Summary
Spherocytosis is one of the most common inherited disorders, yet presents with a wide range of clinical severity. While several genes have been found mutated in patients with spherocytosis, the molecular basis for the variability in severity of haemolytic anaemia is not entirely understood. To identify candidate proteins involved in haemolytic anaemia pathophysiology, we utilized a label-free comparative proteomic approach to detect differences in red blood cells (RBCs) from normal and β-adducin (Add2) knock-out mice. We detected seven proteins that were decreased and 48 proteins that were increased in β-adducin null RBC ghosts. Since haemolytic anaemias are characterized by reticulocytosis, we compared reticulocyte-enriched samples from phenylhydrazine-treated mice with mature RBCs from untreated mice. Among the 48 proteins increased in Add2 knockout RBCs, only 11 were also increased in reticulocytes. Of the proteins decreased in Add2 knockout RBCs, α-adducin showed the greatest intensity difference, followed by SLC9A1, the sodium-hydrogen exchanger previously termed NHE1. We verified these mass spectrometry results by immunoblot. This is the first example of SLC9A1deficiency in haemolytic anaemia and suggests new insights into the mechanisms leading to fragile RBCs.
Keywords: red blood cell, reticulocyte, adducin, SLC9A1, label-free proteomics
Spherocytosis is one of the most common inherited disorders with an incidence of 1:2500–1:5000 in populations of Northern European descent. These anaemias arise from abnormalities in the red blood cell (RBC) membrane skeleton, a network of proteins organized around a hexagonal array of spectrin tetramers crosslinked by short actin filaments (Bennett & Gilligan, 1993). The membrane skeleton is crucial to the RBC, providing both support and flexibility as cells move rapidly through the circulation and traverse narrow capillaries (Chasis & Mohandas, 1986; Mohandas & Chasis, 1993).
Defects in membrane skeleton proteins cause mild to severe haemolytic anaemia (Mohandas & Gallagher, 2008) and even hydrops fetalis (Birkenmeier & Barker, 2004). Most clinical cases of hereditary haemolytic anaemia due to membrane skeleton defects have been attributed to defects in ankyrin, spectrin, band 3, and protein 4·2 (Iolascon et al, 2003). However, fundamental issues remain unresolved surrounding the pathophysiology of these disorders. First, not all clinical cases are accounted for by known membrane skeleton defects. Secondly, the clinical variability of currently known RBC mutations suggests the presence of genetic modifiers of disease severity and outcomes (Iolascon et al, 2003). A deeper understanding of RBC proteins involved in haemolytic anaemia pathophysiology would clarify these issues.
Mouse models have contributed significantly to our understanding of haemolytic anaemia (Peters & Barker, 2001; Eber & Lux, 2004). Targeted disruptions or spontaneous mutations have been described in the genes for many of the well-studied components of the RBC membrane skeleton [for e.g., spectrin (Bodine et al, 1984), ankyrin (White et al, 1990), band 3 (Peters et al, 1996; Southgate et al, 1996), protein 4·2 (Peters et al, 1999)]. These models differ in disease severity and in their secondary effects (Birkenmeier & Barker, 2004). Thus, they represent a useful tool for identifying modifiers of hereditary haemolytic anaemia severity. The β-adducin knockout (Add2-KO) mouse is a model of compensated haemolytic anaemia (Gilligan et al, 1999). Adducin is present in human RBCs as a mixture of α/β heterodimers (Gardner & Bennett, 1986) while mouse RBCs also have the γ subunit at low levels (Gilligan et al, 1999; Muro et al, 2000). Add2-KO RBCs are completely deficient in β-adducin with reduced levels of α-adducin and increased levels of γ-adducin (Gilligan et al, 1999). Similar to patients with hereditary spherocytosis, RBCs from β-adducin null mice are osmotically fragile, spherocytic, and dehydrated compared with wild type. Finally, the Add2-KO and other mouse models have been under-utilized in that normal versus disease protein differences have not been globally and comprehensively analysed. This report is the first such comprehensive comparison and reveals new insights into the pathophysiology of haemolytic anaemia.
Past studies of protein changes in the RBC have been hampered by the limitations of methods for studying complex systems and limitations in detecting low abundance protein components. Newly developed mass spectrometry technology represents a promising alternative for analysing hereditary haemolytic anaemias. The RBC is particularly amenable to quantitative mass spectrometry due to its relatively limited protein complexity [approximately 600 proteins (Pasini et al, 2006)]. Interest has grown in label-free comparative approaches that would circumvent the need for metabolic labelling or chemical labels (i.e., ICAT, iTRAQ). The low level of RBC protein complexity makes it an attractive candidate for recently developed label-free quantitative approaches using shotgun liquid chromatography tandem mass spectrometry (LC-MS/MS). This approach uses information in LC-MS/MS spectra to identify the peptides present in a protease digest of a complex mixture and to estimate differences in abundance of peptides from their intensities in LC-MS spectra.
Here we demonstrate the usefulness of label-free analysis to comprehensively detect protein differences in RBCs from normal and Add2-KO mice. We have used the recently developed software package CRAWDAD (Chromatographic Retention Time Alignment and Warping for Differential Analysis of LC-MS Data) (Finney et al, 2008) to find significant changes in abundance between biological replicates of normal and Add2-KO mice. CRAWDAD was also used to identify differences in the protein composition of reticulocytes versus mature RBCs which enabled us to filter the results from the Add2-KO mice for proteins likely to be increased from reticulocytes. Comparative proteomics reveals differences in protein abundance which span a broad range of cellular processes, including a significant deficiency of SLC9A1 [solute carrier family 9 (sodium/hydrogen exchanger), member 1 (previously termed NHE1; gene symbol Slc9a1] in the Add2-KO RBC ghosts. Some of the detected differences represent candidate proteins involved in pathophysiology of hereditary haemolytic anaemia.
Materials and methods
Animals
Mice were bred at The Jackson Laboratory (Bar Harbor, ME, USA) in climate-controlled rooms (12-h light/dark cycle) and provided with acidified water and chow (NIH 5K52) ad libitum. The β-adducin null mice have been fully backcrossed (>20 generations) onto the C57BL/6J strain. All studies were performed on adult mice 6 weeks of age or older. Procedures were approved by the Jackson Laboratory Animal Care and Use Committee and by the Institutional Animal Care and Use Committee at the University of Washington.
Protein isolation from β-adducin (Add2) knock-out mice RBC samples
Whole blood samples (0·3–0·9 ml) were obtained from independent biological replicates of each genotype via cardiac puncture of anesthetized mice. Samples were collected into microfuge tubes containing 10% EDTA (approximately 55 μl per 0·3 ml blood) and mixed by inversion. All subsequent sample processing was standardized so as to minimize pre-analytical variability in the RBC proteome. Blood samples were passed through cellulose acetate (2 ml resin bed in 10 cc syringe) to remove contaminating white blood cells and platelets (Beutler et al, 1976). Fractions containing RBCs were pooled and washed three times with phosphate-buffered saline (PBS; 10 mmol/l NaCl, 155 mmol/l KCl, 10 mmol/l glucose, 1 mmol/l MgCl2, 2·5 mmol/l KHPO4, pH 7·4) to remove any remaining platelets and contaminating serum proteins. Purified RBCs were lysed hypotonically by adding 1 ml cold lysis buffer (5 mmol/l NaHPO4, 1 mmol/l EDTA, pH 7·4) and vortexing. Lysates were incubated on ice for 5 min with occasional vortexing and then centrifuged at 13·8 g, 4°C for 15 min. The haemoglobin-rich supernatant was removed by aspiration and the pellet (known as the RBC ghost) was resuspended in 1 ml lysis buffer followed by centrifugation. This was repeated for a total of five washes to remove residual haemoglobin. The final pellet was resuspended in lysis buffer to an approximate total volume of 100 μl. Protein concentrations were determined using the BioRad Protein Assay (BioRad Laboratories, Hercules CA, USA). Samples were aliquoted and stored at −80°C.
Protein isolation from reticulocyte-enriched samples
Whole blood samples enriched for reticulocytes were generated by treating healthy C57BL/6J mice with phenylhydrazine for 5 d (Chui et al, 1980). Nine d from the start of injections, independent whole blood samples were collected via cardiac puncture from the phenylhydrazine-treated mice and matched untreated controls. Reticulocyte counts were determined via microscopic examination of methylene blue stained samples. Red blood cells were purified from whole blood and RBC ghost protein isolated as described above.
Sample preparation for mass spectrometry analysis
Samples were first solubilized by mixing 1:1 with 0·4% RapiGest (Waters Corporation, Milford, MA, USA) in 50 mmol/l Ammonium Bicarbonate pH 7·8 and boiling at 99°C for approximately 2 min. Samples were then allowed to cool at room temperature for 2 min and centrifuged for 1 min at 8·2 g. Dithiothreitol was added to 5 mmol/l followed by mixing and incubation at 60°C for 30 min. Samples were cooled to room temperature, adjusted to 15 mmol/l Iodoacetamide, incubated in the dark at room temperature for 30 min, and then adjusted to 1 mmol/l CaCl2. Samples were proteolysed by adding trypsin (sequencing grade; Promega, Madison, WI, USA) at a ratio of 1:20 enzyme:protein (i.e., 1 μg trypsin: 20 μg protein) and incubating for 4 h with shaking at 37°C. Protein digests were adjusted to 100 mmol/l HCl, incubated at 37°C for 45 min, and centrifuged at 14000 rpm at 4°C for 10 min to remove any precipitates. The samples were then stored at −20°C.
Sample analysis on linear ion trap tandem mass spectrometer
Initially, Add2-KO RBC samples were comprehensively profiled for their constituent proteins using a Linear Ion-Trap Mass Spectrometer (ThermoElectron, Waltham, MA, USA). A chromatography column was prepared by pulling a 75 μm internal dimension (ID) fused silica capillary to a 5 μm ID tip using a Sutter Instruments P-2000 CO2 laser-based micropipette puller. The capillary was then packed with 40 cm of Jupiter Proteo C12 reverse-phase chromatography material (4 μm particle size, 90 Å pore size) using an in-house constructed pressure bomb and compressed helium gas. The column was then placed inline between a Surveyor high performance liquid chromatography (HPLC)/Autosampler and the mass spectrometer. For each sample, three replicate runs were performed, loading 0·5 μg per run of protein onto the microcapillary column using a conventional binary HPLC and autosampler. HPLC separation consisted of a gradient between Buffer A (95% water, 5% acetonitrile and 0·1% formic acid) and Buffer B (20% water, 80% acetonitrile, and 0·1% formic acid). The HPLC gradient was held constant for 15 min at 95% A during the loading of the sample from the autosampler to the microcapillary column. The buffer conditions were then changed from 5% B to 35% B over 80 min, followed by a 5-min wash at 80% B. The column was then re-equilibrated in 95% A for 20 min. As molecules were eluted from the microcapillary column, they were electrosprayed directly into the mass spectrometer using a distal 2·2 kV spray voltage. MS scans were acquired in profile mode, and up to 5 MS/MS scans were acquired with data-dependent acquisition per MS scan to annotate any MS changes detected.
For label-free comparative proteomic analysis, samples were profiled on a hybrid linear ion trap fourier-transform ion cyclotron resonance (FTICR) mass spectrometer (LTQ-FT Ultra, ThermoElectron). The high resolution of this instrument enables it to detect larger numbers of peptides, thus allowing for the identification of protein differences between samples based on an increased number of peptides. Sample runs were as described above except for the use of a Nano Acquity HPLC (Waters Corparation) prior to loading onto a 30 cm reverse phase column. HPLC separation consisted of a gradient between Buffer A (95% water and 0·1% formic acid) and Buffer B (100% acetonitrile and 0·1% formic acid). The HPLC gradient was held constant for 15 min at 95% A during the loading of the sample from the autosampler to the microcapillary column. The buffer conditions were then changed from 5% B to 35% B over 140 min, followed by a 15-min wash at 80% B. The column was then re-equilibrated in 95% A for 25 min.
Peptide and protein identification
For peptide and protein identification, fragmentation spectra acquired for replicate runs were pooled and searched using SEQUEST against a database containing Mus musculus sequences (IPI mouse release 3.43) and common contaminants. Data were searched using a precursor ion mass tolerance of ±3 Da with no enzyme specificity. To control the false discovery rate, search results from a shuffled decoy database were used to assign a q-value (Storey & Tibshirani, 2003) to each spectrum identification using the program Percolator, which uses a semi-supervised machine learning algorithm (Kall et al, 2007). Spectra matching to peptides with a q-value ≤0·01 were retained.
Label-free differential analysis
Protein differences between normal and β-adducin null RBCs were detected using the label-free differential analysis program CRAWDAD (Finney et al, 2008). Data sets for each genotype consisted of three technical replicates for each of two biological replicates for a total six μLC-MS runs. MS1 scans were binned on equally-spaced 1/24 m/z and 0·01 min intervals. Runs from these analyses were truncated to include only the signal-rich retention time regions between 35 and 115 min. μLC-MS runs were aligned and warped in the retention time domain to a common template run so that signals at a given m/z were compared at an equal retention time across all runs. The replicate runs from the two biological classes were compared at each (m/z, RT) bin using a t-test (independent variances between groups), and a ‘difference region’ feature defined when the P-value of the t-test was at or below 0·05 over at least 0·25′ (12 interpolated scans).
CRAWDAD associated MS/MS peptide identifications with difference regions in software when an MS/MS event from any run was within the retention time window of a difference region and within ±1 m/z of the difference region m/z bin for LTQ data. For LTQ-FT data, a more stringent mapping was done by limiting the mappings to those where the difference region's m/z corresponds to one of isotopes M+0 through M+2 of the identified peptide. Peptides were considered differentially expressed if mapped to a MS difference region as described above. MS/MS associations were made at a Percolator q-value of 0·01 or less. Differentially expressed peptides were organized into a hierarchical list grouped by proteins and the sample in which their relative abundance increased (see Supporting information tables for list of peptide differences). While it is likely that the measured ion current ratios will reflect the appropriate molar ratios of the respective peptides between samples, without validating the linear response for each analyte throughout the entire intensity range these data should be conservatively considered semi-quantitative or simply “different” unless demonstrated otherwise.
Immunoblotting to confirm differences revealed by mass spectrometry
Samples of RBC ghosts were prepared from three mice for each genotype (wild-type [wt] or Add2 knockout mice) using the method described above. RBC ghosts were solubilized with Laemmli sample buffer and stored at −80°C. For the Add1 knockout and Spnasph–4J RBC ghosts, samples were prepared as described (Robledo et al, 2010). RBC ghost proteins were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to nitrocellulose by semi-dry electroblotting. Nitrocellulose blots were blocked with 4% bovine serum albumin (BSA) before primary antibody was added. Blots were incubated at 4°C overnight or longer. After washing with PBS, blots were incubated with horseradish peroxidase conjugated to protein A (BioRad Laboratories) for one hour at room temperature, washed again, incubated with enhanced chemiluminescent reagents, and exposed to X-ray film. Rabbit polyclonal antibody to SLC9A1 was purchased from Santa Cruz Biotechnology (sc28758).
Results
RBC ghost proteome
The wild-type (wt) and Add2-KO murine RBC ghost proteomes were qualitatively profiled on the lower resolution LTQ-ion trap utilizing using six reverse-phase MS/MS runs per genotype. In total, 572 proteins were identified for the wt RBC ghost using SEQUEST and postprocessing with Percolator (peptide spectrum match q-value <0·005, ≥1 peptide per protein). Likewise, 710 proteins were identified for the Add2-KO RBC ghost (see supplementary Tables SV and SVI for complete lists of proteins identified). The difference in proteins identified (572 in wild-type versus 710 in Add2-KO) is due in part to the increased number of reticulocytes in the Add2-KO mouse (Gilligan et al, 1999).
Protein differences detected between wt and Add2-KO RBCs
MS data generated on the lower resolution LTQ-ion trap was initially used for label-free comparative analysis. This data set consisted of a total of six technical replicates per genotype (three technical replicates for each of two biological replicates). CRAWDAD alignment of the data is demonstrated in Fig 1 (see panel A). Successful comparison depends upon accurate chromatographic alignments of replicate runs, shown here for wt versus Add2-KO RBC ghosts. This analysis resulted in the identification of 38 protein differences, but the majority of the differences were based on a single peptide (data not shown). Thus, a second set of wt and Add2-KO RBC samples were profiled on a higher resolution LTQ-ion trap (LTQ-FT Ultra, see Materials and methods). A complete list of proteins identified in these samples is presented in supplementary data (see Tables SVII and VIII).
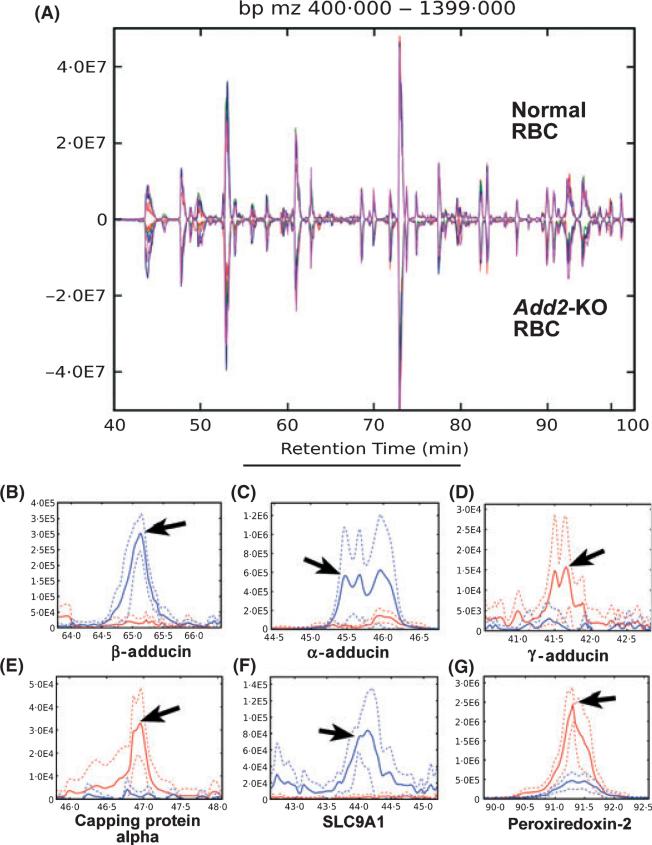
Fig 1.
Detection of red blood cell protein differences. (A) Chromatographic alignments of replicate runs for normal and Add2-KO red blood cells. Alignments of six technical replicate μLC-MS runs for normal (above) and Add2-KO red blood cells (below) shown in a base peak plot (ion intensity vs. retention time). Individual replicates are shown in distinct colours for each genotype. (B–G) Differentially expressed features revealed by chromatographic alignment. CRAWDAD detection of difference region for beta adducin peptide TRWLNTPNTYLRVNVADEVQR (B), alpha adducin peptide YFDRVDENNPEYLR (C), gamma adducin peptide IELQKVLGPSCK (D), capping protein (actin filament) muscle Z-line alpha 1 peptide EASDPQPEDVDGGLKSWR (E), SLC9A1peptide IGSDPLAYEPK (F), and peroxiredoxin-2 peptide KEGGLGPLNIPLLADVTK (G). In B – G, aligned replicate μLC-MS runs from the normal and Add2-KO RBC series are shown in blue and red, respectively. Mean intensity values are shown in solid lines and ± SD in dashed lines. Difference regions for each peptide indicated with arrows.
Examples of peptide differences detected between wt and Add2-KO RBC ghosts by CRAWDAD are shown in Fig 1B–G. These difference regions in the chromatograms are shown in blue for wt and red for the Add2-KO RBC ghosts. Panel B shows a peptide from β-adducin, missing in the knockout as expected. Panel C shows a peptide from α-adducin, the partner for β-adducin, also expected to be missing as shown by previous immunoblots (Gilligan et al, 1999). Panel D shows a peptide from γ-adducin, which we previously demonstrated by immunoblot to be increased in the Add2-KO RBC ghosts (Gilligan et al, 1999). Panel E shows a peptide from capping protein alpha, previously shown by immunoblot to be increased in the Add2-KO RBCs (Porro et al, 2004). Panel F shows a peptide from SLC9A1, the sodium hydrogen exchanger, demonstrated here for the first time to be greatly decreased in the Add2-KO RBC ghosts. Panel G shows a peptide for peroxiredoxin-2, shown here to be increased in Add2-KO RBC ghosts.
The higher resolution data set also consisted of a total of six technical replicates per genotype (three technical replicates for each of two biological replicates). Importantly, the LTQ-FT data allowed for detection of protein differences based on an increased number of peptides. Table I shows the list of proteins that were found to be decreased in Add2-KO RBC ghosts compared to wt with a P value of 0·05 or less, an intensity difference of two-fold or greater, and a peptide cutoff of two or more. For each protein, the denoted intensity difference represents an average based on the fold difference of all the peptide differences detected by CRAWDAD. The number of peptides listed is the number of peptides for that protein that showed an intensity difference. A complete list of peptide differences detected by CRAWDAD is presented in supplemental Table SX.
Table I.
Proteins decreased in β-adducin knock-out RBC's.
| No. | Protein* | Intensity Difference† | No. of peptides | ID |
|---|---|---|---|---|
| 1 | Adducin 2 (Beta)‡ | –79·8 | 8 | IPI00323122.1 |
| 2 | Adducin 1 (Alpha)‡ | –78·1 | 8 | IPI00225322.1 |
| 3 | Solute carrier family 9 (sodium/hydrogen exchanger), member 1 | –28·6 | 2 | IPI00380785.1 |
| 4 | Ubiquitin carboxyl-terminal hydrolase 5‡ | –10·6 | 3 | IPI00113214.1 |
| 5 | Epb4.9 Dematin‡ | –8·7 | 14 | IPI00125328.3 |
| 6 | Sodium/potassium-transporting ATPase subunit alpha-1 | –3·0 | 3 | IPI00311682.5 |
| 7 | Ubiquitin C‡ | –2·6 | 7 | IPI00755916.3 |
Intensity difference significant at P < 0·05.
Fold difference by which the intensity in Add2 KO exceeds that in wild-type; negative values denote proteins found to be more abundant in wild-type than Add2 KO.
Intensity difference significant at P < 0·01.
Deficiency of SLC9A1 in Add2 KO RBC ghosts
CRAWDAD detected 7 proteins that were decreased in Add2-KO RBC ghosts (see Table I). As expected, β-adducin is decreased the most, with an intensity difference of 79·8-fold followed by its binding partner, α-adducin, with a nearly identical intensity difference of 78·1-fold. The third most decreased protein is the sodium/hydrogen exchanger (SLC9A1) encoded by Slc9a1. The intensity difference for this protein is 28·6-fold and based upon two peptides. SLC9A1 is an integral membrane protein with twelve membrane-spanning domains and the peptides that were consistently detected by mass spectrometry are derived from the cytoplasmic tail of the protein, so it is likely that the integral membrane domains were not accessible to proteolysis in our preparation.
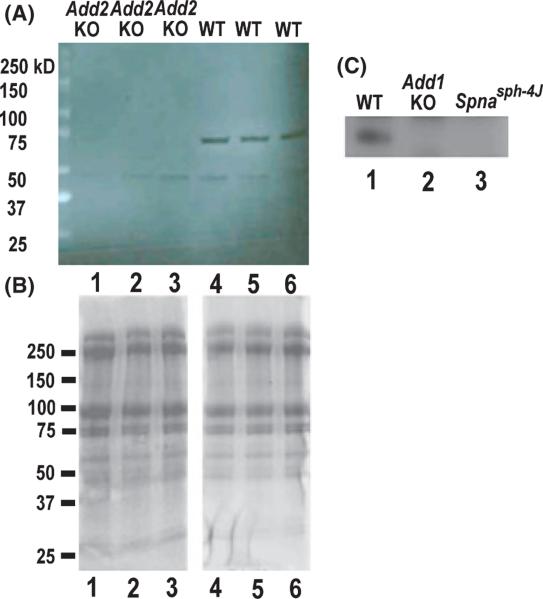
Previous studies have demonstrated by immunoblot the decrease of α-adducin when β-adducin was deleted (Gilligan et al, 1999; Muro et al, 2000). However, no studies have examined the expression of SLC9A1 in inherited haemolytic anaemia. Thus, we used immunoblotting to examine RBC ghosts from wt and Add2-KO mice for the level of SLC9A1 protein. A prominent band with a relative mobility of approximately 75 kDa was detected in samples from independent wild-type mice but not in samples from Add2-KO mice (Fig 2A, lanes 1–3 vs. 4–6). Coomassie blue staining demonstrates equal protein loading for wt and Add2-KO RBC ghosts (Fig 2B). We also examined SLC9A1 in RBC ghost samples from the Add1 knockout mouse (Robledo et al, 2008) and the Spnasph–4J (Robledo et al, 2010) mouse (spontaneous mutation of α-spectrin). SLC9A1 was detected in a wild-type control sample but not in Add1-KO or Spnasph–4J RBC ghosts (Fig 2C). These RBC ghosts have been shown previously to be deficient in β-adducin (Robledo et al, 2010), so this result is consistent with the lack of β-adducin being associated with lack of SLC9A1.
Fig 2.
Deficiency of SLC9A1 in β-adducin (Add2) KO, α-adducin (Add1) KO, and Spnasph–4J RBC ghosts. (A) RBC ghost protein samples from independent Add2-KO mice (lanes 1–3) and wild-type mice (lanes 4–6) were probed with SLC9A1 antibody as described in methods. Wild-type ghosts have a prominent band at approximately 75 kDa, but this band is missing in the Add2-KO ghosts. (B) Coomassie blue stained gel to demonstrate protein loading for Add2-KO and wild-type ghosts. (C) RBC ghost samples from wild-type (lane 1), Add1-KO (lane 2), and Spnasph–4J (lane 3) mice probed with SLC9A1 antibody. Again, the prominent band is seen in wt ghosts, but not Add1-KO or Spnasph–4J ghosts.
Four other proteins were decreased in Add2-KO RBC ghosts compared to wt at the cutoffs listed above (Table I). The intensity differences were much lower than the intensity difference for SLC9A1, and thus we have not pursued confirmation of these protein differences by immunoblot.
Proteins that are increased in Add2-KO RBCs
CRAWDAD detected 46 proteins that were increased in Add2-KO RBC ghosts at a P-value of 0·05 or less, two peptides or more per protein, and an intensity difference of two-fold or greater (see supplemental Table SI). Some of these proteins (listed in italics) were also increased in reticulocytes (see below) and may be representative of the increased number of reticulocytes in Add2-KO mice compared to wt mice. Nonetheless, several of the increased proteins have been demonstrated by immunoblot in previous studies. We demonstrated increased γ-adducin (Add3) in our initial description of the Add2 KO mice (Gilligan et al, 1999). Porro et al (2004) demonstrated increased capping protein and suggested that this reflected compensation for the lack of adducin to cap actin filaments in the Add2 KO RBCs. They also demonstrated increased tropomodulin. Finding the same proteins high on our list of increased proteins confirms the reliability of this method.
Other proteins on this list with lower intensity differences, such as band 4·1, spectrin, band 7, ankyrin, p55, and band 3, have not been demonstrated to be increased by immunoblot, suggesting that immunoblotting may not be as sensitive as mass spectrometry in detecting differences in protein abundance. Other proteins may be contaminants from platelets or leucocytes, such as platelet factor 4 precursor, filamin, villin, and α-actin. Proteins that may represent physiological differences in the Add2-KO RBCs include ATPase, Ca++ transporting, plasma membrane 4, pyruvate kinase, peroxiredoxin-2, and bisphosphoglycerate mutase.
Protein differences detected between reticulocyte and mature RBCs
As discussed earlier, haemolytic anaemias are characterized by reticulocytosis. As expected, we found more proteins in Add2-KO RBC ghosts compared to wt, partly due to the increased reticulocyte count (4·3% vs. 2%). To guide the interpretation of wild-type versus Add2-KO protein differences, we performed reticulocyte control studies using samples generated by phenylhydrazine treatment of mice with a genetic background identical to the Add2-KO mice (C57BL/6J). Phenylhydrazine treatment of mice is known to result in profound reticulocytosis (Chui et al, 1980). Nine d from the start of injections, whole blood samples were collected and found to have a reticulocyte count of 62%. Three independent reticulocyte-enriched samples and three independent matched untreated control samples were profiled on the higher resolution LTQ-FT. The complete list of proteins identified is presented in Tables SVIII and SIX.
Label-free analysis identified 66 protein differences in total. Among these, 45 proteins were increased and 21 proteins were decreased in the reticulocyte-enriched samples (Tables SII and SIII). Many of the proteins increased in reticulocytes relate to known RBC developmental processes, such as haem biosynthesis and protein synthesis. In fact, the loss of transferrin receptor and ribosomal proteins is a hallmark difference between reticulocytes and mature RBCs. Thus, the phenylhydrazine treatment was successful in generating a sample enriched in reticulocyte-related proteins. While many of these proteins are normally considered to be mitochondrial, cytoplasmic, or located in the endoplasmic reticulum, they have been previously identified in murine reticulocyte-rich membrane ghosts analysed by two-dimensional (2DE)-PAGE (Prenni et al, 2007). Among the proteins increased in Add2-KO RBCs, only 11 were also found increased in reticulocytes (see Table SI). A complete list of peptide differences detected by CRAWDAD for the reticulocyte control studies is presented in supplemental Table SXI.
Discussion
We have performed comprehensive protein profiling of RBC ghosts from wt and Add2-KO mice. In addition to the expected membrane skeleton proteins and integral membrane proteins, a surprising number of associated proteins were found. This is consistent with published proteomic analyses of human RBC ghosts, where proteins involved in translation, adhesion, protein repair, protein degradation, glycolysis, and cellular defense were identified (Kakhniashvili et al, 2004; Pasini et al, 2006). Many of these proteins were classically believed to reside in the RBC cytosol, but evidence now suggests functional interactions with the membrane skeleton apparatus. For example, the glycolytic enzymes phosphofructokinase, glyceraldehyde 3-phosphate dehydrogenase, pyruvate kinase, and lactate dehydrogenase were consistently detected in our RBC ghost preparations. This association is corroborated by immunostaining of intact RBCs in which these enzymes are found to localize along the periphery and may result in a molecular crowding effect that facilitates catalysis (Campanella et al, 2005). Furthermore, subsequent studies have identified binding sites on the transmembrane protein band three for aldolase, glyceraldehyde 3-phosphate dehydrogenase, and phosphofructokinase (Campanella et al, 2008).
We used the label-free analysis program CRAWDAD to globally detect protein differences between wild-type and Add2-KO RBCs. We detected seven proteins that were decreased and 48 proteins with a greater abundance in Add2-KO RBC ghosts. As haemolytic anaemias are characterized by reticulocytosis, we performed control studies in which reticulocyte-enriched samples from phenylhydrazine-treated mice were compared with mature RBCs from matched untreated controls. Our label-free analysis identified 47 proteins that were increased and 21 proteins that were decreased in the RBC ghosts from reticulocyte-enriched samples. A recent study by Liu et al (2010) examined a selected set of RBC proteins by immunoblot, comparing a reticulocyte-enriched sample to mature RBCs. While their approach is limited to proteins with available antibodies, their results are largely confirmed and expanded by our global proteomic approach.
The ATPase Ca++ transporting plasma membrane four was significantly increased. Activity of the calcium pump has been reported to decrease steeply throughout the lifespan of the red blood cell (Lew et al, 2003) and interactions with cytoskelatal proteins have recently been suggested (Vanagas et al, 2007). Thus, increasing its presence may help ameliorate the loss in life span for the fragile Add2-KO spherocyte. Alternatively, its increase may be a consequence of altered functional interactions with the membrane skeleton. The enzymes bisphosphoglycerate mutase, peroxiredoxin-2, and pyruvate kinase were also increased. Bisphosphoglycerate mutase, which is found only in RBCs and placental cells (Pritlove et al, 2006), plays a major role in regulating the oxygen affinity of haemoglobin through the synthesis of 2,3-bisphosphoglycerate (Rose, 1980). Changes in its abundance may be an attempt by the cell to reduce oxygen loading of haemoglobin via increased bisphosphoglycerate mutase activity. Mice deficient in peroxiredoxin-2 have significantly reduced haematocrits with morphologically abnormal RBCs that contain higher levels of reactive oxygen species and highly oxidized proteins (Lee et al, 2003). Peroxiredoxin-2 has been found in the RBC membrane fractions of hereditary spherocytosis patients but not in controls (Rocha et al, 2008). More recently, peroxiredoxin-2 expression and net content was shown to be increased in RBCs from mouse models of β-thalassemia that are characterized by oxidative damage to the membrane (Matte et al, 2010). Pyruvate kinase deficiency is a common cause of hereditary nonspherocytic anaemia and has been found to alter RBC survival (Aizawa et al, 2005). It is thought to play an important role as an antioxidant during RBC development (Aisaki et al, 2007). Increases in pyruvate kinase and the antioxidant peroxiredoxin-2 may help to combat increased oxidative stress in a morphologically altered Add2-KO RBC. Nonetheless, these changes in non-membrane skeleton proteins represent potential compensatory mechanisms that may collectively reduce the severity of the β-adducin defect.
SLC9A1 was greatly decreased in Add2-KO RBCs with the highest detected intensity difference after the β- and α-adducins (28·6-fold, see Table I and Fig 1). SLC9A1 is an ubiquitously expressed regulator of cell volume and pH (Alexander & Grinstein, 2006). Its activity has been associated with many cellular functions, such as adhesion, migration, proliferation, and apoptosis. SLC9A1 has 12 highly conserved transmembrane domains and a large carboxy-terminal cytoplasmic domain that is phosphorylated by several kinases and binds calmodulin. Targeted disruption of SLC9A1 in mice causes ataxia, growth retardation, and seizures (Bell et al, 1999). The deficiency of SLC9A1 in Add2-KO RBC ghosts was confirmed by immunoblot. Two additional mouse strains that have reduced β-adducin [Add1-KO (Robledo et al, 2008) and Spnasph–4J, an α-spectrin mutant (Robledo et al, 2010)] also showed deficiency of SLC9A1 by immunoblot. The mechanism for deficiency of SLC9A1 in adducin-deficient RBC ghosts will be determined in future studies. We do not expect this to be simply a result of membrane loss because other integral membrane proteins were not decreased in the Add2-KO RBC ghosts. A recent report by Salomao et al (2010) demonstrated aberrant protein sorting during erythroblast enucleation in cultured cells from mice with haemolytic anaemia. Aberrant protein sorting may be the mechanism by which adducin-deficient RBCs lose SLC9A1. Another possibility is that adducin has a role in targeting SLC9A1 to the plasma membrane during erythropoiesis and, without adducin, SLC9A1 does not reach the plasma membrane efficiently.
This is the first example of SLC9A1 deficiency in haemolytic anaemia and suggests new mechanisms leading to fragile RBCs. Cellular pH is crucial to protein stability and function. Without proper maintenance of pH, RBC proteins may lose their normal interactions and ability to regulate cell shape. SLC9A1 also plays an important role in maintaining cell volume and adducin-deficient RBCs are smaller and dehydrated (Gilligan et al, 1999). The mechanism for dehydration and reduced volume is likely to involve the deficiency of SLC9A1. The deficiency of SLC9A1 in adducin-targeted mice may explain additional phenotypic changes observed in these mice, such as abnormal learning and memory formation in the Add2-targeted mice (Rabenstein et al, 2005) and hydrocephaly in the Add1-targeted mice (Robledo et al, 2010). Finally, our finding of SLC9A1 deficiency in association with adducin deficiency may provide a clue to the role of adducin polymorphisms in hypertension in humans (Bianchi et al, 2005).
Supplementary Material
Acknowledgements
We thank Gennifer Merrihew (University of Washington), Babette Gwynn (Jackson Labs), and Mark Tsang (University of Washington) for assistance in sample preparation. We thank Doug Bolgiano (Puget Sound Blood Center) for statistical assistance. This work was supported by National Institutes of Health (grants 5F32HL079771-02, J.M.W.; P41RR011823-14, M.J.M.; and HL088468 and HL075714, L.L.P.). G.L.F. gratefully acknowledges training grant support from NHGRI grant T32-HG00035. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Author contributions
JMW, GLF, AJL, RFR, and DMG performed experiments. JMW, GLF, ER, MJM, LLP, and DMG designed the research and analysed the data. JMW and DMG wrote the paper.
Conflict of interest
The authors declare no competing financial interests.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table SI. Proteins increased in beta-adducin knock-out RBCs.
Table SII. Proteins decreased in reticulocytes.
Table SIII. Proteins increased in reticulocytes.
Table SIV. Proteins identified in wild-type RBCs on LTQ.
Table SV. Proteins identified in beta-adducin KO RBCs on LTQ.
Table SVI. Proteins identified in wild-type RBCs on LTQ-FT.
Table SVII. Proteins identified in beta-adducin KO RBCs on LTQ-FT.
Table SVIII. Proteins identified in RBCs from untreated mice on LTQ-FT.
Table SIX. Proteins identified in RBCs from phenylhydrazine treated mice LTQ-FT.
Table SX. Peptides for protein differences identified in beta-adducin KO RBCs.
Table SXI. Peptides for protein differences identified in RBCs from phenylhydrazine treated mice.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aisaki K, Aizawa S, Fujii H, Kanno J, Kanno H. Glycolytic inhibition by mutation of pyruvate kinase gene increases oxidative stress and causes apoptosis of a pyruvate kinase defi cient cell line. Experimental Hematology. 2007;35:1190–1200. doi: 10.1016/j.exphem.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Aizawa S, Harada T, Kanbe E, Tsuboi I, Aisaki K, Fujii H, Kanno H. Ineffective erythropoiesis in mutant mice with deficient pyruvate kinase activity. Experimental Hematology. 2005;33:1292–1298. doi: 10.1016/j.exphem.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Alexander RT, Grinstein S. Na+/H+ exchangers and the regulation of volume. Acta Physiologica (Oxford) 2006;187:159–167. doi: 10.1111/j.1748-1716.2006.01558.x. [DOI] [PubMed] [Google Scholar]
- Bell SM, Schreiner CM, Schultheis PJ, Miller ML, Evans RL, Vorhees CV, Shull GE, Scott WJ. Targeted disruption of the murine Nhe1 locus induces ataxia, growth retardation, and seizures. American Journal of Physiology. 1999;276:C788–C795. doi: 10.1152/ajpcell.1999.276.4.C788. [DOI] [PubMed] [Google Scholar]
- Bennett V, Gilligan DM. The spectrin-based membrane skeleton and micron-scale organization of the plasma membrane. Annual Review of Cell Biology. 1993;9:27–66. doi: 10.1146/annurev.cb.09.110193.000331. [DOI] [PubMed] [Google Scholar]
- Beutler E, West C, Blume KG. The removal of leukocytes and platelets from whole blood. Journal of Laboratory and Clinical Medicine. 1976;88:328–333. [PubMed] [Google Scholar]
- Bianchi G, Ferrari P, Staessen JA. Adducin polymorphism: detection and impact on hypertension and related disorders. Hypertension. 2005;45:331–340. doi: 10.1161/01.HYP.0000156497.39375.37. [DOI] [PubMed] [Google Scholar]
- tBirkenmeier CS, Barker JE. Hereditary haemolytic anaemias: unexpected sequelae of mutations in the genes for erythroid membrane skeletal proteins. Journal of Pathology. 2004;204:450–459. doi: 10.1002/path.1636. [DOI] [PubMed] [Google Scholar]
- Bodine D.M.t., Birkenmeier CS, Barker JE. Spectrin deficient inherited hemolytic anemias in the mouse: characterization by spec-trin synthesis and mRNA activity in reticulocytes. Cell. 1984;37:721–729. doi: 10.1016/0092-8674(84)90408-2. [DOI] [PubMed] [Google Scholar]
- Campanella ME, Chu H, Low PS. Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2402–2407. doi: 10.1073/pnas.0409741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella ME, Chu H, Wandersee NJ, Peters LL, Mohandas N, Gilligan DM, Low PS. Characterization of glycolytic enzyme interactions with murine erythrocyte membranes in wild-type and membrane protein knockout mice. Blood. 2008;112(9):3900–3906. doi: 10.1182/blood-2008-03-146159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasis JA, Mohandas N. Erythrocyte membrane deformability and stability: two distinct membrane properties that are independently regulated by skeletal protein associations. Journal of Cell Biology. 1986;103:343–350. doi: 10.1083/jcb.103.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui DH, Patterson M, Bayley ST. Unequal alpha and beta globin mRNA in reticulocytes of normal and mutant f/f fetal mice. British Journal of Haematology. 1980;44:431–439. doi: 10.1111/j.1365-2141.1980.tb05913.x. [DOI] [PubMed] [Google Scholar]
- Eber S, Lux SE. Hereditary spherocytosis–defects in proteins that connect the membrane skeleton to the lipid bilayer. Seminars in Hematology. 2004;41:118–141. doi: 10.1053/j.seminhematol.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Finney GL, Blackler AR, Hoopmann MR, Canterbury JD, Wu CC, MacCoss MJ. Label-free comparative analysis of proteomics mixtures using chromatographic alignment of high-resolution muLC-MS data. Analytical Chemistry. 2008;80:961–971. doi: 10.1021/ac701649e. [DOI] [PubMed] [Google Scholar]
- Gardner K, Bennett V. A new erythrocyte membrane-associated protein with calmodulin binding activity. Identification and purification. Journal of Biological Chemistry. 1986;261:1339–1348. [PubMed] [Google Scholar]
- Gilligan DM, Lozovatsky L, Gwynn B, Brugnara C, Mohandas N, Peters LL. Targeted disruption of the beta adducin gene (Add2) causes red blood cell spherocytosis in mice. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:10717–10722. doi: 10.1073/pnas.96.19.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iolascon A, Perrotta S, Stewart G. Red blood cell membrane defects. Reviews in Clinical and Experimental Hematology. 2003;1:22–56. [PubMed] [Google Scholar]
- Kakhniashvili DG, Bulla LA, Jr, Goodman SR. The human erythrocyte proteome: analysis by ion trap mass spectrometry. Molecular & Cellular Proteomics : MCP. 2004;3:501–509. doi: 10.1074/mcp.M300132-MCP200. [DOI] [PubMed] [Google Scholar]
- Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nature Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- Lee TH, Kim SU, Yu SL, Kim SH, Park DS, Moon HB, Dho SH, Kwon KS, Kwon HJ, Han YH, Jeong S, Kang SW, Shin HS, Lee KK, Rhee SG, Yu DY. Peroxiredoxin II is essential for sustaining life span of erythrocytes in mice. Blood. 2003;101:5033–5038. doi: 10.1182/blood-2002-08-2548. [DOI] [PubMed] [Google Scholar]
- Lew VL, Daw N, Perdomo D, Etzion Z, Book-chin RM, Tiffert T. Distribution of plasma membrane Ca2+ pump activity in normal human red blood cells. Blood. 2003;102:4206–4213. doi: 10.1182/blood-2003-06-1787. [DOI] [PubMed] [Google Scholar]
- Liu J, Guo X, Mohandas N, Chasis JA, An X. Membrane remodeling during reticulocyte maturation. Blood. 2010;115:2021–2027. doi: 10.1182/blood-2009-08-241182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matte A, Low PS, Turrini F, Bertoldi M, Campanella ME, Spano D, Pantaleo A, Siciliano A, De Franceschi L. Peroxiredoxin-2 expression is increased in beta-thalassemic mouse red cells but is displaced from the membrane as a marker of oxidative stress. Free Radical Biology & Medicine. 2010;49:457–466. doi: 10.1016/j.freeradbiomed.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohandas N, Chasis JA. Red blood cell deformability, membrane material properties and shape: regulation by transmembrane, skeletal and cytosolic proteins and lipids. Seminars in Hematology. 1993;30:171–192. [PubMed] [Google Scholar]
- Mohandas N, Gallagher PG. Red cell membrane: past, present, and future. Blood. 2008;112:3939–3948. doi: 10.1182/blood-2008-07-161166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro AF, Marro ML, Gajovic S, Porro F, Luzzatto L, Baralle FE. Mild spherocytic hereditary elliptocytosis and altered levels of alpha- and gamma-adducins in beta-adducindeficient mice. Blood. 2000;95:3978–3985. [PubMed] [Google Scholar]
- Pasini EM, Kirkegaard M, Mortensen P, Lutz HU, Thomas AW, Mann M. In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood. 2006;108:791–801. doi: 10.1182/blood-2005-11-007799. [DOI] [PubMed] [Google Scholar]
- Peters L, Barker J. Spontaneous and targeted mutations in erythrocyte membrane skeleton genes: mouse models of hereditary spherocytosis. In: Zon LI, editor. Clinical Medicine. Oxford Press; New York: 2001. pp. 582–608. [Google Scholar]
- Peters LL, Shivdasani RA, Liu SC, Hanspal M, John KM, Gonzalez JM, Brugnara C, Gwynn B, Mohandas N, Alper SL, Orkin SH, Lux SE. Anion exchanger 1 (band 3) is required to prevent erythrocyte membrane surface loss but not to form the membrane skeleton. Cell. 1996;86:917–927. doi: 10.1016/s0092-8674(00)80167-1. [DOI] [PubMed] [Google Scholar]
- Peters LL, Jindel HK, Gwynn B, Korsgren C, John KM, Lux SE, Mohandas N, Cohen CM, Cho MR, Golan DE, Brugnara C. Mild spherocytosis and altered red cell ion transport in protein 4.2-null mice. Journal of Clinical Investigation. 1999;103:1527–1537. doi: 10.1172/JCI5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porro F, Costessi L, Marro ML, Baralle FE, Muro AF. The erythrocyte skeletons of beta-adducin deficient mice have altered levels of tropomyosin, tropomodulin and EcapZ. FEBS Letters. 2004;576:36–40. doi: 10.1016/j.febslet.2004.08.057. [DOI] [PubMed] [Google Scholar]
- Prenni JE, Avery AC, Olver CS. Proteomics: a review and an example using the reticulocyte membrane proteome. Veterinary Clinical Pathology/American Society for Veterinary Clinical Pathology. 2007;36:13–24. doi: 10.1111/j.1939-165x.2007.tb00176.x. [DOI] [PubMed] [Google Scholar]
- Pritlove DC, Gu M, Boyd CA, Randeva HS, Vatish M. Novel placental expression of 2,3-bisphosphoglycerate mutase. Placenta. 2006;27:924–927. doi: 10.1016/j.placenta.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Rabenstein RL, Addy NA, Caldarone BJ, Asaka Y, Gruenbaum LM, Peters LL, Gilligan DM, Fitzsimonds RM, Picciotto MR. Impaired synaptic plasticity and learning in mice lacking beta-adducin, an actin-regulating protein. The Journal of Neuroscience. 2005;25:2138–2145. doi: 10.1523/JNEUROSCI.3530-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo RF, Ciciotte SL, Gwynn B, Sahr KE, Gilligan DM, Mohandas N, Peters LL. Targeted deletion of alpha-adducin results in absent beta- and gamma-adducin, compensated hemolytic anemia, and lethal hydrocephalus in mice. Blood. 2008;112:4298–4307. doi: 10.1182/blood-2008-05-156000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo RF, Lambert AJ, Birkenmeier CS, Cirlan MV, Cirlan AF, Campagna DR, Lux SE, Peters LL. Analysis of novel sph (spherocytosis) alleles in mice reveals allele-specific loss of band 3 and adducin in alpha-spectrin-deficient red cells. Blood. 2010;115:1804–1814. doi: 10.1182/blood-2009-07-232199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha S, Vitorino RM, Lemos-Amado FM, Castro EB, Rocha-Pereira P, Barbot J, Cleto E, Ferreira F, Quintanilha A, Belo L, Santos-Silva A. Presence of cytosolic peroxiredoxin 2 in the erythrocyte membrane of patients with hereditary spherocytosis. Blood Cells, Molecules & Diseases. 2008;41:5–9. doi: 10.1016/j.bcmd.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Rose ZB. The enzymology of 2,3-bisphosphoglycerate. Advances in Enzymology and Related Areas of Molecular Biology. 1980;51:211–253. doi: 10.1002/9780470122969.ch5. [DOI] [PubMed] [Google Scholar]
- Salomao M, Chen K, Villalobos J, Mohandas N, An X, Chasis JA. Hereditary spherocytosis and hereditary elliptocytosis: aberrant protein sorting during erythroblast enucleation. Blood. 2010;116:267–269. doi: 10.1182/blood-2010-02-264127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southgate CD, Chishti AH, Mitchell B, Yi SJ, Palek J. Targeted disruption of the murine erythroid band 3 gene results in spherocytosis and severe haemolytic anaemia despite a normal membrane skeleton. Nature Genetics. 1996;14:227–230. doi: 10.1038/ng1096-227. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods in Molecular Biology. 2003;224:149–157. doi: 10.1385/1-59259-364-X:149. [DOI] [PubMed] [Google Scholar]
- Vanagas L, Rossi RC, Caride AJ, Filoteo AG, Strehler EE, Rossi JP. Plasma membrane calcium pump activity is affected by the membrane protein concentration: evidence for the involvement of the actin cytoskeleton. Biochimica et Biophysica Acta. 2007;1768:1641–1649. doi: 10.1016/j.bbamem.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RA, Birkenmeier CS, Lux SE, Barker JE. Ankyrin and the hemolytic anemia mutation, nb, map to mouse chromosome 8: presence of the nb allele is associated with a truncated erythrocyte ankyrin. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:3117–3121. doi: 10.1073/pnas.87.8.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.