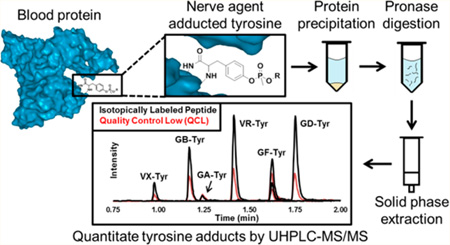
Abstract
This work describes a new specific, sensitive, and rapid stable isotope dilution method for the simultaneous detection of the organophosphorus nerve agents (OPNAs) tabun (GA), sarin (GB), soman (GD), cyclosarin (GF), VR, VX, and VM adducts to tyrosine (Tyr). Serum, plasma, and lysed whole blood samples (50 µL) were prepared by protein precipitation followed by digestion with Pronase. Specific Tyr adducts were isolated from the digest by a single solid phase extraction (SPE) step, and the analytes were separated by reversed-phase ultra high performance liquid chromatography (UHPLC) gradient elution in less than 2 min. Detection was performed on a triple quadrupole tandem mass spectrometer using time-triggered selected reaction monitoring (SRM) in positive electrospray ionization (ESI) mode. The calibration range was characterized from 0.100–50.0 ng/mL for GB– and VR– Tyr and 0.250–50.0 ng/mL for GA–, GD–, GF–, and VX/VM–Tyr (R2 ≥ 0.995). Inter- and intra-assay precision had coefficients of variation of ≤17 and ≤10%, respectively, and the measured concentration accuracies of spiked samples were within 15% of the targeted value for multiple spiking levels. The limit of detection was calculated to be 0.097, 0.027, 0.018, 0.074, 0.023, and 0.083 ng/mL for GA–, GB–, GD–, GF–, VR–, and VX/VM–Tyr, respectively. A convenience set of 96 serum samples with no known nerve agent exposure was screened and revealed no baseline values or potential interferences. This method provides a simple and highly specific diagnostic tool that may extend the time postevent that a confirmation of nerve agent exposure can be made with confidence.
The first confirmed large-scale use of organophosphorus nerve agents (OPNAs) in the 1984 Iraq–Iran conflict1 emphasized the need for sensitive diagnostic biomarkers and methods for detection of OPNA exposure.2 Since then, there have been several additional large-scale uses of OPNAs including the sarin (GB) gas attacks in Matsumoto in 1994,3 Tokyo in 1995,4 and Syria in 2013.5 While measurement of environmental samples plays a key role in investigations of alleged production or use of chemical weapons, detection of nerve agent biomarkers in clinical samples can provide critical evidence of human exposure. Such analyses may be crucial to separating the “worried-well” from exposed individuals in the case of a large exposure event.
Nerve agents are rapidly metabolized in vivo and exist partially as free agent (for a short time after exposure), partially as degradation products or metabolites, and partially as covalent adducts to macromolecules.6–8 Measurement of cholinesterase (ChE) activity in blood by the Ellman assay is widely used as a preliminary test to diagnose exposure to OPNAs.9 Unfortunately, this assay cannot differentiate between exposure to Schedule 1 nerve agents prohibited by the Chemical Weapons Convention (CWC) and exposure to commonly used organophosphorus (OP) pesticides. Additionally, reliance upon the Ellman assay requires routine measurement of baseline activity values to account for intra- and inter-individual activity variations.10 More specific methods based on the detection of nerve agent hydrolysis products such as O-alkyl methylphosphonic acid have also been developed for urine11,12 and blood samples;13 however, samples must be collected shortly after exposure because these metabolites are rapidly eliminated from the body.12
Fluoride reactivation has proven to be a highly sensitive method for detecting OPNA exposure and works by displacing the adducted OP moiety from acetylcholinesterase (AChE), butyrylcholinesterase (BChE), human serum albumin (HSA), and fibrous tissue with fluoride ions in turn regenerating the fluorinated agent.14–16 Unfortunately, loss of the agent defining O-alkyl group from cholinesterases through an enzyme mediated hydrolysis process known as “aging” eliminates the possibility of refluoridation and makes the detection of rapidly aging OPNA adducts such as soman (GD) unlikely.14 Likewise, displacement of the OP adduct from BChE and AChE after oxime therapy reduces the effectiveness of this otherwise sensitive diagnostic approach.17
Detecting OPNAs adducted to macromolecules in blood is an alternative approach with many advantages. Most notably, the extended lifetime relative to that of hydrolysis products or free agent makes protein adducts ideal for retrospective detection of nerve agent exposure.18 Methods have been developed to detect OPNAs adducted to the catalytic serine (Ser) residue of BChE (Ser-198)18–21 and tyrosine (Tyr-411) of HSA.17,22–24 While BChE has proven to be an excellent biomarker protein for assessing OPNA exposure,6,25,26 the aging process that can occur removes the structural identity of the adducted nerve agent. Conversely, OPNA adducts to HSA do not “age” and persist following oxime therapy.27–29
Proteins other than HSA have also been reported to contain OPNA–Tyr adducts; however, the high concentration of HSA, even after depletion, makes these additional protein biomarkers difficult to detect at low OPNA exposure levels.27 A method has been established for the determination of GD and tabun (GA) adducts in guinea pigs 7 days following exposure and subsequent oxime treatment.17,22,24 GB, VX, VR, and cyclosarin (GF) have also been reported to covalently bind to Tyr-411 of HSA.17,22,28 Even under conditions that would typically cause clinical samples to age and nerve agent specificity to be lost, analysis of Tyr adducts allows identification of the specific nerve agents of exposure.29
This work reports a newly developed sensitive and accurate method for the simultaneous detection of GA, GB, GD, GF, VR, and VX/VM adducted to Tyr in total protein. Pronase digestion followed by solid phase extraction (SPE) and ultrahigh pressure liquid chromatography-isotope dilution tandem mass spectrometry (UHPLC-MS/MS) was used to isolate OPNA–Tyr adducts from human serum and plasma that was exposed to OPNAs in vitro. Proteins were precipitated from plasma, serum, or whole lysed blood by the addition of acetone, and the resulting dried pellet was digested with Pronase. The OPNA-modified Tyr was purified by a single solid phase extraction (SPE) step, separated by UHPLC, and detected on a triple quadrupole tandem mass spectrometer by time function selected reaction monitoring (SRM) in positive electrospray ionization (ESI) mode in under 2 min. Quantitation was performed using isotope dilution calculations with a labeled analog for each analyte.
EXPERIMENTAL SECTION
Reagents and Supplies
The following materials were purchased from Fisher Scientific (Hanover Park, IL): ammonium bicarbonate, HPLC-grade methanol, Optima-grade acetone, and LC-MS-grade formic acid. Protease Type XIV from Streptomyces griseus (Pronase) was purchased from Sigma-Aldrich (P5147, St. Louis, MO). HPLC-grade acetonitrile was purchased from Tedia (Fairfield, OH). Strata SDB-L 96-well (50 mg) SPE plates were purchased from Phenomenex (Torrance, CA). Cibacron Blue beads were purchased from Pierce Biotechnology (Rockford, IL). Synthetically prepared native and isotopically labeled Tyr adducts (≥95% purity) were obtained from Battelle Memorial Institute (Columbus, OH) and the Defence Science and Technology Laboratory (Porton Down, U.K.). Isotopically labeled GA–Tyr was enriched with D5-ethyl, and GB-, GD-, GF-, VR-, and VX–Tyr were enriched with 13CD3 at the phosphomethyl position. A convenience set of 96 serum samples was purchased from Tennessee Blood Services (Memphis, TN) to assess background levels of each biomarker. Additionally, pooled plasma for preparing inhibited quality control (QC) materials and whole blood samples for determining method compatibility were purchased from Tennessee Blood Services. Pooled serum was purchased from Bioreclamation Inc. (Westbury, NY). Plasma and serum pools were screened by the vendors, in accordance with FDA regulations, to be free of Hepatitis B, Hepatitis C, Syphilis, and HIV. This study used de-identified blood acquired from commercial sources, and thus, the work did not meet the definition of human subjects as specified in 45 CFR 46.102 (f).
Preparation of Calibrators and Quality Controls
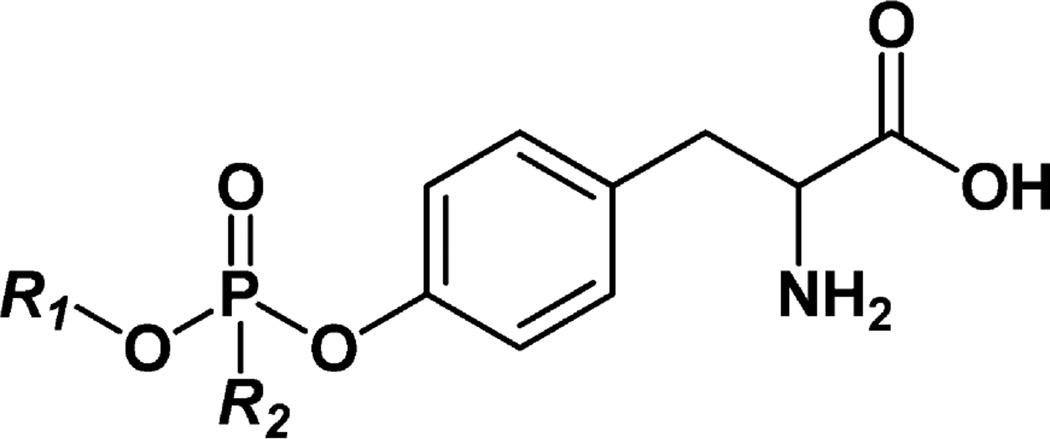
Synthetic GA–, GB–, GD–, GF–, VR–, and VX/VM–Tyr standards (Figure 1) were analyzed for amino acid content by Midwest Biotech (Fishers, IN), and purity estimates were adjusted accordingly. Individual stock solutions for each native and isotopically labeled standard (1.00 mg/mL) were prepared in HPLC-grade water and stored at −20 °C. The native stock solutions were combined and diluted to prepare eight calibrators at a final concentration for each of 0.100, 0.250, 0.500, 1.00, 5.00, 10.0, 25.0, and 50.0 ng/mL in HPLC-grade water. The isotopically labeled standard stock solutions were combined to prepare a single internal standard (ISTD) solution at a final concentration for each of 10 ng/mL in HPLC-grade water.
Figure 1.
Structures of organophosphorus nerve agent–tyrosine adducts. GA–Tyr, R1 = ethyl and R2 = dimethylamino; GB–Tyr, R1 = isopropyl and R2 = methyl; GD–Tyr, R1 = pinacolyl and R2 = methyl; GF–Tyr, R1 = cyclohexyl and R2 = methyl; VR–Tyr, R1 = isobutyl and R2 = methyl; VX/VM–Tyr, R1 = ethyl and R2 = methyl. VX and VM adducts to tyrosine cannot be differentiated.
Three QC samples were prepared: QC low (QCL), QC high (QCH), and QC digest (QCD). The QCL and QCH samples were prepared by spiking 1.00 mg/mL native OPNA–Tyr stock solutions into HPLC-grade water. The QCD samples were prepared by pooling previously characterized GA- and GB-spiked serum with GD- and GF-spiked plasma to monitor for the completeness of digestion. Plasma spiked with V-series agents was intentionally not included in the QCD pool due to the already low concentrations of VR– and VX–Tyr in undiluted plasma. The concentrations of OPNA–Tyr adducts were characterized for each QC sample as part of the method validation in a set of 20 different experiments.
Preparation of Agent-Spiked Materials for use as Quality Control Samples
A single pool of plasma was divided into seven parts and spiked with nerve agent so that each part was exposed to a single nerve agent or blank (isopropyl alcohol) at the Edgewood Chemical and Biological Center (Edgewood, MD). Specifically, 20.0 µL of a 1.00 mg/mL solution of GA, GB, GD, GF, VR, or VX nerve agent in isopropyl alcohol was added to 1.00 L of plasma at room temperature (RT) and gently shaken for 20 min for a final concentration of 20.0 ng/mL nerve agent in plasma. At the Battelle Memorial Institute, a pool of plasma (1.00 L) was spiked with 100 µL of a VM nerve agent stock (825 µg/mL in plasma) and stirred for 1 h at RT for a final concentration of 75.0 ng/mL VM in plasma. In a similar fashion, Battelle divided and spiked a pool of serum with a serum stock solution of GA (877 µg/mL) or GB (1050 µg/mL) to afford final concentrations of 1580 and 396 ng/mL in serum, respectively. All plasma and serum materials were assayed for BChE activity using a modified Ellman assay9 before and after spiking, and screened for free/excess agent prior to storage at −80 °C.
Sample Preparation
Plasma, serum, or whole lysed blood (50 µL) was transferred to a 2 mL conical bottom 96-well plate, and acetone (300 µL) was added for protein precipitation. The samples were sealed and centrifuged at 3000g for 5 min at 20 °C to pelletize proteins. Supernatant was removed by aspiration, and the remaining pellet was allowed to air-dry for 5 min at room temperature. The dried pellet was reconstituted in 400 µL of ammonium bicarbonate (50 mM, pH 7.8) with mixing. ISTD solution (20 µL) was added to all processed wells. Calibrators (50 µL) were spiked into wells containing reconstituted pellets formed from blank plasma for the purpose of matrix matching. To ensure homogeneity between calibrators, QCs, and unknowns, HPLC-grade water (50 µL) was added to all unknowns, reference materials, and digest controls. Pronase solution (100 µL of 10 mg/mL in 50 mM ammonium bicarbonate, pH 7.8) was added to each well. The plate was sealed with adhesive foil and incubated at 50 °C for 1.5 h with intermittent mixing. The entire volume (~570 µL) of the sample digest was added to a Strata SDB-L 96-well SPE plate that was first conditioned with methanol (1 mL) and then water (1 mL). The plate was washed with 10% methanol in water (2 × 1 mL). The Tyr adducts were eluted with methanol (500 µL) into a clean 2 mL 96-well receiving plate. All samples were evaporated to dryness under nitrogen at 50 °C using a Porvair Ultravap (Porvair Sciences, Leatherhead, U.K.). The dried samples were reconstituted in HPLC-grade water (75 µL), transferred to an Eppendorf 96-well PCR plate, and heat sealed with pierceable foil.
UHPLC-MS/MS
The chromatographic system consisted of an Agilent 1290 binary pump, refrigerated autosampler, and temperature controlled column compartment (Santa Clara, CA). Separation was performed on an Acquity HSS-T3, 1.8 µm, 50 × 1.0 mm analytical column (Waters, Milford, MA) at 60 °C. Water with 0.1% formic acid (mobile phase A, MPA) and acetonitrile with 0.1% formic acid (mobile phase B, MPB) made up the binary mobile phase, which was delivered at a flow rate of 250 µL/min. Following a 6 s needle wash in MPB, samples were injected (5 µL) on column with initial conditions set to 98% MPA and 2% MPB. A linear gradient was used to reach 40% MPB at 2 min followed by an immediate return to initial conditions for column regeneration and a total run time of 2.5 min. The extra-column volume of this system was found to be 10 µL, while the gradient delay volume was determined to be 105 µL in loop mainpass mode and 65 µL in loop bypass mode. To reduce the gradient delay volume and potential for loop carryover, the autosampler state was set to bypass mode at 12 s (10 × injection volume, 50 µL) following sample injection.
The UHPLC eluent was directed to an Agilent 6460 triple quadrupole mass analyzer with an ESI source operating in positive ionization mode. Data were acquired with Mass Hunter 6.0 in dynamic SRM mode monitoring the two most selective and sensitive transitions for the native Tyr adducts (quantitation and confirmation transitions) and a single transition for the labeled Tyr adducts. Ionization source parameters were optimized by post column infusion of a 100 ng/mL solution of VR–Tyr in HPLC-grade water at a flow rate of 5 µL/min. During the infusion experiment, the LC pump was set to deliver 23% MPB at 245 µL/min corresponding to the approximate solvent composition in source at the elution time of VR–Tyr, factoring in gradient delay, column void, and post column volume. The optimized source settings were found to be 200 °C sheath gas at 11 L/min, 300 °C drying gas at 8 L/min, nebulizer gas at 60 psi, nozzle voltage of 1000 V, and capillary voltage of 3500 V. Optimized parameters for the fragmentor voltage, collision energy (CE), and cell accelerator (CA) voltage were independently collected for each native and labeled Tyr adduct (Table 1). It is important to note that the nerve agents VX and VM differ in structure only by the leaving group and are indistinguishable from one another once adducted to proteins through phosphorylation. For this reason, the same mass spectrometer settings were used to identify transitions for VX–Tyr and VM–Tyr.
Table 1.
Optimized ESI-MS/MS Parameters for Detection of Native and Labeled Nerve Agent Tyrosine Adducts
| compd | precursor ion (m/z) | product ion (m/z) | fragmentor (V) | collision energy (V) | cell accelerator (V) | |
|---|---|---|---|---|---|---|
| GA–Tyr | quant | 317.1 | 243.1 | 100 | 15 | 3.0 |
| GA–Tyr | conf | 317.1 | 198.1 | 100 | 7.0 | 2.0 |
| D5-GA–Tyr | ISTD | 322.1 | 243.1 | 100 | 15 | 3.0 |
| GB–Tyr | quant | 302.1 | 214.1 | 100 | 15 | 4.5 |
| GB–Tyr | conf | 302.1 | 260.1 | 100 | 2.0 | 3.0 |
| 13CD3-GB–Tyr | ISTD | 306.1 | 218.1 | 100 | 15 | 4.5 |
| GD–Tyra | quant | 260.1 | 214.1 | 110 | 8.0 | 4.5 |
| GD–Tyra | conf | 260.1 | 136.1 | 110 | 23 | 7.5 |
| 13CD3-GD–Tyra | ISTD | 264.1 | 218.1 | 110 | 8.0 | 4.5 |
| GF–Tyr | quant | 342.1 | 214.1 | 70 | 20 | 4.5 |
| GF–Tyr | conf | 342.1 | 260.1 | 70 | 3.0 | 3.0 |
| 13CD3-GF–Tyr | ISTD | 346.1 | 218.1 | 70 | 20 | 4.5 |
| VR–Tyr | quant | 316.1 | 214.1 | 100 | 14 | 4.5 |
| VR–Tyr | conf | 316.1 | 260.1 | 100 | 2.0 | 3.0 |
| 13CD3-VR–Tyr | ISTD | 320.1 | 218.1 | 100 | 14 | 4.5 |
| VX/VM–Tyrb | quant | 288.1 | 214.1 | 110 | 17 | 4.5 |
| VX/VM–Tyrb | conf | 288.1 | 242.1 | 110 | 7.0 | 3.5 |
| 13CD3-VX/VM–Tyrb | ISTD | 292.1 | 218.1 | 110 | 17 | 4.5 |
Optimized for source fragmentation, [M + H]+ not observed.
The VX/VM–Tyr transitions are used to detect both VX–Tyr and VM–Tyr.
Calibration curves were constructed by plotting the response ratio (analyte area divided by internal standard area) versus the expected calibrator concentration. Linear regression was used to fit the data with 1/x weighting as determined by residual analysis. The origin was not forced through zero. The peak area ratio of the quantitation (quant) to confirmation (conf) transitions were used to calculate the confirmation ion ratio (CIR), a value established for each adduct and used to confirm detection.
Method Validation
Data from 20 replicate calibration curves and QC samples were evaluated to assess accuracy, precision, linearity, lowest reportable limit (LRL), and limits of detection (LOD) for each analyte. A maximum of two runs were prepared and analyzed per day over the span of two months by three separate analysts. Accuracy and inter-assay precision were calculated using the 0.250, 2.50, and 25.0 ng/mL calibrators. Intra-assay precision was calculated on five individual preparations of each QC level within a single batch. The LOD was calculated from the standard deviation of the concentration of the lowest four calibrators per analyte, as described by Taylor.30 Method ruggedness was investigated for five digestion parameters, which were not normalized by isotope dilution and therefore expected to have the greatest impact on method variance. Temperature, time, Pronase concentration, digestion buffer concentration, and pH were separately evaluated against the validated conditions using the QCD or QCL samples at values ±20% of the final conditions.
Matrix Effects and Extraction Recovery
Matrix effects and extraction recovery were evaluated by a pre/post-SPE spiking experiment performed in triplicate. Blank serum was processed through the precipitation and digestion steps and split into two samples prior to SPE. The resulting two identical blank digests were identified as the pre- and postspiked samples. Prior to SPE, the prespike sample was spiked with 50 µL of the 1.00 ng/mL standard, and the postspike sample was spiked with 50 µL of water. Both samples were extracted by SPE, and the eluent was removed by evaporation. The prespike extract was reconstituted in 50 µL of water and 50 µL of the ISTD solution. The postspike extract was reconstituted in 50 µL of the 1.00 ng/mL standard and 50 µL of the ISTD solution. These samples were analyzed and compared to a matrix-free solution containing 50 µL of the 1.00 ng/mL standard and 50 µL of the ISTD solution. The average of the response ratio of the prespike was compared to that of the postspike to assess extraction recovery:
The average of the response ratio of the postspike was compared to that of the matrix-free solution to assess the matrix effect in terms of ionization efficiency:
This process was used to evaluate various conditioning, loading, washing, and elution steps on several SPE phases during method development.
Safety Considerations
Nerve agent adducts to Tyr are not expected to pose a risk greater than that of free Tyr. Universal safety precautions for handling biological samples should be strictly adhered to at all times when handling blood products. Nerve agents should only be handled in an assurity laboratory.
RESULTS AND DISCUSSION
Sample Preparation Optimization
There are several approaches to enrich HSA from serum or plasma;31 however, these techniques are semiselective processes that require proper internal standards to account for losses in recovery of the protein.32 Due to the lack of an isotopically labeled nerve agent adduct to HSA, total protein was digested without enrichment. The internal standard solution was added to each sample at the earliest possible step (following the removal of the protein precipitation solvent) and was present during all remaining steps, including digestion. The calibrator solutions were added to blank matrix at the same step, ensuring normalized quantitation by matrix matching and isotope dilution.
This reported method digests total protein and not explicitly HSA nerve agent adducts. The difference between samples containing total protein and samples enriched for HSA was evaluated by processing G-series nerve-agent-spiked plasma using the two approaches (Table 2). In brief, nerve-agent-spiked samples were either enriched with HSA using Cibacron Blue prior to Pronase digestion, as described by Andacht et al.,32 or treated using the precipitation method, described here, without enrichment. When compared, plasma spiked with GB, GD, and GF had similar Tyr adduct values for HSA-enriched and total protein samples, suggesting HSA as the primary source of the OPNA–Tyr adducts measured. Interestingly, plasma spiked with GA showed greater differences between the two sample preparations. The value calculated for GA–Tyr was within the characterized method limits for both preparations, but the value for HSA-enriched sample was significantly lower and more variable than that for samples prepared by total protein precipitation. This finding is consistent with observations of the GA–Tyr adduct being acid labile, perhaps similar to reported acid hydrolysis of GA adducts to cholinesterases,33–35 as the HSA enrichment protocol incorporates trichloroacetic acid to precipitate the protein following enrichment with Cibacron Blue.
Table 2.
Comparison between the Digestion of Total Protein and Albumin Enriched Plasma Exposed to G-Series Nerve Agents
| calcd concn (ng/mL) (% CV) | |||
|---|---|---|---|
| agent-spiked plasma |
total protein digest |
albumin-enriched digest |
difference (%)a |
| GA | 3.24 (10.9) | 1.02 (47.1) | 104 |
| GB | 4.54 (5.10) | 4.23 (13.0) | 7.04 |
| GD | 7.48 (8.56) | 6.44 (13.1) | 15.0 |
| GF | 3.82 (7.50) | 3.75 (13.7) | 1.66 |
The difference (%) was calculated as 100 × the absolute value of [(total protein digest concentration − albumin-enriched digest concentration)/(average of total protein and albumin-enriched digests)].
Significant attention was given to the optimization of conditions and the ruggedness evaluation of the digestion because protein adducted with isotopically labeled nerve agent was not available and a nonspecific protease was used. To optimize digestion conditions, temperature, pH, digestion time, Pronase concentration, and buffer concentrations were varied ±20% using the QCD materials to identify the most reproducible conditions. Temperature, pH, Pronase concentration, and buffer concentration had insignificant influence on measured OPNA–Tyr adducts when varied ±20%; however, digestion time had significant impact on the calculated values, particularly for GA–Tyr. The GA–Tyr values decreased as the digestion time increased beyond 1 h, despite the use of basic pH conditions, which have previously been shown to reduce degradation of GA adducts.34 After 2 h of digestion, the GA–Tyr concentration was nearly 3-fold lower than those collected after 1 h at 50 °C. The values for GB–, GD–, and GF–Tyr increased beyond 1 h of digestion and plateaued at 1.5 h at 50 °C. A digestion time of 1.5 h was chosen for the robustness of the majority of Tyr adducts in the method. Despite the sacrifice in robustness for the digestion of GA–Tyr, the characterized QCD values were within acceptable limits of precision (CV ≤ 20%).
The effect of sample integrity and composition on the sensitivity of the OPNA–Tyr assay was also evaluated. Unexposed serum, plasma, and lysed whole blood samples were spiked with internal standard and calibrators to make a matrix-based calibration curve, then digested and extracted according to the validated method. As expected and recently demonstrated for the OPNA–BChE adduct method,36 there was no statistically significant difference in measured OPNA–Tyr values between serum and plasma. Lysed whole blood was analyzed to simulate samples where serum or plasma could not be harvested. The calibration curves were compared for consistency, response, and detection limit and were found to be indistinguishable between matrices. The uniformity of response in different matrices permits the preparation and quantitation of a variety of sample matrices in a single batch against a calibration curve constructed in any of the three matrices.
This high-throughput method allows for the accurate quantitation of clinical samples without sacrificing sensitivity. Using the reported method, 96 samples can be processed and ready for analysis in less than 4 h and analyzed by UHPLC-MS/MS in a single batch in less than 4 h. Because sample preparation of a second set of 96 samples can be performed in parallel with instrumental analysis of the first, up to 480 samples can be prepared and analyzed in 24 h by a single instrument. This level of throughput is defined by the simplicity of the sample preparation and the short UHPLC analysis time.
Matrix Effects and Extraction Recovery
OPNA–Tyr reference standard spiked matrix materials were used to evaluate SPE chemistries for matrix effects. Ionization efficiency and extraction recovery were both considered in the overall assessment of sensitivity and performance of the method. Polymer- and silica-ligand sorbent beds composed of silica, cyanopropyl, mixed mode anion exchange, mixed mode cation exchange, phenyl, styrene divinylbenzene, pentafluorophenyl, and porous graphitic carbon were all assessed under conditions appropriate to the volume of sample and their mode(s) of extraction. The Strata SDB-L, a styrene divinylbenzene sorbent specifically designed for the purification of aromatic and hydrophobic compounds, provided the most consistent recovery and ionization data and proved to be a good all-around sorbent for OPNA–Tyr adducts (Table 3). The ionization efficiency generally increased with the composition of acetonitrile introduced during the gradient separation. This decrease in ionization efficiency for the early eluting compounds is consistent with reversed phase chromatography and desolvation of mobile phase with a high aqueous composition. VX/VM–Tyr adducts had the lowest recovery and were typically lost due to partial breakthrough during the wash steps of the extraction. Efforts to improve the recovery of VX/VM–Tyr resulted in higher recovery but also lower ionization efficiency and a net overall decrease in sensitivity for many of the other adducts.
Table 3.
Retention Time, Ionization Efficiency, and Extraction (SPE) Recovery of OPNA–Tyrosine Adducts
| compd | retention time (min) (CV) |
ionization efficiency (%) |
SPE recovery (%) |
|---|---|---|---|
| VX–Tyr, VM–Tyr | 0.993 (1.01%) | 56.0 | 37.8 |
| GB–Tyr | 1.18 (0.840%) | 66.7 | 64.9 |
| GA–Tyr | 1.26 (0.814%) | 53.4 | 89.9 |
| VR–Tyr | 1.43 (0.741%) | 64.0 | 74.8 |
| GF–Tyr | 1.64 (0.720%) | 88.5 | 67.8 |
| GD–Tyr | 1.77 (0.670%) | 92.1 | 69.9 |
UHPLC-MS/MS Optimization
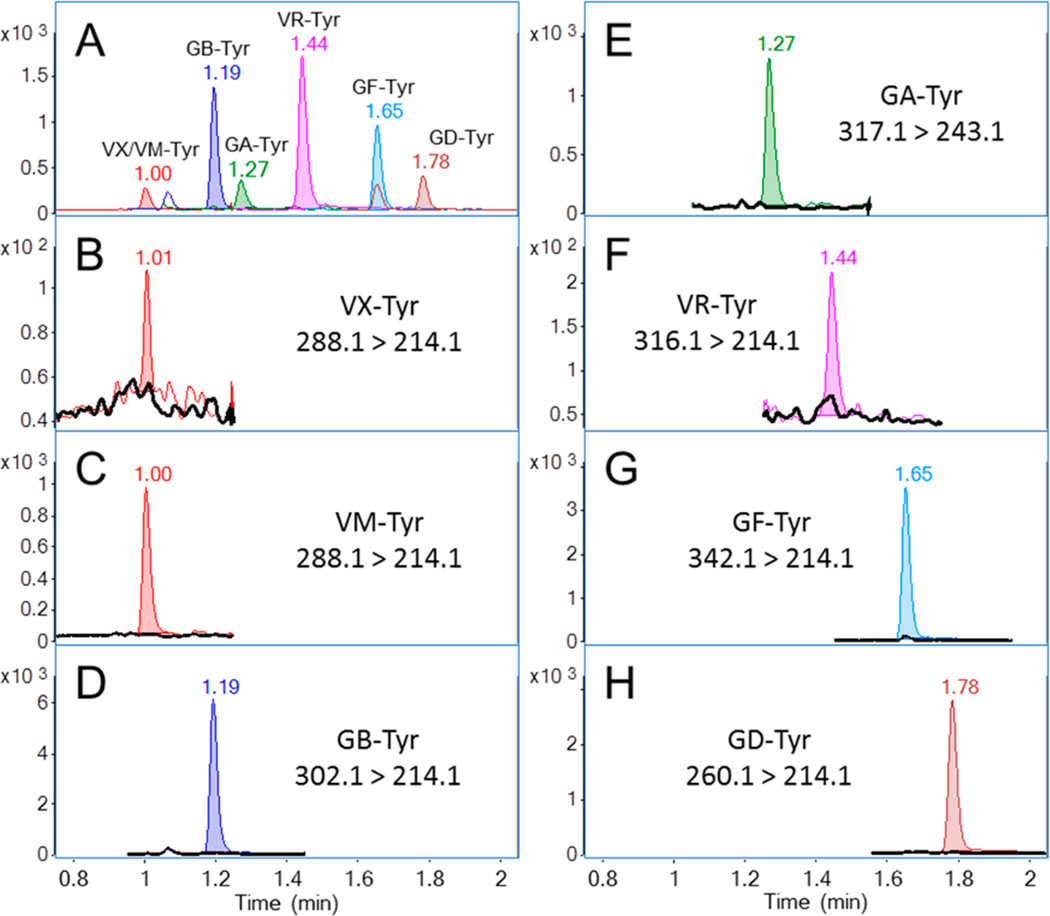
The Atlantis-T3 column was selected after careful review of several separation phases and modes. A scouting gradient and linear solvent strength model were used to simplify chromatographic development.37 Gradient elution was chosen due to the large fraction of run time occupied between VX– and GD–Tyr. A separation producing a gradient retention factor (k*) of approximately 10 was required to produce baseline resolution of GA–Tyr from closely eluting low abundance mass interferences endogenous to serum. The final gradient time of 2 min and end point of 40% MPB was effective at producing a well-resolved chromatogram. The 30 s taken by the UHPLC autosampler to draw, wash, and load the next sample allowed approximately 4 column volumes of initial mobile phase conditions to re-equilibrate the column prior to injection. The total time from injection to injection was 2.5 min. Figure 2A shows an example of the resulting chromatographic separation on the 1.00 ng/mL calibration standard. The retention time for each Tyr adduct was recorded during the two-month validation at each calibration and QC level, and the average retention time and precision are reported (Table 3). There was little variation in retention times, even when multiple column and mobile phase lots were used.
Figure 2.
Overlay of extracted ion chromatograms of the OPNA–Tyr adducts. (A) Blank plasma spiked with 1.00 ng/mL synthetic standards. (B–H, black trace) Blank plasma and (colored trace) detected tyrosine adduct following separate and independent addition of corresponding nerve agent to the blanks.
The high separation efficiency afforded by UHPLC demanded very short MS/MS dwell times per transition to obtain the 10–15 data points per peak we desired for quantitation. While the instrument was capable of acquiring acceptable data under these conditions, the signal-to-noise ratio was increased by utilizing the “Dynamic MRM” time function acquisition feature. The mass spectrometer was set to acquire transition-specific signal during a 30 s window centered on the compound’s expected retention time.
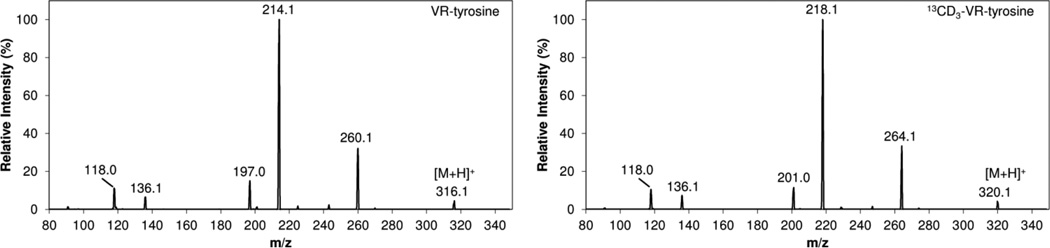
The MS/MS fragmentation for GA–, GB–, GD–, GF–, and VX–Tyr adducts have been previously reported.17,24 The product ion scans for VR– and 13CD3-VR–Tyr were collected at collision energies of 5, 15, and 25, and the resulting spectra were summed for the native and labeled compounds (Figure 3). The fragmentation for VR–Tyr is consistent with GB–, GD–, and GF–Tyr adducts with the collision induced disassociation (CID) of the nerve agent defining O-alkyl group resulting in m/z 260.1 and the additional loss of HCO2H to yield m/z 214.1 as the two most abundant product ions. Fragmentation of VX/VM–Tyr also yielded m/z 260.1 and m/z 214.1; however, the loss of HCO2H to m/z 242.1 was more abundant than m/z 260.1 and thus used for confirmation. For GA–Tyr, the less abundant product ion of m/z 198.0 was chosen for confirmation instead of m/z 226.117 due to background contamination in the matrix.
Figure 3.
Product ion scans for VR– and 13CD3-VR–Tyrosine.
In-source fragmentation resulting in the loss of the O-alkyl group was observed for GB–, GF–, GD–, VR–, and VX/VM–Tyr and increased with the stability of the O-alkyl leaving group. The most pronounced in-source fragmentation occurred for GF– and GD–Tyr. Decreasing the fragmentor voltage to 70 was sufficient to minimize the loss of cyclohexanol from GF–Tyr so the [M + H]+ ion of m/z 342.1 could be used for precursor selection of GF–Tyr. However, the expected GD–Tyr [M + H]+ ion of m/z 344.1 was not observed, even at low fragmentor voltage, indicating instrument-related variation.17,24 Instead, the fragmentor voltage was increased to promote full in-source fragmentation loss of pinacolyl alcohol, and the resulting m/z 260.1 was used as the GD–Tyr precursor ion (Table 1). The CID of m/z 260.1 resulted in loss of HCO2H to yield m/z 214.1 and further loss of HPO3 to yield m/z 136.1. These MS/MS transitions had no background contamination and were used for quantitation and confirmation of GD–Tyr. Additionally, the MS/MS transitions from this in-source fragmentation may prove to be useful as a general screen for O-alkyl methylphosphonate–Tyr adducts since GB–, GF–, VR– and VX/VM–Tyr were all detected in the same transitions and can be identified by retention time.
Linearity, Accuracy, Precision, and Limits of Detection
Validation data was collected from 20 processed calibration curves that were recorded over a period of two months (Table 4). Calibration curves were linear for all Tyr adducts with a coefficient of determination value (R2) ≥0.995 over a range of 0.100–50.0 ng/mL for GB– and VR–Tyr and a range of 0.250–50.0 ng/mL for GA–, GD–, GF–, VX–, and VM–Tyr. The mean accuracies for all Tyr adducts were 99.3–115% at 0.250 ng/mL, 94.9–100% at 2.50 ng/mL, and 98.5–101% at 25.0 ng/mL. The inter-assay precision on the same samples was 6.16–17.3, 4.52–8.11, and 3.30–7.70%, respectively. The three different QC samples were prepared and analyzed with each calibration curve, and the mean concentration and precision values were characterized. The inter-assay precision was 6.96–13.6% for QCL, 3.78–8.92% for QCH, and 7.48–17.2% for QCD. The intra-assay precision was evaluated using five simultaneous preparations of QCL, QCH, and QCD samples and was found to be between 1.54 and 9.72% for all adducts and all concentrations (Table 5). As expected, GA–Tyr was the most variable adduct measured for the calibrators and QC samples, regardless of concentration, although the precision was still within acceptable limits (CV ≤ 20%). The LOD values were calculated using the Taylor method of evaluating the standard deviation at known concentrations. The LOD values reported were calculated using the quantitation ion (Table 4) and are comparable to those reported for GB–, GD–, and VX–Tyr in rat plasma;24 however, this is the first report of limits of detection for GA–, GF–, and VR–Tyr. The LRL was chosen so that it was above the LOD with the confirmation ion reliably detected, and a CIR could be established for each adduct. The CIRs were calculated to be 1.10 (±0.137), 1.31 (±0.128), 6.94 (±0.502), 0.867 (±0.085), 1.55 (±0.151), and 0.692 (±0.078) for GA–, GB–, GD–, GF–, VR–, and VX/VM–Tyr adducts, respectively.
Table 4.
Accuracy, Inter-Assay Precision, Reporting Limits, and Detection Limits of OPNA–Tyr Adduct Calibrators (n = 20)
| compd | expected concn (ng/mL) |
calcd concn (ng/mL) |
accuracy of mean (%) |
precision (% CV) |
LRL (ng/mL)a |
URL (ng/mL)b |
LOD (ng/mL)c |
|---|---|---|---|---|---|---|---|
| GA–Tyr | 0.250 | 0.257 | 103 | 17.3 | 0.250 | 0.250 | 0.097 |
| 2.50 | 2.40 | 96.0 | 8.11 | ||||
| 25.0 | 25.1 | 101 | 7.70 | ||||
| GB–Tyr | 0.250 | 0.248 | 99.3 | 10.7 | 0.100 | 50.0 | 0.027 |
| 2.50 | 2.37 | 94.9 | 4.52 | ||||
| 25.0 | 25.4 | 101 | 3.45 | ||||
| GD–Tyr | 0.250 | 0.285 | 114 | 6.16 | 0.250 | 50.0 | 0.018 |
| 2.50 | 2.50 | 100 | 4.82 | ||||
| 25.0 | 25.2 | 101 | 3.63 | ||||
| GF–Tyr | 0.250 | 0.288 | 115 | 11.8 | 0.250 | 50.0 | 0.074 |
| 2.50 | 2.44 | 97.5 | 7.31 | ||||
| 25.0 | 24.6 | 98.5 | 5.39 | ||||
| VR–Tyr | 0.250 | 0.251 | 101 | 9.47 | 0.100 | 50.0 | 0.023 |
| 2.50 | 2.40 | 95.9 | 5.74 | ||||
| 25.0 | 24.9 | 99.4 | 3.30 | ||||
| VX–Tyr, VM–Tyr | 0.250 | 0.254 | 101 | 15.5 | 0.250 | 50.0 | 0.083 |
| 2.50 | 2.50 | 100 | 6.09 | ||||
| 25.0 | 25.0 | 100 | 5.37 |
Lowest reportable limit.
Upper reportable limit.
Limit of detection calculated by Taylor method.
Table 5.
Inter-Assay (n = 20) and Intra-Assay (n = 5) Precision of Quality Control Materials
| inter-assay precision | intra-assay precision | ||||
|---|---|---|---|---|---|
| compd | QC material |
calcd concn (ng/mL) |
% CV | calcd concn (ng/mL) |
% CV |
| GA–Tyr | QCLa | 1.03 | 13.6 | 0.948 | 7.86 |
| QCHb | 13.6 | 8.92 | 15.1 | 7.67 | |
| QCDc | 0.663 | 17.2 | 0.510 | 9.61 | |
| GB–Tyr | QCL | 1.01 | 9.30 | 0.923 | 3.59 |
| QCH | 14.0 | 3.78 | 14.1 | 1.77 | |
| QCD | 0.894 | 8.56 | 0.862 | 5.44 | |
| GD–Tyr | QCL | 1.03 | 6.96 | 1.05 | 2.22 |
| QCH | 14.0 | 5.49 | 14.8 | 3.26 | |
| QCD | 1.47 | 7.48 | 1.49 | 4.05 | |
| GF–Tyr | QCL | 1.00 | 7.58 | 0.953 | 3.78 |
| QCH | 14.1 | 7.91 | 14.9 | 5.51 | |
| QCD | 1.14 | 11.1 | 1.17 | 6.13 | |
| VR–Tyr | QCL | 0.985 | 9.35 | 0.935 | 3.96 |
| QCH | 13.9 | 4.09 | 14.7 | 1.54 | |
| VX–Tyr, VM–Tyr | QCL | 1.12 | 14.6 | 0.950 | 9.72 |
| QCH | 13.7 | 4.36 | 13.7 | 2.08 | |
QCL = low concentration synthetic OP–Tyr spike used for quality control.
QCH = high concentration synthetic OP–Tyr spike used for quality control..
QCD = G-series nerve agents individually spiked into separate plasma or serum and pooled for quality control of the digestion. Due to the low concentration of VR– and VX–Tyr measured in plasma spiked with VR and VX, these materials were not sued in the preparation of the QCD pool.
Stability
The synthetic OPNA–Tyr standards were stored in water at −20 °C for 9 months with no signs of degradation. Similarly, pooled plasma and serum samples independently spiked with GA, GB, GD, GF, VR, VX, and VM were stored at −80 °C with no signs of degradation in 9 months of testing. Initially, prepared samples were reconstituted in MPA following the removal of the extraction solvents; however, the signal intensity for GA–Tyr in MPA reconstituted samples decreased dramatically and was undetectable after 3 days stored on the instrument at 4 °C. Removal of the 0.1% formic acid from the reconstitution solvent eliminated the poor short-term stability of the extracted GA–Tyr samples.
It has been reported that adducts to HSA are stable and do not lose the agent defining O-alkyl side chain22,38 in a process similar to aging on cholinesterase.39,40 Still, there has been little discussion of the fate of HSA nerve agent adducts when exposed to conditions that would otherwise produce an aged cholinesterase adduct. As a model, GB was added to serum (396 ng/mL) which was then divided into two pools. One pool was stored at −80 °C, and the second was held at 40 °C for 96 h. As previously shown, these conditions are suitable for producing an aged methylphosphonate (MeP) adduct on BChE.41 The samples were characterized (n ≥ 8) yielding the result of 11.8 (±1.14) ng/mL GB–Tyr for the material stored at −80 °C and 2.37 (±0.194) ng/mL for the material stored at 40 °C. Evidence of O-alkyl side chain loss from the GB–Tyr adduct was not observed when compared to a standard of MeP–Tyr; however, the 80% reduction in GB–Tyr at a storage temperature of 40 °C is an important finding. The distinction between O-alkyl hydrolysis and overall adduct loss must be considered when attempting to measure Tyr adducts in samples that have not been collected, transported, or stored properly. Furthermore, it is likely that adduct loss will also occur in vivo, prior to specimen collection. A more detailed study of the degradation kinetics for additional OP–Tyr adducts is currently underway.
Nerve Agent Spiked Materials and Convenience Samples
Various OPNA spiked materials were characterized for Tyr adducts (n = 10) using the validated method (Table 6). Figure 2B–H contains representative chromatograms for plasma samples before and after the addition of nerve agents. The plasma samples were spiked with nerve agent so that complete BChE inhibition was not reached. These materials were used to assess the detection and reporting limit for nerve agents adducted to Tyr in terms of BChE percent inhibition. For the G-series agents, the extrapolated lower reporting limit is equivalent to approximately 2–5% BChE inhibition; although, based on the detection limits, it may be possible to identify nerve agent adducts to Tyr below 2% BChE inhibition. Approximately 10–20% of the spiked nerve agent was detected as a Tyr adduct for the G-series agents. Assuming that the measured Tyr adducts are predominately from HSA and that the normal range of HSA is 35–55 mg/mL in plasma,42 these values represent an approximately 0.001–0.004% molar adduction to HSA for GA, GB, GD, and GF.
Table 6.
Concentrations of OPNA–Tyr Adducts in Plasma after Spiking with Nerve Agent
| agent spiked into plasma |
BChE inhibition (%)a |
final spike in plasma (nM) |
measured OP– Tyr adduct (nM) (±std dev) |
calcd agent spike bound to tyrosine (%) |
|---|---|---|---|---|
| GA | 67.1 | 123 | 10.2 (±1.10) | 8.29 |
| GB | 88.1 | 143 | 15.0 (±0.76) | 10.5 |
| GD | 87.8 | 110 | 21.7 (±1.86) | 19.7 |
| GF | 76.9 | 111 | 11.2 (±0.848) | 10.1 |
| VR | 53.3 | 74.9 | 0.443 (±0.063) | 0.591 |
| VX | 92.1 | 74.9 | 0.625 (±0.278) | 0.834 |
| VM | 99.0 | 313 | 10.4 (±1.73) | 3.32 |
[1 − (prespike BChE activity/postspike BChE activity)] × 100.
As shown in Table 6, there is a clear difference in reactivity with Tyr between the G-series and the V-series agents with less than 1% of the spike of VX and VR binding to Tyr. The low reactivity of VX to HSA (Figure 2B) has previously been reported17 and suggested to be related to the nature of the leaving group. The reactivity of VR to Tyr is similar to that of VX (Figure 2F); however, while VM appears to be more reactive with Tyr than the other V-series investigated (Figure 2C), the direct comparison of the reactivity of VM to VX and VR with tyrosine should be qualified by confounding factors such as the different spiking conditions and concentrations used during in vitro preparations of these materials. As a whole, the V-series adducts are still much lower than observed for the G-series adducts.
To assess background and possible interferences, commercially obtained serum samples with no expectation of nerve agent exposure were prepared and analyzed. No peaks corresponding to any of the OPNA–Tyr adducts measured by the reported method were detected in the 96 convenience set samples studied. The selected SRM transitions and their ratio to one another coupled with the high-efficiency chromatographic separation provide excellent selectivity for the OPNA–Tyr adducts.
CONCLUSION
A new method for the detection of GA, GB, GD, GF, VR, VX, and VM exposure through the measurement of Tyr adducts in blood products has been developed and validated which may extend the time postevent that the exact OPNA of exposure may be identified in clinical specimens. The simplified sample preparation provides high accuracy and precision, low background interference, and low limits of detection using only 50 µL of whole lysed blood, plasma, or serum without the need for HSA enrichment. Rapid UHPLC-MS/MS analysis coupled with high throughput sample preparation allows up to 480 samples to be processed and analyzed per day per single instrument. Synthetic calibrators and OPNA spiked QC materials have been shown to be stable for at least 9 months when stored at −20 °C and −80 °C, respectively. The method has been used to characterize clinical samples exposed to live nerve agent in vitro and to assess the effect of sample aging conditions for the GB–Tyr adduct. Additionally, promoting in-source fragmentation may allow the described method to be readily adapted for use as a general screen for exposure to prohibited CWC Schedule 1 nerve agents containing an O-alkyl methylphosphonate group.
Acknowledgments
This work was supported by the Centers for Disease Control and Prevention, the Defense Threat and Reduction Agency (11-005-12430), and the Oak Ridge Institute for Science and Education. The authors would like to thank Ms. Chariety Sapp of the CDC’s Incident Response Laboratory (IRL) for dispensing convenience set samples prior to analysis. The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. Use of trade names is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention, the Public Health Service, or the U.S. Department of Health and Human Services.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.United Nations Security Council. Report of the Specialists Appointed by the Secretary-General to Investigate Allegations by the Islamic Republic of Iran Concerning the Use of Chemical Weapons. United Nations: New York: 1984. [PubMed] [Google Scholar]
- 2.Black RM, Clarke RJ, Read RW, Reid MTJ. J. Chromatogr. A. 1994;662:301–321. doi: 10.1016/0021-9673(94)80518-0. [DOI] [PubMed] [Google Scholar]
- 3.Morita H, Yanagisawa N, Nakajima T, Shimizu M, Hirabayashi H, Okudera H, Nohara M, Midorikawa Y, Mimura S. Lancet. 1995;346:290–293. doi: 10.1016/s0140-6736(95)92170-2. [DOI] [PubMed] [Google Scholar]
- 4.Nagao M, Takatori T, Matsuda Y, Nakajima M, Iwase H, Iwadate K. Toxicol. Appl. Pharmacol. 1997;144:198–203. doi: 10.1006/taap.1997.8110. [DOI] [PubMed] [Google Scholar]
- 5.United Nations Mission to Investigate Allegations of the Use of Chemical Weapons in the Syrian Arab Republic. Report on the Alleged Use of Chemical Weapons in the Ghouta Area of Damascus on 21 August 2013. The Hague, The Netherlands: Organisation for the Prohibition of Chemical Weapons; 2013. [Google Scholar]
- 6.Black RM. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2010;878:1207–1215. doi: 10.1016/j.jchromb.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Black RM, Read RW. Arch. Toxicol. 2013;87:421–437. doi: 10.1007/s00204-012-1005-1. [DOI] [PubMed] [Google Scholar]
- 8.Black RM. J. Anal. Toxicol. 2008;32:2–9. doi: 10.1093/jat/32.1.2. [DOI] [PubMed] [Google Scholar]
- 9.Worek F, Mast U, Kiderlen D, Diepold C, Eyer P. Clin. Chim. Acta. 1999;28:73–90. doi: 10.1016/s0009-8981(99)00144-8. [DOI] [PubMed] [Google Scholar]
- 10.Ellman GL, Courtney KD, Andres V, Featherstone RM. Biochem. Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- 11.Mawhinney DB, Hamelin EI, Fraser R, Silva SS, Pavlopoulos AJ, Kobelski RJ. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2007;852:235–243. doi: 10.1016/j.jchromb.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 12.Riches J, Morton I, Read RW, Black RM. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2005;816:251–258. doi: 10.1016/j.jchromb.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 13.Hamelin EI, Schulze ND, Shaner RL, Coleman RM, Lawrence RJ, Crow BS, Jakubowski EM, Johnson RC. Anal. Bioanal. Chem. 2014;406:5195–5202. doi: 10.1007/s00216-014-7702-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams TK, Capacio BR, Smith JR, Whalley CE, Korte WD. Drug Chem. Toxicol. 2005;27:77–91. doi: 10.1081/dct-120027901. [DOI] [PubMed] [Google Scholar]
- 15.Heilbronn E. Acta Chem. Scand. 1965;19:1333–1346. doi: 10.3891/acta.chem.scand.19-1333. [DOI] [PubMed] [Google Scholar]
- 16.Heilbronn E. Biochem. Pharmacol. 1965;14:1363–1373. doi: 10.1016/0006-2952(65)90120-6. [DOI] [PubMed] [Google Scholar]
- 17.Williams NH, Harrison JM, Read RW, Black RM. Arch. Toxicol. 2007;81:627–639. doi: 10.1007/s00204-007-0191-8. [DOI] [PubMed] [Google Scholar]
- 18.Fidder A, Hulst AG, Noort D, de Ruiter R, van der Schans M, Benschop HP, Langenberg JP. Chem. Res. Toxicol. 2002;15:582–590. doi: 10.1021/tx0101806. [DOI] [PubMed] [Google Scholar]
- 19.Knaack JS, Zhou Y, Abney CW, Jacob JT, Prezioso SM, Hardy K, Lemire SW, Thomas J, Johnson RC. Anal. Chem. 2012;84:9470–9477. doi: 10.1021/ac302301w. [DOI] [PubMed] [Google Scholar]
- 20.Pantazides BG, Watson CM, Carter MD, Crow BS, Perez JW, Blake TA, Thomas JD, Johnson RC. Anal. Bioanal. Chem. 2014;406:5187–5194. doi: 10.1007/s00216-014-7718-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sporty JL, Lemire SW, Jakubowski EM, Renner JA, Evans RA, Williams RF, Schmidt JG, van der Schans MJ, Noort D, Johnson RC. Anal. Chem. 2010;82:6593–6600. doi: 10.1021/ac101024z. [DOI] [PubMed] [Google Scholar]
- 22.Black RM, Harrison JM, Read RW. Arch. Toxicol. 1999;73:123–126. doi: 10.1007/s002040050596. [DOI] [PubMed] [Google Scholar]
- 23.Peeples ES, Schopfer LM, Duysen EG, Spaulding R, Voelker T, Thompson CM, Lockridge O. Toxicol. Sci. 2005;83:303–312. doi: 10.1093/toxsci/kfi023. [DOI] [PubMed] [Google Scholar]
- 24.Bao Y, Liu Q, Chen J, Lin Y, Wu B, Xie J. J. Chromatogr. A. 2012;1229:164–171. doi: 10.1016/j.chroma.2012.01.032. [DOI] [PubMed] [Google Scholar]
- 25.Nigg HN, Knaak JB. Rev. Environ. Contam. Toxicol. 2000;163:29–111. doi: 10.1007/978-1-4757-6429-1_2. [DOI] [PubMed] [Google Scholar]
- 26.Aurbek N, Thiermann H, Eyer F, Eyer P, Worek F. Toxicology. 2009;259:133–139. doi: 10.1016/j.tox.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Ding S-J, Carr J, Carlson JE, Tong L, Xue W, Li Y, Schopfer LM, Li B, Nachon F, Asojo O, Thompson CM, Hinrichs SH, Masson P, Lockridge O. Chem. Res. Toxicol. 2008;21:1787–1794. doi: 10.1021/tx800144z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.John H, Breyer F, Thumfart JO, Hochstetter H, Thiermann H. Anal. Bioanal. Chem. 2010;398:2677–2691. doi: 10.1007/s00216-010-4076-y. [DOI] [PubMed] [Google Scholar]
- 29.Read RW, Riches JR, Stevens JA, Stubbs SJ, Black RM. Arch. Toxicol. 2010;84:25–36. doi: 10.1007/s00204-009-0473-4. [DOI] [PubMed] [Google Scholar]
- 30.Taylor JK. Quality Assurance of Chemical Measurements. Chelsea, MI: Lewis Publishers; 1987. [Google Scholar]
- 31.Young CL, Woolfitt AR, McWilliams LG, Moura H, Boyer AE, Barr JR. Proteomics. 2005;5:4973–4979. doi: 10.1002/pmic.200402081. [DOI] [PubMed] [Google Scholar]
- 32.Andacht TM, Pantazides BG, Crow BS, Fidder A, Noort D, Thomas JD, Blake TA, Johnson RC. J. Anal. Toxicol. 2014;38:8–15. doi: 10.1093/jat/bkt088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carletti E, Li H, Li B, Ekstrom F, Nicolet Y, Loiodice M, Gillon E, Froment M-T, Lockridge O, Schopfer LM, Masson P, Nachon F. J. Am. Chem. Soc. 2008;130:16011–16020. doi: 10.1021/ja804941z. [DOI] [PubMed] [Google Scholar]
- 34.Barak D, Ordentlich A, Kaplan D, Barak R, Mizrahi D, Kronman C, Segall Y, Velan B, Shafferman A. Biochemistry. 2000;39:1156–1161. doi: 10.1021/bi992009n. [DOI] [PubMed] [Google Scholar]
- 35.Elhanany E, Ordentlich A, Dgany O, Kaplan D, Segall Y, Barak R, Velan B, Shafferman A. Chem. Res. Toxicol. 2001;14:912–918. doi: 10.1021/tx0100542. [DOI] [PubMed] [Google Scholar]
- 36.Pantazides BG, Watson CM, Carter MD, Crow BS, Perez JW, Blake TA, Thomas JD, Johnson RC. Anal. Bioanal. Chem. 2014;406:5187–5194. doi: 10.1007/s00216-014-7718-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snyder LR, Dolan JW. High-Performance Gradient Elution: The Practical Application of the Linear-Solvent-Strength Model. Hoboken, NJ: Wiley-Interscience; 2007. [Google Scholar]
- 38.Li B, Schopfer LM, Hinrichs SH, Masson P, Lockridge O. Anal. Biochem. 2007;361:263–272. doi: 10.1016/j.ab.2006.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berry WK, Davies DR. Biochem. J. 1966;100:572–576. doi: 10.1042/bj1000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Worek F, Aurbek N, Koller M, Becker C, Eyer P, Thiermann H. Biochem. Pharmacol. 2007;73:1807–1817. doi: 10.1016/j.bcp.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 41.Carter MD, Crow BS, Pantazides BG, Watson CM, Thomas JD, Blake TA, Johnson RC. Anal. Chem. 2013;85:11106–11111. doi: 10.1021/ac4029714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen Y, Jacobs JM, Camp DG, Fang R, Moore RJ, Smith RD. Anal. Chem. 2004;76:1134–1144. doi: 10.1021/ac034869m. [DOI] [PubMed] [Google Scholar]