Abstract
Background
Clinically relevant polymorphisms often demonstrate population-specific allele frequencies. Central and South America remain largely uncategorized in the context of pharmacogenomics.
Materials & methods
We assessed 15 polymorphisms from 12 genes (ABCB1 3435C>T, ABCG2 Q141K, CYP1B1*3, CYP2C19*2, CYP3A4*1B, CYP3A5*3C, ERCC1 N118N, ERCC2 K751Q, GSTP1 I105V, TPMT 238G>C, TPMT 460G>A, TPMT 719A>G, TYMS TSER, UGT1A1*28 and UGT1A1 −3156G>A) in 81 Peruvian and 95 Mexican individuals.
Results
Six polymorphism frequencies differed significantly between the two populations: ABCB1 3435C>T, CYP1B1*3, GSTP1 I105V, TPMT 460G>A, UGT1A1*28 and UGT1A1 −3156G>A. The pattern of observed allele frequencies for all polymorphisms could not be accurately estimated from any single previously studied population.
Conclusion
This highlights the need to expand the scope of geographic data for use in pharmacogenomics studies.
Keywords: genotype, Hispanic, Mexico, Peru, pharmacogenomics, polymorphism, population
Clinically relevant DNA polymorphisms have significantly variable allele frequencies among different world populations [1–7]. However, many areas of the world are under-represented in current pharmacogenomics research [8,9]. Organizations such as the Pharmacogenetics for Every Nation Initiative are attempting to redress this balance by collecting country and region-specific data on pharmacogenomically relevant polymorphisms to ensure rational medication selection not based on Western European data [2,8–13].
Hispanic populations represent an important challenge for pharmacogenomics. Mexico is the 11th most populous nation in the world, and Mexican–Americans comprised 10.9% of the US population as of 2010 [14]. With over 30 million people of Mexican descent living in North America, this population is too large to be ignored by researchers. Approximately 60% of Mexicans classify themselves as Mestizo, with an ancestry that includes both Amerindians and Europeans. Another 30% are of Amerindian descent and 9% self-classify as white [15]. A study of Texas Hispanics (90% Mexican–Americans) calculated the degree of admixture to be approximately 36% Amerindian and 64% European [16]. Such a significant non-European genetic contribution to the Mexican population makes inferences based on European–American pharmacogenomic data problematic [17].
Peru has a population of approximately 30.15 million people. The major ethnic groups are 45% Amerindian, 37% Mestizo (mixed Amerindian and white) and 15% white, with the remaining 3% made up from Japanese, Chinese and African ethnicities [18]. A study of 25 regions of Peru found different levels of European ancestry depending on location [19]. As with the Mexican population this admixture provides a distinct population [20–22] that does not allow for assumptions from existing data for any one ethnic group.
Even generalizations across Hispanic groups are not viable. Both admixture levels and disease prevalence vary across Central and South America. For example, in New Jersey, where the majority of the Hispanic population is of Puerto Rican descent, the relative genetic contributions of Europeans, Amerindians and Africans are approximately 85%, 9% and 6%, respectively [16]. This variation in admixture correlates with real clinical problems. Puerto Ricans have the highest asthma rates in the US while Mexican–Americans have the lowest, and a meta-analysis has shown that Hispanics with African ancestry are at risk for more severe asthma in both Mexican–American and Puerto Rican American populations [23].
There is a paucity of pharmacogenomics data for Hispanic populations. Indeed, the majority of Central and South America are largely uncharacterized in the pharmacogenomics literature [24]. Previous studies have shown that Mexican–Americans have lower frequencies of the CYP2D6*4 alleles than European populations, but similar CYP2E1 and CYP3A4 enzyme activity [24–26]. Significant differences between Mexican–American and Spanish populations have also been observed for the CYP2C9*2 polymorphism, with the Spanish population carrying at least double the amount of CYP2C9*2 alleles (16% vs 8% for Mexican–Americans, 7% for Mexican-Mestizos and 1% for Mexican-Tepehuanos) [27].
Even within genes assumptions cannot be made about allele frequencies based on previously studied populations. In the MTHFR gene, two commonly studied polymorphisms (677 C>T and 1298A>C) showed inconsistent frequencies within the Mexican population. MTHFR 677C>T had the highest reported frequency in Mexicans (58%), significantly different from European (36.1–47.3%) and West African (9%) populations and was also high in Peruvians (46%) [28]. In contrast, the MTHFR 1298 A>C polymorphism had one of the lowest recorded frequencies in Mexico (14.7%), similar to West African populations (13.9%) and significantly differing from Europeans (28–36%) [29]. In Amerindian Peruvians the MTHFR 1298A>C frequency was rare (1.5%) [30]. The ITPA polymorphism, P32T, putatively responsible for toxicity from azathioprine therapy [31] is present at a similar frequency in Peruvians (1%) and Mexicans (2%) [32], which is significantly lower than other world populations studied for this polymorphism (5–19%) [32].
Additionally, frequencies of polymorphisms and haplotypes for the warfarin pathway genes CYP2C9 and VKORC1 highlight that differences also occur between Peruvian and Mexican populations. The warfarin high-dose predictive allele VKORC1 Asp36Tyr was not found at all in Peruvians [33]. For warfarin low dose prediction, 45% of Mexicans carried the CYP2C9/VKORC1 genotype combination compared with only 28% of Peruvians [6]. For this combination the Peruvians were most similar to the African populations (22–23%) despite the low incidence of African admixture [20–22], and the Mexicans were closer to the Caucasian population (55%) [6].
To further elucidate the pharmacogenomic similarities and/or differences within and between Hispanic and other world populations, we have assessed the allele frequencies of key polymorphisms in ABCB1, ABCG2, CYP1B1, CYP2C19, CYP3A4, CYP3A5, ERCC1, ERCC2, GSTP1, TPMT, TYMS and UGT1A1 in Mexican and Peruvian individuals. These genes were selected because they are involved in the transport, metabolism, or are the target for at least 76 systemic drugs from the WHO Essential Medicines List [34], and the specific alleles have previously been identified as clinically relevant in more than one population. Whilst studies on polymorphisms in these genes in many populations have previously been reported (Supplementary Tables 1–3; see online at: www.futuremedicine.com/doi/suppl/10.2217/pgs.15.10), this represents the first comparison of multiple pharmacogenomically relevant polymorphisms in Mexicans and Peruvians.
Materials & methods
Population samples
Genotyping was performed on genomic DNA from 81 healthy unrelated Peruvian volunteers (35 female, 44 male, 2 unknown) recruited as controls in a TB Vitamin D study [35], and 95 healthy unrelated Mexican individuals (50 female, 45 male) from the Coriell Institute [36]. The Mexican individuals were from Los Angeles, CA and defined as individuals with at least three Mexico-born grandparents [37]. This study was approved by the Washington University Human Studies Committee.
Genotyping
Genotypes for ABCB1 3435C>T, ABCG2 Q141K, CYP1B1*3, CYP2C19*2, CYP3A4*1B, CYP3A5*3C, ERCC1 N118N, ERCC2 K751Q, GSTP1 I105V, TPMT 238G>C, TPMT 460G>A, TPMT 719A>G, UGT1A1 −3156G>A and UGT1A1*28 were determined using PCR and Pyrosequencing® methodology as previously described [38–41]. The TYMS TSER polymorphism was assayed using PCR and agarose gel electrophoresis with primers and conditions as previously described [42].
Analysis
Hardy–Weinberg equilibrium was assessed using HWSIM [43]. TPMT variant allele frequencies were combined to determine the overall frequency of high-risk variants in the populations. Pairwise linkage (D′) analysis for UGT1A1 was performed using the Polymorphism and Haplotype Analysis Suite [44]. Significant differences between Mexican and Peruvian genotype frequencies were assessed with χ2 analysis using Statistica (StatSoft Inc. Tulsa, OK).
Results
All genotype frequencies were in Hardy–Weinberg equilibrium. Variant allele frequencies for all 15 polymorphisms in the Peruvian and Mexican populations were not universally similar to each other (Table 1) or any one population previously studied (Supplementary Tables 1–3). Six polymorphisms differed significantly between the Peruvian and Mexican populations in this study: ABCB1 3435C>T, CYP1B1*3, GSTP1 I105V, TPMT 460G>A, UGT1A1*28 and UGT1A1 −3156G>A (Table 1). Combined frequency of TPMT variant alleles was 0.1 in the Peruvian population and 0.06 in the Mexican population (predominantly TPMT*3A, an allele containing a combination of 719G>A and 460G>A for both populations). TPMT*2A (238G>C) was not observed in either population. Pairwise linkage analysis for UGT1A1*28 and −3156G>A indicated that the two alleles were in tighter linkage in the Mexican population (D′ = 0.97; p < 0.001) than the Peruvian population (D′ = 0.86; p < 0.001).
Table 1.
Genotype and variant allele frequencies for polymorphisms in the Peruvian and Mexican populations.
| Gene | Polymorphism | rs number | Population | n | Homozygous wild-type | Heterozygous | Homozygous variant | Variant allele frequency |
|---|---|---|---|---|---|---|---|---|
| ABCB1 | 3435C>T† | rs1045642 | Peruvian | 73 | 39 | 29 | 5 | 0.27 |
| Mexican | 92 | 25 | 45 | 22 | 0.48 | |||
|
| ||||||||
| ABCG2 | Q141K | rs2231142 | Peruvian | 79 | 49 | 24 | 6 | 0.23 |
| Mexican | 93 | 61 | 27 | 5 | 0.20 | |||
|
| ||||||||
| CYP1B1 | *3‡ | rs1056836 | Peruvian | 70 | 49 | 19 | 2 | 0.16 |
| Mexican | 86 | 50 | 28 | 8 | 0.26 | |||
|
| ||||||||
| CYP2C19 | *2 | rs4244285 | Peruvian | 80 | 69 | 10 | 1 | 0.08 |
| Mexican | 94 | 75 | 18 | 1 | 0.11 | |||
|
| ||||||||
| CYP3A4 | *1B | rs2740574 | Peruvian | 74 | 67 | 6 | 1 | 0.05 |
| Mexican | 91 | 83 | 8 | 0 | 0.04 | |||
|
| ||||||||
| CYP3A5 | *3C | rs776746 | Peruvian | 77 | 1 | 18 | 58 | 0.87 |
| Mexican | 94 | 1 | 32 | 61 | 0.82 | |||
|
| ||||||||
| ERCC1 | N118N | rs11615 | Peruvian | 78 | 45 | 30 | 3 | 0.23 |
| Mexican | 91 | 46 | 36 | 9 | 0.30 | |||
|
| ||||||||
| ERCC2 | K751Q | rs13181 | Peruvian | 78 | 57 | 20 | 1 | 0.14 |
| Mexican | 95 | 68 | 23 | 4 | 0.16 | |||
|
| ||||||||
| GSTP1 | I105V† | rs1695 | Peruvian | 65 | 36 | 23 | 6 | 0.27 |
| Mexican | 93 | 23 | 49 | 21 | 0.49 | |||
|
| ||||||||
| TPMT | 238G>C (*2) | rs1800462 | Peruvian | 80 | 80 | 0 | 0 | 0.00 |
| Mexican | 94 | 94 | 0 | 0 | 0.00 | |||
|
| ||||||||
| TPMT | 460G>A (*3B)‡ | rs1800460 | Peruvian | 81 | 68 | 12 | 1 | 0.09 |
| Mexican | 84 | 78 | 6 | 0 | 0.04 | |||
|
| ||||||||
| TPMT | 719A>G (*3C) | rs1142345 | Peruvian | 76 | 65 | 10 | 1 | 0.08 |
| Mexican | 92 | 84 | 8 | 0 | 0.04 | |||
|
| ||||||||
| TYMS | TSER§ | rs34743033 | Peruvian | 73 | 8 | 34 | 31 | 0.66¶ |
| Mexican | 91 | 14 | 34 | 42 | 0.65¶ | |||
|
| ||||||||
| UGT1A1 | *28‡,# | rs3064744 | Peruvian | 78 | 21 | 37 | 19 | 0.48 |
| Mexican | 89 | 35 | 41 | 11 | 0.36 | |||
|
| ||||||||
| UGT1A1 | −3156G>A‡ | rs10929302 | Peruvian | 77 | 21 | 38 | 18 | 0.48 |
| Mexican | 94 | 42 | 40 | 12 | 0.34 | |||
Genotype frequencies significantly differ at p<0.001.
Genotype frequencies significantly differ at p<0.05.
TYMS TSER*4 allele frequency = 0 in Peruvians and 0.005 in Mexicans.
Frequency of TYMS TSER*3 allele.
(TA)8TAA repeat allele frequency = 0.01 for both populations.
Discussion
The Peruvian and Mexican populations are both highly admixed and this is reflected in the lack of similarity to any one commonly genotyped population for the polymorphisms assessed in this study (Supplementary Tables 1–3). Even in this small-scale study, we found that the Mexican and Peruvian populations did not consistently demonstrate similar allele frequencies to each other or to other South American countries. This is understandable taking into account the differences in admixture between the two populations [15,18]. Of particular note is that previous studies in these populations also yielded differences in allele frequencies [6]. We postulate that the admixture in these populations makes basing assumptions on allele frequencies, even within countries, from existing pharmacogenomics data inaccurate. Even resources such as the 1000 Genomes Project [45,46], which uses family trios of LA Mexican samples and Peruvian family trios from Lima, do not accurately reflect the same allele frequencies found here. Consequently, any one source is not a viable surrogate for an estimate of the likelihood of a clinically relevant polymorphism occurring in an individual in Peruvian or Mexican populations. It is important to not make assumptions based on previously published allele frequencies without further assessment of admixture and population comparisons within different regions of each country.
Mexicans born in Los Angeles demonstrated allele frequencies with the most similarities to previously studied European populations, specifically for ABCB1, CYP1B1, CYP2C19, CYP3A4 and TYMS (Supplementary Tables 1–3). However, both Mexicans and Peruvians were closer in allele frequencies to Asian populations for ERCC1 N118N. UGT1A1 polymorphisms in Mexicans were similar to African population allele frequencies. The frequency of GSTP1 I105V in Mexicans was one of the highest reported globally. Peru tracked with Asian populations for ERCC1, African populations for ABCB1 and UGT1A1, Central/South American for CYP2C19, and European for CYP3A4 and CYP3A5 polymorphisms. GSTP1 I105V in Peruvians was similar to both European and Asian reported frequencies (Supplementary Table 3). For ABCG2 Q141K and ERCC2 K751Q Mexico and Peru had similar allele frequencies (Table 1); however, these were unrelated to previously reported populations apart from Pacific Islanders for ABCG2 (Supplementary Tables 1 & 3), highlighting that using existing population frequencies to represent different world populations would be erroneous.
Peru had one of the lowest reported frequencies of CYP2C19*2, a polymorphism important for predicting adverse events from clopidogrel [47], whereas the TPMT combined *2, *3A, *3B and *3C alleles demonstrated one of the highest frequencies (10%) in Peru of all the populations previously studied (1–11%; Supplementary Table 1). In this instance the Peruvian population was most similar to the Ghanaian and Bulgarian populations (11 and 8%, respectively) [48,49] and least similar to Asian populations (1–2%) [50,51]. This polymorphism has been assessed in several South American populations (Figure 1) [52–59]. The allele frequency in Mexicans (6%; Supplementary Table 1) was closest to Bolivians (6%) [52] and almost half of the frequency in Peruvians. The Peruvian TPMT data suggests caution when using azathioprine in Peruvian populations. Taking into account the low frequency of the clinically relevant ITPA polymorphism [32], the TPMT alleles are a likely cause of toxicity from azathioprine in up to one tenth of Peruvians. Genotyping should be considered prior to therapy selection in this population.
Figure 1. TPMT allele frequency (combined *2, *3A, *3B and *3C) distribution in North and South America [52–59].
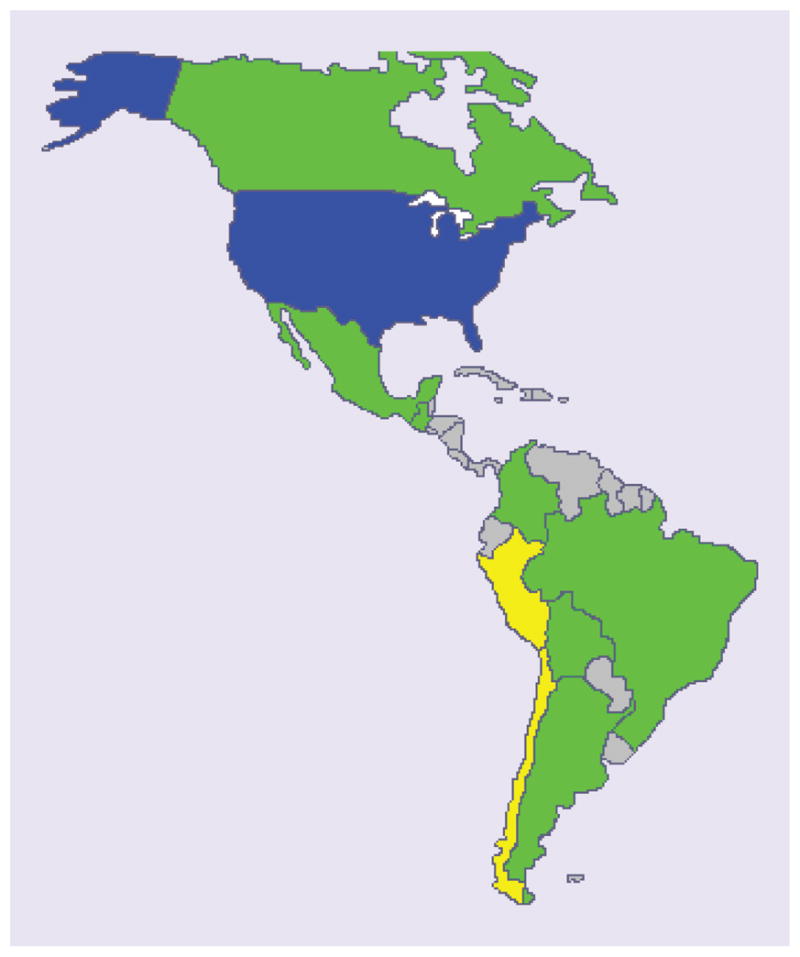
USA Caucasian (blue) represents the reference frequency (0.04) [56]. Green = between 0.5 and 2× the reference frequency, yellow = allele frequencies greater than 2× the reference frequency, gray = unknown frequency. Allele frequencies are listed in Supplementary Table 1.
Likewise, UGT1A1 polymorphisms in Peru were among the highest previously reported (48% for both UGT1A1*28 and −3156G>A; Table 1), suggesting the Peruvian population would be at increased risk from drugs such as irinotecan, where these polymorphisms have shown significant associations with life-threatening toxicities [60]. As genotyping is suggested in the US prior to irinotecan dose selection, this should also be considered in Peruvian populations where available. A dose reduction in UGT1A1*28 patients could significantly reduce the impact and incidence of severe irinotecan-related toxicity.
The UGT1A1 −3156G>A polymorphism is in tight linkage with the UGT1A1*28 polymorphism in multiple populations [61,62]. However, in populations where the linkage between the two is diminished, as seen in the Peruvian population (D′ = 0.86), it may be important to genotype both variants to get an accurate profile of UGT1A1 activity. This difference in linkage highlights the need to comprehensively screen target populations before selecting haplotype tagSNPs to use as markers for association studies. Using public resources such as the International HapMap Project [63], which identified haplotypes in four populations (European Caucasian, Chinese, Japanese and Yoruba) to pre-select tagSNPs would likely miss-represent haplotypes in populations where the linkage disequilibrium blocks, and population-specific polymorphisms and mutations across genes/chromosomes are uncharacterized. Indeed, screening for causative mutations in open-angle glaucoma identified mutations in MYOC exon 3 in Peruvians that had previously been missed by sequencing other populations [64], supporting the need for caution when making assumptions based on previously published data.
Conclusion
This pilot study supports previous studies [17] that it is not possible to estimate the frequency of pharmacogenomic variants in Hispanic populations based on existing information from other populations. Nor can data from one Hispanic country be used to inform medical decision-making in another Hispanic country. The population admixture in Mexico [15] and Peru [18] has likely played an evolutionary role in diversifying allele frequencies and linkage disequilibrium, and caution should be exercised when making assumptions even within populations from different regions of the country [19]. Obviously, there are limitations with such a small sample set lacking in family cohorts and without a detailed population structure analysis on each sample. However, the lack of trend toward other commonly studied populations is clear even from a small-scale study. As the Hispanic population is a significant component of the World’s population [14], it should not be omitted from future large-scale genomics projects. The data presented here suggest that prior genotype testing of individuals should be considered before prescribing common therapeutics, such as azathioprine and irinotecan, in Mexican and Peruvian individuals.
Future perspective
Currently, drug development is mainly based on data from European-derived reference populations. The frequencies of key polymorphisms in the Peruvian and Mexican populations could not be predicted from knowledge of any one commonly studied ethnic group. This study demonstrates that it is essential to take into account a range of world populations when deriving global strategies for optimizing therapy regimens. For example, in the Peruvian population the high frequency of TPMT variant alleles (Figure 1) suggest an elevated toxicity risk from thiopurines. Genotyping every patient for panels of polymorphisms before any drug therapy will not be on the horizon in the majority of countries for some time [8,9,11]. However, polymorphism frequency cannot be extrapolated from similar countries or ethnic groups. Even within countries, population admixture can present with widely different genotype frequencies [27,65–67]. Knowledge of the frequency of functional polymorphisms in the patient’s representative population or subpopulation, compared with the frequencies observed in the reference populations used in drug development, is vital for enhancing the ability to make appropriate therapeutic decisions at a national level.
Executive summary.
Background
Differences in allele frequencies and linkage disequilibrium are common between populations.
The majority of the pharmacogenomic literature centers around Caucasian, Asian and African populations.
Pharmacogenomic data for Hispanic countries are severely lacking in published literature.
Methods
Genotyping of 15 pharmacogenomically relevant polymorphisms was performed on unrelated individuals of Peruvian and Mexican descent.
Results
Peruvians and Mexicans showed significantly different allele frequencies for ABCB1 3435C>T, CYP1B1*3, GSTP1 I105V, TPMT 460G>A, UGT1A1*28 and UGT1A1 −3156G>A.
UGT1A1 linkage disequilibrium was lower in Peruvians (D′ = 0.86) than Mexicans (D′ = 0.97).
Discussion
No consistent pattern could be identified that could predict either Peruvian or Mexican allele frequency distribution for any one polymorphism.
Conclusion
The admixture of the populations is likely a causative factor in the divergent allele frequencies and linkage disequilibrium.
Caution when dispensing medication with known pharmacogenomic influence on drug efficacy and toxicity should be exerted for Hispanic populations in the absence of individual genotype testing.
Future perspective
Ultimately frequencies of all clinically relevant polymorphisms will be archived and medical decisions will be informed by, at the least, population-based risk. However, this is still a long way from current practice.
Acknowledgments
The efforts of D Berg and F Arenas, and the technical support of JB Phu, D Sara, RM Engen, HD Kannall, TJ Scott-Horton and MR Minton are greatly appreciated.
Footnotes
Financial & competing interests disclosure
This work was supported in part by the NIH Pharmacogenetics Research Network (GM63340), the Pharmacogenetics for Every Nation Initiative and the Global Research Training Fund of the Fogarty International Center of the NIH. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
•• of considerable interest
- 1.Kidd KK, Pakstis AJ, Yun L. An historical perspective on the world-wide distribution of allele frequencies at the human dopamine D4 receptor locus. Hum Genet. 2014;133(4):431–433. doi: 10.1007/s00439-013-1386-0. [DOI] [PubMed] [Google Scholar]
- 2••.Stathias V, Sotiris GR, Karagiannidis I, et al. Exploring genomic structure differences and similarities between the Greek and European HapMap populations: implications for association studies. Ann Hum Genet. 2012;76(6):472–483. doi: 10.1111/j.1469-1809.2012.00730.x. Study cautioning the use of generalizations when inferring genotype data in different populations. [DOI] [PubMed] [Google Scholar]
- 3.Engen RM, Marsh S, Van Booven DJ, McLeod HL. Ethnic differences in pharmacogenetically relevant genes. Curr Drug Targets. 2006;7(12):1641–1648. doi: 10.2174/138945006779025446. [DOI] [PubMed] [Google Scholar]
- 4.Cavaco I, Piedade R, Gil JP, Ribeiro V. CYP2C8 polymorphism among the Portuguese. Clin Chem Lab Med. 2006;44(2):168–170. doi: 10.1515/CCLM.2006.030. [DOI] [PubMed] [Google Scholar]
- 5.King CR, Yu J, Freimuth RR, McLeod HL, Marsh S. Interethnic variability of ERCC2 polymorphisms. Pharmacogenomics J. 2005;5(1):54–59. doi: 10.1038/sj.tpj.6500283. [DOI] [PubMed] [Google Scholar]
- 6.Marsh S, King CR, Porche-Sorbet RM, Scott-Horton TJ, Eby CS. Population variation in VKORC1 haplotype structure. J Thromb Haemost. 2006;4(2):473–474. doi: 10.1111/j.1538-7836.2006.01759.x. [DOI] [PubMed] [Google Scholar]
- 7.Lee MY, Mukherjee N, Pakstis AJ, et al. Global patterns of variation in allele and haplotype frequencies and linkage disequilibrium across the CYP2E1 gene. Pharmacogenomics J. 2008;8(5):349–356. doi: 10.1038/tpj.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marsh S, Van Booven DJ, McLeod HL. Global pharmacogenetics: giving the genome to the masses. Pharmacogenomics. 2006;7(4):625–631. doi: 10.2217/14622416.7.4.625. [DOI] [PubMed] [Google Scholar]
- 9.Marsh S. Pharmacogenetics: global clinical markers. Pharmacogenomics. 2008;9(4):371–373. doi: 10.2217/14622416.9.4.371. [DOI] [PubMed] [Google Scholar]
- 10.Mitropoulos K, Innocenti F, Van Schaik RH, et al. Institutional Profile: Golden Helix Institute of Biomedical Research: interdisciplinary research and educational activities in pharmacogenomics and personalized medicine. Pharmacogenomics. 2012;13(4):387–392. doi: 10.2217/pgs.12.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Roederer MW, Sanchez-Giron F, Kalideen K, et al. Pharmacogenetics and rational drug use around the world. Pharmacogenomics. 2011;12(6):897–905. doi: 10.2217/pgs.11.17. Description of the work of the PGENI organization. [DOI] [PubMed] [Google Scholar]
- 12.Roederer MW, McLeod HL. Applying the genome to national drug formulary policy in the developing world. Pharmacogenomics. 2010;11(5):633–636. doi: 10.2217/pgs.10.55. [DOI] [PubMed] [Google Scholar]
- 13.Mitropoulos K, Johnson L, Vozikis A, Patrinos GP. Relevance of pharmacogenomics for developing countries in Europe. Drug Metabol Drug Interact. 2011;26(4):143–146. doi: 10.1515/DMDI.2011.028. [DOI] [PubMed] [Google Scholar]
- 14.US Census Website. www.census.gov.
- 15.CIA World Factbook: Mexico. www.cia.gov.
- 16.Bertoni B, Budowle B, Sans M, Barton SA, Chakraborty R. Admixture in Hispanics: distribution of ancestral population contributions in the Continental United States. Hum Biol. 2003;75(1):1–11. doi: 10.1353/hub.2003.0016. [DOI] [PubMed] [Google Scholar]
- 17.Moreno-Estrada A, Gignoux CR, Fernandez-Lopez JC, et al. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science. 2014;344(6189):1280–1285. doi: 10.1126/science.1251688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CIA World Factbook: Peru. www.cia.gov.
- 19.Sandoval JR, Salazar-Granara A, Acosta O, et al. Tracing the genomic ancestry of Peruvians reveals a major legacy of pre-Columbian ancestors. J Hum Genet. 2013;58(9):627–634. doi: 10.1038/jhg.2013.73. [DOI] [PubMed] [Google Scholar]
- 20.Callegari-Jacques SM, Tarazona-Santos EM, Gilman RH, et al. Autosome STRs in native South America-Testing models of association with geography and language. Am J Phys Anthropol. 2011;145(3):371–381. doi: 10.1002/ajpa.21505. [DOI] [PubMed] [Google Scholar]
- 21.Pereira L, Zamudio R, Soares-Souza G, et al. Socioeconomic and nutritional factors account for the association of gastric cancer with Amerindian ancestry in a Latin American admixed population. PLoS ONE. 2012;7(8):e41200. doi: 10.1371/journal.pone.0041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scliar MO, Gouveia MH, Benazzo A, et al. Bayesian inferences suggest that Amazon Yunga Natives diverged from Andeans less than 5000 ybp: implications for South American prehistory. BMC Evol Biol. 2014;14(1):174. doi: 10.1186/s12862-014-0174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pino-Yanes M, Thakur N, Gignoux CR, et al. Genetic ancestry influences asthma susceptibility and lung function among Latinos. J Allergy Clin Immunol. 2014;135(1):228–235. doi: 10.1016/j.jaci.2014.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Cuautle-Rodriguez P, Llerena A, Molina-Guarneros J. Present status and perspective of pharmacogenetics in Mexico. Drug Metabol Drug Interact. 2014;29(1):37–45. doi: 10.1515/dmdi-2013-0019. Comprehensive review of the status of pharmacogenomics research in Mexico. [DOI] [PubMed] [Google Scholar]
- 25.Poland RA, Lin KM, Nuccio C, Wilkinson GR. Cytochrome P450 2E1 and 3A activities do not differ between Mexicans and European Americans. Clin Pharmacol Ther. 2002;72(3):288–293. doi: 10.1067/mcp.2002.127398. [DOI] [PubMed] [Google Scholar]
- 26.Mendoza R, Wan YJ, Poland RE, et al. CYP2D6 polymorphism in a Mexican American population. Clin Pharmacol Ther. 2001;70(6):552–560. doi: 10.1067/mcp.2001.120675. [DOI] [PubMed] [Google Scholar]
- 27.Dorado P, Sosa-Macias MG, Penas-Lledo EM, et al. CYP2C9 allele frequency differences between populations of Mexican-Mestizo, Mexican-Tepehuano, and Spaniards. Pharmacogenomics J. 2011;11(2):108–112. doi: 10.1038/tpj.2010.29. [DOI] [PubMed] [Google Scholar]
- 28.Williams MA, Sanchez SE, Zhang C, Bazul V. Methylenetetrahydrofolate reductase 677 C-->T polymorphism and plasma folate in relation to pre-eclampsia risk among Peruvian women. J Matern Fetal Neonatal Med. 2004;15(5):337–344. doi: 10.1080/14767050410001680037. [DOI] [PubMed] [Google Scholar]
- 29.Gueant-Rodriguez RM, Gueant JL, Debard R, et al. Prevalence of methylenetetrahydrofolate reductase 677T and 1298C alleles and folate status: a comparative study in Mexican, West African, and European populations. Am J Clin Nutr. 2006;83(3):701–707. doi: 10.1093/ajcn.83.3.701. [DOI] [PubMed] [Google Scholar]
- 30.Monsalve MV, Salzano FM, Rupert JL, et al. Methylenetetrahydrofolate reductase (MTHFR) allele frequencies in Amerindians. Ann Hum Genet. 2003;67(Pt 4):367–371. doi: 10.1046/j.1469-1809.2003.00027.x. [DOI] [PubMed] [Google Scholar]
- 31.Marinaki AM, Ansari A, Duley JA, et al. Adverse drug reactions to azathioprine therapy are associated with polymorphism in the gene encoding inosine triphosphate pyrophosphatase (ITPase) Pharmacogenetics. 2004;14:181–187. doi: 10.1097/00008571-200403000-00006. [DOI] [PubMed] [Google Scholar]
- 32.Marsh S, King CR, Ahluwalia R, McLeod HL. Distribution of ITPA P32T alleles in multiple world populations. J Hum Genet. 2004;49(10):579–581. doi: 10.1007/s10038-004-0183-y. [DOI] [PubMed] [Google Scholar]
- 33.Shahin MH, Cavallari LH, Perera MA, et al. VKORC1 Asp36Tyr geographic distribution and its impact on warfarin dose requirements in Egyptians. Thromb Haemost. 2013;109(6):1045–1050. doi: 10.1160/TH12-10-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.WHO Essential Medicines List. www.who.int/selection_medicines/list/en/
- 35.Roth DE, Soto G, Arenas F, et al. Association between vitamin D receptor gene polymorphisms and response to treatment of pulmonary tuberculosis. J Infect Dis. 2004;190(5):920–927. doi: 10.1086/423212. [DOI] [PubMed] [Google Scholar]
- 36.Coriell Institute. https://catalog.coriell.org/1/NIGMS.
- 37.Llerena A, Dorado P, O’Kirwan F, Jepson R, Licinio J, Wong ML. Lower frequency of CYP2C9*2 in Mexican–Americans compared with Spaniards. Pharmacogenomics J. 2004;4(6):403–406. doi: 10.1038/sj.tpj.6500278. [DOI] [PubMed] [Google Scholar]
- 38.Marsh S, King CR, Garsa AA, McLeod HL. Pyrosequencing of clinically relevant polymorphisms. Methods Mol Biol. 2005;311:97–114. doi: 10.1385/1-59259-957-5:097. [DOI] [PubMed] [Google Scholar]
- 39.Saeki M, Saito Y, Jinno H, et al. Comprehensive UGT1A1 genotyping in a Japanese population by pyrosequencing. Clin Chem. 2003;49(7):1182–1185. doi: 10.1373/49.7.1182. [DOI] [PubMed] [Google Scholar]
- 40.Oliveira E, Marsh S, Van Booven DJ, Amorim A, Prata MJ, McLeod HL. Pharmacogenetically relevant polymorphisms in Portugal. Pharmacogenomics. 2007;8(7):703–712. doi: 10.2217/14622416.8.7.703. [DOI] [PubMed] [Google Scholar]
- 41.Garsa AA, McLeod HL, Marsh S. CYP3A4 and CYP3A5 genotyping by Pyrosequencing. BMC Med Genet. 2005;6(1):19. doi: 10.1186/1471-2350-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horie N, Aiba H, Oguro K, Hojo H, Takeishi K. Functional analysis and DNA polymorphism of the tandemly repeated sequences in the 5′-terminal regulatory region of the human gene for thymidylate synthase. Cell Struct Funct. 1995;20(3):191–197. doi: 10.1247/csf.20.191. [DOI] [PubMed] [Google Scholar]
- 43.Hardy–Weinberg software. http://krunch.med.yale.edu/hwsim.
- 44.Schaid DJ, Mcdonnell SK, Wang L, Cunningham JM, Thibodeau SN. Caution on pedigree haplotype inference with software that assumes linkage equilibrium. Am J Hum Genet. 2002;71(4):992–995. doi: 10.1086/342666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.100 Genomes Project: Peru. https://catalog.coriell.org.
- 46.1000 Genomes Project: Mexico. https://catalog.coriell.org.
- 47.Holmes MV, Perel P, Shah T, Hingorani AD, Casas JP. CYP2C19 genotype, clopidogrel metabolism, platelet function, and cardiovascular events: a systematic review and meta-analysis. JAMA. 2011;306(24):2704–2714. doi: 10.1001/jama.2011.1880. [DOI] [PubMed] [Google Scholar]
- 48.Ameyaw MM, Collie-Duguid ES, Powrie RH, Ofori-Adjei D, McLeod HL. Thiopurine methyltransferase alleles in British and Ghanaian populations. Hum Mol Genet. 1999;8(2):367–370. doi: 10.1093/hmg/8.2.367. [DOI] [PubMed] [Google Scholar]
- 49.Indjova D, Atanasova S, Shipkova M, Armstrong VW, Oellerich M, Svinarov D. Phenotypic and genotypic analysis of thiopurine s-methyltransferase polymorphism in the bulgarian population. Ther Drug Monit. 2003;25(5):631–636. doi: 10.1097/00007691-200310000-00013. [DOI] [PubMed] [Google Scholar]
- 50.Kumagai K, Hiyama K, Ishioka S, et al. Allelotype frequency of the thiopurine methyltransferase (TPMT) gene in Japanese. Pharmacogenetics. 2001;11(3):275–278. doi: 10.1097/00008571-200104000-00012. [DOI] [PubMed] [Google Scholar]
- 51.Chang JG, Lee LS, Chen CM, et al. Molecular analysis of thiopurine S-methyltransferase alleles in South-east Asian populations. Pharmacogenetics. 2002;12(3):191–195. doi: 10.1097/00008571-200204000-00003. [DOI] [PubMed] [Google Scholar]
- 52.Lu HF, Shih MC, Hsueh SC, Chen CM, Chang JY, Chang JG. Molecular analysis of the thiopurine S-methyltransferase alleles in Bolivians and Tibetans. J Clin Pharm Ther. 2005;30(5):491–496. doi: 10.1111/j.1365-2710.2005.00640_1.x. [DOI] [PubMed] [Google Scholar]
- 53.Dubinsky MC, Lamothe S, Yang HY, et al. Pharmacogenomics and metabolite measurement for 6-mercaptopurine therapy in inflammatory bowel disease. Gastroenterology. 2000;118(4):705–713. doi: 10.1016/s0016-5085(00)70140-5. [DOI] [PubMed] [Google Scholar]
- 54.Larovere LE, De Kremer RD, Lambooy LH, De Abreu RA. Genetic polymorphism of thiopurine S-methyltransferase in Argentina. Ann Clin Biochem. 2003;40(Pt 4):388–393. doi: 10.1258/000456303766477039. [DOI] [PubMed] [Google Scholar]
- 55.Isaza C, Henao J, Lopez AM, Cacabelos R. Allelic variants of the thiopurine methyltransferase (TPMT) gene in the Colombian population. Methods Find Exp Clin Pharmacol. 2003;25(6):423–429. doi: 10.1358/mf.2003.25.6.769646. [DOI] [PubMed] [Google Scholar]
- 56.Hon YY, Fessing MY, Pui CH, Relling MV, Krynetski EY, Evans WE. Polymorphism of the thiopurine S-methyltransferase gene in African-Americans. Hum Mol Genet. 1999;8(2):371–376. doi: 10.1093/hmg/8.2.371. [DOI] [PubMed] [Google Scholar]
- 57.Boson WL, Romano-Silva MA, Correa H, Falcao RP, Teixeira-Vidigal PV, De Marco L. Thiopurine methyltransferase polymorphisms in a Brazilian population. Pharmacogenomics J. 2003;3(3):178–182. doi: 10.1038/sj.tpj.6500175. [DOI] [PubMed] [Google Scholar]
- 58.Farfan MJ, Salas C, Canales C, et al. Prevalence of TPMT and ITPA gene polymorphisms and effect on mercaptopurine dosage in Chilean children with acute lymphoblastic leukemia. BMC Cancer. 2014;14:299. doi: 10.1186/1471-2407-14-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garrido C, Santizo VG, Mullers P, et al. Frequency of thiopurine S-methyltransferase mutant alleles in indigenous and admixed Guatemalan patients with acute lymphoblastic leukemia. Med Oncol. 2013;30(1):474. doi: 10.1007/s12032-013-0474-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst. 2007;99(17):1290–1295. doi: 10.1093/jnci/djm115. [DOI] [PubMed] [Google Scholar]
- 61.Sai K, Saeki M, Saito Y, et al. UGT1A1 haplotypes associated with reduced glucuronidation and increased serum bilirubin in irinotecan-administered Japanese patients with cancer. Clin Pharmacol Ther. 2004;75(6):501–515. doi: 10.1016/j.clpt.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 62.Innocenti F, Undevia SD, Iyer L, et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J Clin Oncol. 2004;22(8):1382–1388. doi: 10.1200/JCO.2004.07.173. [DOI] [PubMed] [Google Scholar]
- 63.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mendoza-Reinoso V, Patil TS, Guevara-Fujita ML, et al. Novel and known MYOC exon 3 mutations in an admixed Peruvian primary open-angle glaucoma population. Mol Vision. 2012;18:2067–2075. [PMC free article] [PubMed] [Google Scholar]
- 65.Estrela RC, Ribeiro FS, Carvalho RS, et al. Distribution of ABCB1 polymorphisms among Brazilians: impact of population admixture. Pharmacogenomics. 2008;9(3):267–276. doi: 10.2217/14622416.9.3.267. [DOI] [PubMed] [Google Scholar]
- 66.Sosa-Macias M, Dorado P, Alanis-Banuelos RE, Llerena A, Lares-Asseff I. Influence of CYP2D6 deletion, multiplication, −1584C-->G, 31G-->A and 2988G-->a gene polymorphisms on dextromethorphan metabolism among Mexican tepehuanos and mestizos. Pharmacology. 2010;86(1):30–36. doi: 10.1159/000314334. [DOI] [PubMed] [Google Scholar]
- 67.Sosa-Macias M, Llerena A. Cytochrome P450 genetic polymorphisms of Mexican indigenous populations. Drug Metabol Drug Interact. 2013;28(4):193–208. doi: 10.1515/dmdi-2013-0037. [DOI] [PubMed] [Google Scholar]


