, Breen G, St Clair D, Caesar S, Gordon-Smith K, Jones L, Fraser C, Green EK, Frozeva D, Jones IR, Kirov G, Moskvina V, Nikolov I, O’Donovan MC, Owen MJ, Collier DA, Elkin A, Williamson R, Young AH, Ferrier IN, Stefansson K, Stefansson H, Porgeirsson P, Steinberg S, Gustafsson Ó, Bergen SE, Nimgaonkar V, Hultman C, Landén M, Lichtenstein P, Sullivan P, Schalling M, Osby U, Backlund L, Frisén L, Langstrom N, Jamain S, Leboyer M, Etain B, Bellivier F, Petursson H, Sigurđsson E, Müller-Mysok B, Lucae S, Schwarz M, Fullerton JM, Schofield PR, Martin N, Montgomery GW, Lathrop M, Óskarsson H, Bauer M, Wright A, Mitchell PB, Hautzinger M, Reif A, Kelsoe JR, Purcell SM. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4 . Nature Genetics. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW, Ripke S, Lee PH, Neale B, Nurnberger JI, Santangelo S, Sullivan PF, Perlis RH, Purcell SM, Fanous A, Neale MC, Rietschel M, Schulze TG, Thapar A, Anney R, Buitelaar JK, Farone SV, Hoogendijk WJ, Levinson DF, Lesch KP, Riley B, Schachar R, Sonuga-Barke E, Absher D, Agartz I, Akil H, Amin F, Andreassen OA, Anjorin A, Arking D, Asherson P, Azevedo MH, Backlund L, Badner JA, Banaschewski T, Barchas JD, Barnes MR, Bass N, Bauer M, Bellivier F, Bergen SE, Berrettini W, Bettecken T, Biederman J, Binder EB, Black DW, Blackwood DH, Bloss CS, Boehnke M, Boomsma DI, Breen G, Breuer R, Buccola NG, Bunner WE, Burmeister M, Buxbaum JD, Byerley WF, Sian C, Cantor RM, Chakravarti A, Chambert K, Chicon S, Cloniger CR, Collier DA, Cook E, Coon H, Corvin A, Coryell WH, Craig DW, Craig IW, Curtis D, Czamara D, Daly M, Datta S, Day R, De Geus EJ, Degenhardt F, Devlin B, Srdjan D, Doyle AE, Duan J, Dudbridge F, Edenberg HJ, Elkin A, Etain B, Farmer AE, Ferreira MA, Ferrier IN, Flickinger M, Foroud T, Frank J, Franke B, Fraser C, Freedman R, Freimer NB, Friedl M, Frisén L, Gejman PV, Georgieva L, Gershon ES, Giegling I, Gill M, Gordon SD, Gordon-Smith K, Green EK, Greenwood TA, Gross M, Grozeva D, Guan W, Gurling H, Gustafsson Ó, Hakonarson H, Hamilton SP, Hamshere ML, Hansen TF, Hartmann AM, Hautzinger M, Heath AC, Henders AK, Herms S, Hickie IB, Hipolito M, Hoefels S, Holmans PA, Holsboer F, Hottenga JJ, Hultman CM, Ingason A, Ising M, Jamain S, Jones EG, Jones L, Jones I, Jung-Ying T, Kahler A, Kandaswamy R, Keller MC, Kelsoe JR, Kennedy JL, Kenny E, Kim Y, Kirov GK, Knowles JA, Kohli MA, Koller DL, Konte B, Korszun A, Krasucki R, Kuntsi J, Phoenix K, Landén M, Langstrom N, Lathrop M, Lawrence J, Lawson WB, Leboyer M, Lencz T, Lesch KP, Lewis CM, Li J, Lichtenstein P, Lieberman JA, Lin D, Liu C, Lohoff FW, Loo SK, Lucae S, MacIntyre D, Madden PA, Magnusson P, Mahon PB, Maier W, Malhotra AK, Mattheisen M, Matthews K, Mattingsdal M, McCarroll S, McGhee KA, McGough JJ, McGrath PJ, McGuffin P, McInnis MG, McIntosh A, McKinney R, McClean AW, McMahon FJ, McQuillin A, Medeiros H, Medland SE, Meier S, Melle I, Meng F, Middeldorp CM, Middleton L, Vihra M, Mitchell PB, Montgomery GW, Moran J, Morken G, Morris DW, Moskvina V, Mowry BJ, Muglia P, Mühleisen TW, Muir WJ, Müller-Myhsok B, Myers RM, Nelson SF, Nievergelt CM, Nikolovq I, Nimgaonkar V, Nolen WA, Nöthen MM, Nwulia EA, Nyholt DR, O'Donovan MC, O'Dushlaine C, Oades RD, Olincy A, Olsen L, Ophoff RA, Osby U, Óskarsson H, Owen MJ, Palotie A, Pato MT, Pato CN, Penninx BP, Pergadia ML, Petursson H, Pickard BS, Pimm J, Piven J, Porgeirsson P, Posthuma D, Potash JB, Propping J, Puri V, Quested D, Quinn EM, Rasmussen HB, Raychaudhuri S, Rehnström K, Reif A, Rice J, Rossin L, Rothenberger A, Rouleau G, Ruderfer D, Rujescu D, Sanders AR, Schalling M, Schatzberg AF, Schftner WA, Schellenberg G, Schofield PR, Schork NJ, Schumacher J, Schwarz MM, Scolnick E, Scott LJ, Shi J, Shillling PD, Shyn SI, Sigurdsson E, Silverman JM, Sklar P, Slager SL, Smalley SL, Smit JH, Smith EN, Sonuga-Barke E, St Clair D, State M, Stefansson K, Stefansson H, Steffans M, Steinberg S, Steinhausen HC, Strauss J, Strohmaier J, Stroup TS, Sutcliffe J, Szatmari P, Szelinger S, Thirumalai S, Thompson RC, Tozzi F, Treutlein J, Uhr M, van den Oord EJ, Van Grootheest G, Vieland V, Vincent JB, Visscher PM, Watson SJ, Weissman MM, Werge T, Wienker TF, Willemsen G, Williamson R, Witt SH, Wray NR, Wright A, Xu W, Young AH, Zammit S, Zandi PP, Zhang P, Zitman FG, Zöllner S, Craddock N, Kendler K. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013 doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Publishing Group. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Wegmann D, Trynka G, Gutierrez-Achury J, Do R, Voight BF, Kraft P, Chen R, Kallberg HJ, Kurreeman FA Diabetes Genetics Replication and Meta-analysis Consortium; Myocardial Infarction Genetics Consortium. Kathiresan S, Wijmenga C, Gregersen PK, Alfredsson L, Siminovitch KA, Worthington J, de Bakker PI, Raychaudhuri S, Plenge RM. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nature Genetics. 2012;44:483–489. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Bi J, Chan G, Oslin D, Farrer L, Gelernter J, Kranzler HR. Improved methods to identify stable, highly heritable subtypes of opioid use and related behaviors. Addict Behav. 2012;37:1138–1144. doi: 10.1016/j.addbeh.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Bi J, Kranzler HR. Multi-view singular value decomposition for disease subtyping and genetic associations. BMC Genet. 2014;15:73. doi: 10.1186/1471-2156-15-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F, Isono M, Nabika T, Katsuya T, Sugiyama T, Yamaguchi S, Kobayashi S, Ogihara T, Yamori Y, Fujioka A, Kato N. Confirmation of ALDH2 as a major locus of drinking behavior and of its variants regulating multiple metabolic phenotypes in a Japanese population. Circ J. 2011;75:911–918. doi: 10.1253/circj.cj-10-0774. [DOI] [PubMed] [Google Scholar]
- Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1999;48:677–681. [PMC free article] [PubMed] [Google Scholar]
- Thorleifsson G, Magnusson KP, Sulem P, Walters GB, Gudbjartsson DF, Stefansson H, Jonsson T, Jonasdottir A, Jonasdottir A, Stefansdottir G, Masson G, Hardarson GA, Petursson H, Arnarsson A, Motallebipour M, Wallerman O, Wadelius C, Gulcher JR, Thorsteinsdottir U, Kong A, Jonasson F, Stefansson K. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science. 2007;317:1397–1400. doi: 10.1126/science.1146554. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nöthen MM, Kiefer F, Spanagel R, Mann K, Rietschel M. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True WR, Xian H, Scherrer JF, Madden PA, Bucholz KK, Heath AC, Eisen SA, Lyons MJ, Goldberg J, Tsuang M. Common genetic vulnerability for nicotine and alcohol dependence in men. Arch Gen Psychiatry. 1999;56:655. doi: 10.1001/archpsyc.56.7.655. [DOI] [PubMed] [Google Scholar]
- Verhulst B, Neale MC, Kendler KS. The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol Med. 2014;45:1061–1072. doi: 10.1017/S0033291714002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, McGue M, Miller MB, Hicks BM, Iacono WG. Three mutually informative ways to understand the genetic relationships among behavioral disinhibition, alcohol use, drug use, nicotine use/dependence, and their co-occurrence: Twin biometry, GCTA, and genome-wide scoring. Behav Genet. 2013;43:97–107. doi: 10.1007/s10519-013-9584-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Foroud T, Hinrichs AL, Le NX, Bertelsen S, Budde JP, Harari O, Koller DL, Wetherill L, Agrawal A, Almasy L, Brooks AI, Bucholz K, Dick D, Hesselbrock V, Johnson EO, Kang S, Kapoor M, Kramer J, Kuperman S, Madden PA, Manz N, Martin NG, McClintick JN, Montgomery GW, Nurnberger JI, Jr, Rangaswamy M, Rice J, Schuckit M, Tischfield JA, Whitfield JB, Xuei X, Porjesz B, Heath AC, Edenberg HJ, Bierut LJ, Goate AM. A genome-wide association study of alcohol-dependence symptom counts in extended pedigrees identifies C15orf53. Molecular Psychiatry. 2013;18:1218–1224. doi: 10.1038/mp.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Jia P, Wolfinger RD, Chen X, Zhao Z. Gene set analysis of genome-wide association studies: Methodological issues and perspectives. Genomics. 2011;98:1–8. doi: 10.1016/j.ygeno.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetherill L, Kapoor M, Agrawal A, Bucholz K, Koller D, Bertelsen SE, Le N, Wang JC, Almasy L, Hesselbrock V, Kramer J, Nurnberger JI, Jr, Schuckit M, Tischfield JA, Xuei X, Porjesz B, Edenberg HJ, Goate AM, Foroud T. Family-based association analysis of alcohol dependence criteria and severity. Alcohol Clin Exp Res. 2013;38:354–366. doi: 10.1111/acer.12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nature Genetics. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Aliev F, Webb BT, Kendler KS, Williamson VS, Edenberg HJ, Agrawal A, Kos MZ, Almasy L, Nurnberger JI, Jr, Schuckit MA, Kramer JR, Rice JP, Kuperman S, Goate AM, Tischfield JA, Porjesz B, Dick DM. Using genetic information from candidate gene and genome-wide association studies in risk prediction for alcohol dependence. Addiction Biology. 2014;19:708–721. doi: 10.1111/adb.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Li C, Kranzler HR, Farrer LA, Zhao H, Gelernter J. Exploring the genetic architecture of alcohol dependence in African-Americans via analysis of a genomewide set of common variants. Hum Genet. 2014;133:617–624. doi: 10.1007/s00439-013-1399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Association of haplotypic variants in DRD2 ANKK1 TTC12 and NCAM1 to alcohol dependence in independent case control and family samples. Hum Mol Genet. 2007;16:2844–2853. doi: 10.1093/hmg/ddm240. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Haplotypic variants in DRD2 ANKK1 TTC12, and NCAM1 are associated with comorbid alcohol and drug dependence. Alcohol Clin Exp Res. 2008;32:2117–2127. doi: 10.1111/j.1530-0277.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nature Genetics. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. American Journal of Human Genetics. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, Malison RT, Li CS, Wang F, Zhang XY, Deng HW, Krystal JH, Zhang F, Luo X. Genome-wide association study of alcohol dependence implicates KIAA0040 on chromosome 1q. Neuropsychopharmacology. 2012;37:557–566. doi: 10.1038/npp.2011.229. [DOI] [PMC free article] [PubMed] [Google Scholar]