Abstract
Sphingolipids are bioactive lipid effectors, which are involved in the regulation of various cellular signaling pathways. Sphingolipids play essential roles in controlling cell inflammation, proliferation, death, migration, senescence, metastasis and autophagy. Alterations in sphingolipid metabolism has been also implicated in many human cancers. Macroautophagy (referred to here as autophagy) is a form of nonselective sequestering of cytosolic materials by double membrane structures, autophagosomes, which can be either protective or lethal for cells. Ceramide, a central molecule of sphingolipid metabolism is involved in the regulation of autophagy at various levels, including the induction of lethal mitophagy, a selective autophagy process to target and eliminate damaged mitochondria. In this review, we focused on recent studies with regard to the regulation of autophagy, in particular lethal mitophagy, by ceramide, and aimed at providing discussion points for various context-dependent roles and mechanisms of action of ceramide in controlling mitophagy.
Keywords: Ceramide, sphingolipids, mitophagy, tumor suppression
1. Introduction
Sphingolipids are membrane lipids with important functions in regulating membrane fluidity and subdomain structures. Advances in sphingolipid research suggest that sphingolipids are highly bioactive molecules and have a great impact on cellular signaling and disease pathogenesis [1]. Ceramide is one of the central molecules of sphingolipid metabolism and plays a key role in the regulation of various cellular functions, including cell proliferation, death, migration, and senescence [2]. Ceramide is intimately involved in cancer pathogenesis; alterations in its metabolism are involved in controlling cancer initiation, progression, and/or response of cancer cells to chemotherapeutic agents and radiation. Endogenous levels of ceramide, especially C18-ceramide, are suppressed in many types of tumor tissues compared to non-cancerous counterparts. Additionally, ceramide levels increase upon exposure of cancer cells to stress-causing agents, such as cytokines, anticancer drugs, and radiation, leading to cancer cell death and tumor suppression [2, 3]. Ceramide-mediated tumor suppression is regulated by various mechanisms such as apoptosis, necroptosis, lethal autophagy, and mitophagy [4-8].
Mitophagy is a form of autophagy that results in selective degradation of mitochondria through the autophagic machinery [9]. Ceramide plays a key role in the regulation of general autophagy at many levels from initiation to formation of autophagosomes [10]. Recently, we have begun to better understand ceramide’s role in inducing selective lethal mitophagy. Several recent studies showed the ability of endogenous or exogenous ceramides to cause cell death after accumulating in the mitochondria [11-13]. Our recent study showed that mitochondrial ceramide acts as a receptor for LC3-II [14]. Ceramide binding directly to LC3-II protein leads to the recruitment of autophagosomes to damaged mitochondria, resulting in lethal mitophagy [14]. This review will focus on the roles and mechanism of action of ceramide in the regulation of mitophagy and tumor suppression.
2. Metabolism and biological roles of ceramide
a. Structure and metabolism of ceramide
Ceramide is a bioactive sphingolipid with a peculiar structure. It is composed of a sphingosine backbone that is esterified to a fatty acyl chain via an amide linkage at carbon 3 [1, 2]. The variety in the length of the fatty acyl chain generates many different ceramides, such as C14-to C26-ceramides. A trans-double bond between carbons 4 and 5 in the sphingosine backbone is required for its biological activity, as the loss of the double bond generates dihydro-ceramide [1].
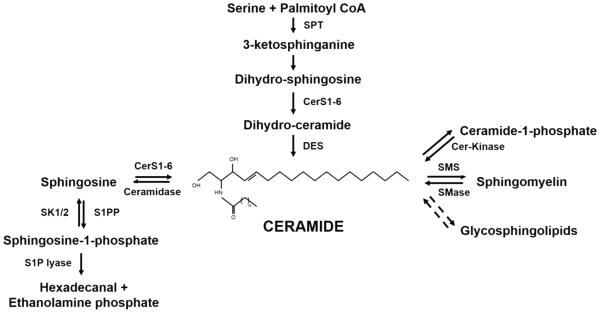
Ceramide lies at the center of sphingolipid metabolism: acting as a substrate for the generation of more complex sphingolipids, or as a product of the breakdown of complex sphingolipid molecules (Figure 1). As a substrate, ceramide is converted to sphingomyelin, ceramide-1-phosphate, hexosylceramides, and other complex glycosphingolipids and gangliosides [1]. As a product, ceramide can be generated by the breakdown of these more complex sphingolipids by specialized enzymes such as sphingomyelinases (SMAse), which hydrolyze sphingomyelin (SM), and cerebrosidases which hydrolyze hexosylceramides [15].
Fig. 1.
Ceramide metabolic pathways. Ceramide lies at the center of sphingolipid metabolism. Ceramide de novo generation starts with a condensation reaction involving Serine and Palmitoyl CoA by the enzyme Serine Palmitoyl CoA Transferase (SPT), to generate 3-ketosphinganine, which is then converted to sphinganine or dihydrosphingosine. Then, Ceramide Synthases (CerS1-6) transfer a fatty acyl CoA to the amino group yielding dihydroceramide which gets desaturated to ceramide by Dihydro-ceramide Desaturase enzyme (DES). Ceramide can be generated back from Sphingosine-1-phosphate with the help of S1P phosphatase (S1PP) and ceramide synthase (salvage pathway) or from Sphingomyelin by Sphingomyelinase (SMase), from Glycosphingolipids, or from Ceramide-1-phosphate by Ceramide kinase (Cer-Kinase). As a metabolic outlet for ceramide, it is converted to sphingosine by the action of ceramidase. Sphingosine kinases 1 and 2 (SK1/2) phosphorylate sphingosine to sphingosine-1-phosphate that can be further degraded by S1P lyase to hexadecanal and ethanolamine phosphate.
De novo generation of ceramide begins with the condensation of serine and palmitoylCoA to form 3-ketosphinganine, and then dihydrosphingosine (sphinganine) [16], involving multiple metabolic enzymes. This is followed by the action of ceramide synthases 1-6 (CerS1-6) (also known as dihydro-ceramide synthases) that esterify the dihydrosphingosine to generate dihydroceramide [17]. Ceramide is then generated by the action of (dihydroceramide)-desaturase that irreversibly inserts a double bond between carbons 4 and 5 [18].
Ceramide catabolism is regulated mainly by ceramidases, which cleave ceramide to generate sphingosine. Sphingosine then gets phosphorylated by sphingosine kinases 1 or 2 (SphK1 or SphK2) to produce sphingosine 1-phosphate (S1P). S1P can be lysed to hexadecanal and ethanolaminephosphate by S1P lyase or dephosphorylated by S1P phosphatases[19]. Hence, endogenous levels of ceramide increase through de novo synthesis, activation of sphingomyelinases, or decreased clearance through the inhibition of glucosylceramide synthase (GCS), sphingomyelin synthase (SMS), or ceramidase (CDase) [20].
Sphingolipid metabolism is highly compartmentalized in cells, as the subcellular localization of different sphingolipid molecules and the metabolic enzymes play key roles in this process [1]. For example, CerS1-6 enzymes are localized in the endoplasmic reticulum (ER) where de novo synthesis of ceramide occurs. For the synthesis of SM, ceramide is transported from the ER to the Golgi apparatus by ceramide transporter protein (CERT) in a non-vesicular fashion [21, 22]. Similarly, for the synthesis of glucosylceramide (GlcCer), ceramide is transported to the Golgi by Fabb2 transporter [22]. Recently, a role for a lipid transporter protein GLTPD1 for ceramide 1-phosphate, referred to as CPTP, has been discovered, which plays a critical role in the regulation of inflammation [23]. Ceramide is also found in the mitochondria where it can be generated by neutral sphingomyelinase (N-SMase) in response to increased reactive oxygen species (ROS) production [1]. Ceramide accumulation in the mitochondria can lead to ceramide stress-induced mitochondrial fragmentation, and decrease in ATP production [14]. In lysosomes, ceramide is mainly produced by acid sphingomyelinase (A-SMase), and in the plasma membrane, ceramide is localized within lipid rafts, which are specialized membrane microdomains that regulate various signaling pathways [24].
b. Biological functions of Ceramide Synthases 1-6
Ceramide Synthases 1-6 (CerS1-6) were first identified in yeast as longevity assurance gene 1 (LAG1). LAG1 regulates longevity in yeast in a way that its deletion prolongs the replicative lifespan [17]. The mouse homologue of LAG1 is LASS1, also known as the upstream of growth and differentiation factor 1 (UOG1), which was discovered to specifically regulate the synthesis of C18-ceramide [25, 26]. There are at least six mammalian LASS proteins that are currently known as CerS1-6 [17]. All of the CerS enzymes share a domain required for their enzymatic activity to generate ceramide called TLC domain (TRAM, LAG1, and CLN8 homology), which is made up of five predicted transmembrane helices [27, 28]. CerS2-6 also contain a HOX domain which CerS1 lacks [29, 30].
CerS1-6 exhibit some specificity for the generation of endogenous ceramides with different fatty acid chain lengths [31, 32]. For example, CerS1 mainly generates ceramide with an 18-carbon containing fatty acid chain (C18-ceramide), CerS4 generates both C18-ceramide and C20-ceramide, and CerS5 and CerS6 generate mainly C16-ceramide, and to a lesser extent, C12- and C14-ceramides. CerS2 and CerS3 are known to generate very long chain C22-24- and C26-ceramides, respectively [1-3, 7, 15, 33].
Different species of ceramide with distinct fatty acyl chain lengths have diverse biological functions (Table 1). For example, CerS1 and CerS6, generating C18-ceramide and C16-ceramide respectively, have opposing roles in cell death and proliferation in head and neck cancer cells and tumors. CerS1/C18-ceramide axis leads to cancer cell death and decreases head and neck tumor growth [34-36]. Increased levels of serum C18-ceramide act as a potential biomarker to monitor patients’ response to chemotherapy [37]. On the other hand, C16-ceramide promotes head and neck cancer cell proliferation and its increased serum levels associate with a positive lymph node status in breast cancer patients [38, 39]. On the other hand, there are studies showing that C16-ceramide is pro-apoptotic, whereas C24-ceramide protects from cell death [40, 41]. Thus, overall, distinct roles of ceramides appear to be context dependent, especially that knockout mice for multiple CerS enzymes exhibit different phenotypes: mice that express a mutant CerS1 (toppler mice) exhibit neurological disorders associated mainly with alterations in Purkinjee cells; mice with genetic loss of CerS2 results in liver damage; mice with CerS6 knockout exhibit neurological/behavioral alterations; and CerS4 knockout mice exhibit severe alopecia with alterations in sebaceous glands and sebum contents [42-43,204]. The demonstration of the distinct roles of CerS-generated ceramides in the regulation of cell death is also consistent with studies performed in Drosophila and C. elegans that showed the distinct roles of CerS-generated ceramides in these organisms [45-47]. Moreover, recent evidence suggests that changes in the carbon length of the sphingoid base of ceramides with 18 versus 16 carbons play roles in inducing survival autophagy versus cell death in cardiomyocytes [48].
Table 1.
Diverse biological roles of Ceramide Synthases 1-6.
| Ceramide Synthase isoform |
Ceramide product | Biological Role |
|---|---|---|
| Ceramide Synthase 1 | C18-ceramide | |
|
| ||
| Ceramide Synthase 2 | C22-24- ceramide | |
|
| ||
| Ceramide Synthase 3 | Ultra long chain ceramides >C26 |
|
|
| ||
| Ceramide Synthase 4 | C18- and C20-ceramide |
|
|
| ||
| Ceramide Synthase 5 | C12-, C14., and C16-ceramide |
|
|
| ||
| Ceramide Synthase 6 | C12-, C14-, and C16-
ceramide |
|
c. Cancer suppressing role of ceramide
Ceramide induces a variety of anti-proliferative responses such as programmed cell death, cell cycle arrest, senescence, and differentiation. Indeed, studies show that ceramide is involved in apoptosis, necroptosis, lethal autophagy, and mitophagy, all of which decrease cancer cell viability [14, 49-51]. Exogenously supplied ceramides (C2-C18-ceramides), also have anti-proliferative activities when added to cells in culture [11, 12, 52]. Thus, ceramide metabolism has a significant role in suppressing cancer progression, and ceramide is emerging as a tumor suppressor lipid.
Ceramide is generated during stress conditions such as hypoxia, growth factor withdrawal, hyperthermia, and DNA damage which then mediates cell death [53]. In addition, there is an increase in ceramide levels during the activation of extrinsic apoptosis via FAS/FAS ligand pathway or tumor necrosis factor (TNF)-alpha pathway [54]. Ionizing radiation causes ceramide formation by activating A-SMase while androgen ablation in prostate cancer cells induces de novo generation of ceramide, leading to cell death [55-57].
One mechanism by which ceramide leads to cell death is by activating protein phosphatases PP1 and PP2A [58]. PP2A is a tumor suppressor protein that acts as a phosphatase to regulate signaling of many targets including Akt, c-Myc, and Bcr-Abl [59, 60]. Ceramide binds to the biological inhibitor of PP2A, I2PP2A or SET oncoprotein, leading to PP2A reactivation. PP2A can then dephosphorylate and inactivate several anti-apoptotic proteins such as Bcl-2 and AKT, or c-Myc. [50, 61]. Interestingly, SET/I2PP2A oncoprotein preferentially associates with endogenous C18-ceramide compared with other ceramide species [62]. This specificity for binding to a specific species of ceramide is also evident in CERT binding preferentially to C16- and C18-ceramides but not very long chain ceramides [63]. Ceramide interacts with another phosphatase, PP1, which inactivates the pro-apoptotic protein Bid [54]. Moreover, ceramide-PP1 is involved in the regulation of retinoblastoma protein (RB), a tumor suppressor protein that plays an important role in cell cycle regulation. Ceramide treatment can dephosphorylate and activate RB leading to cell cycle arrest in cancer cells [64]. In addition, ceramide generated in lysosomes by A-SMase activates cathepsin D by inducing autocatalytic proteolysis, resulting in Bid cleavage and caspase activation [65, 66].
Another mechanism that associates with the tumor suppressor roles of ceramide is its regulation of telomere length by acting as an upstream regulator of telomerase. Studies showed that ceramide accumulation in lung cancer cells inhibits telomerase expression by inactivating c-Myc transcription factor, which is an activator of the human telomerase reverse transcriptase (hTERT) promoter, via increased ubiquitination and proteasome mediated degradation. The inactivation of telomerase prevents the cancer cell from elongating the telomeric ends of the chromosomes after each replication cycle, leading eventually to cell death [67, 68].
Ceramide is also involved in pathways that lead to quiescence and senescence in cancer cells. For example, ceramide inactivates cyclin dependent kinase 2 (CDK2), and upregulates the expression of CDK inhibitors p21 and p27 in Wi-38 fibroblasts and nasopharyngeal carcinoma cells, respectively [69-71]. In addition, exogenous supply of ceramide to fibroblasts cultured at low passage induces the biochemical and morphological features of senescence [1, 72]. By inducing senescence, ceramide helps in the suppression of key mitogenic pathways, leading to tumor suppression [73].
Because of these pro-death characteristics, ceramide analogues or mimetics have the potential to act as anti-cancer agents. Endogenous accumulation of ceramide might also be beneficial to suppress tumor growth. One example is the group of ceramide analogues, Ceramidoids or pyridinium ceramides, which preferentially accumulate in the mitochondria/nuclei, and suppress the tumor growth of lung, breast, and head and neck squamous cell carcinomas [52, 74]. The mitochondrial accumulation of pyridinium ceramides results in mitochondrial permeability transition and either caspase-dependent apoptosis or mitophagy [14, 74]. Another example is the group of glucosylceramide synthase inhibitors (PPMP and PPPP) that lead to the accumulation of ceramide, which decrease glucosylceramide generation, and suppress solid tumor growth [75, 76]. Ceramide can also be delivered exogenously in PEGylated nanoliposomes. Studies using these liposomes show that this delivery method of ceramide decreases phosphorylation of AKT, stimulates the activity of caspase 3/7, and prevents the growth of breast cancer cells in vitro and in vivo [77, 78].
3. General autophagy and its regulation by ceramide
a. Progression of autophagy
Autophagy, or self-eating in Greek, describes the mechanism utilized by the cell to self-digest internal organelles and misfolded proteins using lysosomal hydrolytic enzymes [79]. Autophagy starts with the formation of cup shaped structures called phagophores, also known as isolation membranes, which will elongate to engulf organelles and other cytoplasmic components. The maturation of the phagophores leads to the formation of an autophagosome that fuses with lysosomes for the formation of autophagolysosomes [80]. It is at this stage that lysosomal enzymes start the digestion process and the rate of breakdown of the cellular components is referred to as lysosomal flux. Autophagy plays a critical role in physiology and cellular homeostasis, allowing the cells to recycle nutrients from digested organelles at times of starvation and remove aberrantly folded proteins [81-84]. Dysregulation in autophagy has been implicated in various human diseases such as neurodegenerative disorders, cardiovascular diseases, and cancer [84-89].
The discovery of autophagy genes (Atg) in yeast had a great impact on our understanding of the mechanisms of autophagy process. Most of the Atg genes are conserved in humans and play various roles including autophagy initiation, autophagosome formation and maturation [90-92]. During initiation, Atg1 forms a complex with ULK (Unc-51 like kinase) that integrates inputs from mTOR signaling [87, 93-95]. During autophagosome formation, Atg9 allows for membrane addition and retrieval to and from sites of autophagosome biogenesis [83, 96], while Atg6 (or Beclin 1 in mammals) forms a multimeric complex with Atg14, Vps34/PI3kinase, and Vps15 [94, 96]. In addition, during autophagosome formation, Atg12 is activated by the enzyme Atg7 (E1-like ubiquitin ligase) and then transferred to Atg10 (E2-like ubiquitin ligase) to be conjugated to Atg5 (E3-like ubiquitin ligase), forming an autophagosomal precursor. Finally, Atg8 is required during the maturation phase [93, 95, 97, 100, 101].
LC3, mammalian homologue of Atg8, which plays important roles for the maturation of autophagosomes, has three isoforms: LC3A, LC3B and LC3C. GABARAP and GATE16 are also mammalian homologues of Atg8 [102]. LC3 is synthesized in the cell in its cytosolic form, LC3-I, with a carboxy terminal glycine (Gly120) [102]. During autophagy, LC3 is cleaved by Atg4 protease and then activated by Atg7 to be transferred to Atg3 (E2-like ubiquitin ligase) [80, 103]. This allows the Gly120 residue on the carboxyl terminal to be conjugated to phosphatidylethanolamine (PE) to form LC3-II. This allows its docking to the membranes, leading to membrane closure and formation of a mature autophagosome [104]. This sets LC3-II as a well-established marker of autophagosomes.
b. Autophagy paradox: cell survival or death regulation
Autophagy was initially discovered as a mechanism occurring at a low rate to remove protein aggregates and damaged organelles that are otherwise toxic for the cell [81]. In addition, autophagy is considered as a vital process during metabolic stress or nutrient deprivation, in which the degradation of organelles provides macromolecules and nutrients that maintain energy production and the basic cellular functions [79, 82, 99]. Mechanistically, several reports show that upon cell starvation, LC3-I is modified to LC3-II to promote autophagy [80]. In vivo, GFP-LC3 localizes in punctate structures in heart and skeletal muscle tissues in transgenic mice when the animals were under fasting conditions [102, 105]. These functions of autophagy provide pro-survival and cyto-protective mechanisms.
Recently, autophagy was found to be a pro-death mechanism especially if it occurs for a sustained period of time with a high intensity. Cells with sustained upregulation of autophagic activity became atrophic with loss of vital organelles and cellular functions [106]. This suggests that over-activated autophagy can lead to cell death when associated with major elimination of essential organelles. Moreover, studies indicate that autophagy can lead to cell death via its ability to degrade cyto-protective proteins such as catalase, an anti-oxidant enzyme [107]. Autophagy can also result in cell death by upregulating apoptosis or necroptosis [108-110]. One particular example is the case of Atg5, which during autophagy can translocate to the mitochondria to induce mitochondrial membrane depolarization and caspase dependent cell death [111]. However, autophagic cell death, also known as lethal autophagy, or autosis, can be achieved independent of apoptosis or necroptosis. This type of cell death is rescued by suppression of autophagy by pharmacological or genetic approaches, and involves the action of autophagy genes and lysosomal flux during the death process [106, 108, 112-114].
c. Autophagy in cancer
There are several lines of evidence supporting that autophagy is a tumor-suppressor mechanism: Cancer cells have increased oxidative and metabolic stress that cause chromosomal abnormalities, DNA strand breaks, and gene mutations. Autophagy helps in scavenging reactive oxygen species by removing the damaged organelles, thus preventing the genetic abnormalities that might otherwise lead to oncogene activation or tumor suppressor gene inactivation [115-118]. Some cancer cells suppress autophagy as a mechanism to avoid the quality control during oxidative stress, DNA damage, and genetic instability [119]. One example to illustrate this is the role of autophagy in the turnover of p62 protein. When autophagy is suppressed, p62 protein clearance is prevented, leading to its accumulation, which in turn activates NRF2 (nuclear factor erythroid 2 related factor 2). NRF2 can then translocate to the nucleus, where it activates an anti-oxidant and pro-survival response [117, 120-122].
Some of the Atg proteins act as tumor suppressor genes. Beclin 1 (Atg6) expression is suppressed in malignant breast epithelial cells, and it is monoallelically deleted in 40-70% of prostate, ovarian, and breast cancers. Overexpression of Beclin 1 in breast cancer cells promoted autophagy and inhibited the malignant phenotype [123-125]. In vivo, targeted deletion of Beclin 1 in mice led to early embryonic death. Heterozygous disruption of Beclin 1 resulted in an increased risk of spontaneous tumor development, even though the other allele is intact. This suggests that the pro-autophagic Beclin 1 is a haplo-insufficient tumor suppressor protein [126]. Further studies then highlighted that other pro-autophagic proteins also act as tumor suppressors, such as Atg5 and Bif1 [83, 84, 112]. Autophagy can also be suppressed due to its regulation by signaling pathways that are up- or down-regulated in the cancer cells [112]. For instance, cancer cells with upregulation of PI3K-AKT-mTOR signaling cascade, or downregulation of PTEN activity, will have suppressed autophagy, promoting tumor growth/proliferaiton [83, 127, 128].
As cancer progresses to late stages, autophagy can act as a mechanism to help the cancer cells meet metabolic demands and repair intracellular damages inflicted by the aggressive tumor environment [84, 112, 119]. Pancreatic cancer cell lines demonstrate a high basal rate of autophagy, and upon pharmacological inhibition of autophagy by chloroquine, pancreatic cancer cell growth was diminished mainly due to increased DNA damage and oxidative stress [117, 129]. Additionally, cancer cells expressing the Ras oncogene have a higher basal rate of autophagy, such that Ras expressing Atg5−/− and Atg7−/− cells, have suppressed autophagy levels and showed reduced tumor growth in vivo [130].
The implication of autophagy in cancer pathogenesis gives insight into new pharmacological therapies for cancer. If autophagy is required for the survival of cancer cells in the late stages, then pharmacological inhibition of autophagy can enhance the anti-cancerous effect of chemotherapeutic drugs [84, 112, 112, 117, 120]. For instance, combining vinblastine with C6-ceramide attenuated autophagy and inhibited cancer cell growth in a synergistic fashion [131]. In contrast, if autophagy induction leads to cancer cell death via lethal autophagy, then drugs that induce autophagy will lead to tumor suppression. For example, pyridinium ceramide treatment leads to cancer cell death via in part activating autophagy [11].
d. Role of ceramide in mediating general autophagy
Ceramide is known to induce both survival and lethal autophagy via several mechanisms that are outlined in Figure 2 [4, 132, 133]. One mechanism by which ceramide can induce survival autophagy is by regulating cellular nutrient transporters [134-136]. Transporter proteins are required by cells to move nutrients across the plasma membrane. Since these transporters control the cellular fuel supply, alterations of the expression of the nutrient transporters is one way to affect survival and cell growth. Ceramide was shown to down-regulate the expression of amino acid and nutrient transporters leading to starvation, a state that induces survival autophagy by reducing mTOR signaling or activating AMPK [135, 137]. Survival autophagy was also induced in the context of CerS2 downregulation that dysregulated the normal trafficking of ceramide in the ER, leading to long chain ceramide accumulation, and activation of pro-survival IRE-1 (inositol requiring element 1) to prevent induction of cell death [133].
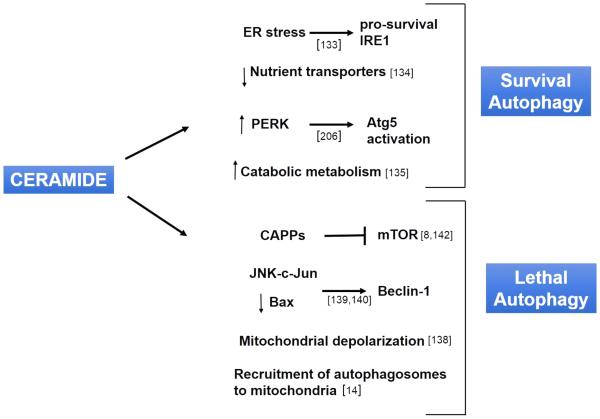
Fig. 2.
Ceramide’s role in survival and lethal autophagy. Ceramide regulates several signaling pathways in autophagy, some of which lead to cytoprotective autophagy and survival while some lead to cell death through lethal autophagy. In the context of survival autophagy, ceramide can induce ER stress that activates the pro-survival IRE1 (inositol-requiring element 1), downregulate nutrient transporters and improves catabolic metabolism, and can result in activation of Atg5 via CD95 and PERK. In the context of lethal autophagy, ceramide can induce ER stress that inhibits mTOR through TRB3, activate Ceramide associated phosphatases (CAPPs) that also inhibit mTOR by inactivating Akt, increase the expression of Beclin-1 by activating JNK-c-Jun axis or by inactivating Bax, leading to loss of mitochondrial membrane potential and activation of BNIP3, and bind to LC3-II to recruit autophagosomes to engulf mitochondria.
Moreover, ceramide is shown to induce lethal autophagy by affecting the expression of pro-autophagic protein Beclin 1 whereby exogenous treatment of cells with C2-ceramide increased Beclin 1 expression and induced lethal autophagy [138]. This is due to the conversion of C2-ceramide to long chain ceramide as the effect was inhibited when de novo ceramide synthesis was blocked using myriocin, the pharmacological inhibitor of SPT [138]. Further evidence indicated that ceramide increases Beclin 1 expression by activating JNK kinase, which in turn activates c-Jun, a known transcription factor for Beclin 1 expression [139]. In addition, JNK activation by ceramide leads to Bcl2 phosphorylation allowing it to dissociate from Beclin 1 [140].. In addition, chemotherapeutic drugs and arsenic trioxide lead to ceramide production that increase Beclin1 expression and promote lethal autophagy [141]. Other drugs such as cannabinoids also lead to ceramide accumulation to induce ER stress, mTOR inhibition via TRB3 (tribbles homolog 3), and lethal autophagy [142].
Ceramide can be hydrolyzed for the generation of S1P, which plays important roles in the regulation of autophagy [19, 143]. When cells were subjected to starvation, the activity of sphingosine kinase 1 (SphK1) increased, leading to increased accumulation of S1P [144]. SphK1 overexpression was able to induce autophagy by inhibiting the mTOR pathway, but unlike the case of ceramide, this mechanism was independent of AKT dephosphorylation [143-145]. In addition, SphK1 downregulation enhanced ER stress and induced autophagy in an mTOR independent fashion [146].
4. Ceramide-mediated mitophagy
a. Mitophagy: selective autophagy of the mitochondria
Autophagy was considered to be a general process whereby the autophagosomes engulf many cytoplasmic elements, including mitochondria, endoplasmic reticulum, and peroxisomes [147]. However, recent findings suggest that autophagy can be selective to a specific organelle. The findings are based on the discovery of specific proteins of the organelles that are required to instigate the autophagy process. These include peroxin 14 of peroxisomes in yeast for pexophagy, the autophagic degradation of peroxisomes; and Uth1p, an outer mitochondrial membrane protein required for selective mitochondrial autophagy, also known as mitophagy [148-150].
Aged and dysfunctional mitochondria are removed from cells to prevent the harm of unhealthy mitochondria, which generate reactive oxygen species, and release pro-apoptotic proteins [151]. This turnover process involves the action of autophagosomes and lysosomal hydrolytic enzymes. The term mitophagy has been suggested to refer to such process that selectively removes the mitochondria by autophagy [151-153].
It has been shown that mitochondria in hepatocytes that had undergone a mitochondrial permeability transition or a depolarization of the mitochondrial membrane potential are selectively removed by autophagosomes [153]. These studies showed that upon loss of mitochondrial membrane potential or during mitochondrial permeability transition, mitochondria are engulfed by GFP-LC3 positive autophagosomes [9, 154]. Photo-damaged mitochondria also recruited GFP-LC3 positive structures to the damaged areas [155]. It is suggested that reactive oxygen species (ROS) act as a signal in damaged mitochondria to recruit the LC3 positive autophagosomes. ROS can activate Atg4B, the protease that is required for LC3-I to be converted to LC3-II [156, 157].
b. Types of mitophagy
It is proposed that there are at least three types of mitophagy depending on the cellular mechanism of sequestration of mitochondria into autophagosomes [152].
Type 1 mitophagy refers to the mitophagy process that occurs during nutrient deprivation. The process occurs in coordination with mitochondrial fission, which starts with the formation of phagophores that enlarge to surround mitochondria to form structures called mitophagosomes. Mitophagosomes are then acidified to activate the hydrolytic enzymes of lysosomes [152, 155, 158].
Type 2 mitophagy refers to the degradation of mitochondria during photo damage. In this case, depolarized mitochondria recruit LC3 positive structures to aggregate onto their surface. These structures then fuse together to sequester the mitochondria into a mitophagosome [159-161]. This type of mitophagy is not coordinated with mitochondrial fission, unlike Type 1. Another difference is that Beclin-1 protein is required for Type 1 mitophagy but not for Type 2. This came from studies showing that Type 1 mitophagy can be prevented by pharmacological inhibition of PI3K using 3-methyladenine or wortmannin. On the other hand, Type 2 mitophagy was shown to be independent of Beclin-1 and PI3K [152].
Type 3 mitophagy, also known as micromitophagy, involves the formation of mitochondria-derived vesicles, which translocate to the lysosomes [152, 162, 163]. The release of mitochondrial derived vesicles depends on oxidative stress in the mitochondria and involves pink 1 and parkin proteins. Micromitophagy does not involve LC3 or Atg5, and it is independent of mitochondrial depolarization or mitochondrial fission [164]. This process allows the cell to selectively remove damaged or oxidized components of the mitochondria without total degradation.
c. Progression of mitophagy and the involvement of mitochondrial fission/fusion
The signaling pathways involved in the progression of mitophagy share many similarities with general autophagy. At baseline, LC3 is dispersed throughout the cytosol, and some LC3 is found in pre-autophagic structures close to the mitochondrial membrane [80]. During mitophagy, LC3 is conjugated to phosphatidylethanolamine (PE), forming LC3-II. During type 1 mitophagy, already existing preautophagic structures enlarge in size to envelope and sequester the mitochondria. This event forms the mitophagosome, which then fuses with a lysosome or a late endosome to form a mitophagolysosome that digests the mitochondrial content [152, 155].
The molecular events contributing to mitophagy initiation were first identified in yeast. Studies showed that there are three yeast proteins participating in mitophagy initiation: outer mitochondrial protein Uth1, intermembrane space protein phosphatase Aup1, and inner membrane protein required for K+/H+ exchange Mdm38p [9]. Atg32 was identified as the main signal to direct autophagosomes to mitochondria after interacting with Atg8 and Atg11 [165, 166, 202]. There are no mammalian homologues for Atg32; however, studies showed that there are some receptors on the mitochondrial outer membrane that signal for the mitophagy process. For instance, optineurin acts as an autophagy receptor in parkin-mediated mitophagy, and FUNDC1 mediates hypoxia induced mitophagy [196,197]. Autophagy receptors can also be lipids in the mitochondrial membrane such as cardiolipin and ceramide [14,198,199].
Moreover, proteins involved in mitochondria fission/fusion are key regulators for the selective elimination of mitochondria in mammalian cells [167]. Mitochondria are dynamic mobile organelles continuously dividing or fusing [147]. The processes of fusion and fission are intrinsic for mitochondrial viability, and they are important in the regulation of calcium homeostasis and the generation of ATP and ROS [168, 169]. In addition, fission and fusion of the mitochondria are important during mitophagy [9]. Fission, or mitochondrial division, involves the translocation of DRP-1 (dynamin related protein 1) to the mitochondria, where it oligomerizes to bind to Fission 1 (Fis1) in the outer mitochondrial membrane [170]. Fusion, a process that fuses two mitochondria together, involves mitofusin 1 and mitofusin 2, located in the outer mitochondrial membrane, and OPA1 (optic atrophy protein 1) located in the inner mitochondrial membrane [171]. OPA1 is processed by mitochondrial peptidase OMA1 and i-AAA protease YME1L and is regulated by mitofusin 1 during inner mitochondrial membrane fusion [200,201].
The role of fission and fusion during the mitophagy process is illustrated in several studies. Cells with a knockdown of DRP-1 had suppressed rates of mitophagy, whereas cells with overexpression of DRP-1 had excessive mitochondrial disappearance [172-174]. In addition, mitochondria going through a round of fusion followed by fission generate two populations of mitochondria: those that re-fuse and are healthy, and those that never re-fuse, have a depolarized membrane potential, and get degraded by mitophagy [173, 175]. The loss of the pro-fusion OPA-1 is a key process for mitophagy, such that OPA-1 overexpression was able to decrease mitophagy [173, 175].
The pro-fission function of DRP-1 makes it an important player during mitophagy. DRP-1 is a cytosolic GTPase with three domains: GTP binding domain, bundle signaling element (BSE), and a stalk that allows for stable dimerization and oligomerization [176]. Upon its activation, DRP-1 translocates to the mitochondria to form dimers and oligomers that are necessary for fission. It is believed that DRP-1 translocation requires adapter proteins, such as mitochondrial fission factor (MFF), mitochondrial elongation factor 1/mitochondrial dynamics proteins of 49 and 51 kDa (MIEF1/MiD49/MiD51), and mitochondrial fission protein Fis1 [177-181]. DRP-1 is regulated by several post-transcriptional modifications such as phosphorylation. There are two sites of phosphorylation for DRP-1: Ser637 and Ser616. DRP-1 is activated when it is phosphorylated by cyclin B1-CDK1 at Ser616 and dephosphorylated by calcineurin at Ser637 [170, 182]. DRP-1 is inactivated when it is phosphorylated by protein kinase A at Ser637 [170, 182]. Another form of regulation of DRP-1 is nitrosylation by nitric oxide in the mitochondria, and deSUMOylation by SENP5 protease, both of which promote DRP-1 activation and dimer formation [182].
Two other proteins, which are associated with Parkinson’s disease, are involved in DRP-1 mediated mitophagy: pink1 and parkin. Pink1 is a serine/threonine protein kinase located inside the mitochondria, while parkin is an E3 ubiquitin protein ligase located mainly in the cytosol [183-186]. It is believed that Parkin and Pink1 promote mitochondrial fission such that silencing their expression leads to mitochondrial defects due to lack of fission [184, 187]. Pink 1 and Parkin also regulate other mitochondrial functions, including mitochondrial biogenesis, mitochondrial transport, and calcium homeostasis [188-192]. In healthy mitochondria, Pink1 is cleaved and exported to the cytosol where it is rapidly degraded by proteasomes [200]. Upon uncoupling or depolarization of the mitochondria, Pink1 is stabilized, and Parkin is translocated from the cytosol to the mitochondria in a Pink 1-dependent fashion. This leads to ubiquitin mediated proteasomal degradation of outer mitochondrial proteins such as mitofusin 1 and mitofusin 2, leading to mitochondrial fragmentation and initiation of mitophagy [177, 184, 193]. DRP-1 is recruited to mitochondrial sites in close proximity of Pink1 and Parkin highlighting their importance in DRP-1-dependent mitophagy. In addition, Parkin can induce mitochondrial fission independent of Pink1 by affecting DRP-1 phosphorylation [184].
Moreover, ROS serve as candidates to initiate mitophagy. Cells supplied exogenously with hydrogen peroxide or superoxide showed evidence of autophagosomal structure formation [166]. It has been shown that upon ROS generation in the mitochondria, the mitochondrial membrane was depolarized, leading to Parkin translocation and initiation of mitophagy [195]. Interestingly, overexpression of the anti-oxidant enzyme superoxide dismutase 2 or pre-treatment with antioxidants prevented ROS-induced mitophagy [194, 195]. This suggests that oxidative stress may be an important signal to initiate mitophagy.
d. Ceramide mediated mitophagy as a tumor suppressor mechanism
There is evidence to support a role for mitophagy in cell survival or death, which appears to be context dependent. Mitophagy promotes cell survival under circumstances where it degrades the mitochondria that are about to activate caspase dependent apoptosis. In this case, disrupting the autophagic and lysosomal processes will prevent survival and lead to apoptosis [152, 153]. On the other hand, when mitophagy occurs excessively or for a sustained period of time, enzymes from the lysosomal flux such as cathepsins can leak to the cytosol where they initiate caspase dependent cell death [79, 83, 152]. Therefore, the functional outcome of mitophagy inducing cell death or survival depends on the intensity and duration of the stress as well as cellular contact [152].
Moreover, mitophagy can serve as a programmed cell death mechanism independent of apoptosis. This mechanism of cell death depends on ceramide synthase 1 (CerS1) and its metabolic product C18-ceramide. Sentelle et al. showed that CerS1 and C18-ceramide selectively induce non-apoptotic lethal mitophagy independent of Bax, Bak, or caspase activity, in head and neck squamous cell carcinoma cells and tumors. Ectopic expression of CerS1 or treatment with C18-pyridinium-ceramide resulted in LC3-II formation, and promoted its direct binding to ceramide on the mitochondrial membranes. This lipid-protein binding then allowed the mitochondria to be targeted by the LC3-II containing autophagosomes. This report was the first to describe the role of ceramide signaling in mediating lethal mitophagy through ceramide-LC3-II binding (Figure 3). Interestingly, endogenous C16-ceramide generated by CerS6 did not show any mitophagy promoting function in these cells. However, treatment with C16-pyridinium ceramide, which accumulates in the mitochondria, induced mitophagy. This suggested that the subcellular localization of endogenous ceramides, and not their fatty acid chain length per se, is of great importance to determine their distinct biological actions during mitophagy [14].
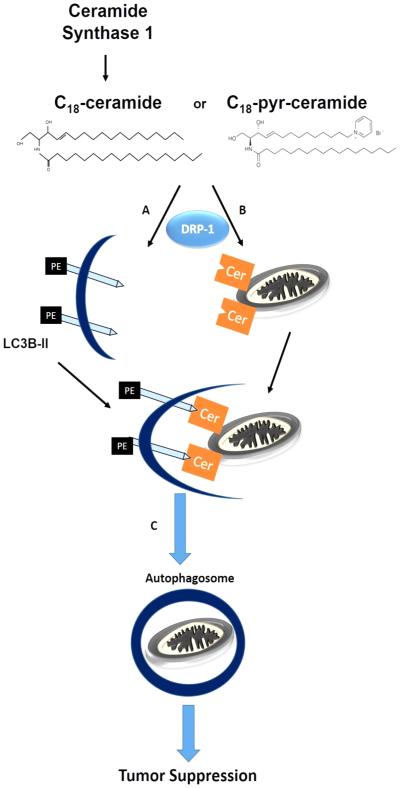
Fig. 3.
Regulation of mitophagy by ceramide. Endogenous generation of C18-ceramide via CerS1 or exogenous treatment by C18-pyridinium-ceramide is followed by two processes: A. conjugation of LC3-I to phosphatidylethanolamine on the carboxy terminal to form LC3-II and B. accumulation of ceramide in the mitochondrial outer membrane. Ceramide in the mitochondrial membrane acts as a receptor to LC3-II by binding to its amino terminal, opposite to where PE is conjugated. This results in C. recruiting the autophagosome to engulf the mitochondria. Lysosomes then fuse with the autophagosomes (D) for hydrolytic degradation of the contents.
The binding of ceramide to LC3-II indicates that ceramide acts as a tumor suppressor lipid that can directly bind proteins. Ceramide is shown to have a higher affinity to the PE-conjugated LC3-II than LC3-I. This interaction was proposed to involve the central hydrophobic domain of LC3 that has structural similarities to the domain of CERT (ceramide transporter protein) that binds C16- and C18-ceramides. Within this hydrophobic domain, the Ile35 and Phe52 residues of LC3-II were required for ceramide binding. Computational docking simulations and molecular modeling suggested that conjugation of LC3-I to phosphatidylethanolamine hides a low-affinity ceramide-binding sites allowing ceramide to bind selectively to the opposite end of the protein [14, 49]. More importantly, although point mutations at the Ile35 and/or Phe52 for conversion to Ala did not prevent ceramide-mediated LC3 lipidation, and inhibition of ceramide binding modulated mitophagy and resulted in resistance to ceramide-mediated tumor suppression. Thus, these data support that ceramide plays a novel receptor role at the mitochondrial membranes to recruit LC3-II-containing autophagosomes to the mitochondria, which have been subjected to DRP-1-mediated fission. These data also suggest that targeting LC3 containing autophagosomes to mitochondria by ceramide at the mitochondrial membranes results in cancer cell death and tumor suppression, which seems to be regulated downstream of DRP1-mediated mitochondrial fission. Interestingly, in the absence of CerS1 overexpression, tumor cells with knockdown of LC3 had a reduced growth in vivo suggesting that at baseline, LC3 mediated autophagy is required for tumor growth in head and neck squamous cell carcinoma tumors [14, 49].
5. Conclusions and future perspectives
Ceramide, as a bioactive sphingolipid, plays key roles in the regulation of general and selective autophagy and/or mitophagy. This role of ceramide is of great importance in tumor biology as in most cases ceramide mediated autophagy leads to cell death and is thus called lethal autophagy or autosis [108, 114]. One example by which ceramide regulates lethal autophagic signaling pathways is its activation of c-Jun through JNK signaling, causing upregulated Beclin 1 expression and autophagic cell death [138]. Exogenous supply of C18-pyridinium ceramide or overexpression of CerS1 resulted in caspase independent mitophagy where ceramide acts as a mitochondrial receptor for LC3-II-containing autophagosomes by interacting directly with LC3-II, recruiting autophagolysosomes to damaged mitochondria [14]. Exogenous ceramide mediated autophagic cell death is believed to involve BNIP3 activation after a reduction in mitochondrial membrane potential [140]. Chemotherapeutic drugs and arsenic trioxide lead to ceramide production that increase Beclin1 expression and promote lethal autophagy [141]. Other drugs such as cannabinoids also lead to ceramide accumulation to induce ER stress, mTOR inhibition via TRB3 (tribbles homolog 3), and lethal autophagy [142]. Amino acid deprivation is known to induce lethal autophagy in a ceramide dependent manner by activating CAPPs (ceramide activated protein phosphatases), which inhibits Akt/mTOR pathway [8]. The knowledge of the role of ceramide in autophagy/mitophagy sets an example to the importance of sphingolipid metabolism and signaling in these cellular mechanisms. Thus, more studies should be invested in this area of research to define the roles and mechanisms of how ceramide and.or other bioactive sphingolipid molecules mediate mitophagy and their relation to mitochondrial dynamics. Ceramide’s role in recruiting autophagosomes specifically to mitochondria gave support to the findings that removal of mitochondria by autophagy can be selective rather than inadvertent. Ceramide acts as a receptor in the mitochondria binding the LC3-II in the autophagosomes to direct them specifically to mitochondria. However, the relationship between ceramide and the fission/fusion machinery is still not clear. DRP-1 is required for ceramide-mediated mitophagy, however the mechanism underlying DRP-1 activation remains unknown. Other studies looking at the interplay of ceramide and Pink1 or Parkin are also important for the field. Importantly, studies should consider the compartmentalized roles of ceramides with different fatty acyl chain lengths, involved in the regulation of the mitophagy process. We expect that there will be key discoveries to dissect the mechanisms of how ceramide regulates lethal mitophagy and tumor suppression during the next few years, as analytical, molecular, pharmacologic and/or genetic tools are now available to define these roles of ceramides/sphingoilipids in various disease models.
Highlights.
This review focuses on the roles and mechanisms of ceramide-induced mitophagy.
Ceramide is a key bioactive sphingolipid molecule, which is involved in the regulation of mitophagy.
Ceramide-mediated mitophagy involves ceramide-LC3B-II binding and Drp1-mediated mitochondrial fission.
Ceramide-induced mitophagy results in cell death and tumor suppression.
5. Acknowledgments
This work was supported by grant support from the National Institutes of Health (CA088932, CA173687 and DE016572). We thank Ms. Raquela Thomas and Ms. Rose Ndeto at the Ogretmen Laboratory for their helpful discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- [1].Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat. Rev. Cancer. 2004;4:604–616. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- [2].Saddoughi SA, Ogretmen B. Diverse functions of ceramide in cancer cell death and proliferation. Adv. Cancer Res. 2013;117:37–58. doi: 10.1016/B978-0-12-394274-6.00002-9. [DOI] [PubMed] [Google Scholar]
- [3].Ponnusamy S, Meyers-Needham M, Senkal CE, Saddoughi SA, Sentelle D, Selvam SP, Salas A, Ogretmen B. Sphingolipids and cancer: ceramide and sphingosine-1-phosphate in the regulation of cell death and drug resistance. Future Oncol. 2010;6:1603–1624. doi: 10.2217/fon.10.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Young MM, Kester M, Wang HG. Sphingolipids: regulators of crosstalk between apoptosis and autophagy. J. Lipid Res. 2013;54:5–19. doi: 10.1194/jlr.R031278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Saddoughi SA, Gencer S, Peterson YK, Ward KE, Mukhopadhyay A, Oaks J, Bielawski J, Szulc ZM, Thomas RJ, Selvam SP, Senkal CE, Garrett-Mayer E, De Palma RM, Fedarovich D, Liu A, Habib AA, Stahelin RV, Perrotti D, Ogretmen B. Sphingosine analogue drug FTY720 targets I2PP2A/SET and mediates lung tumour suppression via activation of PP2A-RIPK1-dependent necroptosis. EMBO Mol. Med. 2013;5:105–121. doi: 10.1002/emmm.201201283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Selvam SP, Ramshesh VK, Peterson YK, Lemasters JJ, Szulc ZM, Bielawski J, Ogretmen B. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat. Chem. Biol. 2012;8:831–838. doi: 10.1038/nchembio.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jiang W, Ogretmen B. Autophagy paradox and ceramide. Biochim. Biophys. Acta. 2014;1841:783–792. doi: 10.1016/j.bbalip.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Taniguchi M, Kitatani K, Kondo T, Hashimoto-Nishimura M, Asano S, Hayashi A, Mitsutake S, Igarashi Y, Umehara H, Takeya H, Kigawa J, Okazaki T. Regulation of autophagy and its associated cell death by "sphingolipid rheostat": reciprocal role of ceramide and sphingosine 1-phosphate in the mammalian target of rapamycin pathway. J. Biol. Chem. 2012;287:39898–39910. doi: 10.1074/jbc.M112.416552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tolkovsky AM. Mitophagy. Biochim. Biophys. Acta. 2009;1793:1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- [10].Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. Sphingolipids in macroautophagy. Methods in Molecular Biology. 2008;445:159–173. doi: 10.1007/978-1-59745-157-4_11. [DOI] [PubMed] [Google Scholar]
- [11].Beckham TH, Lu P, Jones EE, Marrison T, Lewis CS, Cheng JC, Ramshesh VK, Beeson G, Beeson CC, Drake RR, Bielawska A, Bielawski J, Szulc ZM, Ogretmen B, Norris JS, Liu X. LCL124. a cationic analog of ceramide, selectively induces pancreatic cancer cell death by accumulating in mitochondria, J. Pharmacol. Exp. Ther. 2013;344:167–178. doi: 10.1124/jpet.112.199216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hou Q, Jin J, Zhou H, Novgorodov SA, Bielawska A, Szulc ZM, Hannun YA, Obeid LM, Hsu YT. Mitochondrially targeted ceramides preferentially promote autophagy, retard cell growth, and induce apoptosis. J. Lipid Res. 2011;52:278–288. doi: 10.1194/jlr.M012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Senkal CE, Ponnusamy S, Rossi MJ, Sundararaj K, Szulc Z, Bielawski J, Bielawska A, Meyer M, Cobanoglu B, Koybasi S, Sinha D, Day TA, Obeid LM, Hannun YA, Ogretmen B. Potent antitumor activity of a novel cationic pyridinium-ceramide alone or in combination with gemcitabine against human head and neck squamous cell carcinomas in vitro and in vivo. J. Pharmacol. Exp. Ther. 2006;317:1188–1199. doi: 10.1124/jpet.106.101949. [DOI] [PubMed] [Google Scholar]
- [14].Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Selvam SP, Ramshesh VK, Peterson YK, Lemasters JJ, Szulc ZM, Bielawski J, Ogretmen B. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nature Chemical Biology. 2012;8:831–838. doi: 10.1038/nchembio.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Saddoughi SA, Song P, Ogretmen B. Roles of bioactive sphingolipids in cancer biology and therapeutics. Subcell. Biochem. 2008;49:413–440. doi: 10.1007/978-1-4020-8831-5_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Merrill AH, Jr, Wang E, Mullins RE. Kinetics of long-chain (sphingoid) base biosynthesis in intact LM cells: effects of varying the extracellular concentrations of serine and fatty acid precursors of this pathway. Biochemistry. 1988;27:340–345. doi: 10.1021/bi00401a051. [DOI] [PubMed] [Google Scholar]
- [17].Pewzner-Jung Y, Ben-Dor S, Futerman AH. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)?: Insights into the regulation of ceramide synthesis. J. Biol. Chem. 2006;281:25001–25005. doi: 10.1074/jbc.R600010200. [DOI] [PubMed] [Google Scholar]
- [18].Kraveka JM, Li L, Szulc ZM, Bielawski J, Ogretmen B, Hannun YA, Obeid LM, Bielawska A. Involvement of dihydroceramide desaturase in cell cycle progression in human neuroblastoma cells. J. Biol. Chem. 2007;282:16718–16728. doi: 10.1074/jbc.M700647200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Selvam SP, Ogretmen B. Sphingosine kinase/sphingosine 1-phosphate signaling in cancer therapeutics and drug resistance. Handb. Exp. Pharmacol. (216):3–27. doi: 10.1007/978-3-7091-1511-4_1. doi (2013) 3-27. [DOI] [PubMed] [Google Scholar]
- [20].Futerman AH, Hannun YA. The complex life of simple sphingolipids. EMBO Rep. 2004;5:777–782. doi: 10.1038/sj.embor.7400208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- [22].Kumagai K, Yasuda S, Okemoto K, Nishijima M, Kobayashi S, Hanada K. CERT mediates intermembrane transfer of various molecular species of ceramides. J. Biol. Chem. 2005;280:6488–6495. doi: 10.1074/jbc.M409290200. [DOI] [PubMed] [Google Scholar]
- [23].Simanshu DK, Kamlekar RK, Wijesinghe DS, Zou X, Zhai X, Mishra SK, Molotkovsky JG, Malinina L, Hinchcliffe EH, Chalfant CE, Brown RE, Patel DJ. Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids. Nature. 2013;500:463–467. doi: 10.1038/nature12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tirodkar TS, Voelkel-Johnson C. Sphingolipids in apoptosis. Exp. Oncol. 2012;34:231–242. [PubMed] [Google Scholar]
- [25].Venkataraman K, Riebeling C, Bodennec J, Riezman H, Allegood JC, Sullards MC, Merrill Jr AH, Futerman AH. Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine (C18-(dihydro)ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. J. Biol. Chem. 2002;277:35642–35649. doi: 10.1074/jbc.M205211200. [DOI] [PubMed] [Google Scholar]
- [26].Lee SJ. Expression of growth/differentiation factor 1 in the nervous system: conservation of a bicistronic structure. Proc. Natl. Acad. Sci. U. S. A. 1991;88:4250–4254. doi: 10.1073/pnas.88.10.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kageyama-Yahara N, Riezman H. Transmembrane topology of ceramide synthase in yeast. Biochem. J. 2006;398:585–593. doi: 10.1042/BJ20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Spassieva S, Seo JG, Jiang JC, Bielawski J, Alvarez-Vasquez F, Jazwinski SM, Hannun YA, Obeid LM. Necessary role for the Lag1p motif in (dihydro)ceramide synthase activity. J. Biol. Chem. 2006;281:33931–33938. doi: 10.1074/jbc.M608092200. [DOI] [PubMed] [Google Scholar]
- [29].Mesika A, Ben-Dor S, Laviad EL, Futerman AH. A new functional motif in Hox domain-containing ceramide synthases: identification of a novel region flanking the Hox and TLC domains essential for activity. J. Biol. Chem. 2007;282:27366–27373. doi: 10.1074/jbc.M703487200. [DOI] [PubMed] [Google Scholar]
- [30].Teufel A, Maass T, Galle PR, Malik N. The longevity assurance homologue of yeast lag1 (Lass) gene family (review) Int. J. Mol. Med. 2009;23:135–140. [PubMed] [Google Scholar]
- [31].Schulz A, Mousallem T, Venkataramani M, Persaud-Sawin DA, Zucker A, Luberto C, Bielawska A, Bielawski J, Holthuis JC, Jazwinski SM, Kozhaya L, Dbaibo GS, Boustany RM. The CLN9 protein, a regulator of dihydroceramide synthase. J. Biol. Chem. 2006;281:2784–2794. doi: 10.1074/jbc.M509483200. [DOI] [PubMed] [Google Scholar]
- [32].Imgrund S, Hartmann D, Farwanah H, Eckhardt M, Sandhoff R, Degen J, Gieselmann V, Sandhoff K, Willecke K. Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J. Biol. Chem. 2009;284:33549–33560. doi: 10.1074/jbc.M109.031971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J. 2005;390:263–271. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Senkal CE, Ponnusamy S, Rossi MJ, Bialewski J, Sinha D, Jiang JC, Jazwinski SM, Hannun YA, Ogretmen B. Role of human longevity assurance gene 1 and C18-ceramide in chemotherapy-induced cell death in human head and neck squamous cell carcinomas. Mol. Cancer. Ther. 2007;6:712–722. doi: 10.1158/1535-7163.MCT-06-0558. [DOI] [PubMed] [Google Scholar]
- [35].Karahatay S, Thomas K, Koybasi S, Senkal CE, Elojeimy S, Liu X, Bielawski J, Day TA, Gillespie MB, Sinha D, Norris JS, Hannun YA, Ogretmen B. Clinical relevance of ceramide metabolism in the pathogenesis of human head and neck squamous cell carcinoma (HNSCC): attenuation of C(18)-ceramide in HNSCC tumors correlates with lymphovascular invasion and nodal metastasis. Cancer Lett. 2007;256:101–111. doi: 10.1016/j.canlet.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Koybasi S, Senkal CE, Sundararaj K, Spassieva S, Bielawski J, Osta W, Day TA, Jiang JC, Jazwinski SM, Hannun YA, Obeid LM, Ogretmen B. Defects in cell growth regulation by C18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. J. Biol. Chem. 2004;279:44311–44319. doi: 10.1074/jbc.M406920200. [DOI] [PubMed] [Google Scholar]
- [37].Saddoughi SA, Garrett-Mayer E, Chaudhary U, O'Brien PE, Afrin LB, Day TA, Gillespie MB, Sharma AK, Wilhoit CS, Bostick R, Senkal CE, Hannun YA, Bielawski J, Simon GR, Shirai K, Ogretmen B. Results of a phase II trial of gemcitabine plus doxorubicin in patients with recurrent head and neck cancers: serum C(1)(8)-ceramide as a novel biomarker for monitoring response. Clin. Cancer Res. 2011;17:6097–6105. doi: 10.1158/1078-0432.CCR-11-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Senkal CE, Ponnusamy S, Bielawski J, Hannun YA, Ogretmen B. Antiapoptotic roles of ceramide-synthase-6-generated C16-ceramide via selective regulation of the ATF6/CHOP arm of ER-stress-response pathways. FASEB J. 2010;24:296–308. doi: 10.1096/fj.09-135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Senkal CE, Ponnusamy S, Manevich Y, Meyers-Needham M, Saddoughi SA, Mukhopadyay A, Dent P, Bielawski J, Ogretmen B. Alteration of ceramide synthase 6/C16-ceramide induces activating transcription factor 6-mediated endoplasmic reticulum (ER) stress and apoptosis via perturbation of cellular Ca2+ and ER/Golgi membrane network. J. Biol. Chem. 2011;286:42446–42458. doi: 10.1074/jbc.M111.287383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Holliday MW, Jr, Cox SB, Kang MH, Maurer BJ. C22:0- and C24:0-dihydroceramides confer mixed cytotoxicity in T-cell acute lymphoblastic leukemia cell lines. PLoS One. 2013;8:e74768. doi: 10.1371/journal.pone.0074768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sassa T, Suto S, Okayasu Y, Kihara A. A shift in sphingolipid composition from C24 to C16 increases susceptibility to apoptosis in HeLa cells. Biochim. Biophys. Acta. 2012;1821:1031–1037. doi: 10.1016/j.bbalip.2012.04.008. [DOI] [PubMed] [Google Scholar]
- [42].Zhao L, Spassieva SD, Jucius TJ, Shultz LD, Shick HE, Macklin WB, Hannun YA, Obeid LM, Ackerman SL. A deficiency of ceramide biosynthesis causes cerebellar purkinje cell neurodegeneration and lipofuscin accumulation. PLoS Genet. 2011;7:e1002063. doi: 10.1371/journal.pgen.1002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ebel P, Imgrund S, Vom Dorp K, Hofmann K, Maier H, Drake H, Degen J, Dormann P, Eckhardt M, Franz T, Willecke K. Ceramide synthase 4 deficiency in mice causes lipid alterations in sebum and results in alopecia. Biochem. J. 2014;461:147–158. doi: 10.1042/BJ20131242. [DOI] [PubMed] [Google Scholar]
- [44].Pewzner-Jung Y, Brenner O, Braun S, Laviad EL, Ben-Dor S, Feldmesser E, Horn-Saban S, Amann-Zalcenstein D, Raanan C, Berkutzki T, Erez-Roman R, Ben-David O, Levy M, Holzman D, Park H, Nyska A, Merrill AH, Jr, Futerman AH. A critical role for ceramide synthase 2 in liver homeostasis: II. insights into molecular changes leading to hepatopathy. J. Biol. Chem. 2010;285:10911–10923. doi: 10.1074/jbc.M109.077610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mosbech MB, Kruse R, Harvald EB, Olsen AS, Gallego SF, Hannibal-Bach HK, Ejsing CS, Faergeman NJ. Functional loss of two ceramide synthases elicits autophagy-dependent lifespan extension in C. elegans. PLoS One. 2013;8:e70087. doi: 10.1371/journal.pone.0070087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Menuz V, Howell KS, Gentina S, Epstein S, Riezman I, Fornallaz-Mulhauser M, Hengartner MO, Gomez M, Riezman H, Martinou JC. Protection of C. elegans from anoxia by HYL-2 ceramide synthase. Science. 2009;324:381–384. doi: 10.1126/science.1168532. [DOI] [PubMed] [Google Scholar]
- [47].Voelzmann A, Bauer R. Embryonic expression of Drosophila ceramide synthase schlank in developing gut, CNS and PNS. Gene Expr. Patterns. 2011;11:501–510. doi: 10.1016/j.gep.2011.08.006. [DOI] [PubMed] [Google Scholar]
- [48].Russo SB, Tidhar R, Futerman AH, Cowart LA. Myristate-derived d16:0 sphingolipids constitute a cardiac sphingolipid pool with distinct synthetic routes and functional properties. J. Biol. Chem. 2013;288:13397–13409. doi: 10.1074/jbc.M112.428185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jiang W, Ogretmen B. Ceramide stress in survival versus lethal autophagy paradox: ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Autophagy. 2013;9:258–259. doi: 10.4161/auto.22739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Saddoughi SA, Gencer S, Peterson YK, Ward KE, Mukhopadhyay A, Oaks J, Bielawski J, Szulc ZM, Thomas RJ, Selvam SP, Senkal CE, Garrett-Mayer E, De Palma RM, Fedarovich D, Liu A, Habib AA, Stahelin RV, Perrotti D, Ogretmen B. Sphingosine analogue drug FTY720 targets I2PP2A/SET and mediates lung tumour suppression via activation of PP2A-RIPK1-dependent necroptosis. EMBO Mol. Med. 2013;5:105–121. doi: 10.1002/emmm.201201283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Truman JP, Garcia-Barros M, Obeid LM, Hannun YA. Evolving concepts in cancer therapy through targeting sphingolipid metabolism. Biochim. Biophys. Acta. 2014;1841:1174–1188. doi: 10.1016/j.bbalip.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Senkal CE, Ponnusamy S, Rossi MJ, Sundararaj K, Szulc Z, Bielawski J, Bielawska A, Meyer M, Cobanoglu B, Koybasi S, Sinha D, Day TA, Obeid LM, Hannun YA, Ogretmen B. Potent antitumor activity of a novel cationic pyridinium-ceramide alone or in combination with gemcitabine against human head and neck squamous cell carcinomas in vitro and in vivo. J. Pharmacol. Exp. Ther. 2006;317:1188–1199. doi: 10.1124/jpet.106.101949. [DOI] [PubMed] [Google Scholar]
- [53].Pettus BJ, Chalfant CE, Hannun YA. Ceramide in apoptosis: an overview and current perspectives. Biochim. Biophys. Acta. 2002;1585:114–125. doi: 10.1016/s1388-1981(02)00331-1. [DOI] [PubMed] [Google Scholar]
- [54].Chalfant CE, Ogretmen B, Galadari S, Kroesen BJ, Pettus BJ, Hannun YA. FAS activation induces dephosphorylation of SR proteins; dependence on the de novo generation of ceramide and activation of protein phosphatase 1. J. Biol. Chem. 2001;276:44848–44855. doi: 10.1074/jbc.M106291200. [DOI] [PubMed] [Google Scholar]
- [55].Cheng JC, Bai A, Beckham TH, Marrison ST, Yount CL, Young K, Lu P, Bartlett AM, Wu BX, Keane BJ, Armeson KE, Marshall DT, Keane TE, Smith MT, Jones EE, Drake RR, Jr, Bielawska A, Norris JS, Liu X. Radiation-induced acid ceramidase confers prostate cancer resistance and tumor relapse. J. Clin. Invest. 2013;123:4344–4358. doi: 10.1172/JCI64791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Eto M, Bennouna J, Hunter OC, Hershberger PA, Kanto T, Johnson CS, Lotze MT, Amoscato AA. C16 ceramide accumulates following androgen ablation in LNCaP prostate cancer cells. Prostate. 2003;57:66–79. doi: 10.1002/pros.10275. [DOI] [PubMed] [Google Scholar]
- [57].Santana P, Pena LA, Haimovitz-Friedman A, Martin S, Green D, McLoughlin M, Cordon-Cardo C, Schuchman EH, Fuks Z, Kolesnick R. Acid sphingomyelinase-deficient human lymphoblasts and mice are defective in radiation-induced apoptosis. Cell. 1996;86:189–199. doi: 10.1016/s0092-8674(00)80091-4. [DOI] [PubMed] [Google Scholar]
- [58].Chalfant CE, Kishikawa K, Mumby MC, Kamibayashi C, Bielawska A, Hannun YA. Long chain ceramides activate protein phosphatase-1 and protein phosphatase-2A. Activation is stereospecific and regulated by phosphatidic acid. J. Biol. Chem. 1999;274:20313–20317. doi: 10.1074/jbc.274.29.20313. [DOI] [PubMed] [Google Scholar]
- [59].Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, Hahn WC, Stukenberg PT, Shenolikar S, Uchida T, Counter CM, Nevins JR, Means AR, Sears R. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- [60].Salas A, Ponnusamy S, Senkal CE, Meyers-Needham M, Selvam SP, Saddoughi SA, Apohan E, Sentelle RD, Smith C, Gault CR, Obeid LM, El-Shewy HM, Oaks J, Santhanam R, Marcucci G, Baran Y, Mahajan S, Fernandes D, Stuart R, Perrotti D, Ogretmen B. Sphingosine kinase-1 and sphingosine 1-phosphate receptor 2 mediate Bcr-Abl1 stability and drug resistance by modulation of protein phosphatase 2A. Blood. 2011;117:5941–5952. doi: 10.1182/blood-2010-08-300772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Mukhopadhyay A, Saddoughi SA, Song P, Sultan I, Ponnusamy S, Senkal CE, Snook CF, Arnold HK, Sears RC, Hannun YA, Ogretmen B. Direct interaction between the inhibitor 2 and ceramide via sphingolipid-protein binding is involved in the regulation of protein phosphatase 2A activity and signaling. FASEB J. 2009;23:751–763. doi: 10.1096/fj.08-120550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Mukhopadhyay A, Saddoughi SA, Song P, Sultan I, Ponnusamy S, Senkal CE, Snook CF, Arnold HK, Sears RC, Hannun YA, Ogretmen B. Direct interaction between the inhibitor 2 and ceramide via sphingolipid-protein binding is involved in the regulation of protein phosphatase 2A activity and signaling. FASEB J. 2009;23:751–763. doi: 10.1096/fj.08-120550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Muto S, Senda M, Akai Y, Sato L, Suzuki T, Nagai R, Senda T, Horikoshi M. Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4285–4290. doi: 10.1073/pnas.0603762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Alesse E, Zazzeroni F, Angelucci A, Giannini G, Di Marcotullio L, Gulino A. The growth arrest and downregulation of c-myc transcription induced by ceramide are related events dependent on p21 induction, Rb underphosphorylation and E2F sequestering. Cell Death Differ. 1998;5:381–389. doi: 10.1038/sj.cdd.4400358. [DOI] [PubMed] [Google Scholar]
- [65].Heinrich M, Neumeyer J, Jakob M, Hallas C, Tchikov V, Winoto-Morbach S, Wickel M, Schneider-Brachert W, Trauzold A, Hethke A, Schutze S. Cathepsin D links TNF-induced acid sphingomyelinase to Bid-mediated caspase-9 and -3 activation. Cell Death Differ. 2004;11:550–563. doi: 10.1038/sj.cdd.4401382. [DOI] [PubMed] [Google Scholar]
- [66].Heinrich M, Wickel M, Schneider-Brachert W, Sandberg C, Gahr J, Schwandner R, Weber T, Saftig P, Peters C, Brunner J, Kronke M, Schutze S. Cathepsin D targeted by acid sphingomyelinase-derived ceramide. EMBO J. 1999;18:5252–5263. doi: 10.1093/emboj/18.19.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ogretmen B, Schady D, Usta J, Wood R, Kraveka JM, Luberto C, Birbes H, Hannun YA, Obeid LM. Role of ceramide in mediating the inhibition of telomerase activity in A549 human lung adenocarcinoma cells. J. Biol. Chem. 2001;276:24901–24910. doi: 10.1074/jbc.M100314200. [DOI] [PubMed] [Google Scholar]
- [68].Kraveka JM, Li L, Bielawski J, Obeid LM, Ogretmen B. Involvement of endogenous ceramide in the inhibition of telomerase activity and induction of morphologic differentiation in response to all-trans-retinoic acid in human neuroblastoma cells. Arch. Biochem. Biophys. 2003;419:110–119. doi: 10.1016/j.abb.2003.08.034. [DOI] [PubMed] [Google Scholar]
- [69].Lee JY, Bielawska AE, Obeid LM. Regulation of cyclin-dependent kinase 2 activity by ceramide. Exp. Cell Res. 2000;261:303–311. doi: 10.1006/excr.2000.5028. [DOI] [PubMed] [Google Scholar]
- [70].Marchesini N, Luberto C, Hannun YA. Biochemical properties of mammalian neutral sphingomyelinase 2 and its role in sphingolipid metabolism. J. Biol. Chem. 2003;278:13775–13783. doi: 10.1074/jbc.M212262200. [DOI] [PubMed] [Google Scholar]
- [71].Zhu XF, Liu ZC, Xie BF, Feng GK, Zeng YX. Ceramide induces cell cycle arrest and upregulates p27kip in nasopharyngeal carcinoma cells. Cancer Lett. 2003;193:149–154. doi: 10.1016/s0304-3835(03)00050-8. [DOI] [PubMed] [Google Scholar]
- [72].Venable ME, Lee JY, Smyth MJ, Bielawska A, Obeid LM. Role of ceramide in cellular senescence. J. Biol. Chem. 1995;270:30701–30708. doi: 10.1074/jbc.270.51.30701. [DOI] [PubMed] [Google Scholar]
- [73].Marchesini N, Osta W, Bielawski J, Luberto C, Obeid LM, Hannun YA. Role for mammalian neutral sphingomyelinase 2 in confluence-induced growth arrest of MCF7 cells. J. Biol. Chem. 2004;279:25101–25111. doi: 10.1074/jbc.M313662200. [DOI] [PubMed] [Google Scholar]
- [74].Novgorodov SA, Szulc ZM, Luberto C, Jones JA, Bielawski J, Bielawska A, Hannun YA, Obeid LM. Positively charged ceramide is a potent inducer of mitochondrial permeabilization. J. Biol. Chem. 2005;280:16096–16105. doi: 10.1074/jbc.M411707200. [DOI] [PubMed] [Google Scholar]
- [75].Watters RJ, Fox TE, Tan SF, Shanmugavelandy S, Choby JE, Broeg K, Liao J, Kester M, Cabot MC, Loughran TP, Liu X. Targeting glucosylceramide synthase synergizes with C6-ceramide nanoliposomes to induce apoptosis in natural killer cell leukemia. Leuk. Lymphoma. 2013;54:1288–1296. doi: 10.3109/10428194.2012.752485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Lavie Y, Cao H, Volner A, Lucci A, Han TY, Geffen V, Giuliano AE, Cabot MC. Agents that reverse multidrug resistance, tamoxifen, verapamil, and cyclosporin A, block glycosphingolipid metabolism by inhibiting ceramide glycosylation in human cancer cells. J. Biol. Chem. 1997;272:1682–1687. doi: 10.1074/jbc.272.3.1682. [DOI] [PubMed] [Google Scholar]
- [77].Stover T, Kester M. Liposomal delivery enhances short-chain ceramide-induced apoptosis of breast cancer cells. J. Pharmacol. Exp. Ther. 2003;307:468–475. doi: 10.1124/jpet.103.054056. [DOI] [PubMed] [Google Scholar]
- [78].Stover TC, Sharma A, Robertson GP, Kester M. Systemic delivery of liposomal short-chain ceramide limits solid tumor growth in murine models of breast adenocarcinoma. Clin. Cancer Res. 2005;11:3465–3474. doi: 10.1158/1078-0432.CCR-04-1770. [DOI] [PubMed] [Google Scholar]
- [79].Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- [80].Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell. Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N. Engl. J. Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- [85].Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat. Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- [87].Mizumura K, Cloonan SM, Haspel JA, Choi AM. The emerging importance of autophagy in pulmonary diseases. Chest. 2012;142:1289–1299. doi: 10.1378/chest.12-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kirshenbaum LA. Regulation of autophagy in the heart in health and disease. J. Cardiovasc. Pharmacol. 2012;60:109. doi: 10.1097/FJC.0b013e31825f6faa. [DOI] [PubMed] [Google Scholar]
- [90].Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Thumm M. Structure and function of the yeast vacuole and its role in autophagy. Microsc. Res. Tech. 2000;51:563–572. doi: 10.1002/1097-0029(20001215)51:6<563::AID-JEMT6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- [92].Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J. Biol. Chem. 2003;278:51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- [93].Tanida I. Autophagy basics. Microbiol. Immunol. 2011;55:1–11. doi: 10.1111/j.1348-0421.2010.00271.x. [DOI] [PubMed] [Google Scholar]
- [94].Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Tanida I. Mammalian Atg-conjugation systems: key players essential for the formation of autophagosomes. Tanpakushitsu Kakusan Koso. 2006;51:1490–1493. [PubMed] [Google Scholar]
- [96].Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- [97].Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- [98].Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr. Biol. 2012;22:R29–34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- [100].Nemoto T, Tanida I, Tanida-Miyake E, Minematsu-Ikeguchi N, Yokota M, Ohsumi M, Ueno T, Kominami E. The mouse APG10 homologue, an E2-like enzyme for Apg12p conjugation, facilitates MAP-LC3 modification. J. Biol. Chem. 2003;278:39517–39526. doi: 10.1074/jbc.m300550200. [DOI] [PubMed] [Google Scholar]
- [101].Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- [102].Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Tanida I, Ueno T, Kominami E. LC3 and Autophagy. Methods Mol. Biol. 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- [104].Tanida I, Ueno T, Kominami E. Human light chain 3/MAP1LC3 is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J. Biol. Chem. 2004;279:47704–47710. doi: 10.1074/jbc.M407016200. [DOI] [PubMed] [Google Scholar]
- [105].Schworer CM, Shiffer KA, Mortimore GE. Quantitative relationship between autophagy and proteolysis during graded amino acid deprivation in perfused rat liver. J. Biol. Chem. 1981;256:7652–7658. [PubMed] [Google Scholar]
- [106].Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- [107].Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, Baehrecke EH, Lenardo M. Autophagic programmed cell death by selective catalase degradation. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Gozuacik D, Kimchi A. Autophagy and cell death. Curr. Top. Dev. Biol. 2007;78:217–245. doi: 10.1016/S0070-2153(06)78006-1. [DOI] [PubMed] [Google Scholar]
- [109].Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- [110].Levine B, Yuan J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- [112].Choi KS. Autophagy and cancer. Exp. Mol. Med. 2012;44:109–120. doi: 10.3858/emm.2012.44.2.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, Gottlieb E, Green DR, Hengartner MO, Kepp O, Knight RA, Kumar S, Lipton SA, Lu X, Madeo F, Malorni W, Mehlen P, Nunez G, Peter ME, Piacentini M, Rubinsztein DC, Shi Y, Simon HU, Vandenabeele P, White E, Yuan J, Zhivotovsky B, Melino G, Kroemer G. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Liu Y, Shoji-Kawata S, Sumpter RM, Jr, Wei Y, Ginet V, Zhang L, Posner B, Tran KA, Green DR, Xavier RJ, Shaw SY, Clarke PG, Puyal J, Levine B. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc. Natl. Acad. Sci. U. S. A. 2013;110:20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Mah LY, Ryan KM. Autophagy and cancer. Cold Spring Harb Perspect. Biol. 2012;4:a008821. doi: 10.1101/cshperspect.a008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Eskelinen EL. The dual role of autophagy in cancer. Curr. Opin. Pharmacol. 2011;11:294–300. doi: 10.1016/j.coph.2011.03.009. [DOI] [PubMed] [Google Scholar]
- [118].Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Sridhar S, Botbol Y, Macian F, Cuervo AM. Autophagy and disease: always two sides to a problem. J. Pathol. 2012;226:255–273. doi: 10.1002/path.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- [122].Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol. Cell. Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Saito H, Inazawa J, Saito S, Kasumi F, Koi S, Sagae S, Kudo R, Saito J, Noda K, Nakamura Y. Detailed deletion mapping of chromosome 17q in ovarian and breast cancers: 2-cM region on 17q21.3 often and commonly deleted in tumors. Cancer Res. 1993;53:3382–3385. [PubMed] [Google Scholar]
- [124].Gao X, Zacharek A, Salkowski A, Grignon DJ, Sakr W, Porter AT, Honn KV. Loss of heterozygosity of the BRCA1 and other loci on chromosome 17q in human prostate cancer. Cancer Res. 1995;55:1002–1005. [PubMed] [Google Scholar]
- [125].Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Martelli AM, Tazzari PL, Evangelisti C, Chiarini F, Blalock WL, Billi AM, Manzoli L, McCubrey JA, Cocco L. Targeting the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin module for acute myelogenous leukemia therapy: from bench to bedside. Curr. Med. Chem. 2007;14:2009–2023. doi: 10.2174/092986707781368423. [DOI] [PubMed] [Google Scholar]
- [128].Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat. Rev. Clin. Oncol. 2011;8:528–539. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- [129].Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell'antonio G, Mautner J, Tonon G, Haigis M, Shirihai OS, Doglioni C, Bardeesy N, Kimmelman AC. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, Coller HA, Dipaola RS, Gelinas C, Rabinowitz JD, White E. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Adiseshaiah PP, Clogston JD, McLeland CB, Rodriguez J, Potter TM, Neun BW, Skoczen SL, Shanmugavelandy SS, Kester M, Stern ST, McNeil SE. Synergistic combination therapy with nanoliposomal C6-ceramide and vinblastine is associated with autophagy dysfunction in hepatocarcinoma and colorectal cancer models. Cancer Lett. 2013;337:254–265. doi: 10.1016/j.canlet.2013.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Pattingre S, Bauvy C, Levade T, Levine B, Codogno P. Ceramide-induced autophagy: to junk or to protect cells? Autophagy. 2009;5:558–560. doi: 10.4161/auto.5.4.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Spassieva SD, Mullen TD, Townsend DM, Obeid LM. Disruption of ceramide synthesis by CerS2 down-regulation leads to autophagy and the unfolded protein response. Biochem. J. 2009;424:273–283. doi: 10.1042/BJ20090699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Guenther GG, Peralta ER, Rosales KR, Wong SY, Siskind LJ, Edinger AL. Ceramide starves cells to death by downregulating nutrient transporter proteins. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17402–17407. doi: 10.1073/pnas.0802781105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Edinger AL. Starvation in the midst of plenty: making sense of ceramide-induced autophagy by analysing nutrient transporter expression. Biochem. Soc. Trans. 2009;37:253–258. doi: 10.1042/BST0370253. [DOI] [PubMed] [Google Scholar]
- [136].Peralta ER, Edinger AL. Ceramide-induced starvation triggers homeostatic autophagy. Autophagy. 2009;5:407–409. doi: 10.4161/auto.5.3.7809. [DOI] [PubMed] [Google Scholar]
- [137].Guenther GG, Edinger AL. A new take on ceramide: starving cells by cutting off the nutrient supply. Cell. Cycle. 2009;8:1122–1126. doi: 10.4161/cc.8.8.8161. [DOI] [PubMed] [Google Scholar]
- [138].Scarlatti F, Bauvy C, Ventruti A, Sala G, Cluzeaud F, Vandewalle A, Ghidoni R, Codogno P. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J. Biol. Chem. 2004;279:18384–18391. doi: 10.1074/jbc.M313561200. [DOI] [PubMed] [Google Scholar]
- [139].Li DD, Wang LL, Deng R, Tang J, Shen Y, Guo JF, Wang Y, Xia LP, Feng GK, Liu QQ, Huang WL, Zeng YX, Zhu XF. The pivotal role of c-Jun NH2-terminal kinase-mediated Beclin 1 expression during anticancer agents-induced autophagy in cancer cells. Oncogene. 2009;28:886–898. doi: 10.1038/onc.2008.441. [DOI] [PubMed] [Google Scholar]
- [140].Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64:4286–4293. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- [141].Dbaibo GS, Kfoury Y, Darwiche N, Panjarian S, Kozhaya L, Nasr R, Abdallah M, Hermine O, El-Sabban M, de The H, Bazarbachi A. Arsenic trioxide induces accumulation of cytotoxic levels of ceramide in acute promyelocytic leukemia and adult T-cell leukemia/lymphoma cells through de novo ceramide synthesis and inhibition of glucosylceramide synthase activity. Haematologica. 2007;92:753–762. doi: 10.3324/haematol.10968. [DOI] [PubMed] [Google Scholar]
- [142].Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, Vazquez P, Blazquez C, Torres S, Garcia S, Nowak J, Fimia GM, Piacentini M, Cecconi F, Pandolfi PP, Gonzalez-Feria L, Iovanna JL, Guzman M, Boya P, Velasco G. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J. Clin. Invest. 2009;119:1359–1372. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Bedia C, Levade T, Codogno P. Regulation of autophagy by sphingolipids. Current Medicinal Chemistry - Anti-Cancer Agents. 2011;11:844–853. doi: 10.2174/187152011797655131. [DOI] [PubMed] [Google Scholar]
- [144].Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J. Biol. Chem. 2006;281:8518–8527. doi: 10.1074/jbc.M506182200. [DOI] [PubMed] [Google Scholar]
- [145].Lavieu G, Scarlatti F, Sala G, Levade T, Ghidoni R, Botti J, Codogno P. Is autophagy the key mechanism by which the sphingolipid rheostat controls the cell fate decision? Autophagy. 2007;3:45–47. doi: 10.4161/auto.3416. [DOI] [PubMed] [Google Scholar]
- [146].Lepine S, Allegood JC, Park M, Dent P, Milstien S, Spiegel S. Sphingosine-1-phosphate phosphohydrolase-1 regulates ER stress-induced autophagy. Cell Death Differ. 2011;18:350–361. doi: 10.1038/cdd.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Gomes LC, Scorrano L. Mitochondrial morphology in mitophagy and macroautophagy. Biochim. Biophys. Acta. 2013;1833:205–212. doi: 10.1016/j.bbamcr.2012.02.012. [DOI] [PubMed] [Google Scholar]
- [148].De Duve C, Wattiaux R. Functions of lysosomes. Annu. Rev. Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- [149].Veenhuis M, Douma A, Harder W, Osumi M. Degradation and turnover of peroxisomes in the yeast Hansenula polymorpha induced by selective inactivation of peroxisomal enzymes. Arch. Microbiol. 1983;134:193–203. doi: 10.1007/BF00407757. [DOI] [PubMed] [Google Scholar]
- [150].Bolender RP, Weibel ER. A morphometric study of the removal of phenobarbital-induced membranes from hepatocytes after cessation of threatment. J. Cell Biol. 1973;56:746–761. doi: 10.1083/jcb.56.3.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Lemasters JJ. Variants of mitochondrial autophagy: Types 1 and 2 mitophagy and micromitophagy (Type 3) Redox Biol. 2014;2:749–754. doi: 10.1016/j.redox.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- [154].Lemasters JJ, Nieminen AL, Qian T, Trost LC, Elmore SP, Nishimura Y, Crowe RA, Cascio WE, Bradham CA, Brenner DA, Herman B. The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim. Biophys. Acta. 1998;1366:177–196. doi: 10.1016/s0005-2728(98)00112-1. [DOI] [PubMed] [Google Scholar]
- [155].Rodriguez-Enriquez S, Kim I, Currin RT, Lemasters JJ. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2:39–46. doi: 10.4161/auto.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17:422–427. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- [157].Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Kim I, Lemasters JJ. Mitochondrial degradation by autophagy (mitophagy) in GFP-LC3 transgenic hepatocytes during nutrient deprivation. Am. J. Physiol. Cell. Physiol. 2011;300:C308–17. doi: 10.1152/ajpcell.00056.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Aggarwal BB, Quintanilha AT, Cammack R, Packer L. Damage to mitochondrial electron transport and energy coupling by visible light. Biochim. Biophys. Acta. 1978;502:367–382. doi: 10.1016/0005-2728(78)90057-9. [DOI] [PubMed] [Google Scholar]
- [160].Kim I, Lemasters JJ. Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxid. Redox Signal. 2011;14:1919–1928. doi: 10.1089/ars.2010.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Alexandratou E, Yova D, Handris P, Kletsas D, Loukas S. Human fibroblast alterations induced by low power laser irradiation at the single cell level using confocal microscopy. Photochem. Photobiol. Sci. 2002;1:547–552. doi: 10.1039/b110213n. [DOI] [PubMed] [Google Scholar]
- [162].Soubannier V, McLelland GL, Zunino R, Braschi E, Rippstein P, Fon EA, McBride HM. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol. 2012;22:135–141. doi: 10.1016/j.cub.2011.11.057. [DOI] [PubMed] [Google Scholar]
- [163].Soubannier V, Rippstein P, Kaufman BA, Shoubridge EA, McBride HM. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS One. 2012;7:e52830. doi: 10.1371/journal.pone.0052830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, Dorn GW, 2nd, Yin XM. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J. Biol. Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, Dorn GW, 2nd, Yin XM. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J. Biol. Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Szabadkai G, Simoni AM, Chami M, Wieckowski MR, Youle RJ, Rizzuto R. Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Mol. Cell. 2004;16:59–68. doi: 10.1016/j.molcel.2004.09.026. [DOI] [PubMed] [Google Scholar]
- [169].Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- [170].Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, Scorrano L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J. Cell. Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- [172].Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, Barja F, Martinou JC. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS One. 2008;3:e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Arnoult D, Rismanchi N, Grodet A, Roberts RG, Seeburg DP, Estaquier J, Sheng M, Blackstone C. Bax/Bak-dependent release of DDP/TIMM8a promotes Drp1-mediated mitochondrial fission and mitoptosis during programmed cell death. Curr. Biol. 2005;15:2112–2118. doi: 10.1016/j.cub.2005.10.041. [DOI] [PubMed] [Google Scholar]
- [175].Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim. Biophys. Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Frohlich C, Grabiger S, Schwefel D, Faelber K, Rosenbaum E, Mears J, Rocks O, Daumke O. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 2013;32:1280–1292. doi: 10.1038/emboj.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlen P, Tomilin N, Shupliakov O, Lendahl U, Nister M. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 2011;30:2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 2005;16:5077–5086. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Chang CR, Blackstone C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann. N. Y. Acad. Sci. 2010;1201:34–39. doi: 10.1111/j.1749-6632.2010.05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Corti O, Lesage S, Brice A. What genetics tells us about the causes and mechanisms of Parkinson's disease. Physiol. Rev. 2011;91:1161–1218. doi: 10.1152/physrev.00022.2010. [DOI] [PubMed] [Google Scholar]
- [184].Buhlman L, Damiano M, Bertolin G, Ferrando-Miguel R, Lombes A, Brice A, Corti O. Functional interplay between Parkin and Drp1 in mitochondrial fission and clearance. Biochim. Biophys. Acta. 2014;1843:2012–2026. doi: 10.1016/j.bbamcr.2014.05.012. [DOI] [PubMed] [Google Scholar]
- [185].Hatano Y, Li Y, Sato K, Asakawa S, Yamamura Y, Tomiyama H, Yoshino H, Asahina M, Kobayashi S, Hassin-Baer S, Lu CS, Ng AR, Rosales RL, Shimizu N, Toda T, Mizuno Y, Hattori N. Novel PINK1 mutations in early-onset parkinsonism. Ann. Neurol. 2004;56:424–427. doi: 10.1002/ana.20251. [DOI] [PubMed] [Google Scholar]
- [186].Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- [187].Tan JM, Dawson TM. Parkin blushed by PINK1. Neuron. 2006;50:527–529. doi: 10.1016/j.neuron.2006.05.003. [DOI] [PubMed] [Google Scholar]
- [188].Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Pacelli C, De Rasmo D, Signorile A, Grattagliano I, di Tullio G, D'Orazio A, Nico B, Comi GP, Ronchi D, Ferranini E, Pirolo D, Seibel P, Schubert S, Gaballo A, Villani G, Cocco T. Mitochondrial defect and PGC-1alpha dysfunction in parkin-associated familial Parkinson's disease. Biochim. Biophys. Acta. 2011;1812:1041–1053. doi: 10.1016/j.bbadis.2010.12.022. [DOI] [PubMed] [Google Scholar]
- [190].Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson's disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Sandebring A, Dehvari N, Perez-Manso M, Thomas KJ, Karpilovski E, Cookson MR, Cowburn RF, Cedazo-Minguez A. Parkin deficiency disrupts calcium homeostasis by modulating phospholipase C signalling. FEBS J. 2009;276:5041–5052. doi: 10.1111/j.1742-4658.2009.07201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Kuroda Y, Mitsui T, Kunishige M, Shono M, Akaike M, Azuma H, Matsumoto T. Parkin enhances mitochondrial biogenesis in proliferating cells. Hum. Mol. Genet. 2006;15:883–895. doi: 10.1093/hmg/ddl006. [DOI] [PubMed] [Google Scholar]
- [193].Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Lin WJ, Kuang HY. Oxidative stress induces autophagy in response to multiple noxious stimuli in retinal ganglion cells. Autophagy. 2014;10 doi: 10.4161/auto.36076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Wei X, Qi Y, Zhang X, Qiu Q, Gu X, Tao C, Huang D, Zhang Y. Cadmium induces mitophagy through ROS-mediated PINK1/Parkin pathway. Toxicol. Mech. Methods. 2014:1–8. doi: 10.3109/15376516.2014.943444. [DOI] [PubMed] [Google Scholar]
- [196].Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 2014;111:4439–48. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, Huang L, Xue P, Li B, Wang X, Jin H, Wang J, Yang F, Liu P, Zhu Y, Sui S, Chen Q. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–85. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- [198].Chu CT, Bayır H, Kagan VE. LC3 binds externalized cardiolipin on injured mitochondria to signal mitophagy in neurons: implications for Parkinson disease. Autophagy. 2014;10:376–8. doi: 10.4161/auto.27191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Chu CT, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, Qiang Wang KZ, Klein-Seetharman J, Balasubramanian K, Amoscato AA, Borisneko G, Huang Z, Gusdon AM, Cheikhi A, Steer EK, Wang R, Baty C, Watkins S, Bahar I, Bayir H, Kagan VE. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197–205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A. 2004;101:15927–32. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Anand R, Wai T, Baker MJ, Kladt N, Schauss AC, Rugarli E, Langer T. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol. 2014;204:919–29. doi: 10.1083/jcb.201308006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- [203].Krieg P, Rosenberger S, de Juanes S, Latzko S, Hou J, Dick A, Kloz U, van der Hoeven K, Hausser I, Esposito I, Rauh M, Schneider H. Aloxe3 knockout mice reveal a function of epidermal lipoxygenase-3 as hepoxilin synthase and its pivotal role in barrier formation. J Invest Dermatol. 2013;133:172–80. doi: 10.1038/jid.2012.250. [DOI] [PubMed] [Google Scholar]
- [204].Russo SB, Baicu CF, Van Laer A, Geng T, Kasiganesan H, Zile MR, Cowart LA. J Clin Invest. 2012;122:3919–30. doi: 10.1172/JCI63888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Ebel P, Vom Dorp K, Petrasch-Parwez E, Zlomuzica A, Kinugawa K, Mariani J, Minich D, Ginkel C, Welcker J, Degen J, Eckhardt M, Dere E, Dörmann P, Willecke K. Inactivation of ceramide synthase 6 in mice results in an altered sphingolipid metabolism and behavioral abnormalities. J Biol Chem. 2013;288:21433–47. doi: 10.1074/jbc.M113.479907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Park MA, Zhang G, Martin AP, Hamed H, Mitchell C, Hylemon PB, Graf M, Rahmani M, Ryan K, Liu X, Spiegel S, Norris J, Fisher PB, Grant S, Dent P. Vorinostat and sorafenib increase ER stress, autophagy and apoptosis via ceramide-dependent CD95 and PERK activation. Cancer Biol Ther. 2008;10:1648–62. doi: 10.4161/cbt.7.10.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]