Abstract
In tumor cells, two factors are abnormally increased that contribute to metastatic bone disease: Runx2, a transcription factor that promotes expression of metastasis related and osteolytic genes; and IL-11, a secreted osteolytic cytokine. Here, we addressed a compelling question: Does Runx2 regulate IL-11 gene expression? We find a positive correlation between Runx2, IL-11 and TGFβ1, a driver of the vicious cycle of metastatic bone disease, in prostate cancer (PC) cell lines representing early (LNCaP) and late (PC3) stage disease. Further, like Runx2 knockdown, IL-11 knockdown significantly reduced expression of several osteolytic factors. Modulation of Runx2 expression results in corresponding changes in IL-11 expression. The IL-11 gene has Runx2, AP-1 sites and Smad binding elements located on the IL-11 promoter. Here, we demonstrated that Runx2-c-Jun as well as Runx2-Smad complexes upregulate IL-11 expression. Functional studies identified a significant loss of IL-11 expression in PC3 cells in the presence of the Runx2-HTY mutant protein, a mutation that disrupts Runx2-Smad signaling. In response to TGFβ1 and in the presence of Runx2, we observed a 30-fold induction of IL-11 expression, accompanied by increased c-Jun binding to the IL-11 promoter. Immunoprecipitation and in situ co-localization studies demonstrated that Runx2 and c-Jun form nuclear complexes in PC3 cells. Thus, TGFβ1 signaling induces two independent transcriptional pathways - AP-1 and Runx2. These transcriptional activators converge on IL-11 as a result of Runx2-Smad and Runx2-c-Jun interactions to amplify IL-11 gene expression that, together with Runx2, supports the osteolytic pathology of cancer induced bone disease.
Keywords: Runx2, Prostate Cancer, Bone Metastasis, IL-11 Gene
Tumor invasion into bone is associated with secretion of growth factors, cytokines and other signaling molecules by cancer cells that promote the vicious cycle of tumor and metastatic bone disease. Prostate cancer metastatic bone disease results in predominantly painful bone forming osteoblastic bone lesions, but it is now clinically recognized that osteolysis is also a component of the metastatic disease in the bone microenvironment during progression of prostate tumor growth [Morrissey et al., 2013; Sottnik and Keller, 2013]. Recent studies in mouse models have also identified the osteolytic component of prostate cancer tumor growth in bone [Akech et al., 2010; Dutta et al., 2014; Eswaraka et al., 2014; Fradet et al., 2013; Ortiz and Lin, 2012; Zhang et al., 2015]. TGFβ1 and PTHrP are considered the major factors driving this ‘vicious cycle’ of metastatic bone disease [Buijs et al., 2011; Massague, 2012; Weilbaecher et al., 2011]. While cancer cells that secrete PTHrP promote osteoclast differentiation, TGFβ1 stimulates cancer cells to express other osteolytic factors, including multiple matrix metalloproteinases [Dutta et al., 2014]. In the TGFβ1 pathway, phosphorylated receptor-regulated R Smads (Smad2/3) form complexes with the co-Smad Smad4, then, enter the nucleus to bind with other transcription factors, such as Runx2 and AP1 factors to regulate gene expression [Ikushima and Miyazono, 2010; Padua and Massague, 2009; Zaidi et al., 2002]. Similar to breast cancer cells, two deregulated osteoytic factors, the Runx2 transcription factor and the osteolytic cytokine IL-11, are highly expressed in metastatic prostate cancer cells [Akech et al., 2010; Pratap et al., 2008; Zhang et al., 2015]. Both these pathologic activities result in woven bone formation and loss of normal bone during progression of tumor growth.
Several interleukins are also secreted from different tumor cells and among these is the highly osteolytic cytokine IL-11 [Sottnik and Keller, 2013]. IL-11 is a cytokine secreted from mesenchymal, epithelial and tumor cells and it was found to be a valuable biomarker for tumor aggressiveness and prognosis in several cancers [Casimiro et al., 2012; Furuya et al., 2005; Necula et al., 2012; Xiang et al., 2012; Zurita et al., 2004]. IL-11 a plays an important role in inflammation, motility and invasion in cancer and also increases transcription of bone sialoprotein, linked with cancer metastasis to bone [Begley et al., 2008; Lay et al., 2012; Matsumura et al., 2014; Shen et al., 2012]. As an osteolytic factor, IL-11 stimulates the development and/or survival of osteoclast progenitor cells [Calon et al., 2012; McCoy et al., 2013]. IL-11 is one of four highly overexpressed genes in bone metastatic breast cancer cells [Kang et al., 2003] and it was strongly associated with resorptive activity in breast cancer cells; however, no mechanisms were established [Mendoza-Villanueva et al., 2011]. Potentially relevant to the osteoblastic lesions in PC tumors, IL-11 has been reported to have osteogenic properties in normal bone cells [Matsumoto et al., 2012]. Elevated levels of IL-11 and its receptor IL-11Rα have been observed in PC tumors [Campbell et al., 2001; Kamradt et al., 2007]. The cytokine is long known to be induced by TGFβ1 signaling, stimulating IL-11 gene transcription via induction of AP-1 factors [Kang et al., 2003; Matsumoto et al., 2012; Tang et al., 1998a]. However, transcriptional mechanisms contributing to its high level of expression in bone metastatic breast and prostate cells is understudied.
Previous studies have established that Runx2 mediates TGF/BMP signaling to promote metastatic bone disease by PC cells and contributes to expression of genes that promote both osteomimmetic and osteolytic properties of PC cells [Akech et al., 2010; van der Deen et al., 2010; Wan et al., 2012; Yang et al., 2001; Zhang et al., 2015]. In response to TGFβ1 Runx2 recruits Smads to unique subnuclear domains forming Runx2-Smad complexes [Javed et al., 2008; Zaidi et al., 2002; Zhang et al., 2000]. Runx2-Smad is critical in facilitating osteolytic disease resulting from orthotopic growth of human breast and prostate tumor cells in the mouse [Akech et al., 2010; Pratap et al., 2008; Zhang et al., 2015]. Studies have further demonstrated a striking reduction of bone resorption by Runx2 RNAi in vivo in the intratibial model of metastatic bone disease [Akech et al., 2010; Pratap et al., 2008]. The Runx2 transcription factor promotes tumor growth and metastatic bone disease through multiple mechanisms: direct transcriptional regulation of invasion-associated and bone homing genes (e.g., VEGF, MMPs, osteopontin, bone sialoprotein); increased transcription of TGFβ1-induced bone resorbing genes through Runx2-Smad signaling and Runx2-Gli complexes mediating IHH-PTHrP signaling [Pratap et al., 2008] promoting proliferation, motility, immortality of tumor cells and the disruption of normal acini [Leong et al., 2010; Pratap et al., 2009]. These findings showed that Runx2 is highly expressed in breast and prostate cancer cell lines that metastasize to bone and that it plays important roles in supporting the osteolytic disease associated with tumor growth in bone.
In this study, to further understand the observed impact of knockdown of Runx2 in reducing prostate cancer-induced osteolytic disease in vivo [Akech et al., 2010; Zhang et al., 2015], we examined Runx2 regulation of the IL-11 gene. These studies identify for the first time that two TGFβ1 signaling pathways, via Smad co-receptors and induced AP-1, converge on Runx2 through Runx2-Smad and Runx2-c-Jun complexes at SBE and AP-1 sites within the IL-11 proximal promoter. This cooperativity of two distinct Runx2 complexes greatly amplifies IL-11 gene expression in response to TGFβ1. Together, Runx2 and IL-11 are mediating TGFβ1-induced osteolytic disease in prostate cancer.
METHODS
CELL LINES AND CELL CULTURE
Three PC cell lines were used in these studies to model PC progression during tumor growth in bone. LNCaP cells that are lymph node, but not bone, metastatic were used as a control cell line; PC3-L cells that produce mixed osteolytic and osteblastic lesions and have low Runx2 levels; and PC3-H cells that have high Runx2 levels and that exhibit aggressive osteolytic disease in mouse models, followed by mixed osteolytic and osteblastic lesions. Microsatellite analyses carried out by the University of Vermont DNA Analysis Facility were used to identify the genotype as authentic LNCaP and/or PC3 cells [Zhang et al., 2015].
LNCaP cells and PC3-L cells were cultured in RPMI 1640 with 10% FBS, 10 mM non-essential amino acids, 2 mM L-glutamine and 1 mM sodium pyruvate. PC3-H cells were cultured in T-medium with 5% fetal bovine serum (FBS) [Huang et al., 2005]. All media were supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin. Cell culture media and supplements were obtained from Invitrogen, Carlsbad, CA, with the exception of FBS, which was obtained from Atlanta Biologicals, Norcross, GA.
TGFβ1 AND BMP2 TREATMENT
For experiments involving growth factor additions, sub-confluent cell layers were first cultured in 1% charcoal-stripped media (Life Technologies, Carlsbad, CA) for 24 h. Some cultures were treated with the TGFβ inhibitor SB431542 at 5 μM for 1 h pre-incubation prior to TGFβ1 treatment, where indicated. Treatment was for 24 h, with vehicle control (DMSO), porcine TGFβ1 (10 ng/ml), or BMP2 (100 ng/ml) (R&D Systems, Minneapolis, MN). Cells were then harvested for protein detection by Western blot and for mRNA levels by qPCR.
WESTERN BLOT ANALYSIS
Cells were lysed in RIPA buffer (50 mMTris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate) containing 25 mM MG132, EDTA-free cOmplete Protease Inhibitor Cocktail (Roche, Nutley, NJ) and 1mM PMSF. Proteins were resolved by SDS-PAGE and transferred to PVDF membranes (EMD Millipore, Billerica, MA). Membranes were incubated with mouse anti-Runx2 monoclonal (MBL International Corporation, Woburn, MA), rabbit anti-Smad2/3 (Cell Signaling Technology, Danvers, MA), or rabbit anti-phospho-Smad2, rabbit anti-phospho-Smad3, rabbit anti-c-Jun (Cell Signaling Technology, Danvers, MA), rabbit anti-cdk2 polyclonal antibody (Santa Cruz Biotechnology, Dallas, TX). Proteins were detected using Clarity™ Western ECL Substrate (Bio-Rad Laboratories, Hercules, CA).
REVERSE TRANSCRIPTION-QUANTITATIVE PCR (qPCR)
Total RNA was isolated from cells using Trizol reagent according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA) and then further purified using a DNA-Free RNA kit (Zymo Research, Irvine, CA). cDNA was synthesized using Superscript First-Strand Synthesis System (Invitrogen, Carlsbad, CA). qPCR was performed using SYBR Green Master Mix (Bio-Rad, Hercules, CA) and gene-specific primers shown in Table S1 in an ABI Prism 7000 real-time PCR instrument (Life Technologies, Carlsbad, CA). Data were normalized using human glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
IL-11 ELISA ASSAY
Supernatants collected from cell cultures were analyzed for secreted IL-11 expression using the Human IL-11 Quantikine kit (R&D Systems, Minneapolis, MN), according to manufacturer’s instructions. All samples were measured in duplicate and a biological replicate assay was carried out.
siRNA TRANSFECTION
Cells at approximately 60–70% confluency were transfected with 500 nM siRNA targeting Runx2 or with 500 nM non-silencing control siRNA (NS siRNA) using lipofectamine 2000 (Life Technologies, Carlsbad, CA) according to manufacturer’s instructions. “Control” transfections, in which cells were exposed to Lipofectamine 2000 only, were also included where indicated. mRNA and protein knockdown was assessed 48 h after transfection.
ADENOVIRUS INFECTION
For studies involving expression of Runx2 wild-type (WT) and mutant proteins, adenoviral vectors containing Runx2 WT-IRES-GFP, Runx2-HTY-IRES-GFP, and Runx2-ΔC-IRES-GFP, and GFP-only (infection-efficiency control) constructs under the control of the CMV5 promoter were delivered to low Runx2 expressing PC-L cells using previously described methods [Afzal et al., 2005]. Preparation and purification of virus were performed using the Adeno-X Maxi Purification Kit (Clonetech, Mountain View, CA), according to the manufacturer protocol. After infection, cells were washed twice with serum-free medium, incubated for 24 h, and prepared for downstream analysis or cultured for TGFβ1 treatment, as described above.
CHROMATIN IMMUMOPRECIPITATION (ChIP)
For ChIP experiments, approximately 1×107 PC3-H cells with high endogenous Runx2 expression were either single-crosslinked in 1% formaldehyde for 10 min at room temperature (RT) or dual-crosslinked in 2 mM disuccinimidyl glutarate (DSG) for 30 min with agitation at RT, washed with PBS and then treated with 1% formaldehyde for 10 min at RT with rotation. Formaldehyde crosslinking was halted by addition of 2.5 M Glycine to a final concentration of 125 mM. Crosslinked cells were then washed with ice-cold PBS, harvested by scraping, pelleted, and resuspended in nuclei isolation buffer A (50 mM HEPES pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% IGEPAL CA-630. 0.25% Triton-X100, Roche cOmplete protease inhibitor) for10 min at 4°C with constant rotation. Nuclei were then pelleted by centrifugation, washed in washing buffer B (10 mM Tris-HCL pH 8.0, 200 mM NaCl, 1 mM EDTA, 1 mM EGTA, Roche cOmplete protease inhibitor), collected by centrifugation and resuspended in lysis buffer C (10 mM Tris-HCL pH 8.0, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.1% sodium deoxycholate, 0.5% N-laurylsarcosine Roche cOmplete protease inhibitor). Samples were then sonicated for 7 min (formaldehyde crosslinking) or 15 min (dual-crosslinking) using a Covaris S220 focused-ultrasonicator (Covaris, Woburn, MA) to reduce chromatin to an average size of 0.3 kb. Chromatin was then incubated overnight with Runx2 antibody (M-70, Santa Cruz Biotechnology, Dallas, TX), rabbit c-Jun antibody (Cell Signaling Technology, Danvers, MA) or rabbit IgG (EMD Millipore, Billerica, MA) at 4°C followed by Protein A Dynabeads (Life Technologies, Carlsbad, CA). Immune complexes were washed three times with low salt buffer (20 mM Tris-HCl pH 8.0, 1 mM EDTA, 0.1% SDS, 1% Triton-X100, 150 mM NaCl) once with high salt buffer (20 mM Tris-HCl pH 8.0, 1 mM EDTA, 0.1% SDS, 1% Triton-X100, 500 mM NaCl) and once with TEN buffer (50 mM Tris-HCl pH 8.0, 1 mM EDTA, 50 mM NaCl) and eluted into elution buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS) at 55°C for 30 min with agitation. Chromatin was then treated with RNase A for 2 h followed by Proteinase K for 2 h at 55°C. DNA was then recovered by phenol/chloroform/isoamyl alcohol extraction followed by ethanol precipitation. Recovered DNA was then used as a template in qPCR reactions using primer pairs specific for each gene of interest (Table S2).
CO-IMUNOPRECIPITATION
PC3-H cells (TGFβ1 treated or not treated) were scraped into ice-cold PBS and pelleted by centrifugation. Cells were then lysed with lysis buffer (150 mM NaCl, 50 mM Tris, pH 8.0, 1% IGEPAL CA-630, EDTA-free cOmplete protease inhibitor cocktail (Roche, Nutley, NJ), 25 mM MG132) on ice for 20 min and centrifuged to pellet insoluble cellular debris. Runx2 M70 antibody (Santa Cruz Biotechnology, Dallas, TX) or rabbit IgG (EMD Millipore, Billerica, MA) was added to supernatant and incubated at 4°C overnight with agitation. Fifty μl protein A/G PLUS Agarose beads (Santa Cruz Biotechnology, Dallas, TX) were then added and incubated at 4°C for 3 h. Protein-antibody complexes were then collected by centrifugation, beads were washed four times with lysis buffer, resuspended in 2 x SDS sample buffer (Bio-Rad Laboratories, Hercules, CA), containing 5% β-mercaptoethanol and boiled for 5 min. immunoprecipated proteins were separated by SDS-PAGE and transferred to PVDF membranes for Western blot analysis.
IMMUNOFLUORESCENCE MICROSCOPY
Cells were grown on gelatin-coated coverslips and processed for immunofluorescence microscopy as described previously [Young et al., 2007]. Antibodies were diluted in 1% BSA in PBS. The following primary antibodies were used in immunofluorescence experiments: mouse monoclonal RUNX2 (MBL International Corporation, Woburn, MA), goat polyclonal Smad3 (Santa Cruz Biotechnology, Dallas, TX) and rabbit polyclonal c-Jun (described above). Secondary antibodies used were goat anti-mouse-IgG conjugated to Alexa Fluor 488, donkey anti-goat-IgG conjugated to Alexa Fluor 594, and goat anti-mouse-IgG conjugated to Alexa Fluor 594, all from Life Technologies, Carlsbad, CA. ProLong gold antifade reagent with DAPI (Life Technologies, Carlsbad, CA) was added to the cells before they were visualized. The cells were examined by fluorescence microscopy and images were captured and processed using Zen pro2011 (Carl Zeiss Microscopy, Thornwood, NY). Immunofluorescence signals were quantified by using the ImageJ software package (National Institutes of Health, Bethesda, MD).
STATISTICAL ANALYSIS
Data were expressed as the mean ± standard error of the mean (SEM). Statistically significant differences were determined using the Student’s t-test with Welch’s correction. p<0.05 was considered statistically significant.
RESULTS
IL-11 EXPRESSION RELATED TO TGFβ1 AND RUNX2 SIGNALING
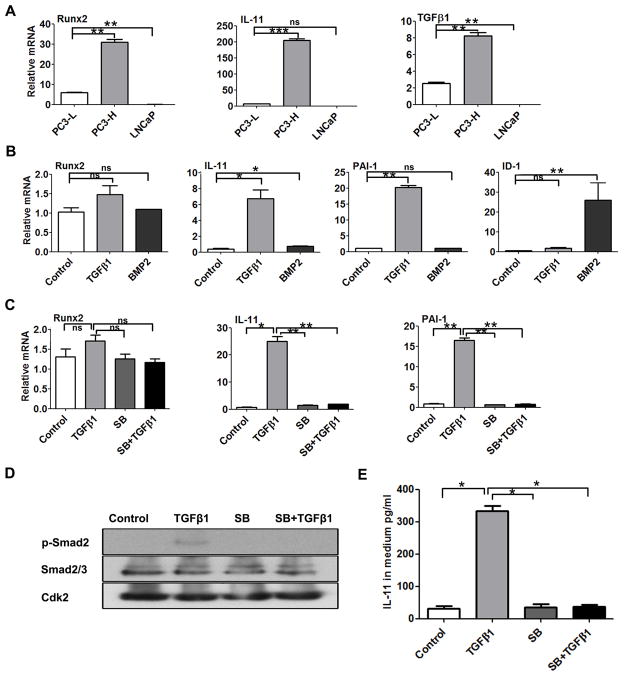
To begin to elucidate mechanisms involving both Runx2 and IL-11 in mediating prostate cancer metastatic disease, we initially characterized the relationship between Runx2 and expression of TGFβ1 and IL-11 among three prostate cancer cell lines, PC3-L, PC3-H, and LNCaP (Fig. 1). qPCR results showed high levels and a positive correlation between Runx2, IL-11 and TGFβ1 expression in PC3-H cells (known to have aggressive osteolytic properties in vivo). In contrast, PC3-L cells have significantly lower Runx2 and TGFβ1 levels and nearly undetectable IL-11 expression compared to PC-3H cells (Fig. 1A). Notably, LNCaP, a non-bone metastatic cell, exhibited no detectable levels of all three genes. Expression of Runx2 is 7-fold, IL-11 ~25-fold, and TGFβ1 4-fold higher in PC3-H than in PC3-L cells. We also confirmed expression of known osteolytic genes in these cells, RANKL and MMP2 (data not shown, as reported in [Dutta et al., 2014]), as well as PTHrP, CSF2, and CTGF (Supplementary Fig. S1). These genes also correlated with IL-11 and Runx2 expression in PC3-H cells.
Fig 1. IL-11 expression is related to TGFβ1 and Runx2 signaling.
(A) Robust expression of Runx2, IL-11 and TGFβ1 in highly osteolytic PC3-H cell line, with low to undetectable expression in PC3-L and LNCaP cells. Plots show relative gene expression of Runx2, IL-11 and TGFβ1 in PC3-L, PC3-H and non-metastatic LNCaP cell cultures determined by qPCR. (B) Response of low Runx2-expressing PC3-L cells (control) to TGFβ1 or BMP2 treatment. Sub-confluent PC3-L cells were cultured in 1% charcoal stripped FBS for 24 h before treatment with DMSO-control, TGFβ1, or BMP2, as indicated. Plots show relative gene expression of TGFβ1-regulated genes, Runx2, IL-11, PAI-1, and ID1. (C) Inhibition of TGFβ1 signaling in PC3-L cells. Sub-confluent PC3-L cells were cultured in 1% charcoal stripped FBS for 24 h before treatment with DMSO-control, TGFβ1, TGFβ1 inhibitor SB431542 (SB),m and SB431542 pre-treatment, followed by TGFβ1 (SB + TGFβ1). Plots show relative gene expression of Runx2, IL-11, and PA-I-1. Data were normalized to GAPDH and are mean ± SEM of n=3 experiments analyzed in duplicate. (D) Western blot analysis of phospho-Smad2, (p-Smad2), Smad2/3, and Cdk2 in PC3-L cells treated as indicated. (E) ELISA analysis of IL-11 levels in culture media from cells treated as indicated. Values are mean ± SEM of n=3 experiments analyzed in duplicate. Data in panels A–C and E were statistically analyzed using the T-test with Welch’s correction: not significant (ns), *p<0.05, **p<0.01, ***p<0.001.
TGFβ1 and BMPs are secreted during bone resorption and increase tumor cell secretion of osteolytic factors and osteoblastic factors that stimulate stromal cells to cause pathologic bone formation. The response of IL-11 and Runx2 to TGFβ1 and BMP2 was examined in PC3-L cells using molecular markers: PAI-1 for TGFβ1 activity and ID1, a target gene of BMP2 stimulation (Fig. 1B). TGFβ1 modestly increased Runx2 expression in PC3-L cells (1.5-fold). In striking contrast, IL-11 gene expression was induced significantly by TGFβ1 (7-fold), although not as high as the PAI-1 marker gene of TGFβ activity (20-fold upregulated). BMP2 treatment had no significant effect on Runx2 or IL-11 compared to non-treated PC3-L cells with the exception of ID1, as expected (Fig. 1B). We find that TGFβ-induced expression of IL-11 and PAI-1 is almost completely inhibited by the TGFβ1 inhibitor SB431542 (in the presence or absence of exogenous TGFβ1 treatment) (Fig. 1C). In contrast, the same inhibitor had no significant effect on Runx2 expression. In addition, Western blot results showed that TGFβ1-induced phosphorylation of its co-receptor Smad2, which was completely inhibited by SB431542 (Fig. 1D). Furthermore, an ELISA assay showed secretion of IL-11 in response to TGFβ1 was prevented by the inhibitor (Fig. 1E). Together these findings show a positive correlation between IL-11, Runx2 and TGFβ1 expression, with robust induction of IL-11, but only a modest increase in Runx2 in response to TGFβ1.
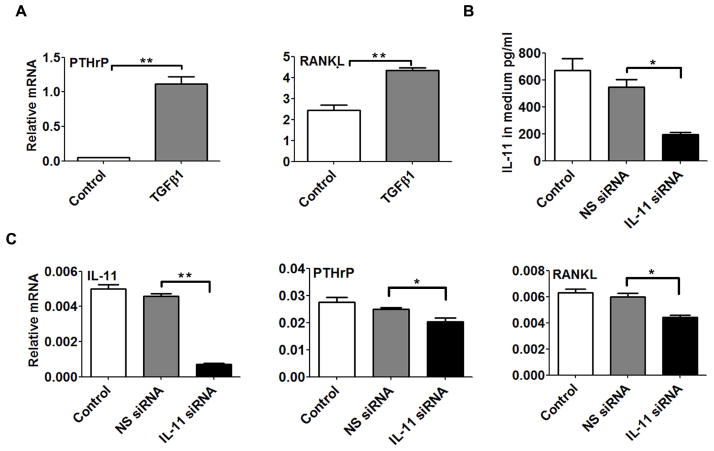
EXPRESSION OF OSTEOLYTIC GENES PTHrP AND RANKL RESPOND TO CHANGES IN IL-11
To further investigate the role of IL-11 in the context of the ‘vicious cycle’ of osteolytic bone disease, we addressed whether changes in IL-11 affected the expression of PTHrP and RANKL. Consistent with the finding that IL-11 is induced by TGFβ1 in PC3-L cells (Fig. 1B), both PTHrP and RANKL expression can also be induced by TGFβ1 in the same cells (Fig. 2A). To determine the relationship between IL-11 expression and these osteolytic factors, IL-11 was selectively inhibited by RNA interference in PC3-H cells, which express high endogenous IL-11 levels (Fig. 2B). Effective knockdown (60–80%) of IL-11 was confirmed by assaying secreted IL-11 (Fig. 2B, left panel) and IL-11 mRNA expression (Fig. 2B, right panel). As a result, the expression of the osteolytic factors PTHrP and RANKL were significantly decreased upon IL-11 depletion (p<0.05) (Fig. 2C). Thus, in support of the hypothesized functional relationship between Runx2 and IL-11, we find that the same osteolytic genes increased by Runx2 transcription, PTHrP and RANKL, are also responsive to IL-11 knockdown. These results suggest a functional cooperation between Runx2 and IL-11 mediated by TGFβ1 in advanced prostate cancer cell lines.
Fig. 2. IL-11 regulates expression of osteolytic genes PTHrP and RANKL in PC3 cells.
(A) Effect of TGFβ1 treatment on osteolytic gene expression in PC3-L cells. Relative MRNA expression of PTHrP and RANKL was determined using qPCR in PC3-L cells treated with DMSO-control or TGFβ1. (B) Secretion of IL-11 protein measured by testing a sample of the culture medium using ELISA following IL-11 knockdown using siRNA (NS siRNA: non-silencing control siRNA). (C) Expression of IL-11, PTHrP and RANKL mRNA following IL-11 knockdown in PC3-H cells compared to controls. Data in panels A and C were normalized to GAPDH and were statistically analyzed using the Student’s t-test with Welch’s correction: not significant (ns), * p<0.05, ** p<0.01.
IL-11 EXPRESSION IS RELATED TO RUNX2-SMAD SIGNALING IN PROSTATE CANCER PC3 CELLS
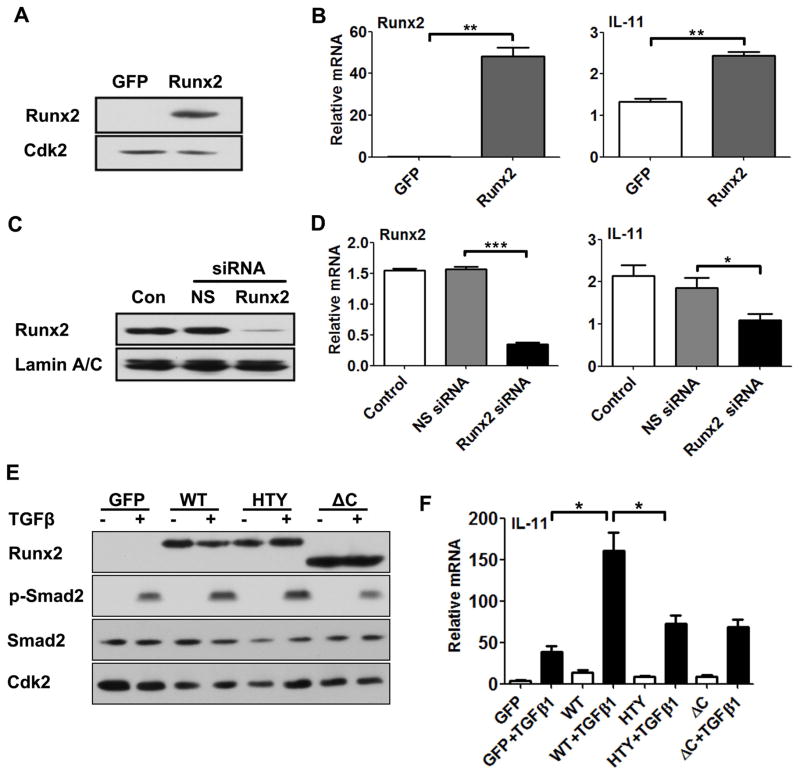
To address the functional relationship between Runx2 and IL-11 gene expression, we first determined if Runx2-regulated IL-11. Runx2 was exogenously expressed in PC3-L cells (having low endogenous Runx2 levels) by adenovirus infection (Fig. 3A). The results showed that Runx2 can activate IL-11 gene expression in PC3-L cells (Fig. 3B). We then selectively inhibited Runx2 expression by RNA interference in PC3-H cells (Fig. 3C). Accordingly, Runx2 protein expression was almost completely depleted, along with an 80% reduction in Runx2 mRNA levels. This knockdown of Runx2 reduced IL-11 gene expression by 40–50%, again supporting a potential role for Runx2 in regulating IL-11 (Fig. 3D).
Fig. 3. Modulation of Runx2 expression and activity alters IL-11 mRNA levels and response to TGFβ1.
A) Ectopic expression of Runx2 in PC3-L cells. PC3-L cells were infected with adenoviral GFP (control) and wildtype Runx2 expression constructs. Western blot analysis is shown, with Cdk2 as a loading control. (B) Gene expression analysis of Runx2 and induced IL-11 in PC3-L cells with overexpressed Runx2 GFP control, relative to GAPDH. (C–D) Runx2 knockdown in PC3-H cells. (C) Western blot analysis of Runx2 protein in nuclear extracts 48 h after mock transfection (Con), and transfection with siRNA targeting (Runx2) or a non-silencing (NS) control siRNA. Lamin A/C was used as loading control. (D) Corresponding qPCR analysis for Runx2 and IL-11 expression. (E–F) Effect of Runx2 mutations on TGFβ1 signaling (E) and IL-11 expression (F) in PC3-L cells. (E) Western blot analysis of Runx2, p-Smad2 and total Smad2 following infection with adenovirus expressing GFP, Runx2 WT, Runx2–HTY and ΔC mutants in the presence (+) and absence (−) of TGFβ1 treatment. Cdk2 was used as internal control. (F) Corresponding analysis of IL-11 gene expression using qPCR. Data in panels B, D and F were normalized to GAPDH. Values are mean ±SEM of n=3 experiments analyzed in duplicate. Data were statistically analyzed using the Student’s t-test with Welch’s correction: *p<0.05, **p<0.01***p<0.001.
Our previous studies showed that Runx2 enhances the TGFβ1 response in cancer cells through formation of a Runx2-SMAD complex (reviewed in Introduction). To determine whether this mechanism is also involved in IL-11 gene regulation, we utilized constructs expressing either wild-type Runx2 or two mutations (Runx2-HTY and Runx2-ΔC) that abrogate Runx2-Smad signaling [Zhang et al., 2015]. The two Runx2 mutant proteins retain DNA binding domain, but are not fully functional [Zhang et al., 2015]. Importantly, TGFβ1 treatment efficiently induced phosphorylation of Smad2 in cells expressing WT Runx2 and the mutant proteins (Fig. 3E). When the Runx2 mutant proteins were overexpressed in the PC3-L cells, IL-11 gene expression was reduced by 50% compared to Runx2 WT (Supplementary Fig. S2 and Fig. 3F). In these same cells, in the presence of exogenous expression of WT Runx2, IL-11 expression increased ~8-fold. However, exogenous expression of Runx2-HTY and −ΔC mutant proteins exhibited a 50% reduction in TGFβ1-stimulated activation of the IL-11 gene when compared with controls (Fig. 3F). These data strongly implicate TGFβ-Runx2-Smad signaling in regulation of IL-11 gene expression in advanced prostate cancer cells, in addition to the known TGFβ-AP1 pathway that increases IL-11 expression in numerous cell types.
RUNX2 AND C-JUN FORM A COMPLEX FOR UPREGULATION OF IL-11 GENE EXPRESSION
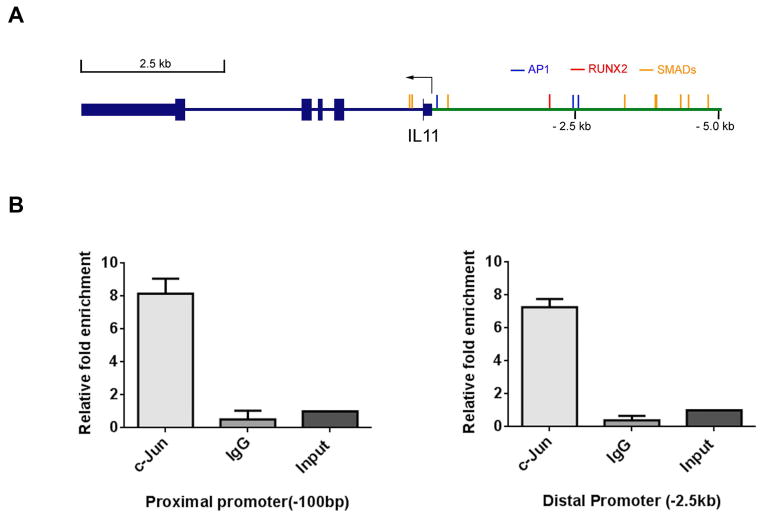
The above findings prompted an examination of direct binding of Runx2 in the IL-11 gene promoter, but only a single Runx2 site resides in the proximal promoter in the vicinity of two previously characterized AP-1 sites (Fig. 4A). Multiple Smad binding elements and several additional AP-1 consensus sites are located within 5 kb of the IL-11 promoter region. Even after optimizing several experimental conditions (e.g., testing of two lots of M-70 Runx2 antibody from Santa Cruz Biotechnology, Dallas, TX, lot #s E2410 and F0112), we could not detect direct Runx2 binding to the IL-11 gene. Supplementary Fig. S3 displays the absence of Runx2 binding at several promoter regions enriched in Smad sites. Therefore, we considered the possibility of a Runx2-c-Jun complex involved in regulating IL-11 gene expression. Several members of the AP-1 family of transcription factors are proven regulators of the IL-11 gene dependent on the cell type [Bamba et al., 2003; Tang et al., 1998b; Tohjima et al., 2003; Yang and Yang, 1994]. Previous studies showed that Runx2 can augment c-Jun activation of the AP-1 binding site in the collagenase-3 promoter [D’Alonzo et al., 2002; Hess et al., 2001] and also regulates PTH [Selvamurugan et al., 1998]. Chromatin immunoprecipitation (ChIP) was therefore pursued with a c-Jun antibody which confirmed c-Jun binding within the proximal, as well as in the distal IL-11 promoter regions (Fig. 4B).
Fig. 4. c-Jun binding in the IL-11 promoter region.
(A) Illustration of the IL-11 gene promoter region (+/− 5 kb). There is one Runx2 binding site ~2.5 kb upstream of the TSS, two nearby AP-1 sites and a third AP-1 site approximately 100 bp upstream of the TSS, as well as eight SMAD binding sites. (B) ChIP assay. Chromatin was immunoprecipitated with c-Jun or control IgG antibody and analyzed using qPCR analysis with primers for the AP-1 sites in the proximal (left) and distal (right) promoter regions.
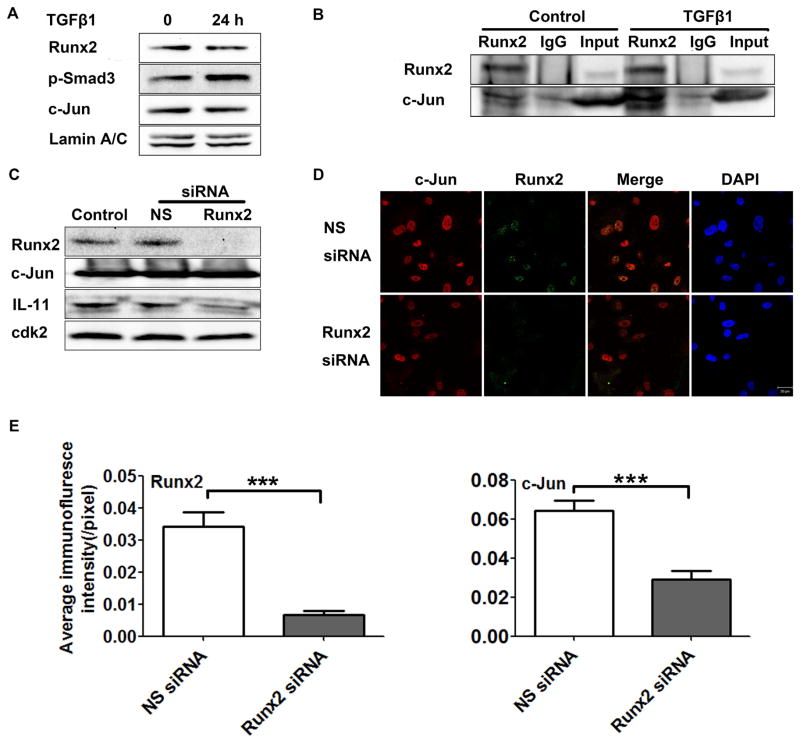
We further interrogated whether Runx2 regulated the IL-11 gene through direct interaction with c-Jun under TGFβ1 stimulation. In response to TGFβ1, protein levels of Runx2 and c-Jun remained unchanged; however, levels of phosphorylated Smad were noticeably increased demonstrating activation of TGFβ1 signaling (Fig. 5A). In PC3-H cells, with and without TGFβ1 treatment, we examined formation of a Runx2-c-Jun protein-protein complex. In Fig. 5B, results of a co-immunoprecipitation show an interaction between Runx2 and c-Jun in PC3-H cells that is amplified in the presence of TGFβ1. Validation that the Runx2-c-Jun complex contributes to IL-11 expression is shown by a decrease in IL-11 protein levels upon Runx2 depletion in the presence of constitutively high c-Jun protein (Fig. 5C). Data derived from immunofluorescence (IF) microscopy studies revealed nuclear co-localization of Runx2 and c-Jun in PC3-H cells, which was diminished by Runx2 knockdown (Fig. 5D). Quantifying the IF intensity corroborated these observations, with the presence of Runx2, as well as c-Jun, significantly decreasing after Runx2 depletion (Fig. 5E). Together these data provide compelling evidence for formation of a nuclear Runx2-c-Jun complex.
Fig. 5. Runx2- and TGFβ1-induced c-Jun form a complex in the nucleus.
(A) Western blot showing expression of Runx2, p-Smad3, and c-Jun in PC3-H cells at the indicated times in the presence or absence of TGFβ1 treatment. Lamin A/C was used as internal control. (B) Runx2 co-immunoprecipitation in PC3-H cells after TGFβ1 treatment. PC3-H cells were treated with TGFβ1 or DMSO-control, then immunoprecipitated using Runx2 or IgG control. The Western blot shown was probed with Runx2 and c-Jun antibodies, as indicated. (C–E) Runx2 silencing in PC3-H cells. (C) Western blot analyses of Runx2, c-Jun, IL-11 protein in non-transfected (control) cells or 48 h after transfection with Runx2 siRNA or a non-silencing (NS) control siRNA. Cdk2 was used as loading control. (D) Representative confocal images 48 h after delivery of Runx2 or a non-silencing (NS) control siRNA (Scale Bar = 20 μm). (E) Quantification of the immunofluorescence signal in panel D calculated using the ImageJ software package. Data are mean ±s.d, calculated using the average intensity of fewer random fields of view (~10 cells/field) in three independent experiments. Statistical analysis (T-test with Welch’s correction) was used to compare the values of cells transfected with NS siRNA or Runx2 siRNA: ***p<0.001.
The summation of our findings supported by biochemical, molecular and in situ cellular approaches, is that a circuitry exists in prostate tumor cells whereby TGFβ1 signals to: 1) AP-1 activity for induced levels of IL-11 by Runx2-c-Jun; and 2) the formation of Runx2-Smad complexes that induce osteolytic factors PTHrP, RANKL. These findings reveal the cooperativity of multiple transcriptional regulators of the IL-11 gene induced by TGFβ1 that contribute to metastatic bone disease.
DISCUSSION
IL-11 is a bone marrow stromal fibroblast-derived cytokine, highly induced by TGFβ1 through AP1 sites in the IL-11 promoter [Bamba et al., 2003; Tang et al., 1998b]. This inflammatory cytokine has long been associated with breast, and prostate and other tumors [Campbell et al., 2001]. Because of its role in invasion and as a secreted factor in breast cancer cells that contribute to osteolytic bone disease, we examined IL-11 functional activity in prostate cancer cells with the goal of determining regulation of the IL-11 gene by Runx2 as a mechanism for increased IL-11 expression in metastatic PC3 cells. In addition, it was also necessary to identify the contribution of IL-11 to osteolytic disease. In this study, we show by IL-11 expression profiles in PC cell lines and by specific knockdown that IL-11 promotes expression of several osteolytic factors (e.g., Runx2, RANKL, CSF2, CTGF, PTHrP, VEGF) which are also genes directly regulated by Runx2-co-regulatory protein complexes. Importantly we have demonstrated that IL-11 induction in prostate cancer cells is mediated through two TGFβ1-induced pathways, Smad and AP-1, resulting in formation of Runx2-Smad and Runx2-c-Jun interactions that, together, amplify expression of IL-11. Therefore, we clearly demonstrate that increased IL-11 expression, together with Runx2, is contributing to augmented expression of osteolytic genes in prostate cancer cells.
The Runx2 transcription factor is involved in mediating the ‘vicious cycle’ of metastatic bone disease [Pratap et al., 2011]. Our key findings have provided evidence for a TGFβ1-Runx2-IL-11 regulatory pathway that affects expression of the osteolytic drivers PTHrP and RANKL [Weilbaecher et al., 2011]. Supporting evidence includes: 1) a positive correlation between IL-11, Runx2 and TGFβ1 in the aggressive PC3-H cell line; 2) the barely detectable IL-11, low TGFβ1 and Runx2 levels in metastatic PC3-L cells that produce mainly osteoblastic lesions; and 3) the near absence of these factors in LNCaP non-bone metastatic cells. Moreover, a synergistic effect of TGFβ1 on Runx2 and IL-11 in promoting osteolytic bone disease was evidenced by the following observations: TGFβ1 has a minimal effect in modifying Runx2 levels but a significant fold induction of IL-11. PC3-L cells exposed to exogenous Runx2 causes IL-11 to become induced 2.5-fold, but importantly, in the presence of both TGFβ1 and Runx2, these cells exhibit in a 30-fold induction of IL-11 compared to either TGFβ1 or Runx2 alone. Furthermore, knockdown of Runx2 WT protein in PC3-H cells or expression of Runx2 mutant proteins that disrupt Runx2-mediated TGFβ1 signaling in PC3-L cells, reduced IL-11 induction by TGFβ1. These in vitro-defined mechanisms are consistent with in vivo studies that demonstrated reduced osteolytic disease by expression of the Runx2-HTY mutant that is not responsive to TGFβ1 signaling [Zhang et al., 2015]. Thus, these data strongly implicate IL-11 as direct target gene of Runx2 and supports the notion of cooperative activities amongst TGFβ1 in prostate cancer cells.
Based on the finding that Runx2 expression increases and knockdown of Runx2 decreases IL-11 in PC3 cells, we performed studies to identify more precisely the mechanism by which Runx2 regulates IL-11. Upon examining Runx2 sites in the IL-11-gene, many were clustered in far distal regions of the gene, but only one consensus sequence occurred in the promoter. The IL-11 gene promoter (a region ≤ −5 kb upstream of the TSS) contains multiple AP-1 sites and Smad binding elements (SBEs). Given the well-established property of Runx2 as a scaffolding protein [Stein et al., 2004], we identified Runx2-Smad and Runx2-c-Jun complexes induced by TGFβ signaling that highly upregulated IL-11 gene expression. Although we demonstrated a functional relationship between Runx2 and IL-11, surprisingly, we did not detect direct binding of Runx2 in the IL-11 gene. This is in contrast to robust c-Jun binding in proximal and distal promoter regions. These results may be a consequence of weak Runx2 interaction at a single site or possibly epitope masking due to the formation of a Runx2 multimeric complex with co-regulatory factors. Large Runx2 complexes can include histone modifying enzymes, co-activators and co-repressors, and other co-regulators associated with cancer, such as AP-1, c-myc, p53, mediators of src signaling, HOXa10, PBX, WWOX, TLE and ETS-1 [Ali et al., 2012; Aqeilan et al., 2007; Gordon et al., 2010; Pratap et al., 2011; Schroeder et al., 2005; Yang et al., 2001; Yang et al., 2010]. Thus, although a Runx2 ChIP could not detect Runx2 binding to IL-11, all other evidences [Bamba et al., 2003] strongly indicate Runx2 regulation via Runx2-Smad complexes at SBEs. For example, mutant Runx2 proteins that disrupt TGFβ1 signaling, significantly reduce IL-11 expression and secretion. We have also demonstrated a Runx2-c-Jun nuclear complex in prostate cancer cells by co-IP and in situ IF, as well as c-Jun binding across the IL-11 promoter. Collectively, our findings of multiple levels of IL-11 gene regulation reveal an integrated network of TGFβ and Runx2 signaling, resulting in increased IL-11 gene expression that contributes to aggressive disease.
Both breast and prostate cancer-induced metastatic bone disease is a complex process involving interactions of tumor cells with stromal cells, osteoblasts, osteoclasts that destroy bone, resulting in disease morbidity and mortality [Sottnik and Keller, 2013; Weilbaecher et al., 2011]. TGFβ Receptor I Kinase Inhibitors are in clinical trials and drugs that inhibit osteoclastic bone resorption are clinically effective in reducing skeletal complications of malignancy, although individual patient variables must be considered [Iranikhah et al., 2014; Mathew and Brufsky, 2014]. There is a compelling need to develop novel therapeutic strategies that can block tumor cell endogenous and secreted factors that deregulate the normal homeostasis of bone [Deng et al., 2014]. From our studies, Runx2 and IL-11, each with far reaching consequences on the skeleton, may represent potential therapeutic targets, if specifically directed to the bone-homing tumor cell to prevent aggressive bone disease.
Supplementary Material
Acknowledgments
Contract grant sponsor: National Institutes of Health; Contract grant numbers: P01 CA082834, P01 CA140043, R37 DE012528
We thank all members of the laboratory for stimulating discussions and support throughout the course of these studies.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no Conflicts of Interest.
Additional Supporting Information may be found in the online version of this article at the publisher’s website.
References
- Afzal F, Pratap J, Ito K, Ito Y, Stein JL, van Wijnen AJ, Stein GS, Lian JB, Javed A. Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J Cell Physiol. 2005;204:63–72. doi: 10.1002/jcp.20258. [DOI] [PubMed] [Google Scholar]
- Akech J, Wixted JJ, Bedard K, van der Deen M, Hussain S, Guise TA, van Wijnen AJ, Stein JL, Languino LR, Altieri DC, Pratap J, Keller E, Stein GS, Lian JB. Runx2 association with progression of prostate cancer in patients: mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene. 2010;29:811–821. doi: 10.1038/onc.2009.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali SA, Dobson JR, Lian JB, Stein JL, van Wijnen AJ, Zaidi SK, Stein GS. A RUNX2-HDAC1 co-repressor complex regulates rRNA gene expression by modulating UBF acetylation. J Cell Sci. 2012;125:2732–2739. doi: 10.1242/jcs.100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqeilan RI, Trapasso F, Hussain S, Costinean S, Marshall D, Pekarsky Y, Hagan JP, Zanesi N, Kaou M, Stein GS, Lian JB, Croce CM. Targeted deletion of Wwox reveals a tumor suppressor function. Proc Natl Acad Sci U S A. 2007;104:3949–3954. doi: 10.1073/pnas.0609783104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamba S, Andoh A, Yasui H, Makino J, Kim S, Fujiyama Y. Regulation of IL-11 expression in intestinal myofibroblasts: role of c-Jun AP-1- and MAPK-dependent pathways. Am J Physiol Gastrointest Liver Physiol. 2003;285:G529–538. doi: 10.1152/ajpgi.00050.2003. [DOI] [PubMed] [Google Scholar]
- Begley LA, Kasina S, MacDonald J, Macoska JA. The inflammatory microenvironment of the aging prostate facilitates cellular proliferation and hypertrophy. Cytokine. 2008;43:194–199. doi: 10.1016/j.cyto.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijs JT, Stayrook KR, Guise TA. TGF-beta in the Bone Microenvironment: Role in Breast Cancer Metastases. Cancer Microenviron. 2011;4:261–281. doi: 10.1007/s12307-011-0075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Cespedes MV, Sevillano M, Nadal C, Jung P, Zhang XH, Byrom D, Riera A, Rossell D, Mangues R, Massague J, Sancho E, Batlle E. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CL, Jiang Z, Savarese DM, Savarese TM. Increased expression of the interleukin-11 receptor and evidence of STAT3 activation in prostate carcinoma. Am J Pathol. 2001;158:25–32. doi: 10.1016/S0002-9440(10)63940-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro S, Luis I, Fernandes A, Pires R, Pinto A, Gouveia AG, Francisco AF, Portela J, Correia L, Costa L. Analysis of a bone metastasis gene expression signature in patients with bone metastasis from solid tumors. Clin Exp Metastasis. 2012;29:155–164. doi: 10.1007/s10585-011-9438-0. [DOI] [PubMed] [Google Scholar]
- D’Alonzo RC, Selvamurugan N, Karsenty G, Partridge NC. Physical interaction of the activator protein-1 factors c-Fos and c-Jun with Cbfa1 for collagenase-3 promoter activation. J Biol Chem. 2002;277:816–822. doi: 10.1074/jbc.M107082200. [DOI] [PubMed] [Google Scholar]
- Deng X, He G, Liu J, Luo F, Peng X, Tang S, Gao Z, Lin Q, Keller JM, Yang T, Keller ET. Recent advances in bone-targeted therapies of metastatic prostate cancer. Cancer Treat Rev. 2014;40:730–738. doi: 10.1016/j.ctrv.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Li J, Lu H, Akech J, Pratap J, Wang T, Zerlanko BJ, FitzGerald TJ, Jiang Z, Birbe R, Wixted J, Violette SM, Stein JL, Stein GS, Lian JB, Languino LR. Integrin alphavbeta6 promotes an osteolytic program in cancer cells by upregulating MMP2. Cancer Res. 2014;74:1598–1608. doi: 10.1158/0008-5472.CAN-13-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaraka J, Giddabasappa A, Han G, Lalwani K, Eisele K, Feng Z, Affolter T, Christensen J, Li G. Axitinib and crizotinib combination therapy inhibits bone loss in a mouse model of castration resistant prostate cancer. BMC Cancer. 2014;14:742. doi: 10.1186/1471-2407-14-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradet A, Sorel H, Depalle B, Serre CM, Farlay D, Turtoi A, Bellahcene A, Follet H, Castronovo V, Clezardin P, Bonnelye E. A new murine model of osteoblastic/osteolytic lesions from human androgen-resistant prostate cancer. PLoS One. 2013;8:e75092. doi: 10.1371/journal.pone.0075092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya Y, Nishio R, Junicho A, Nagakawa O, Fuse H. Serum interleukin-11 in patients with benign prostatic hyperplasia and prostate cancer. Int Urol Nephrol. 2005;37:69–71. doi: 10.1007/s11255-004-6086-1. [DOI] [PubMed] [Google Scholar]
- Gordon JA, Hassan MQ, Saini S, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. Pbx1 represses osteoblastogenesis by blocking Hoxa10-mediated recruitment of chromatin remodeling factors. Mol Cell Biol. 2010;30:3531–3541. doi: 10.1128/MCB.00889-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J, Porte D, Munz C, Angel P. AP-1 and Cbfa/runt physically interact and regulate parathyroid hormone-dependent MMP13 expression in osteoblasts through a new osteoblast-specific element 2/AP-1 composite element. J Biol Chem. 2001;276:20029–20038. doi: 10.1074/jbc.M010601200. [DOI] [PubMed] [Google Scholar]
- Huang WC, Xie Z, Konaka H, Sodek J, Zhau HE, Chung LW. Human osteocalcin and bone sialoprotein mediating osteomimicry of prostate cancer cells: role of cAMP-dependent protein kinase A signaling pathway. Cancer Res. 2005;65:2303–2313. doi: 10.1158/0008-5472.CAN-04-3448. [DOI] [PubMed] [Google Scholar]
- Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- Iranikhah M, Stricker S, Freeman MK. Future of bisphosphonates and denosumab for men with advanced prostate cancer. Cancer Manag Res. 2014;6:217–224. doi: 10.2147/CMAR.S40151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A, Bae JS, Afzal F, Gutierrez S, Pratap J, Zaidi SK, Lou Y, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J Biol Chem. 2008;283:8412–8422. doi: 10.1074/jbc.M705578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamradt J, Jung V, Wahrheit K, Tolosi L, Rahnenfuehrer J, Schilling M, Walker R, Davis S, Stoeckle M, Meltzer P, Wullich B. Detection of novel amplicons in prostate cancer by comprehensive genomic profiling of prostate cancer cell lines using oligonucleotide-based arrayCGH. PLoS One. 2007;2:e769. doi: 10.1371/journal.pone.0000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Lay V, Yap J, Sonderegger S, Dimitriadis E. Interleukin 11 regulates endometrial cancer cell adhesion and migration via STAT3. Int J Oncol. 2012;41:759–764. doi: 10.3892/ijo.2012.1486. [DOI] [PubMed] [Google Scholar]
- Leong DT, Lim J, Goh X, Pratap J, Pereira BP, Kwok HS, Nathan SS, Dobson JR, Lian JB, Ito Y, Voorhoeve PM, Stein GS, Salto-Tellez M, Cool SM, van Wijnen AJ. Cancer-related ectopic expression of the bone-related transcription factor RUNX2 in non-osseous metastatic tumor cells is linked to cell proliferation and motility. Breast Cancer Res. 2010;12:R89. doi: 10.1186/bcr2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A, Brufsky A. Bisphosphonates in breast cancer. Int J Cancer. 2014 doi: 10.1002/ijc.28965. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Kuriwaka-Kido R, Kondo T, Endo I, Kido S. Regulation of osteoblast differentiation by interleukin-11 via AP-1 and Smad signaling. Endocr J. 2012;59:91–101. doi: 10.1507/endocrj.ej11-0219. [DOI] [PubMed] [Google Scholar]
- Matsumura H, Nakayama Y, Takai H, Ogata Y. Effects of interleukin-11 on the expression of human bone sialoprotein gene. J Bone Miner Metab. 2014 doi: 10.1007/s00774-014-0576-8. [DOI] [PubMed] [Google Scholar]
- McCoy EM, Hong H, Pruitt HC, Feng X. IL-11 produced by breast cancer cells augments osteoclastogenesis by sustaining the pool of osteoclast progenitor cells. BMC Cancer. 2013;13:16. doi: 10.1186/1471-2407-13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Villanueva D, Zeef L, Shore P. Metastatic breast cancer cells inhibit osteoblast differentiation through the Runx2/CBFbeta-dependent expression of the Wnt antagonist, sclerostin. Breast Cancer Res. 2011;13:R106. doi: 10.1186/bcr3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey C, Roudier MP, Dowell A, True LD, Ketchanji M, Welty C, Corey E, Lange PH, Higano CS, Vessella RL. Effects of androgen deprivation therapy and bisphosphonate treatment on bone in patients with metastatic castration-resistant prostate cancer: results from the University of Washington Rapid Autopsy Series. J Bone Miner Res. 2013;28:333–340. doi: 10.1002/jbmr.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necula LG, Chivu-Economescu M, Stanciulescu EL, Bleotu C, Dima SO, Alexiu I, Dumitru A, Constantinescu G, Popescu I, Diaconu CC. IL-6 and IL-11 as markers for tumor aggressiveness and prognosis in gastric adenocarcinoma patients without mutations in Gp130 subunits. J Gastrointestin Liver Dis. 2012;21:23–29. [PubMed] [Google Scholar]
- Ortiz A, Lin SH. Osteolytic and osteoblastic bone metastases: two extremes of the same spectrum? Recent Results Cancer Res. 2012;192:225–233. doi: 10.1007/978-3-642-21892-7_11. [DOI] [PubMed] [Google Scholar]
- Padua D, Massague J. Roles of TGFbeta in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- Pratap J, Imbalzano KM, Underwood JM, Cohet N, Gokul K, Akech J, van Wijnen AJ, Stein JL, Imbalzano AN, Nickerson JA, Lian JB, Stein GS. Ectopic runx2 expression in mammary epithelial cells disrupts formation of normal acini structure: implications for breast cancer progression. Cancer Res. 2009;69:6807–6814. doi: 10.1158/0008-5472.CAN-09-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Lian JB, Stein GS. Metastatic bone disease: role of transcription factors and future targets. Bone. 2011;48:30–36. doi: 10.1016/j.bone.2010.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Wixted JJ, Gaur T, Zaidi SK, Dobson J, Gokul KD, Hussain S, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Runx2 transcriptional activation of Indian Hedgehog and a downstream bone metastatic pathway in breast cancer cells. Cancer Res. 2008;68:7795–7802. doi: 10.1158/0008-5472.CAN-08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TM, Jensen ED, Westendorf JJ. Runx2: a master organizer of gene transcription in developing and maturing osteoblasts. Birth Defects Res C Embryo Today. 2005;75:213–225. doi: 10.1002/bdrc.20043. [DOI] [PubMed] [Google Scholar]
- Selvamurugan N, Chou WY, Pearman AT, Pulumati MR, Partridge NC. Parathyroid hormone regulates the rat collagenase-3 promoter in osteoblastic cells through the cooperative interaction of the activator protein-1 site and the runt domain binding sequence. J Biol Chem. 1998;273:10647–10657. doi: 10.1074/jbc.273.17.10647. [DOI] [PubMed] [Google Scholar]
- Shen Z, Seppanen H, Vainionpaa S, Ye Y, Wang S, Mustonen H, Puolakkainen P. IL10, IL11, IL18 are differently expressed in CD14+ TAMs and play different role in regulating the invasion of gastric cancer cells under hypoxia. Cytokine. 2012;59:352–357. doi: 10.1016/j.cyto.2012.04.033. [DOI] [PubMed] [Google Scholar]
- Sottnik JL, Keller ET. Understanding and targeting osteoclastic activity in prostate cancer bone metastases. Curr Mol Med. 2013;13:626–639. doi: 10.2174/1566524011313040012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein GS, Lian JB, van Wijnen AJ, Stein JL, Montecino M, Javed A, Zaidi SK, Young DW, Choi JY, Pockwinse SM. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene. 2004;23:4315–4329. doi: 10.1038/sj.onc.1207676. [DOI] [PubMed] [Google Scholar]
- Tang L, Guo B, van Wijnen AJ, Lian JB, Stein JL, Stein GS, Zhou GW. Preliminary crystallographic study of glutathione S-transferase fused with the nuclear matrix targeting signal of the transcription factor AML-1/CBF-alpha2. J Struct Biol. 1998a;123:83–85. doi: 10.1006/jsbi.1998.4016. [DOI] [PubMed] [Google Scholar]
- Tang W, Yang L, Yang YC, Leng SX, Elias JA. Transforming growth factor-beta stimulates interleukin-11 transcription via complex activating protein-1-dependent pathways. J Biol Chem. 1998b;273:5506–5513. doi: 10.1074/jbc.273.10.5506. [DOI] [PubMed] [Google Scholar]
- Tohjima E, Inoue D, Yamamoto N, Kido S, Ito Y, Kato S, Takeuchi Y, Fukumoto S, Matsumoto T. Decreased AP-1 activity and interleukin-11 expression by bone marrow stromal cells may be associated with impaired bone formation in aged mice. J Bone Miner Res. 2003;18:1461–1470. doi: 10.1359/jbmr.2003.18.8.1461. [DOI] [PubMed] [Google Scholar]
- van der Deen M, Akech J, Wang T, FitzGerald TJ, Altieri DC, Languino LR, Lian JB, van Wijnen AJ, Stein JL, Stein GS. The cancer-related Runx2 protein enhances cell growth and responses to androgen and TGFbeta in prostate cancer cells. J Cell Biochem. 2010;109:828–837. doi: 10.1002/jcb.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, Li ZG, Yingling JM, Yang J, Starbuck MW, Ravoori MK, Kundra V, Vazquez E, Navone NM. Effect of transforming growth factor beta (TGF-beta) receptor I kinase inhibitor on prostate cancer bone growth. Bone. 2012;50:695–703. doi: 10.1016/j.bone.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilbaecher KN, Guise TA, McCauley LK. Cancer to bone: a fatal attraction. Nat Rev Cancer. 2011;11:411–425. doi: 10.1038/nrc3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang ZL, Zeng ZC, Fan J, Tang ZY, Zeng HY. Expression of connective tissue growth factor and interleukin-11 in intratumoral tissue is associated with poor survival after curative resection of hepatocellular carcinoma. Mol Biol Rep. 2012;39:6001–6006. doi: 10.1007/s11033-011-1413-y. [DOI] [PubMed] [Google Scholar]
- Yang J, Fizazi K, Peleg S, Sikes CR, Raymond AK, Jamal N, Hu M, Olive M, Martinez LA, Wood CG, Logothetis CJ, Karsenty G, Navone NM. Prostate cancer cells induce osteoblast differentiation through a Cbfa1-dependent pathway. Cancer Res. 2001;61:5652–5659. [PubMed] [Google Scholar]
- Yang JC, Bai L, Yap S, Gao AC, Kung HJ, Evans CP. Effect of the specific Src family kinase inhibitor saracatinib on osteolytic lesions using the PC-3 bone model. Mol Cancer Ther. 2010;9:1629–1637. doi: 10.1158/1535-7163.MCT-09-1058. [DOI] [PubMed] [Google Scholar]
- Yang L, Yang YC. Regulation of interleukin (IL)-11 gene expression in IL-1 induced primate bone marrow stromal cells. J Biol Chem. 1994;269:32732–32739. [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Sullivan AJ, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Integration of Runx and Smad regulatory signals at transcriptionally active subnuclear sites. Proc Natl Acad Sci U S A. 2002;99:8048–8053. doi: 10.1073/pnas.112664499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Akech J, Browne G, Russell S, Wixted JJ, Stein JL, Stein GS, Lian JB. Runx2-smad signaling impacts the progression of tumor-induced bone disease. Int J Cancer. 2015;136:1321–1332. doi: 10.1002/ijc.29094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Yasui N, Ito K, Huang G, Fujii M, Hanai J, Nogami H, Ochi T, Miyazono K, Ito Y. A RUNX2/PEBP2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc Natl Acad Sci U S A. 2000;97:10549–10554. doi: 10.1073/pnas.180309597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita AJ, Troncoso P, Cardo-Vila M, Logothetis CJ, Pasqualini R, Arap W. Combinatorial screenings in patients: the interleukin-11 receptor alpha as a candidate target in the progression of human prostate cancer. Cancer Res. 2004;64:435–439. doi: 10.1158/0008-5472.can-03-2675. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.