Abstract
Nucleosome assembly following DNA synthesis is critical for maintaining genomic stability. The proteins directly responsible for shuttling newly synthesized histones H3 and H4 from the cytoplasm to the assembly fork during DNA replication comprise the Chromatin Assembly Factor 1 complex (CAF-1). Whereas the diverse functions of the large (CAF-1-p150, CHAF1a) and small (RbAp48, p48) subunits of the CAF-1 complex have been well-characterized in many tissues and extend beyond histone chaperone activity, the contributions of the medium subunit (CAF-1-p60, CHAF1b) are much less well understood. Although it is known that CHAF1b has multiple functional domains (7x WD repeat domain, B-like domain, and a PEST domain), how these components come together to elicit the functions of this protein are still unclear. Here, we review the biology of the CAF-1 complex, with an emphasis on CHAF1b, including its structure, regulation, and function. In addition, we discuss the possible contributions of CHAF1b and the CAF-1 complex to human diseases. Of note, CHAF1b is located within the Down syndrome critical region (DSCR) of chromosome 21. Therefore, we also address the putative contributions of its trisomy to the various manifestations of DS.
Keywords: CAF-1, CHAF1b, Nucleosome, Chaperone
Introduction
The mechanism by which histones are delivered to, and assembled on, newly synthesized DNA was a mystery that began with the first characterization of DNA and histone octamer complexes called “nucleosomes” in 1975 [1, 2]. In 1986, Bruce Stillman and colleagues found that although cytoplasmic extracts from 293T cells promote efficient replication of DNA from SV40 plasmids, it was not until nuclear extracts from 293T cells were added to the reaction that the DNA was able to assemble into novel SV40 mini-chromosomes, complete with nucleosomes [3]. It was this observation that lead Stillman to suggest that, “there may be an active complex that follows the replication machinery along the DNA and promotes nucleosome assembly” [3]. Three years later, in cooperation with Susan Smith, they identified and purified the activity as a heterotrimeric complex consisting of three subunits and named it the “Chromatin Assembly Factor 1” (CAF-1) complex based on its ability to assemble histones onto newly synthesized DNA. Each subunit was named based on its apparent molecular weight following gel electrophoresis: a 150 kDa subunit (CAF-1-p150, CHAF1a), a 60 kDa subunit (CAF-1-p60, CHAF1b), and 48 kDa subunit (CAF-1-p48, RbAp48, p48) [4]. Verreault and colleagues also purified this complex from human cells and revealed that it consisted of each subunit in equal 1:1:1 stoichiometry and that CAF-1 also frequently contained newly synthesized acetylated histone H3/H4 dimers [5]. Drosophila has a similar CAF-1 complex, consisting of a CHAF1a homologue (p180), a CHAF1b homologue (p105/p75), and a p48 homologue (p55) [6]. The CHAF1b homologue p105 is sometimes truncated at its C-terminus, forming a p75 subunit that interacts with CAF-1 and is normally found mutually exclusive of the p105 subunit. Though these two subunits have similar functions, p105 is predominantly expressed during embryogenesis, while p75 dominates after larval formation [7].
CAF-1’s primary identified function is to deliver newly synthesized H3/H4 dimers to the replication fork during the DNA synthesis (S) phase of cell cycle (reviewed in [8]) (Figure 1A). During S-phase, H3 and H4 proteins are newly synthesized in the cytoplasm and form heterotrimeric complexes with histone chaperone Asf1 in a 1:1:1 stoichiometry. Within this complex, Asf1 binds H3 in a manner that precludes its natural homodimerization via a four-helix bundle, thereby preventing H3/H4 heterodimers from becoming tetramers. Following interaction with H2A and DNA in the newly-forming nucleosome, H4 undergoes a conformational change by forming a mini β-sheet, thereby releasing the globular domain of Asf1, and allowing the formation of the heterotetramer [9]. The CAF-1 subunit CHAF1b directly interacts with ASF1a, and while English and colleagues originally proposed that the WD repeat domain of CHAF1b is responsible for maintaining the interaction of ASF1a/b with CAF-1 through the possible formation of a β-propeller structure, Tang and colleagues later found that the WD repeat domain was not required for interaction with ASF1a/b [9, 10].
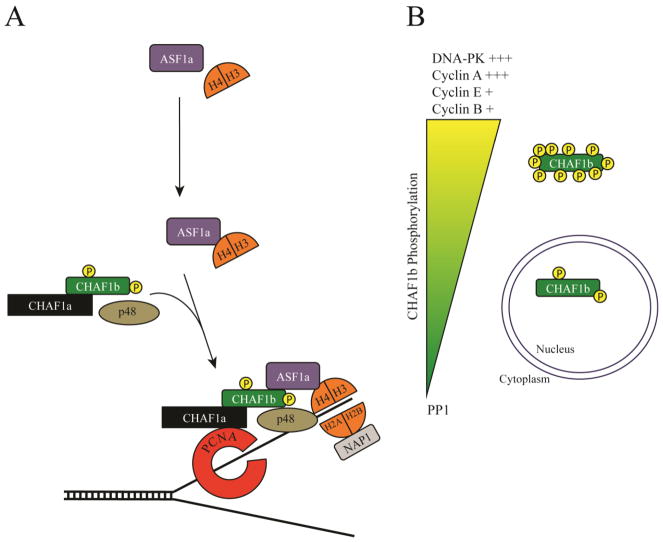
Figure 1.
Schematic of CHAF1b function and regulation. A) ASF1a collects newly synthesized H3/H4 dimers in the cytoplasm. CHAF1b directly interacts with ASF1a through B-like domains. CHAF1b/ASF1a/H3/H4 complex is directed to the replication fork within the nucleus through interactions with other CAF-1 family members and PCNA. CAF-1 deposits H3/H4 heterodimers, driving the first step of nucleosome formation. The complex is jettisoned when NAP1 delivers H2A/H2B. B) CHAF1b is strongly phosphorylated by many different factors including DNA-dependent Protein Kinase (DNA-PK) and Cyclin A, and weakly phosphorylated by Cyclin E and Cyclin B. +’s indicate relative phosphorylation strength due to each factor. CHAF1b is dephosphorylated by Protein Phosphatase 1 (PP1). Hyperphosphorylation is correlated with CHAF1b displacement from chromatin during mitosis.
The association of CAF-1 with newly synthesized histones suggests that it is involved in chaperoning the histones to newly synthesized DNA, as opposed to other histone chaperone complexes that shuttle histones back and forth along pre-existing DNA [3, 11, 12]. Later studies confirmed that CAF-1 has a specific preference for H3 variant H3.1, which is produced primarily during S-phase and is considered to be the replication-dependent H3. The H3 variant H3.2 differs from H3.1 by one amino acid (Cys – Ser, residue 96) and is also produced during DNA replication, though there is no functional evidence showing interactions with CAF-1 [13]. Finally, H3 variant H3.3 is continuously synthesized throughout the lifespan of the cell, its levels are maintained independently of cell cycle regulation, and it can be incorporated into chromatin at any point during cell cycle [14, 15]. However, H3.3 appears to preferentially be found with HIRA instead of CAF-1 [16]. Newly synthesized H3/H4 histone dimers are acetylated in the cytoplasm by a B-type histone acetyltransferase (HAT). This B-type HAT has activity in the cytoplasm, and is known to acetylate lysine residues 5 and 12 on H4 that is not bound in the nucleosome, based on work originally done in yeast and later studied in immortalized HeLa cells and Xenopus eggs [17–20].
During nucleosome formation, CAF-1 is localized to the replication fork in S-phase [21]. This localization allows CAF-1 to facilitate the first step of nucleosome formation on newly synthesized DNA: the deposition of H3/H4 dimers (Figure 1A). shRNA knockdown of either CHAF1a or CHAF1b leads to immediate loss of chromatin chaperoning activity and rapid concomitant degradation of the other subunit, most likely due to the exposure and activation of the PEST domains present in both subunits [22, 23]. Deletion of the CAF-1-p48 subunit was reported to result in minor defects with H3/H4 heterodimer chaperoning, based on failure of RNAi deletion of C. elegans CAF-1-p48 homologue LIN-53 to cause defects in MI motor neuron asymmetry due to improper H3/H4 deposition [24]. However, this observation was only based on one phenotype, and while there may be no other functional homologues of p48 in C. elegans, other species may have homologues that can take on additional roles in CAF-1 activity. Moreover, p48 loss may influence other aspects of CAF-1 chaperone function. A recent study using full-length recombinant human proteins by Kadyrova and colleagues demonstrated that p48 was critical for successful nucleosome formation during DNA-replication-dependent nucleosome packaging [25]. Depletion of CAF-1 in HeLa cells prevents progression through S-phase, activating CHK1, but not CHK2, which is consistent with DNA replication defects, opposed to activating DNA damage response pathways [26]. CAF-1 deletion in yeast is not lethal, but results in significantly longer Ozaki fragments due to incorrect placement of nucleosomes, also suggesting a defect in DNA replication [27]. In addition to Ozaki fragment size changes, yeast lacking components of the CAF-1 complex also exhibit other replicative problems including activation of mitotic checkpoints due to defects in chromosomal centromere and kinetochore formation, especially when combined with additional mutations in Histone Regulatory (HIR) proteins [28, 29].
The CHAF1a subunit
The p150 subunit of human CAF-1 was first cloned by Kaufman et al in 1995, and the human open reading frame plus the promoter region were later sequenced by Dong et al in 2001 [22, 30]. One of the major functions of p150 is in targeting the CAF-1 complex to the replication fork through a direct interaction with Proliferating Cell Nuclear Antigen (PCNA) [31]. PCNA forms a sliding clamp on the DNA, mediated by protein-protein interactions during DNA synthesis, and serves as a scaffold for several different proteins including DNA polymerases and histone chaperones [32–34]. Although CHAF1a has two separate but conserved PCNA interacting peptide regions (PIPs), the second PIP (PIP2), located towards the middle of the protein, is primarily responsible for maintaining the in vivo interaction with PCNA (Figure 2). The two PIPs in CHAF1a have different affinities for PCNA, where the N-terminal PIP1 binds PCNA strongly in in vitro studies, but appears to be unnecessary in a cellular context. The middle PIP2 is required for PCNA binding in an in vivo (cellular) context and differs from PIP1 by a single amino acid change (Q–K) [35]. While a putative PIP2 region was identified in CHAF1b, subsequent experiments did not reveal any evidence of direct binding with PCNA. Ben-shahar and colleagues proposed that the ability for the PIP regions of CHAF1a to interfere with nucleosome activity instead of directly with DNA replication processes allow for CAF-1 to function behind the replication fork, without the hindering other PCNA-mediated processes [35].
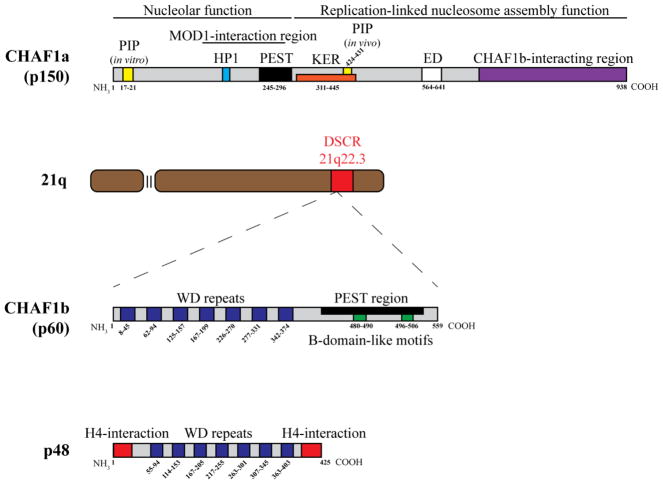
Figure 2.
Diagrams of CAF-1 components. CHAF1a consists of an N-terminal PIP (PCNA Interacting Motif) that has strong activity in vitro, followed by a MOD-1 interacting region, which is required for interaction with heterochromatin. Subsequently, there is also a HP1 (Heterochromatin Protein 1) interacting domain within the MOD-1 region. A PEST domain that drives rapid protein degradation follows. After the PEST domain is a KER domain, which is a highly acidic region thought to facilitate interaction with histones. Within the KER domain is another PIP, which has been experimentally confirmed as the PIP required for in vivo PCNA interaction and nucleosome formation. An ED region and a CHAF1b-interacting region reside in the C-terminus of CHAF1a. The first 300 peptides of CHAF1a are not required for replication-linked nucleosome formation, however they are required for recently discovered novel nucleolar functions. CHAF1b, found within the DSCR of chromosome 21, is one of many WD repeat proteins. CHAF1b consists of 7x WD repeats, followed by a PEST domain as well as two recently identified B-domain-like motifs that facilitate direct interactions with H3/H4 chaperone ASF1a. p48 is also a 7x WD repeat protein that also includes two α-helical domains at the N and C termini that facilitate H4 binding. Known protein domain locations are notated by the amino acid numbers.
In addition to its role in S-phase DNA replication, CHAF1a is also involved in interphase DNA damage repair, specifically during nucleotide excision repair (NER) in an ATP-dependent process involving PCNA [36]. CHAF1a has additional functions in PCNA-independent DNA repair during double strand break (DSB) repair. In DSB repair, CHAF1a is tracked to DSB sites by direct, PCNA-independent, interaction with scaffolding protein 14–3–3ζ and KU70/80 (also known as the DNA-dependent Protein Kinase complex) [37]. This complex is known to recruit the required factors required for DSB repair and facilitates non-homologous end joining and homologous recombination [38]. Hoek et al. postulated that KU70/80 recruits CAF-1 to DNA damage repair sites independently of PCNA because there is a need for new nucleosome assembly without a need for large-scale DNA synthesis [37]. Furthermore, Werner syndrome protein (WRN), a RecQ family DNA helicase, has recently been shown to recruit CHAF1a to sites of DNA damage through direct interactions mediated by its helicase and RQC domains: this complex is promotes DNA repair through homologous recombination [39, 40].
CHAF1a also can bind to Heterochromatin-binding Protein 1 (HP1), allowing the CAF-1 complex to track to heterochromatin via an HP1 binding domain in CHAF1a’s N-terminus that is located within the Mod-interacting region [41, 42]. The N-terminus of CHAF1a also harbors a fragment that is sufficient for maintaining nucleolar chromosome and protein associates, including binding with proliferation antigen Ki67, nucleophosmin, and nucleolin. However, this fragment is unable to bind with CHAF1b, suggesting that this novel nucleolar function is most likely CAF-1-independent, and does not involve histone chaperone activity [43].
Ye and colleagues created a dominant negative CHAF1a protein by deleting the N-terminus, creating a p150C fragment that titrates and sequesters CHAF1b into nonfunctioning complexes. Without CHAF1b present to stabilize it, the remaining endogenous CHAF1a is rapidly degraded due to interactions mediated by its PEST domain (Figure 2). Disruption of CAF-1 with the CHAF1a dominant negative causes stalled replication forks, DSBs, and translocations in U2OS cells, further supporting the role of CHAF1a in maintaining proper nucleosome assembly during DNA synthesis and DNA repair [44].
The p48 subunit
The p48 subunit was first characterized as part of the Retinoblastoma (Rb) family of proteins in yeast based on affinity chromatography experiments and was later sequenced and renamed Retinoblastoma Associated Proteins p46 and p48 (RbAp46/RbAp48) [45, 46]. p48, which consists of 7x WD repeats, has been shown to bind H4 independently of CAF-1 through α-helical domains on its N and C terminus (Figure 2) [47]. It is also tightly associated with the catalytic subunit of human histone deacetylase HDAC1, suggesting it may have a role in facilitating the deacetylation newly synthesized H4 once deposited in the nucleosome [5, 48, 49]. This is in contrast to p48’s predicted function within the yeast histone acetyltransferase (HAT) complex, based on its high degree of conservation with yeast histone chaperone Hat2b. However, subsequent studies have shown that p48 alone has no enzymatic activity [5].
Only a small fraction of cellular p48 is associated with CAF-1, with the majority of the protein present in undetermined large molecular weight complexes [50]. p48 is also found in complex with both H3.1 and H3.3 variants of H3, suggesting that it has additional roles outside of CAF-1-mediated histone chaperone activity [16]. Such roles include functioning within the polycomb repressor complex 2 (PRC2), where p48 assists in facilitating H3K27me3 marks and subsequent repression of the senseless gene and loss of embryonic patterning due to hox gene dysregulation in D. melanogaster [51]. In human cells, p48 interaction with PRC2 is hypothesized to play a role in limiting H3K27me3 deposition, allowing for certain regions of the genome to remain active (reviewed in [52]). In addition to the polycomb repressor complex, p48 has also been identified as playing critical roles within the NuRD remodeling/deacetylase complex and the NURF remodeling complex [53–55]. Mutations of the S. cerevisiae CAC1 (homologous to human CHAF1a) that reduce binding to CAC3 (homologous to human p48) was found to result in telomeric silencing defects [56].
The CHAF1b subunit
CHAF1b was originally discovered as part of the CAF-1 complex [4, 22]. It consists of 7x WD repeats, followed by a B-like region and a PEST domain (Figure 2). Although the WD repeats of CHAF1b were thought to provide the scaffolding necessary to facilitate direct interaction with H3/H4 chaperone ASF1a/b and promote histone chaperone activity, N-terminal deletion experiments removing residues 1–418 of CHAF1b revealed that this domain was not required for binding with ASF1a/b [10]. Rather, it was determined that the C terminus mediates direct binding to ASF1a. The C-terminus of CHAF1b contains a B-like region, first identified in the D. melanogaster CHAF1b homologue p105, and this B-like region is necessary and sufficient for CHAF1b binding to ASF1 [7]. The CHAF1b B-like region forms a β-hairpin that interacts exclusively with the β-sandwich structure of the ASF1a N-terminal core domain. These interactions are further maintained through β-sheet, salt bridge, and van der Waals interactions. Mutation of two arginine residues (R483/R489) to alanine in the CHAF1b B-like region abolishes CHAF1b binding to ASF1a. [10]. CHAF1b also shares homology of its B-like region with HIRA, a replication-independent H3 chaperone that is activated primarily outside of S-phase and has a preference for H3 variant H3.3 [57, 58]. Notably ASF1a binding with HIRA or CHAF1b is mutually exclusive, suggesting HIRA expression may be an indirect method of regulating CHAF1b activity [16]. Additionally, ASF1 is one of two known histone chaperones to have both replication dependent and replication independent functions, due to its interactions with both CHAF1b and HIRA [59]. The other H3/H4 chaperone known to have both replication dependent and independent functions is FACT, although FACT has not been shown to interact with CAF-1 [25, 60, 61]. Finally, in addition to the 7x WD repeats in the N-terminus and the B-like domain in the C-terminus, CHAF1b also has a PEST domain in its C-terminus (Figure 2) [22]. The activity of this PEST domain is further supported by the finding that elimination of CHAF1a by shRNA leads to the subsequent and rapid degradation of CHAF1b [44].
CHAF1b function during S phase
Proper chromatin formation during DNA replication depends on a number of important factors, including a chaperone complex that can assemble the nucleosome. While a strict 1:1:1 stoichiometry of CAF-1 components CHAF1a, CHAF1b, and p48, respectively was initially proposed [62], loss of the p48 subunit was found to cause some defects in CAF-1’s ability to chaperone chromatin to the replication fork [5, 16, 50]. However, these are minor effects when compared to loss of either CHAF1a or CHAF1b, which renders the CAF-1 complex unable to function [22–24].
Loss of either CHAF1a or CHAF1b subunit results in complete loss of CAF-1 function, and is most likely due to two reasons. First, loss of CHAF1b makes CHAF1a unstable due to exposure and subsequent activation of the C-terminal PEST domain [44]. Second, CHAF1b directly interacts with ASF1a/b, the H3/H4 chaperone responsible for maintaining direct contact with H3/H4 heterodimers [10, 63]. Therefore, during S phase, CHAF1b/ASF1a/H3/H4 are in complex with CHAF1a, which in turn is in contact with PCNA at the replication fork. This web of interaction allows the CAF-1 complex to deliver H3/H4 dimers to the DNA ahead of the replication fork (Figure 1A). Evidence for the requirement for CHAF1b in this process is provided by the observation that shRNA-mediated deletion of CHAF1b in replicating HeLa cells leads to induction of programmed cell death within 24 hours. This is due to failure to properly replicate DNA and accumulation of DNA double strand breaks as evidenced by increased levels of phosphorylated H2A variant H2A.X present in the chromatin [64].
CHAF1b functions outside of S phase
CHAF1b has also been shown to be required for NER synthesis following DNA damage [65]. This was first discovered in 1996 when Gaillard and colleagues found that CAF-1 mediates the de novo assembly of nucleosomes onto UV-irradiated plasmids undergoing NER [66]. Following irradiation, CHAF1b is phosphorylated, in an amount that is directly related to the intensity of the UV insult [67]. This phosphorylation drives CHAF1b to form complexes with CHAF1a and PCNA, which then localize to sites of DNA damage [67]. The CHAF1b-ASF1a interaction, which is independent of CHAF1b’s phosphorylation [63], is responsible for bringing H3.1 to the nucleosome during NER. This ability to function independently of its phosphorylation status is critical for CHAF1b’s role in NER.
Recent studies by Zhu and colleagues have shown that Chaf1b is required for ubiquitinated H2A (uH2A) recruitment to damage foci following UV-induced DNA damage [68]. However, these observations may result from the loss of an intact CAF-1 complex, thereby globally reducing H3/H4 deposition at the sites of DNA damage, and subsequently reducing uH2A recruitment. However, it is possible that loss of CHAF1b (subsequently driving the loss of function of the CAF-1 complex) in this system could be globally reducing H3/H4 deposition into nucleosomes at the sites of DNA damage. This could also explain the loss of uH2A deposition observed following CHAF1b shRNA knockdown and UV irradiation.
CHAF1b is also the primary chaperone of a group of histone-like proteins called protamines, which are small highly basic arginine-rich linker proteins that share a high degree of homology with histone linker protein H1 [69]. Protamines mediate a very high degree of chromatin compaction, functionally protecting sperm DNA from physical damage and mutagenesis [70]. In fact, protamines are able to condense the DNA down to a size that is almost 200-fold decreased from its original state, allowing for half the genome to fit inside a spermatid. In mammals, protamines do not directly replace histones in the testes, but operate via interaction with intermediary proteins called Transition Proteins 1 and 2 (TNP1 and TNP2) [70]. A novel, histone-independent chaperone function of CHAF1b was recently elucidated in D. melanogaster testes. Here, the CHAF1b homologue p75 is required for deposition of protamines to the DNA during the late canoe phase of spermatogenesis [71]. These protamines completely replace traditional histones in the mature sperm [72]. While D. melanogaster CHAF1a homologue p180 is responsible for tracking of protamines to the spermatid DNA chromatin, CHAF1b homologue p75 is responsible for their deposition onto the DNA [71]. As of yet, there are no functional studies in mammalian systems to determine if CHAF1b cooperates or can functionally replace TNP1 and TNP2 in spermatogenesis.
CHAF1b regulation
Little is known about how CHAF1b is regulated. Most of the known post-translational modifications of CHAF1b are phosphorylation events that occur throughout the body of the protein (Figure 1B). Smith and colleagues, while developing monoclonal antibodies targeted to CHAF1a and CHAF1b, found that both CHAF1a and CHAF1b can be phosphorylated [73]. Although phosphorylation does not affect CHAF1b’s ability to interact with ASF1a (and concomitantly H3/H4) [63], phosphorylation levels of CHAF1b correlate with cellular localization: hyperphosphorylation of CHAF1b during mitosis correlates with chromosomal displacement and inactivation, suggesting that nucleosome assembly (and CAF-1 activity) is most likely controlled by reversible phosphorylation events [50] (Figure 1B).
There are two families of kinases responsible for the phosphorylation of CHAF1b: Cyclin-Dependent Kinases (CDKs), and the DNA-dependent Protein Kinase (DNA-PK). Keller and colleagues demonstrated that Cyclin A/CDK2, Cyclin E/CDK2, and Cyclin B/CDK1, but not Cyclin D/CDK2, phosphorylate CHAF1b in vitro during DNA synthesis. These data suggest that regulation and activation of CHAF1b by phosphorylation is tightly associated with DNA synthesis checkpoints [74]. Protein phosphatase 1 is responsible for dephosphorylation of CHAF1b (Figure 1B) [74]. It should be noted, however, that the functional consequences of these phosphorylation events have not been determined in any metazoan system.
Nucleosome assembly by CAF-1 outside of S phase is not affected by Cyclin or protein phosphatase activity [74], suggesting that there is a parallel mechanism regulating CHAF1b that occurs during DSB repair and NER. In these situations, additional nuclear factors that could phosphorylate and regulate CHAF1b are part of the DNA-dependent Protein Kinases (DNA-PK) complex, consisting of KU70/80 and DNA-PK, identified as p350, and reviewed in 1996 by Lees-Miller [38, 75, 76]. Later, this work was confirmed by Hoek et al, who showed that, while CHAF1b can be phosphorylated by Cyclin E and Cyclin B, the strongest phosphorylation events were mediated by Cyclin A and DNA-PK [37].
CHAF1b in Down syndrome
CHAF1b was first mapped to the Down syndrome critical region (DSCR) on human chromosome 21 in 1996 by Blouin and colleagues, using EcoRI and PstI-digested pools of HC21-specific cosmids. In their work, they suggested that the imbalance of CHAF1b levels compared to the other CAF-1 components could contribute to the clinical manifestations of DS [77]. Later that year, Katsanis and colleagues also mapped the gene encoding CHAF1b to human chromosome 21q22.2, and similarly speculated that the over-expression of CHAF1b disrupted the 1:1:1 stoichiometry of the CAF-1 complex and could cause nucleosome assembly defects, leading to some characteristics of DS [78]. Of note, although adults with DS have a lower overall incidence of solid tumors as compared to the population without DS, children with DS have a higher incidence of blood cancers including acute lymphoblastic leukemia (ALL) and acute megakaryoblastic leukemia (AMKL). Studies have shown that up to 95% of malignancies in children with DS are of the blood, which is a significantly higher rate when compared to the population without DS, in which leukemia only makes up 34% of childhood malignancies [79]. It is notable that CHAF1b levels are elevated in AMKL in the group of patients with Down syndrome [80]. This observation suggests that the elevation in CHAF1b is most likely due to trisomy 21.
CHAF1b is a marker for poor prognosis in many human malignancies
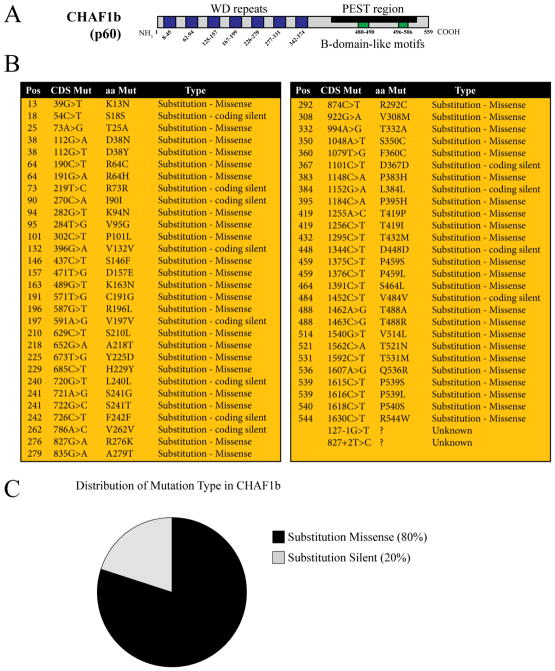
CHAF1b, as a critical component for successful DNA replication and chromatin formation, has a significant role in proliferative tissues. The Sanger Institute’s Catalogue of Somatic Mutations in Cancer (COSMIC) reports 59 identified mutations in CHAF1b that are in some way related to human malignancies [81]. These mutations occur throughout the body of the gene and consist exclusively of missense (app. 80%) and silent (app. 20%) mutations (Figure 3). There are no identified insertions, deletions, or truncations in CHAF1b related to human disease.
Figure 3.
Mutations in CHAF1b found in human disease. A) Schematic of CHAF1b protein domains with amino acid locations notated. B) Table of known mutations of CHAF1b involved in human malignancies. Pos (position), CDS mutant (Coding Strand Mutant), aa Mut (Amino Acid Mutant), Type. C) Relative distribution of known CHAF1b mutations. Only Substitution Missense and Substitution Silent are listed because there are no known insertions, deletions, or truncations. Information was obtained through analysis on the Catalogue of Somatic Mutations in Cancer (COSMIC).
While CHAF1b protein levels are elevated during embryogenesis, most likely due to the high demand for cellular replication during development, healthy terminally differentiated adult tissues express very low levels of CHAF1b. This has been confirmed in studies performed with both brain and skin tissues [82, 83]. However, during malignant tumor development, CHAF1b levels become significantly elevated and may serve as a prognostic marker and predictor of metastasis. In high-grade glioma, CHAF1b was identified as part of a 4-gene signature highly correlated with metastasis along with three other DNA-damage repair proteins (CHAF1b, PDLIM4, EDNRB, and HJURP) [84]. Patients with elevated CHAF1b levels can be expected to survive an average of 14 months, compared to patients expressing low levels of CHAF1b who can survive an average of 23.5 months with the disease [82]. While CHAF1b is expressed at a very low levels in healthy skin melanocytes, elevated CHAF1b in melanomas correlates to malignant phase: mid expression levels of CHAF1b are found in radial growth phase melanoma, and highest CHAF1b levels are found in vertical growth phase melanomas [83]. Similar patterns are found in salivary gland and prostatic cancer tumors. CHAF1b expression level can even be used to accurately predict future metastasis in these tumors [85, 86]. Interestingly, the expression levels of the other components of CAF-1 complex (CHAF1a, p48) are not elevated in the same tumors that CHAF1b is overexpressed, suggesting that CHAF1b levels have an association with metastasis that may be independent of traditional CAF-1 function. Though this correlation has been observed in tumors, no functional studies have been performed to determine the possible contributions of CHAF1b to tumor growth or metastasis.
CHAF1b is also overexpressed in breast, endometrial, renal, and cervical carcinomas, and this is a prognostic marker of poor clinical outcome [87]. It should be noted that the most commonly found solid tumors in DS patients corresponds to the same tumors that overexpress CHAF1b as a poor prognosis marker, including digestive tract tumors, melanomas, and tumors of the female genital organs (endometrial and cervical tumors) [79, 88]. Finally, a recent study of consanguineous families with neurological disorders found that a CHAF1b missense mutation (A496G) was common in members with ADHD, marking one of the first possible genetic mutations associated with the disorder [89]. This is the same mutation identified by Nakano and colleagues as causing loss of asymmetry during MI neuron differentiation in C. elegans, further confirming a possible link between CHAF1b mutations and neurological pathologies [24].
Some patients undergoing allogeneic hematopoietic cell transplant (HCT) as part of the treatment for acute myeloid leukemia (AML) develop an allogeneic antibody reaction against specific allogeneic antigens. Recently, Wadia and colleagues reported such an antibody response against CHAF1b along with another nuclear protein Nucleolar and Spindle Associated Protein (NuSAP) [90]. While this effect appears to be specific to AML patients undergoing HCT treatment, the mechanisms remain unknown [90].
Concluding Remarks
A significant challenge in understanding the functional roles of CHAF1b is that it is difficult to tease out putative functions outside of the CAF-1 complex. The dogma regarding CHAF1b had been that it is a histone chaperone that complexes with CHAF1a and p48 to form CAF-1, whose sole responsibility is chaperoning newly synthesized histone H3 and H4 dimers as part of the CAF-1 complex to replicating DNA and the subsequent assembly of chromatin. While this is a very important role, recent evidence suggests that CHAF1b has additional functions beyond DNA replication. We now know that CHAF1b is a multifaceted protein with several newly emerging functions. For example, in addition to the role of CAF-1 as a histone chaperone, CHAF1b is also a critical protamine chaperone in the testes, and even integrates into the chromatin with protamines. While CHAF1b plays significant roles in DNA repair and replication, it is also a component responsible for maintaining cell fate decisions in some processes such as neuronal differentiation, though this is capacity could also be due to global CAF-1 complex function [24]. Finally, according to some recent studies, CHAF1b is a major factor in driving metastasis in many different human tumors, and elevated CHAF1b protein levels can be used to accurately predict whether or not these tumors will metastasize. Future studies will continue to shed light on the roles and functions of CHAF1b and may even lead to new therapeutic insights.
Highlights.
CAF-1 complex is conserved in yeast, humans, and plants.
CAF-1 is essential for nucleosome formation on newly replicating DNA
The CAF-1 complex participates in both DNA replication and repair
CAF-1 consists of three subunits, CHAF1a, CHAF1b, and p48
The CHAF1b subunit is overexpressed in many tumor types
Acknowledgments
This review is supported in part by a grant from the NCI (CA101774) to JDC. AV is supported by a training grant from the NCI (T32CA080621).
Abbreviations
- CAF-1
Chromatin Assembly Factor-1
- CHAF1a
Chromatin Assembly Factor-1a
- CHAF1b
Chromatin Assembly Factor-1b
- ASF1a
Anti-silencing Factor 1a
- HIRA
Histone Regulatory homologue A
- PCNA
Proliferating Cell Nuclear Antigen
- PIP
PCNA Interacting Peptide
- DS
Down syndrome
- DSCR
Down syndrome Critical Region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Germond JE, Hirt B, Oudet P, Gross-Bellark M, Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oudet P, Gross-Bellard M, Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975;4:281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- 3.Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- 4.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 5.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 6.Kamakaka RT, Bulger M, Kaufman PD, Stillman B, Kadonaga JT. Postreplicative chromatin assembly by Drosophila and human chromatin assembly factor 1. Molecular and cellular biology. 1996;16:810–817. doi: 10.1128/mcb.16.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyler JK, Collins KA, Prasad-Sinha J, Amiott E, Bulger M, Harte PJ, Kobayashi R, Kadonaga JT. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Molecular and cellular biology. 2001;21:6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nature structural & molecular biology. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Y, Poustovoitov MV, Zhao K, Garfinkel M, Canutescu A, Dunbrack R, Adams PD, Marmorstein R. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nature structural & molecular biology. 2006;13:921–929. doi: 10.1038/nsmb1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz-Carrillo A, Wangh LJ, Allfrey VG. Processing of newly synthesized histone molecules. Science. 1975;190:117–128. doi: 10.1126/science.1166303. [DOI] [PubMed] [Google Scholar]
- 12.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin SG, Zweidler A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature. 1977;266:273–275. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- 14.Brown DT, Wellman SE, Sittman DB. Changes in the levels of three different classes of histone mRNA during murine erythroleukemia cell differentiation. Molecular and cellular biology. 1985;5:2879–2886. doi: 10.1128/mcb.5.11.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Molecular cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 16.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 17.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Current biology : CB. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 18.Imhof A, Wolffe AP. Purification and properties of the Xenopus Hat1 acetyltransferase: association with the 14-3-3 proteins in the oocyte nucleus. Biochemistry. 1999;38:13085–13093. doi: 10.1021/bi9912490. [DOI] [PubMed] [Google Scholar]
- 19.Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. The Journal of biological chemistry. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 20.Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 21.Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Experimental cell research. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- 22.Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 23.Lee SB, Ou DS, Lee CF, Juan LJ. Gene-specific transcriptional activation mediated by the p150 subunit of the chromatin assembly factor 1. The Journal of biological chemistry. 2009;284:14040–14049. doi: 10.1074/jbc.M901833200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakano S, Stillman B, Horvitz HR. Replication-coupled chromatin assembly generates a neuronal bilateral asymmetry in C. elegans. Cell. 2011;147:1525–1536. doi: 10.1016/j.cell.2011.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadyrova LY, Rodriges Blanko E, Kadyrov FA. Human CAF-1-dependent nucleosome assembly in a defined system. Cell cycle. 2013;12:3286–3297. doi: 10.4161/cc.26310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoek M, Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proceedings of the National Academy of; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharp JA, Franco AA, Osley MA, Kaufman PD. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes & development. 2002;16:85–100. doi: 10.1101/gad.925302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krude T. Chromatin assembly: the kinetochore connection. Current biology : CB. 2002;12:R256–258. doi: 10.1016/s0960-9822(02)00786-8. [DOI] [PubMed] [Google Scholar]
- 30.Dong H, Lin W, Zhang CK, Xiong H, Fu G, Jin WR, Chen R, Chen Z, Qi ZT, Huang GM. Genomic sequence and expression analyses of human chromatin assembly factor 1 p150 gene. Gene. 2001;264:187–196. doi: 10.1016/s0378-1119(01)00335-3. [DOI] [PubMed] [Google Scholar]
- 31.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 32.Kuriyan J, O’Donnell M. Sliding clamps of DNA polymerases. Journal of molecular biology. 1993;234:915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- 33.Maga G, Stucki M, Spadari S, Hubscher U. DNA polymerase switching: I. Replication factor C displaces DNA polymerase alpha prior to PCNA loading. Journal of molecular biology. 2000;295:791–801. doi: 10.1006/jmbi.1999.3394. [DOI] [PubMed] [Google Scholar]
- 34.Maga G, Jonsson ZO, Stucki M, Spadari S, Hubscher U. Dual mode of interaction of DNA polymerase epsilon with proliferating cell nuclear antigen in primer binding and DNA synthesis. Journal of molecular biology. 1999;285:259–267. doi: 10.1006/jmbi.1998.2314. [DOI] [PubMed] [Google Scholar]
- 35.Rolef Ben-Shahar T, Castillo AG, Osborne MJ, Borden KL, Kornblatt J, Verreault A. Two fundamentally distinct PCNA interaction peptides contribute to chromatin assembly factor 1 function. Molecular and cellular biology. 2009;29:6353–6365. doi: 10.1128/MCB.01051-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moggs JG, Grandi P, Quivy JP, Jonsson ZO, Hubscher U, Becker PB, Almouzni G. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Molecular and cellular biology. 2000;20:1206–1218. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoek M, Myers MP, Stillman B. An analysis of CAF-1-interacting proteins reveals dynamic and direct interactions with the KU complex and 14-3-3 proteins. The Journal of biological chemistry. 2011;286:10876–10887. doi: 10.1074/jbc.M110.217075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lees-Miller SP. The DNA-dependent protein kinase, DNA-PK: 10 years and no ends in sight. Biochemistry and cell biology = Biochimie et biologie cellulaire. 1996;74:503–512. doi: 10.1139/o96-054. [DOI] [PubMed] [Google Scholar]
- 39.Jiao R, Harrigan JA, Shevelev I, Dietschy T, Selak N, Indig FE, Piotrowski J, Janscak P, Bohr VA, Stagljar I. The Werner syndrome protein is required for recruitment of chromatin assembly factor 1 following DNA damage. Oncogene. 2007;26:3811–3822. doi: 10.1038/sj.onc.1210150. [DOI] [PubMed] [Google Scholar]
- 40.Pietrobon V, Freon K, Hardy J, Costes A, Iraqui I, Ochsenbein F, Lambert SA. The chromatin assembly factor 1 promotes Rad51-dependent template switches at replication forks by counteracting D-loop disassembly by the RecQ-type helicase Rqh1. PLoS biology. 2014;12:e1001968. doi: 10.1371/journal.pbio.1001968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Molecular cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 42.Quivy JP, Gerard A, Cook AJ, Roche D, Almouzni G. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nature structural & molecular biology. 2008;15:972–979. doi: 10.1038/nsmb.1470. [DOI] [PubMed] [Google Scholar]
- 43.Smith CL, Matheson TD, Trombly DJ, Sun X, Campeau E, Han X, Yates JR, 3rd, Kaufman PD. A separable domain of the p150 subunit of human chromatin assembly factor-1 promotes protein and chromosome associations with nucleoli. Molecular biology of the cell. 2014;25:2866–2881. doi: 10.1091/mbc.E14-05-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Molecular cell. 2003;11:341–351. doi: 10.1016/s1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 45.Qian YW, Lee EY. Dual retinoblastoma-binding proteins with properties related to a negative regulator of ras in yeast. The Journal of biological chemistry. 1995;270:25507–25513. doi: 10.1074/jbc.270.43.25507. [DOI] [PubMed] [Google Scholar]
- 46.Qian YW, Wang YC, Hollingsworth RE, Jr, Jones D, Ling N, Lee EY. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- 47.Zhang W, Tyl M, Ward R, Sobott F, Maman J, Murthy AS, Watson AA, Fedorov O, Bowman A, Owen-Hughes T, El Mkami H, Murzina NV, Norman DG, Laue ED. Structural plasticity of histones H3–H4 facilitates their allosteric exchange between RbAp48 and ASF1. Nature structural & molecular biology. 2013;20:29–35. doi: 10.1038/nsmb.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 49.Song JJ, Garlick JD, Kingston RE. Structural basis of histone H4 recognition by p55. Genes & development. 2008;22:1313–1318. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marheineke K, Krude T. Nucleosome assembly activity and intracellular localization of human CAF-1 changes during the cell division cycle. The Journal of biological chemistry. 1998;273:15279–15286. doi: 10.1074/jbc.273.24.15279. [DOI] [PubMed] [Google Scholar]
- 51.Anderson AE, Karandikar UC, Pepple KL, Chen Z, Bergmann A, Mardon G. The enhancer of trithorax and polycomb gene Caf1/p55 is essential for cell survival and patterning in Drosophila development. Development. 2011;138:1957–1966. doi: 10.1242/dev.058461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vizan P, Beringer M, Ballare C, Di Croce L. Role of PRC2-associated factors in stem cells and disease. The FEBS journal. 2014 doi: 10.1111/febs.13083. [DOI] [PubMed] [Google Scholar]
- 53.Martinez-Balbas MA, Tsukiyama T, Gdula D, Wu C. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes & development. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & development. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krawitz DC, Kama T, Kaufman PD. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Molecular and cellular biology. 2002;22:614–625. doi: 10.1128/MCB.22.2.614-625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- 58.Rai TS, Adams PD. Lessons from senescence: chromatin maintenance in non-proliferating cells. Biochimica et biophysica acta. 2013;1819:322–331. [PubMed] [Google Scholar]
- 59.Nakatani Y, Ray-Gallet D, Quivy JP, Tagami H, Almouzni G. Two distinct nucleosome assembly pathways: dependent or independent of DNA synthesis promoted by histone H3.1 and H3.3 complexes. Cold Spring Harbor symposia on quantitative biology. 2004;69:273–280. doi: 10.1101/sqb.2004.69.273. [DOI] [PubMed] [Google Scholar]
- 60.Tan BC, Chien CT, Hirose S, Lee SC. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. The EMBO journal. 2006;25:3975–3985. doi: 10.1038/sj.emboj.7601271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 62.Krude T, Knippers R. Nucleosome assembly during complementary DNA strand synthesis in extracts from mammalian cells. The Journal of biological chemistry. 1993;268:14432–14442. [PubMed] [Google Scholar]
- 63.Mello JA, Sillje HH, Roche DM, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO reports. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nabatiyan A, Krude T. Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis. Molecular and cellular biology. 2004;24:2853–2862. doi: 10.1128/MCB.24.7.2853-2862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nature structural & molecular biology. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 66.Gaillard PH, Martini EM, Kaufman PD, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 67.Martini E, Roche DM, Marheineke K, Verreault A, Almouzni G. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. The Journal of cell biology. 1998;143:563–575. doi: 10.1083/jcb.143.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu Q, Wani G, Arab HH, El-Mahdy MA, Ray A, Wani AA. Chromatin restoration following nucleotide excision repair involves the incorporation of ubiquitinated H2A at damaged genomic sites. DNA repair. 2009;8:262–273. doi: 10.1016/j.dnarep.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Braun RE. Packaging paternal chromosomes with protamine. Nature genetics. 2001;28:10–12. doi: 10.1038/ng0501-10. [DOI] [PubMed] [Google Scholar]
- 70.Caldwell KA, Handel MA. Protamine transcript sharing among postmeiotic spermatids. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:2407–2411. doi: 10.1073/pnas.88.6.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doyen CM, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Rathke C, Renkawitz-Pohl R, Verrijzer CP. Subunits of the histone chaperone CAF1 also mediate assembly of protamine-based chromatin. Cell reports. 2013;4:59–65. doi: 10.1016/j.celrep.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 72.Eirin-Lopez JM, Frehlick LJ, Ausio J. Protamines, in the footsteps of linker histone evolution. The Journal of biological chemistry. 2006;281:1–4. doi: 10.1074/jbc.R500018200. [DOI] [PubMed] [Google Scholar]
- 73.Smith S, Stillman B. Immunological characterization of chromatin assembly factor I, a human cell factor required for chromatin assembly during DNA replication in vitro. The Journal of biological chemistry. 1991;266:12041–12047. [PubMed] [Google Scholar]
- 74.Keller C, Krude T. Requirement of Cyclin/Cdk2 and protein phosphatase 1 activity for chromatin assembly factor 1-dependent chromatin assembly during DNA synthesis. The Journal of biological chemistry. 2000;275:35512–35521. doi: 10.1074/jbc.M003073200. [DOI] [PubMed] [Google Scholar]
- 75.Carter T, Vancurova I, Sun I, Lou W, DeLeon S. A DNA-activated protein kinase from HeLa cell nuclei. Molecular and cellular biology. 1990;10:6460–6471. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lees-Miller SP, Chen YR, Anderson CW. Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Molecular and cellular biology. 1990;10:6472–6481. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blouin JL, Duriaux-Sail G, Chen H, Gos A, Morris MA, Rossier C, Antonarakis SE. Mapping of the gene for the p60 subunit of the human chromatin assembly factor (CAF1A) to the Down syndrome region of chromosome 21. Genomics. 1996;33:309–312. doi: 10.1006/geno.1996.0199. [DOI] [PubMed] [Google Scholar]
- 78.Katsanis N, Fisher EM. The gene encoding the p60 subunit of chromatin assembly factor I (CAF1P60) maps to human chromosome 21q22.2, a region associated with some of the major features of Down syndrome. Human genetics. 1996;98:497–499. doi: 10.1007/s004390050246. [DOI] [PubMed] [Google Scholar]
- 79.Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukaemia and solid tumours in individuals with Down’s syndrome. Lancet. 2000;355:165–169. doi: 10.1016/S0140-6736(99)05264-2. [DOI] [PubMed] [Google Scholar]
- 80.Malinge S, Bliss-Moreau M, Kirsammer G, Diebold L, Chlon T, Gurbuxani S, Crispino JD. Increased dosage of the chromosome 21 ortholog Dyrk1a promotes megakaryoblastic leukemia in a murine model of Down syndrome. The Journal of clinical investigation. 2012;122:948–962. doi: 10.1172/JCI60455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, Kok CY, Jia M, De T, Teague JW, Stratton MR, McDermott U, Campbell PJ. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic acids research. 2015;43:D805–811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Tayrac M, Saikali S, Aubry M, Bellaud P, Boniface R, Quillien V, Mosser J. Prognostic significance of EDN/RB, HJURP, p60/CAF-1 and PDLI4, four new markers in high-grade gliomas. PloS one. 2013;8:e73332. doi: 10.1371/journal.pone.0073332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mascolo M, Vecchione ML, Ilardi G, Scalvenzi M, Molea G, Di Benedetto M, Nugnes L, Siano M, De Rosa G, Staibano S. Overexpression of Chromatin Assembly Factor-1/p60 helps to predict the prognosis of melanoma patients. BMC cancer. 2010;10:63. doi: 10.1186/1471-2407-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Tayrac M, Aubry M, Saikali S, Etcheverry A, Surbled C, Guenot F, Galibert MD, Hamlat A, Lesimple T, Quillien V, Menei P, Mosser J. A 4-gene signature associated with clinical outcome in high-grade gliomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:317–327. doi: 10.1158/1078-0432.CCR-10-1126. [DOI] [PubMed] [Google Scholar]
- 85.Staibano S, Mascolo M, Mancini FP, Kisslinger A, Salvatore G, Di Benedetto M, Chieffi P, Altieri V, Prezioso D, Ilardi G, De Rosa G, Tramontano D. Overexpression of chromatin assembly factor-1 (CAF-1) p60 is predictive of adverse behaviour of prostatic cancer. Histopathology. 2009;54:580–589. doi: 10.1111/j.1365-2559.2009.03266.x. [DOI] [PubMed] [Google Scholar]
- 86.Staibano S, Mascolo M, Rocco A, Lo Muzio L, Ilardi G, Siano M, Pannone G, Vecchione ML, Nugnes L, Califano L, Zamparese R, Bufo P, De Rosa G. The proliferation marker Chromatin Assembly Factor-1 is of clinical value in predicting the biological behaviour of salivary gland tumours. Oncology reports. 2011;25:13–22. [PubMed] [Google Scholar]
- 87.Polo SE, Theocharis SE, Grandin L, Gambotti L, Antoni G, Savignoni A, Asselain B, Patsouris E, Almouzni G. Clinical significance and prognostic value of chromatin assembly factor-1 overexpression in human solid tumours. Histopathology. 2010;57:716–724. doi: 10.1111/j.1365-2559.2010.03681.x. [DOI] [PubMed] [Google Scholar]
- 88.Hasle H. Pattern of malignant disorders in individuals with Down’s syndrome. The Lancet Oncology. 2001;2:429–436. doi: 10.1016/S1470-2045(00)00435-6. [DOI] [PubMed] [Google Scholar]
- 89.Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, Hijazi H, Alshammari M, Aldahmesh MA, Salih MA, Faqeih E, Alhashem A, Bashiri FA, Al-Owain M, Kentab AY, Sogaty S, Al Tala S, Temsah M, Tulbah M, Aljelaify RF, Alshahwan SA, Seidahmed MZ, Alhadid AA, Aldhalaan H, AlQallaf F, Kurdi W, Alfadhel M, Babay Z, Alsogheer M, Kaya N, Al-Hassnan ZN, Abdel-Salam GM, Al-Sannaa N, Al Mutairi F, El Khashab HY, Bohlega S, Jia X, Nguyen HC, Hammami R, Adly N, Mohamed JY, Abdulwahab F, Ibrahim N, Naim EA, Al-Younes B, Meyer BF, Hashem M, Shaheen R, Xiong Y, Abouelhoda M, Aldeeri AA, Monies DM, Alkuraya FS. Accelerating Novel Candidate Gene Discovery in Neurogenetic Disorders via Whole-Exome Sequencing of Prescreened Multiplex Consanguineous Families. Cell reports. 2014 doi: 10.1016/j.celrep.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 90.Wadia PP, Coram M, Armstrong RJ, Mindrinos M, Butte AJ, Miklos DB. Antibodies specifically target AML antigen NuSAP1 after allogeneic bone marrow transplantation. Blood. 2010;115:2077–2087. doi: 10.1182/blood-2009-03-211375. [DOI] [PMC free article] [PubMed] [Google Scholar]