Abstract
Hypoxia is a central component of the tumor microenvironment and represents a major source of therapeutic failure in cancer therapy. Recent work has provided a wealth of evidence that noncoding RNAs and, in particular, microRNAs, are significant members of the adaptive response to low oxygen in tumors. All published studies agree that miR-210 specifically is a robust target of hypoxia-inducible factors, and the induction of miR-210 is a consistent characteristic of the hypoxic response in normal and transformed cells. Overexpression of miR-210 is detected in most solid tumors and has been linked to adverse prognosis in patients with soft-tissue sarcoma, breast, head and neck, and pancreatic cancer. A wide variety of miR-210 targets have been identified, pointing to roles in the cell cycle, mitochondrial oxidative metabolism, angiogenesis, DNA damage response, and cell survival. Additional microRNAs seem to be modulated by low oxygen in a more tissue-specific fashion, adding another layer of complexity to the vast array of protein-coding genes regulated by hypoxia.
Keywords: Hypoxia, microRNA, Cancer, Biomarker, miR-210, Mitochondria, Apoptosis, Metabolism
10.1 Introduction
Tissue hypoxia is a dynamic feature of virtually all solid tumors (Semenza 2010a). The adaptive response to low oxygen encompasses complex biochemical and cellular processes, such as energy metabolism, cell survival and proliferation, angiogenesis, adhesion, and motility (Ruan et al. 2009). These, in turn, shape the natural history of cancer and constitute a major source of therapeutic failure in oncology (Brown and Giaccia 1998). During the past two decades, clinical research and animal models have provided strong evidence that tumors with extensive low oxygen tension are more likely exhibit poor prognosis (Vaupel and Mayer 2007). Therefore, a deep understanding of cellular adaptation to oxygen deprivation is key for developing more efficient therapeutic strategies (Wilson and Hay 2011).
Cells react to hypoxia in part via a transcriptional program orchestrated by an oxygen-monitoring machinery that is centered around the hypoxia-inducible factors (HIFs) (Wang and Semenza 1993; Wang et al. 1995; Wang and Semenza 1995). HIFs are heterodimers consisting of an oxygen-sensitive α-subunit (HIFα) and a constitutively expressed β-subunit (HIF1β, also called aryl hydrocarbon receptor nuclear translocator). Among the three homologous HIFα genes, HIF-1α, HIF-2α, and HIF-3α, the functions of HIF-1α and HIF-2α are best characterized. Under normoxic conditions, HIFα is hydroxylated by prolyl-4-hydroxylases, targeting it for proteasomal destruction mediated by the von Hippel-Lindau (VHL) protein-containing E3 ubiquitin ligase (Ivan et al. 2001; Jaakkola et al. 2001). When oxygen becomes limiting, decreased prolyl-4-hydroxylase activity leads to HIFα stabilization and heterodimerization with the β-subunit, followed by translocation to the nucleus and activation of hundreds of target genes (Semenza 2012). The protein-coding components of the HIF-mediated response are discussed in detail elsewhere in this book.
10.2 Noncoding RNA: Wide Roles in Physiology and Pathology
While protein coding sequences account for only approximately 1.1 % of the entire human genome (Venter et al. 2001), during the past decade it has become apparent that the vast majority of the noncoding sequences are, in fact, actively transcribed (Djebali et al. 2012). Given the long history of evolution that has shaped the human genome, it is unlikely that these transcripts are results of “background transcriptional noise.” During the past decade, largely because of rapid advancements in microarray and next-generation sequencing technologies, the “dark matter” of the human genome has been found to encode tens of thousands of noncoding RNAs (ncRNAs). This is a highly heterogeneous superfamily, including diverse entities such as ribosomal RNAs, small nucleolar RNAs, small nuclear RNAs, transfer RNA, small interfering RNAs, piwi-associated RNAs, microRNA (miRNA), unusually small RNA, and long ncRNAs. Despite failing to be translated into proteins, it already has been demonstrated that a small percentage of ncRNAs exhibit important biological functions, and many more are suspected to do so (Hüttenhofer et al. 2005). By far the best-characterized ncRNAs, both in general as well as in the particular context of hypoxia, are miRNAs, which are the main focus of this chapter.
10.2.1 MicroRNAs
miRNAs are single-stranded, small ncRNA molecules (~22 nucleotides in length) that regulate gene expression by inhibiting messenger RNA (mRNA) translation or by facilitating cleavage of the target mRNA (Valencia-Sanchez et al. 2006). Our understanding of miRNA biology was relatively slow to emerge. The first miRNA, lin-4, was discovered in Caenorhabditis elegans in 1993 (Lee et al. 1993; Wightman et al. 1993), followed by the second miRNA, let-7, 7 years later (Reinhart et al. 2000). The finding that let-7 is well conserved in a wide range of animal species (Pasquinelli et al. 2000) spurred an accelerated expansion of miRNA discovery that is still ongoing. To date more than 2,200 miRNAs have been identified in the human genome (miRBase Release 19, http://www.mirbase.org), and at least one-third of all protein-encoding genes are now predicted to be regulated by miRNAs (Lewis et al. 2005). miRNAs are widely recognized as important regulators in developmental, physiological, and pathological settings, including cell growth, differentiation, metabolism, viral infection, and tumorigenesis (Bushati and Cohen 2007). In fact, one would be hard pressed to name a biomedical field that has not been affected in one way or another by miRNA research.
Genes encoding miRNAs are initially transcribed by RNA polymerase II as part of much longer primary transcripts (pri-miRNAs) (Lee et al. 2002) that typically contain the cap structure and the poly(A) tails (Lee et al. 2004). This feature predicts the presence of a wealth of pri-miRNAs alongside mRNA in most whole transcriptome databases. In the second step, pri-miRNAs are processed by the nuclear RNase III Drosha, leading to ~70 nucleotide hairpin-shaped intermediates, called precursor miRNAs (pre-miRNAs). Pre-miRNAs are subsequently exported out of the nucleus and cleaved by the cytoplasmic RNase III Dicer into a short miRNA duplex. One strand of this short-lived duplex is degraded, while the other strand is retained as mature miRNA and incorporated into the RNA-induced silencing complex (RISC), an RNA-protein complex with proteins from the Argonaute family (Schwarz et al. 2003).
The mature miRNA guides the RISC to recognize its target mRNA based on sequence complementarity, most important between the “seed region” of mature miRNAs, nucleotides 2–8, and the 3′ untranslated regions (UTRs) of their target genes, which generally leads to translation inhibition and/or mRNA degradation (Djuranovic et al. 2011, 2012). Because a perfect sequence complementarity is usually only required between the seed region of a miRNA and the 3′ UTR of its target mRNA, a single miRNA can theoretically regulate multiple mRNAs (often hundreds) (Fig. 10.1). Conversely, the 3′ UTR of a given mRNA may contain several miRNA recognition sequences. This relative lack of specificity poses significant challenges for the miRNA research field, in particular in identifying biologically meaningful miRNA targets.
Fig. 10.1.
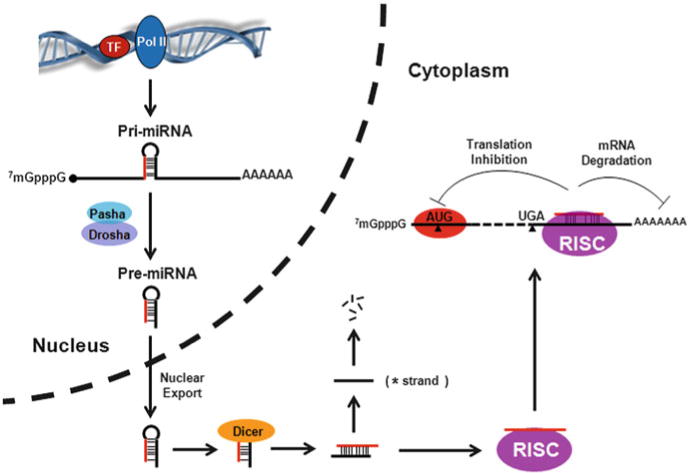
Schematic view of microRNA (miRNA) biogenesis and action. RNA polymerase II (Pol II) transcribes genes that encode miRNAs into primary miRNAs, which usually have a 5′ cap structure and a 3′ poly(A) tail as protein-coding messenger RNAs (mRNAs). The pri-miRNA is first processed in the nucleus by the microprocessor by two partners, Pasha and Drosha, into precursor miRNAs (pre-miRNAs) that are approximately 70 nucleotides in length and have a stem-loop structure. The pre-miRNA is then exported into cytoplasm and further processed by a type III RNA endonuclease Dicer to generate a mature miRNA duplex (~22 nucleotides in length). The sense strand of the miRNA duplex is then loaded into the RNA-induced silencing complex (RISC), whereas the complementary “star” strand (*) of the miRNA duplex is degraded. The RISC regulates gene expression through the inhibition of RNA translation or degradation of target mRNA by base pairing the “seed region” of an miRNA to the 3′ untranslated region of its target mRNA. This figure is modified from Huang et al. (2010)
While highly meaningful for normal cell function, miRNAs have been investigated in depth in most pathological settings, with cancer arguably leading the way. Deregulated miRNA expression has been demonstrated in virtually all neoplasms investigated. It is interesting to note that different cancer types tend to exhibit specific miRNA signatures (Lu et al. 2005; Calin and Croce 2006), including cancers of the colon (Michael et al. 2003), breast (Iorio et al. 2005), brain (Ciafre et al. 2005), liver (Murakami et al. 2006), and lung (Yanaihara et al. 2006). Although the elucidation of the mechanisms behind the specific shifts of profiles in tumors remains a work in progress, recent data on miRNA responses to microenvironmental stresses and oncogenic alterations have provided critical clues.
During the past 6 years multiple reports have demonstrated that miRNAs are involved in the hypoxic response and contribute to the repression of specific genes under low oxygen conditions (Donker et al. 2007; Kulshreshtha et al. 2007a; Camps et al. 2008; Fasanaro et al. 2008; Giannakakis et al. 2008; Huang et al. 2009; Gee et al. 2010). The next section summarizes the current knowledge about the involvement of miRNAs in cellular hypoxic response, discusses challenges for elucidation of their biological functions, and speculates on potential opportunities for cancer diagnosis, prognosis, and treatment.
10.3 Hypoxia-Regulated miRNAs: The New Paradigm of Cellular Response to a Hypoxic Microenvironment
Several groups from diverse fields embarked on genome-wide miRNA profiling, with the goal to identify hypoxia-regulated miRNAs in a variety of cellular contexts. Expression of several dozen mature miRNAs, including miR-210, -21, -23, -24, -26, -103/107, and -181, was found to be induced under hypoxic conditions (Kulshreshtha et al. 2007b; Camps et al. 2008; Fasanaro et al. 2008; Crosby et al. 2009; Huang et al. 2009; Sarkar et al. 2010; Chen et al. 2012). Although these miRNA were reported in at least two publications, relatively limited overall overlap was noticed among these studies. With the caveat that different technologies were employed by different groups, this variability suggests a cell-specific component of miRNA induction and maturation. This in turn may contribute to the well-recognized variations in the magnitude of coding gene responses in hypoxia. Moreover, a large number of mature miRNAs were found to be downregulated in hypoxia (Kulshreshtha et al. 2007a), and their role may be to de-repress expression of specific genes in low oxygen conditions.
In sharp contrast, hypoxic-induced miR-210 stands out as the only miRNA agreed on by all the studies to date (Huang et al. 2010). This is drastically different from the case of classic protein-coding genes in which a plethora of mRNAs with diverse functions are induced by hypoxia, with relatively good overlap between different cell types (Denko et al. 2003). According to most groups that investigated the hypoxic regulation of miR-210, it seems to be a rather specific HIF-1 target (Camps et al. 2008; Crosby et al. 2009; Huang et al. 2009; Kim et al. 2009). However, this specificity is not absolute because HIF-2-dependent regulation of miR-210 also has been reported (Zhang et al. 2009). As is the case for the classic genes, HIF-1 directly binds to a hypoxia-responsive element (HRE) on the proximal miR-210 promoter (Huang et al. 2009). When the miR-210 core promoter is compared across species, this HRE site is highly conserved, indicating the importance of hypoxia/HIFs in regulating miR-210 expression during evolution. Consistent with this observation, the induction of mouse miR-210 under hypoxia is dependent on HIF-1α (Crosby et al. 2009). This highly conserved HRE was recently confirmed to be the functional HRE that is responsible for the robust induction of mouse miR-210 expression under hypoxic conditions (Cicchillitti et al. 2012).
10.3.1 miR-210: A Mirror of HIF Activity with Clinical Implications
miR-210 is upregulated in most solid tumors investigated to date, and its levels generally correlate with a negative clinical outcome (Kulshreshtha et al. 2007b; Porkka et al. 2007; Zhang et al. 2007; Fasanaro et al. 2008; Foekens et al. 2008; Lawrie et al. 2008; Gee et al. 2010; Rothe et al. 2011; Hong et al. 2012). Moreover, miR-210 levels are correlated with a gene expression signature of hypoxia (Camps et al. 2008; Huang et al. 2009), suggesting that its overexpression in tumors is the direct consequence of decreased oxygen tension in the microenvironment. In addition to tissue expression, in our laboratory, we observed a significant increase of circulating miR-210 in patients with pancreatic cancer compared to healthy controls (Ho et al. 2010). This is consistent with our knowledge of miR-210’s responsiveness to hypoxia, since pancreatic adenocarcinomas are usually highly hypoxic (Koong et al. 2000).
Whether miR-210 independently increases tumor aggressiveness and/or decreases the response to therapy is still a matter of debate, although there is preliminary evidence to suggest such an effect (Camps et al. 2008). An emerging paradigm is that miR-210 expression is an accurate readout of HIF-1 activity in vivo, as it is in vitro (Fasanaro et al. 2008; Huang et al. 2009; Devlin et al. 2011).
One particular disease that is connected closely with the HIF pathway is clear-cell renal cell carcinomas (ccRCCs), which is commonly associated with the inactivation of the VHL tumor suppressor gene (Presti et al. 1991; Brugarolas 2007). Mutations and loss of heterozygosity of the VHL gene have been found in 57 % and 98 % of sporadic renal cell carcinoma cases, respectively (Gnarra et al. 1994). The VHL tumor suppressor gene product functions as the adaptor subunit of the E3 ubiquitin ligase complex that targets hydroxylated HIF-1α and HIF-2α for ubiquitination and subsequent degradation by the 26S proteasome (Ivan et al. 2001; Jaakkola et al. 2001). Given its close relationship with HIF, it is not surprising that miR-210 is particularly overexpressed in ccRCCs (Juan et al. 2010; White et al. 2011; Redova et al. 2012). In addition, elevated levels of circulating miR-210 have been found in patients with ccRCC compared to healthy controls (Zhao et al. 2013). Although the origin of circulating miRNAs remains a much-debated subject, the existence of high-level miR-210 in circulation in these patients suggests that miR-210 may serve as a novel biomarker for noninvasive detection of highly hypoxic cancers.
While our own work has focused on the emerging roles of miR-210 in tumors, the impact of this miRNA most likely extends well beyond cancer biology, most notably in cardiac cerebrovascular diseases (Semenza 2010b), cardiac hypertrophy and failure (van Rooij et al. 2006; Thum et al. 2007; Greco et al. 2012), transient focal brain ischemia (Jeyaseelan et al. 2008), limb ischemia (Jeyaseelan et al. 2008; Pulkkinen et al. 2008), ischemic wounds (Biswas et al. 2010), acute myocardial infarction (Bostjancic et al. 2009), atherosclerosis obliterans (Li et al. 2011), and preeclampsia (Pineles et al. 2007; Zhu et al. 2009; Enquobahrie et al. 2011).
10.3.2 miR-210 Targets: A Growing and Diverse List
Identification of biologically relevant targets is an essential step toward understanding the functions of miR-210. A frequently employed approach begins with computational prediction using a growing number of programs available online. These are based on algorithms that search for complementarity between 3′ UTR sequences of annotated coding genes and the seed region sequence of the miRNA (Bartel 2009). Among the most popular programs employed to this end are miRanda (Betel et al. 2008), TargetScan (Lewis et al. 2003), Pictar (Krek et al. 2005), PITA (Kertesz et al. 2007), MicroCosm (Griffiths-Jones et al. 2008), and Dianalab (Maragkakis et al. 2009). Drawing the line after several years of experience with these resources, none stands out as the most accurate predictor of real targets. To further complicate matters, the lists of candidates generated by these programs usually exhibit limited overlap. This seems to be generally true for most miRNAs investigated and is exemplified by a proteomic study comparing the accuracy of different computational predictions of miR-223 targets (Baek et al. 2008). Since the miRNA seed region only consists of 6 or 7? nucleotides, false-positive prediction is a major limitation of this approach. Any given miRNA, including miR-210, may be predicted to regulate hundreds, if not thousands, of coding genes. When the search is extended to the 5′ UTR and the coding region, the number of targets is expected to be even higher. Finally, Fasanaro et al. (2012) have provided recent experimental evidence for a “seedless” target of miR-210 on the basis of complementarity between sequences of the ROD1 (regulator of differentiation 1) gene and 10 consecutive bases in the central portion of miR-210. It is interesting that two widely employed algorithms, PicTar and TargetScan, predict relatively few targets for human miR-210, and, conversely, most of the experimentally validated targets are not predicted by any of these programs with a high score. It is also becoming increasingly apparent that “seed” binding is not always sufficient, as other features of the surrounding sequences can affect binding efficacy (Lewis et al. 2005). In conclusion, while computational predictions still represent powerful tools in the search for targets, they suffer from clear limitations, and exclusive reliance on them can lead to long lists of limited use.
In the particular case of miR-210, a number of genes appearing on these lists have been confirmed, but the success was largely due to the addition of a strong experimental arm. Such confirmed target genes are involved in cell proliferation, mitochondrial metabolism, DNA repair, chromatin remodeling, and cell migration (Camps et al. 2008; Fasanaro et al. 2008; Giannakakis et al. 2008; Pulkkinen et al. 2008; Chan et al. 2009; Crosby et al. 2009; Mizuno et al. 2009; Chen et al. 2010; Qin et al. 2010).
A widely used approach for identification of miRNA targets is a combination of expression profiling following miRNA manipulation using mimics and antagomirs, followed by expression profiling and comparisons with the results of computational predictions. This is a feasible strategy because directing mRNA degradation is a major mechanism that miRNAs use to downregulate the corresponding target genes (Lim et al. 2005; Guo et al. 2010). This effect is usually robust enough to be detected by microarray analysis or whole transcriptome RNA sequencing (RNA-Seq). For example, both Zhang et al. (2009) and Puissegur et al. (2011) began their search by identifying transcripts that were down-modulated upon forced miR-210 expression in colorectal and lung adenocarcinoma cancer cell lines, respectively. The downregulated genes, which also contained a predicted miR-210 binding site, were analyzed further to confirm that MNT (Zhang et al. 2009), NDUFA4, and SDHD (Puissegur et al. 2011) are bona fide miR-210 targets.
However, because miRNA frequently regulates its target gene by inhibiting protein translation rather than altering mRNA abundance (Selbach et al. 2008), complete reliance on mRNA profiling is likely to miss authentic miRNA targets. An ideal sensitive and robust proteomic approach should identify miRNA targets directly when combined with computational prediction. However, lack of sufficient sensitivity, heavy bias for abundant proteins, and high costs still preclude the broader application of this promising technology.
A recent approach has emerged that, at least in part, allows us to overcome the above-mentioned limitations. miRNAs is known to recruit the mRNAs of their target genes to the RISC complex, of which Argonaute family proteins, especially Argonaute 2, are essential components (Kawamata and Tomari 2010). Argonaute protein immunoprecipitation (miRNP-IP) methods have been developed that capture the mRNAs recruited to the complex. This pull-down can be followed by microarray or RNA-Seq and help identify targets that are regulated by both translational blockade or message degradation (Karginov et al. 2007). miRNP-IP can “freeze miRNAs in action” and thus greatly reduce the number of nonspecific targets and the secondary effect that are commonly seen in other approaches. Our groups have successfully pursued this approach in cells overexpressing miR-210 as part of more integrative strategies to identify targets (Fasanaro et al. 2009; Huang et al. 2009). In one of the studies we used a combination of computational prediction, proteomic, transcriptomic, and miRNP-IP approaches in human umbilical vein endothelial cells overexpressing miR-210 (Fasanaro et al. 2009). Proteomic profiling identified 11 downregulated proteins, whereas transcriptome profiling identified 51 transcripts that are induced upon miR-210 knockdown and downregulated by miR-210 overexpression. Despite the fact that 42 of the 62 genes were enriched with miR-210 seed-complementary sequences in their 3′ UTRs, surprisingly few were predicted by the miRNA target identification algorithms listed above. To distinguish between direct and indirect targets, analysis of the miRNP-IP content was analyzed by quantitative reverse transcriptase polymerase chain reaction, revealing that 16 potential targets were enriched in the RISC. An ncRNA (XIST) involved in X chromosome inactivation was identified in the miRNP-IP experiment, indicating a previously unsuspected layer of interaction between ncRNAs. This finding lends support to the recently proposed competing endogenous RNA theory (Salmena et al. 2011).
Using a similar miRNP-IP strategy, we also compared mRNAs enriched in the RISC after exposing immortalized human breast MCF10A cells to low oxygen for 24 hours? versus normoxic controls (Huang et al. 2009). More than 200 mRNAs were enriched, and several algorithms predicted that 50 of these were direct miR-210 targets. Three of five randomly selected genes from the list of 50 genes were confirmed as bona fide miR-210 targets by functional assays.
It is notable that there are few targets in common between these two studies, leading to the hypothesis that miR-210 regulates different sets of target genes in these two cell types. This is not necessarily surprising: comprehensive proteomic studies indicated that miRNAs act as rheostats by performing fine-scale adjustments to the output of hundreds of proteins (Baek et al. 2008; Selbach et al. 2008). Thus, only minor changes at the protein and/or mRNA levels are expected for the majority of miRNA targets, which translate into low fold changes (e.g., 1.2–1.5) in target expression when measured by microarray or RNA-Seq. This inherent variability of our experimental systems to measure such small changes may account at least in part for the discrepancy between our studies.
The current paradigm of miRNA function states that they suppress target gene expression via inhibiting protein translation, degrading mRNA, or both (Bartel 2004). Several reports have provided the first evidence for exceptions to this rule, identifying genes that are upregulated by miRNA transduction (Vasudevan et al. 2007; Place et al. 2008). Although no direct evidence for this has been provided for miR-210, upregulated gene expression was observed in our study (Fasanaro et al. 2009). At this point there is no evidence that this represents more than secondary waves of regulation.
We anticipate that improved understanding of miRNA function will be followed by the increased accuracy of predicting miRNA targets. Moreover, with the lowering cost of next-generation sequencing, the combination of computational tools and advanced experimental approaches will provide a more complete identification of physiologically relevant miR-210 targets. Figure 10.2 summarizes experimentally validated miR-210 targets and their potential regulatory functions. Some of the better-validated miR-210 targets are reviewed below, and their function in hypoxic response and cancer biology are discussed.
Fig. 10.2.
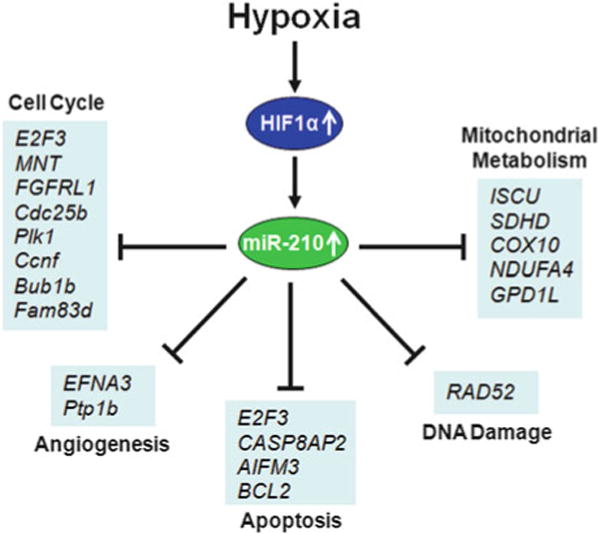
Experimentally identified micro RNA target genes implicated in cancer biology. Genes that have been experimentally identified as miR-210 targets can be classified into five major functional categories: mitochondrial metabolism, cell cycle control, angiogenesis, apoptosis, and DNA damage repair, indicating a complex functional network regulated by miR-210 in the hypoxic microenvironment in which tumor cells reside
10.3.3 miR-210 Regulates Mitochondrial Metabolism and Oxidative Stress
Under normoxic conditions, mitochondria are the “energy factories” of a cell; they generate the majority of adenosine triphosphate through the oxidative phosphorylation pathway using oxygen as an electron acceptor. However, when the oxygen supply is limited, cells switch to glycolysis to produce adenosine triphosphate (the Pasteur effect). During this process, HIF-1 not only upregulates expression of most glycolytic enzymes but also downregulates mitochondrial respiration and biogenesis (Zhang et al. 2007; Denko 2008). Results from several groups have demonstrated that hypoxic induction of miR-210 significantly contributes to this metabolic shift by downregulating the activity of mitochondrial electron transport chains (ETCs). A well-accepted branch of this mechanism is targeting by miR-210 of the iron-sulfur cluster scaffold proteins (ISCU) (Chan et al. 2009; Fasanaro et al. 2009; Chen et al. 2010; Favaro et al. 2010). One of the most consistent miR-210 targets, ISCU is part of an ancient mechanism that catalyzes the assembly of iron-sulfur clusters that are critical for enzymes, such as aconitase, that function in the tricarboxylic acid cycle, as well as for the function of mitochondrial ETC complexes I, II, and III (Tong and Rouault 2006). The role of ISCU as a metabolic regulator has strong backing from clinical data: its mutations are associated with hereditary lactic acidosis characterized by myopathy and exercise intolerance (Mochel et al. 2008).
Further strengthening the case for ISCU as a biologically relevant target, its levels are inversely correlated with miR-210 in multiple tumor data sets (Favaro et al. 2010). In contrast to a poor prognosis predicted by higher levels of miR-210, the expression of ISCU is predictive of a favorable prognosis, at least in breast cancer, indicating that ISCU is regulated by miR-210 in vivo.
While ISCU itself is not a primarily mitochondrial protein, several integral components of the mitochondrial ETC have been found to be miR-210 targets: NADH dehydrogenase (ubiquinone) 1α subcomplex 4 (NDUFA4) (Giannakakis et al. 2008), succinate dehydrogenase complex, subunit D (SDHD) (Puissegur et al. 2011), and cytochrome C oxidase assembly homolog 10 (COX10) (Chen et al. 2010). NDUFA4 was initially considered to be part of ETC complex I until recent work demonstrated that it actually resides in complex IV (Balsa et al. 2012). SDHD is a subunit of complex II and a well-documented tumor suppressor gene product (Baysal et al. 2000; Gottlieb and Tomlinson 2005). Targeting of this gene by miR-210 provides additional support to a protumorigenic role for miR-210. COX10, which encodes a heme A:farnesyltransferase, is another recently discovered target of miR-210. Although COX10 is not a structural subunit of COX (mitochondrial ETC complex IV), it is required for the expression of a functional COX complex (Antonicka et al. 2003). To summarize, by targeting multiple ETC components and regulators, miR-210 may act as a key HIF-1 effector in attenuating oxygen consumption in hypoxia. One unanswered question is whether miR-210 plays biological roles in anoxia, where decreasing oxygen consumption is not a factor.
Another intriguing target predicted by several programs and experimentally confirmed is glycerol-3-phosphate dehydrogenase 1-like (GPD1L) (Fasanaro et al. 2009). GPD1L is highly homologous to glycerol-3-phosphate dehydrogenases that transfer electrons from cytoplasmic NADH to the mitochondrial ETC (Bunoust et al. 2005). Thus, GPD1L may also be involved in the Pasteur effect by regulating NAD+-to-NADH ratios (Liu et al. 2010). Recent insights from Kelly et al. (2011) identified a feedback mechanism based on GPD1L repression, which in turn inactivates HIF prolyl hydroxylase activity, leading to stabilization of HIF-1α. Therefore, miR-210 may be present in or at both downstream and upstream of HIF-1α signaling. Puissegur et al. (2011) also showed that high levels of miR-210 participate to stabilize HIF-1α during hypoxia.
The generation of mitochondrial reactive oxygen species (ROS) is a consequence of electron leakage during electron transport (Murphy 2009). Increased ROS production has been reported in hypoxia, potentially as a result of ETC dysfunction (Guzy and Schumacker 2006); however, whether cell reoxygenation during the ROS assay contributes to this increase is still being debated. We reported that miR-210 increases oxidative stress at least in part by ISCU suppression in normoxic MCF7 cells (Chan et al. 2009; Favaro et al. 2010). However, in hypoxia, and in other cell types, the effects of miR-210 are still a subject of controversy. We observed hypoxic induction of ROS in cancer cell lines, and an miR-210 antagonist reversed this effect to almost normoxic levels (Favaro et al. 2010). Conversely, Chan et al. (2009), working on endothelial cells, did not detect any change in ROS production after exposing the cells to hypoxia and noted a burst of ROS when miR-210 was blocked. This discrepancy is sure to promote additional investigation and may reflect underlying differences between normal versus cancer cells. In addition, miR-210 may exhibit differential effects on various ROS species, a hypothesis that needs to be addressed in future studies.
Aside from its robust response to oxygen deprivation, the HIF pathway plays an increasingly clear role in the Warburg effect (Kim and Dang 2006). HIF is also induced in tumors as part of oncogenic signaling networks, even in the absence of hypoxia (Zundel et al. 2000), so it is tempting to speculate that elevated miR-210 may contribute to the Warburg effect in these tumors by helping to stabilize HIF-1α to promote aerobic glycolysis. Expression of miR-210 is frequently elevated in cancers, including glioblastomas (Malzkorn et al. 2009), melanomas (Satzger et al. 2009; Zhang et al. 2009), ccRCCs (Juan et al. 2010; White et al. 2011), lung (Miko et al. 2009; Raponi et al. 2009), pancreatic cancers (Greither et al. 2009), and breast cancers (Camps et al. 2008; Foekens et al. 2008). However, whether elevated miR-210 expression within the tumor can occur outside of the hypoxic areas remains unclear at this stage, and future laser-capture, microdissection-based analyses may shed critical light on this dilemma.
10.3.4 miR-210 as a Regulator of Angiogenesis
Angiogenesis is a complex, multistep process that normally occurs during embryonic development and rarely in adults. Exceptions include normal repair processes such as wound healing and pathological settings such as in tumor growth and ischemic disorders (Semenza 2003). Tumor growth is highly dependent on the formation of neovessels to establish nutrient and oxygen supplies for cell viability and proliferation. Imbalance between oxygen consumption by tumor cells with high metabolic activities (Gatenby and Gillies 2004) and oxygen delivery by dysfunctional vasculature (Brown and Giaccia 1998) leads to hypoxia and stimulates compensatory angiogenesis (Liu et al. 1995).
Multiple miRNAs seem to be part of the various steps of the angiogenic response, either as positive or negative regulators (Wang and Olson 2009; Wu et al. 2009). On the basis of its involvement in the hypoxic response, it is hardly surprising that miR-210 has been investigated as a candidate angiogenic regulator. Consistent with this hypothesis, miR-210 expression was found to correlate closely with vascular endothelial growth factor (VEGF) expression, hypoxia, and angiogenesis in patients with breast cancer (Foekens et al. 2008). Transduction of miR-210 in human umbilical vein endothelial cells using miRNA mimics functionally stimulates the formation of capillary-like structures as well as VEGF-induced cell migration (Fasanaro et al. 2008; Lou et al. 2012). Conversely, inhibiting miR-210 using the corresponding antagomir blocks both tubulogenesis and VEGF-mediated endothelial chemotaxis. The receptor tyrosine kinase ligand Ephrin-A3 (EFNA3) was identified as a candidate mediator for these effects (Fasanaro et al. 2008), consistent with the knowledge that ephrin ligands and their receptors are important in the development of the cardiovascular system and in vascular remodeling (Kuijper et al. 2007). Overall, EFNA3 seems to be one of the most consistently reported miR-210 target genes (Fasanaro et al. 2008; Fasanaro et al. 2009; Greither et al. 2009; Huang et al. 2009). To complicate matters, regulation of EFNA3 is more complex when ischemic responses are examined (Pulkkinen et al. 2008). Contrary to the expectation of miR210-mediated repression, EFNA3 was present at higher levels in mouse hippocampus after ischemia. It is interesting that EFNA3 transcription is also induced by hypoxia (Fasanaro et al. 2008), suggesting that miR-210 fine-tunes primarily EFNA3 protein translation. Thus, abundance of the EFNA3 protein may reflect the balance between hypoxic induction of mRNA and repression of miR-210, which conceivably varies in different pathological contexts.
The tyrosine phosphatase PTP1B was identified as another miR-210 target that promotes angiogenesis and inhibits apoptosis after myocardial infarction in a mouse model (Hu et al. 2010). PTP1B is documented to negatively regulate activation of the VEGF receptor 2 and stabilize cell-cell adhesions through reducing tyrosine phosphorylation of vascular endothelial cadherin (Nakamura et al. 2008). Thus, by inhibiting EFNA3 and PTP1B, both negative regulators of angiogenesis, miR-210 could promote angiogenesis.
An interesting recent study also suggests positive feedback between VEGF and miR-210. CD34+ cells in umbilical cord blood expanded in VEGF-containing medium upregulated miR-210 expression, and when these cells were transplanted into the ischemic hind limbs of mice tissue perfusion/capillary density were significantly improved, whereas an miR-210 inhibitor abolished the effect (Alaiti et al. 2012).
While the above-mentioned studies were not conducted in the context of cancer, they may nevertheless shed new light on possible roles of miR-210 in tumor angiogenesis. It will be of great interest to investigate whether feedback involving hypoxia, miR-210, and VEGF also occurs in tumors in vivo. This may have implications for developing strategies to increase the efficacy of anti-VEGF therapy, for example, by adding miR-210 inhibitors.
10.3.5 miR-210 Regulation of DNA Damage Response
Genome integrity is challenged by a variety of stresses, including radiation, mutagens, ROS, ultraviolet light, and chemo-or radiotherapeutic agents. Cellular responses to DNA damage involve a complex network of processes that detect and repair genomic lesions. miRNAs have been demonstrated to participate in these processes (Simone et al. 2009; Landau and Slack 2011; Wan et al. 2011). Zhang et al. (2011) provided direct evidence that more than 20 % of examined miRNAs are significantly induced upon DNA damage. While it is not robustly induced by irradiation, miR-210 nevertheless seems to be relevant for this complex process because it targets RAD52 (Crosby et al. 2009; Fasanaro et al. 2009), a key component in the homologous recombination–mediated repair of double-strand breaks (Benson et al. 1998; Shinohara and Ogawa 1998). Suppression of RAD52 by miR-210 may provide an additional mechanism to help explain compromised homologous recombination repair activity in hypoxic cells (Bindra et al. 2007). Consistent with this hypothesis, forced expression of miR-210 was found to lead to double-strand DNA breaks in cultured cells (Faraonio et al. 2012).
10.3.6 miR-210 Regulation of Apoptosis
Cellular stresses, including hypoxia, are well known triggers of programmed cells death, a process also called apoptosis. Evasion from apoptotic responses is critical for tumor progression: transformed cells need to overcome the adverse conditions present in their microenvironment (Hanahan and Weinberg 2011). Thus, it is not surprising that miR-210 has been investigated for possible effects on apoptotic responses. In general, the available evidence suggests an anti-apoptotic role of miR-210 in a variety of cell types. On the one hand, overexpression of miR-210 can protect cells from apoptosis (Kulshreshtha et al. 2007a; Kim et al. 2009; Hu et al. 2010; Mutharasan et al. 2011; Nie et al. 2011); on the other hand, downregulation of miR-210 during hypoxia promotes apoptosis (Cheng et al. 2005; Kulshreshtha et al. 2007b; Fasanaro et al. 2008; Gou et al. 2012; Liu et al. 2012; Yang et al. 2012). While many gaps remain in our understanding of this process, several relevant targets have been identified to help explain such effect: E2F3 (Gou et al. 2012), Ptp1b (Hu et al. 2010), caspase-8-associated protein-2 (CASP8AP2) (Kim et al. 2009), and apoptosis-inducing factor, mitochondrion-associated 3 (AIFM3) (Yang et al. 2012). However, the caveat is that, apart from CASP8AP2 (Kim et al. 2012), none of the other genes has been verified to mediate miR-210’s anti-apoptotic function by an independent study. In a recent report, although AIFM3 was found to be an miR-210 target, its overexpression failed to overcome the cytoprotective effects of the miRNA, suggesting that cooperation with other targets may be necessary (Mutharasan et al. 2011). Despite the evidence of an anti-apoptotic role cited above, a recent report suggested that miR-210 may also exhibit a pro-apoptotic function, at least in hypoxic neuroblastoma cells, by targeting the anti-apoptotic gene BCL2 (Chio et al. 2013).
In summary, evidence for miR-210-mediated regulation of apoptosis in hypoxia is emerging for various cell types. However, it is still premature to state that this miRNA represents a major protector against hypoxia-induced cell death.
10.3.7 miR-210 Effects on the Cell Cycle
In many cell types, extended exposure to hypoxia leads to downregulation of a large number of cell cycle genes, including cyclins and other positive regulators of cell cycle transition (Hammer et al. 2007), while other cells tend to proliferate better under low oxygen conditions (Krick et al. 2005). One of the better-characterized miR-210 targets that belong to cell cycle control is E2F3, a promoter of G 1/S transition (Lees et al. 1993; Leone et al. 1998). E2F3 was first reported as an miR-210 target in ovarian cancer (Giannakakis et al. 2008). Later, several independent studies confirmed that E2F3 was an miR-210 target in various cell types, including ccRCC (Nakada et al. 2011), keratinocytes and in a murine model of ischemic wounds (Biswas et al. 2010), and HEK293 cells (Fasanaro et al. 2009). Despite these findings, the relative contribution of E2F3 to tumor cell cycle alterations in a hypoxic microenvironment remains largely unknown. In addition to E2F3, fibroblast growth factor receptor-like 1 (FGFRL1) also was identified as an miR-210 target involved in cell cycle control (Tsuchiya et al. 2011), consistent with our earlier observation that FGFRL1 is robustly repressed by miR-210 (Huang et al. 2009). De-repression of FGFRL1 by using anti-miR-210 accelerates cell cycle progression, whereas overexpression of miR-210 leads to cell cycle arrest in the G1/G0 and G2/M phases (Tsuchiya et al. 2011). The effects of miR-210 on the cells cycle may, in fact, be significantly broader, including a group of mitosis-related genes, such as Plk1, Cdc25B, cyclin F (CCNF), Bub1B, and Fam83D (He et al. 2013). Whether all these represent direct targets or more indirect responders downstream of the genes discussed above remains unclear.
As is the case with hypoxic cell death, under some circumstances miR-210 may promote cell cycle progression, for example, by downregulating MNT (Zhang et al. 2009), a member of the Myc/MAX/MAD network with a basic-Helix-Loop-Helix-zipper domain. MNT functions as an antagonist of c-Myc and represses Myc target genes by binding the E box DNA sequence (CANNTG) after forming heterodimers with MAX (Hurlin et al. 1997; Meroni et al. 1997). As HIF-1 regulates cell proliferation and metabolism in part by interacting with c-Myc (Gordan et al. 2007), miR-210 may fine-tune the latter under hypoxic conditions to regulate cell cycle progression.
10.3.8 miR-210 as a Candidate Cancer Biomarker
Past studies have demonstrated that miRNAs are frequently dysregulated in human cancers (Ventura and Jacks 2009). Tumor-specific miRNA expression signatures can distinguish between normal and malignant tissues as well as classify cancer subtypes (Garzon et al. 2009). When used to classify poorly differentiated tumors, miRNA expression profiling outperformed mRNA expression profiling (Lu et al. 2005), pointing toward considerable potential as a biomarkers. Expression of miR-210 has been associated with poor clinical outcome in soft-tissue sarcoma, breast, head and neck, and pancreatic tumors (Camps et al. 2008; Foekens et al. 2008; Greither et al. 2009; Gee et al. 2010; Greither et al. 2011; Rothe et al. 2011; Hong et al. 2012; Toyama et al. 2012). However, whether high miR-210 only serves as an indicator of tumor hypoxia or actively promotes a more aggressive disease remains unclear (Huang et al. 2010).
The status of miR-210 expression in tumors may have therapeutic implications. Preliminary in vitro evidence was provided by Chen et al. (2010), who reported that overexpressing miR-210 rendered cells significantly more susceptible to killing by 3-bromo-pyruvate, an inhibitor of the glycolytic pathway. Molecules of this class, such as 2-deoxyglucose and dichloroacetate, have been considered promising therapeutic agents; however, they have yet to fulfill their promise in clinical settings. Therefore, miR-210 may help identify subsets of patients who can benefit from such agents in the future.
Recent publications report that miRNAs are exceptionally stable and can be readily detected in the systemic circulation and other body fluids of healthy subjects and patients with malignant diseases (Chen et al. 2008; Gilad et al. 2008; Lawrie et al. 2008; Mitchell et al. 2008; Taylor and Gercel-Taylor 2008; Weber et al. 2010). It has been suggested that the high stability of miRNAs may be partially attributed to the exosomal miRNA packaging (Valadi et al. 2007). The release of exosomes into the extracellular environment provides an opportunity to use exosome components in body fluids as a proxy to monitor molecular events occurring in tumor cells (Iguchi et al. 2010). Pilot studies assessing the use of circulating miRNAs as cancer biomarkers have attracted broad interest in the field and to date at least 79 miRNAs have been reported as plasma or serum biomarker candidates for solid and hematologic tumors (Allegra et al. 2012). miR-210 was found to be increased in the serum of patients with diffuse large B-cell lymphoma (Lawrie et al. 2008), ccRCC (Zhao et al. 2013), and pancreatic cancer (Wang et al. 2009; Ho et al. 2010). It is interesting that hypoxia has been demonstrated to promote the release of exosomes from cultured breast cancer cells (King et al. 2012); therefore, one can speculate that the elevated levels of circulating miR-210 may directly reflect the hypoxic state of tumor cells.
Circulating miR-210 levels were also correlated with sensitivity to trastuzumab (a human epidermal growth factor receptor 2 monoclonal antibody), tumor presence, and lymph node metastases in patients with breast cancer (Jung et al. 2012). This provides proof of the concept that plasma miR-210 may also be used to monitor response to anticancer therapies (Cortez et al. 2011).
10.3.9 miR-210: A Viable Cancer Therapeutic Target?
Recent development of anti-miRNA agents such as locked nucleic acids or peptide nucleic acids represent significant steps for the therapeutic targeting of miRNAs in vivo (Stenvang et al. 2008; Lanford et al. 2010; Fabbri et al. 2011a, b; Gambari et al. 2011; Iorio and Croce 2012). It is conceivable that inactivation of miRNAs involved in hypoxic adaptation, in combination with other anticancer agents, may be a viable strategy to target a tumor compartment that poses significant therapeutic challenges. At present, efficient delivery of miRNA-related reagents to solid tumors, and in particular to poorly perfused areas, remains a significant hurdle. However, rapid advances in nanoparticle-based nucleic acid delivery (Tabernero et al. 2013) are providing realistic expectations that such limitations eventually will be overcome.
10.4 Concluding Remarks
On the basis of the experimental evidence summarized in this chapter, miR-210 plays complex roles in the cellular responses to hypoxia and in cancer biology. Given the diversity of genes that seem to respond as bona fide miR-210 targets – some with opposing effects on specific cellular functions – a simple and universal model of miR-210 function will be challenging to develop using exclusively in vitro approaches. Answering some of the key questions regarding miR-210 functions will require data from more sophisticated systems such as genetic inactivation of the corresponding locus in animal models.
Acknowledgments
This work is supported in part by funding from the National Institutes of Health (NIH 1R01 CA155332-01 to M.I.) and the American Cancer Society (M.I., X.H.). X.H. is a Liz Tilberis Scholar of the Ovarian Cancer Research Fund.
Contributor Information
Mircea Ivan, Email: mivan@iupui.edu, Department of Medicine, Indiana University, 980 W. Walnut Street Walther Hall, Room C225, Indianapolis, IN 46202, USA; Department of Microbiology and Immunology, Indiana University, 980 W. Walnut Street Walther Hall, Room C225, Indianapolis, IN 46202, USA.
Xin Huang, Email: huangx2@upmc.edu, Magee-Womens Research Institute, Department of Obstetrics, Gynecology & Reproductive Sciences, University of Pittsburgh School of Medicine, B407, 204 Craft Ave, Pittsburgh, PA 15213, USA; Women’s Cancer Research Center, University of Pittsburgh Cancer Institute, B407, 204 Craft Ave, Pittsburgh, PA 15213, USA.
References
- Alaiti MA, Ishikawa M, et al. Up-regulation of miR-210 by vascular endothelial growth factor in ex vivo expanded CD34+ cells enhances cell-mediated angiogenesis. J Cell Mol Med. 2012;16(10):2413–2421. doi: 10.1111/j.1582-4934.2012.01557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegra A, Alonci A, et al. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review) Int J Oncol. 2012;41(6):1897–1912. doi: 10.3892/ijo.2012.1647. [DOI] [PubMed] [Google Scholar]
- Antonicka H, Leary SC, et al. Mutations in COX10 result in a defect in mitochondrial heme a biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum Mol Genet. 2003;12(20):2693–2702. doi: 10.1093/hmg/ddg284. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsa E, Marco R, et al. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16(3):378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal BE, Ferrell RE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287(5454):848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- Benson FE, Baumann P, et al. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391(6665):401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, et al. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(suppl 1):D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindra R, Crosby M, et al. Regulation of DNA repair in hypoxic cancer cells. Cancer Metastasis Rev. 2007;26(2):249–260. doi: 10.1007/s10555-007-9061-3. [DOI] [PubMed] [Google Scholar]
- Biswas S, Roy S, et al. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci. 2010;107(15):6976–6981. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostjancic E, Zidar N, et al. MicroRNA microarray expression profiling in human myocardial infarction. Dis Markers. 2009;27(6):255–268. doi: 10.3233/DMA-2009-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Giaccia AJ. The unique physiology of solid tumors: opportunities (and problems) for cancer therapy. Cancer Res. 1998;58(7):1408–1416. [PubMed] [Google Scholar]
- Brugarolas J. Renal-cell carcinoma – molecular pathways and therapies. N Engl J Med. 2007;356(2):185–187. doi: 10.1056/NEJMe068263. [DOI] [PubMed] [Google Scholar]
- Bunoust O, Devin A, et al. Competition of electrons to enter the respiratory chain: a new regulatory mechanism of oxidative metabolism in Saccharomyces cerevisiae. J Biol Chem. 2005;280(5):3407–3413. doi: 10.1074/jbc.M407746200. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. MicroRNA functions. Annu Rev Cell Dev Biol. 2007;23(1):175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Camps C, Buffa FM, et al. hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14(5):1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- Chan SY, Zhang YY, et al. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10(4):273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ba Y, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Chen Z, Li Y, et al. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29(30):4362–4368. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- Chen H-Y, Lin Y-M, et al. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012;72(14):3631–3641. doi: 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- Cheng AM, Byrom MW, et al. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33(4):1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio C-C, Lin J-W, et al. MicroRNA-210 targets antiapoptotic Bcl-2 expression and mediates hypoxia-induced apoptosis of neuroblastoma cells. Arch Toxicol. 2013;87(3):459–468. doi: 10.1007/s00204-012-0965-5. [DOI] [PubMed] [Google Scholar]
- Ciafre SA, Galardi S, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Cicchillitti L, Di Stefano V, et al. Hypoxia-inducible factor 1-a induces miR-210 in normoxic differentiating myoblasts. J Biol Chem. 2012;287(53):44761–44771. doi: 10.1074/jbc.M112.421255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez MA, Bueso-Ramos C, et al. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby ME, Kulshreshtha R, et al. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69(3):1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8(9):705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- Denko NC, Fontana LA, et al. Investigating hypoxic tumor physiology through gene expression patterns. Oncogene. 2003;22(37):5907–5914. doi: 10.1038/sj.onc.1206703. [DOI] [PubMed] [Google Scholar]
- Devlin C, Greco S, et al. miR-210: more than a silent player in hypoxia. IUBMB Life. 2011;63(2):94–100. doi: 10.1002/iub.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, et al. A parsimonious model for gene regulation by miRNAs. Science. 2011;331(6017):550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, et al. MiRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336(6078):237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donker RB, Mouillet JF, et al. The expression of Argonaute2 and related microRNA biogenesis proteins in normal and hypoxic trophoblasts. Mol Hum Reprod. 2007;13(4):273–279. doi: 10.1093/molehr/gam006. [DOI] [PubMed] [Google Scholar]
- Enquobahrie DA, Abetew DF, et al. Placental microRNA expression in pregnancies complicated by preeclampsia. Am J Obstet Gynecol. 2011;204(2):178 e112–121. doi: 10.1016/j.ajog.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri E, Brognara E, et al. MiRNA therapeutics: delivery and biological activity of peptide nucleic acids targeting miRNAs. Epigenomics. 2011a;3(6):733–745. doi: 10.2217/epi.11.90. [DOI] [PubMed] [Google Scholar]
- Fabbri E, Manicardi A, et al. Modulation of the biological activity of microRNA-210 with peptide nucleic acids (PNAs) ChemMedChem. 2011b;6(12):2192–2202. doi: 10.1002/cmdc.201100270. [DOI] [PubMed] [Google Scholar]
- Faraonio R, Salerno P, et al. A set of miRNAs participates in the cellular senescence program in human diploid fibroblasts. Cell Death Differ. 2012;19(4):713–721. doi: 10.1038/cdd.2011.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanaro P, D’Alessandra Y, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand ephrin-A3. J Biol Chem. 2008;283(23):15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanaro P, Greco S, et al. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem. 2009;284(50):35134–35143. doi: 10.1074/jbc.M109.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanaro P, Romani S, et al. ROD1 is a seedless target gene of hypoxia-induced miR-210. PLoS One. 2012;7(9):e44651. doi: 10.1371/journal.pone.0044651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro E, Ramachandran A, et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and Krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One. 2010;5(4):e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foekens JA, Sieuwerts AM, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci. 2008;105(35):13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambari R, Fabbri E, et al. Targeting microRNAs involved in human diseases: a novel approach for modification of gene expression and drug development. Biochem Pharmacol. 2011;82(10):1416–1429. doi: 10.1016/j.bcp.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Garzon R, Calin GA, et al. MicroRNAs in cancer. Annu Rev Med. 2009;60(1):167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Gee HE, Camps C, et al. hsa-miR-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer. 2010;116(9):2148–2158. doi: 10.1002/cncr.25009. [DOI] [PubMed] [Google Scholar]
- Giannakakis A, Sandaltzopoulos R, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7(2):255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad S, Meiri E, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnarra JR, Tory K, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7(1):85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, et al. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12(2):108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5(11):857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- Gou D, Ramchandran R, et al. miR-210 has an antiapoptotic effect in pulmonary artery smooth muscle cells during hypoxia. Am J Physiol Lung Cell Mol Physiol. 2012;303(8):L682–L691. doi: 10.1152/ajplung.00344.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco S, Fasanaro P, et al. MicroRNA dysregulation in diabetic ischemic heart failure patients. Diabetes. 2012;61(6):1633–1641. doi: 10.2337/db11-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greither T, Grochola LF, et al. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2009;126(1):73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- Greither T, Würl P, et al. Expression of microRNA 210 associates with poor survival and age of tumor onset of soft-tissue sarcoma patients. Int J Cancer. 2011;130(5):1230–1235. doi: 10.1002/ijc.26109. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, et al. MiRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(suppl 1):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, et al. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzy RD, Schumacker PT. Oxygen sensing by mitochondria at complex III: the paradox of increased reactive oxygen species during hypoxia. Exp Physiol. 2006;91(5):807–819. doi: 10.1113/expphysiol.2006.033506. [DOI] [PubMed] [Google Scholar]
- Hammer S, To KK, et al. Hypoxic suppression of the cell cycle gene CDC25A in tumor cells. Cell Cycle. 2007;6(15):1919–1926. doi: 10.4161/cc.6.15.4515. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- He J, Wu J, et al. MiR-210 disturbs mitotic progression through regulating a group of mitosis-related genes. Nucleic Acids Res. 2013;41(1):498–508. doi: 10.1093/nar/gks995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho AS, Huang X, et al. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl Oncol. 2010;3(2):109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Yang J, et al. High expression of miR-210 predicts poor survival in patients with breast cancer: a meta-analysis. Gene. 2012;507(2):135–138. doi: 10.1016/j.gene.2012.07.025. [DOI] [PubMed] [Google Scholar]
- Hu S, Huang M, et al. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122(Suppl 1):S124–S131. doi: 10.1161/CIRCULATIONAHA.109.928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ding L, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35(6):856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Le Q-T, et al. MiR-210 – micromanager of the hypoxia pathway. Trends Mol Med. 2010;16(5):230–237. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlin PJ, Queva C, et al. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev. 1997;11(1):44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- Hüttenhofer A, Schattner P, et al. Non-coding RNAs: hope or hype? Trends Genet. 2005;21(5):289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Iguchi H, Kosaka N, et al. Versatile applications of microRNA in anti-cancer drug discovery: from therapeutics to biomarkers. Curr Drug Discov Technol. 2010;7(2):95–105. doi: 10.2174/157016310793180648. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4(3):143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, et al. Targeting of HIF-alpha to the von hippel-lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Jeyaseelan K, Lim KY, et al. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39(3):959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- Juan D, Alexe G, et al. Identification of a MicroRNA panel for clear-cell kidney cancer. Urology. 2010;75(4):835–841. doi: 10.1016/j.urology.2009.10.033. [DOI] [PubMed] [Google Scholar]
- Jung EJ, Santarpia L, et al. Plasma microRNA 210 levels correlate with sensitivity to Trastuzumab and tumor presence in breast cancer patients. Cancer. 2012;118(10):2603–2614. doi: 10.1002/cncr.26565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Conaco C, et al. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci. 2007;104(49):19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35(7):368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Souza AL, et al. A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol Cell Biol. 2011;31(13):2696–2706. doi: 10.1128/MCB.01242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, et al. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kim JW, Dang CV. Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66(18):8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- Kim HW, Haider HK, et al. Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. J Biol Chem. 2009;284(48):33161–33168. doi: 10.1074/jbc.M109.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HW, Mallick F, et al. Concomitant activation of miR-107/PDCD10 and hypoxamir-210/Casp8ap2 and their role in cytoprotection during ischemic preconditioning of stem cells. Antioxid Redox Signal. 2012;17(8):1053–1065. doi: 10.1089/ars.2012.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King HW, Michael MZ, et al. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12(1):421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koong AC, Mehta VK, et al. Pancreatic tumors show high levels of hypoxia. Int J Radiat Oncol Biol Phys. 2000;48(4):919–922. doi: 10.1016/s0360-3016(00)00803-8. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krick S, Hanze J, et al. Hypoxia-driven proliferation of human pulmonary artery fibroblasts: cross-talk between HIF-1alpha and an autocrine Angiotensin system. FASEB J. 2005;19(7):857–859. doi: 10.1096/fj.04-2890fje. [DOI] [PubMed] [Google Scholar]
- Kuijper S, Turner CJ, et al. Regulation of angiogenesis by Eph–ephrin interactions. Trends Cardiovasc Med. 2007;17(5):145–151. doi: 10.1016/j.tcm.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, et al. Regulation of microRNA expression: the hypoxic component. Cell Cycle. 2007a;6(12):1426–1431. [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, et al. A MicroRNA signature of hypoxia. Mol Cell Biol. 2007b;27(5):1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Slack FJ. MicroRNAs in mutagenesis, genomic instability, and DNA repair. Semin Oncol. 2011;38(6):743–751. doi: 10.1053/j.seminoncol.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie CH, Gal S, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, et al. The C. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, et al. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21(17):4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees JA, Saito M, et al. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol. 1993;13(12):7813–7825. doi: 10.1128/mcb.13.12.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G, DeGregori J, et al. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12(14):2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, et al. Prediction of mammalian MicroRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, et al. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are MicroRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li T, Cao H, et al. Identification of miR-130a, miR-27b and miR-210 as serum biomarkers for atherosclerosis obliterans. Clin Chim Acta. 2011;412(1–2):66–70. doi: 10.1016/j.cca.2010.09.029. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cox SR, et al. Hypoxia regulates vascular endothelial growth factor gene expression in endothelial cells: identification of a 5′ enhancer. Circ Res. 1995;77(3):638–643. doi: 10.1161/01.res.77.3.638. [DOI] [PubMed] [Google Scholar]
- Liu M, Liu H, et al. Reactive oxygen species originating from mitochondria regulate the cardiac sodium channel. Circ Res. 2010;107(8):967–974. doi: 10.1161/CIRCRESAHA.110.220673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Han Y, et al. Synthetic miRNA-mowers targeting miR-183-96-182 cluster or miR-210 inhibit growth and migration and induce apoptosis in bladder cancer cells. PLoS One. 2012;7(12):e52280. doi: 10.1371/journal.pone.0052280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou YL, Guo F, et al. miR-210 activates notch signaling pathway in angiogenesis induced by cerebral ischemia. Mol Cell Biochem. 2012;370(1–2):45–51. doi: 10.1007/s11010-012-1396-6. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Malzkorn B, Wolter M, et al. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2009;20(3):539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37(suppl 2):W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroni G, Reymond A, et al. Rox, a novel bHLHZip protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box and acts as a transcriptional repressor. EMBO J. 1997;16(10):2892–2906. doi: 10.1093/emboj/16.10.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael MZ, O’Connor SM, et al. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–891. [PubMed] [Google Scholar]
- Miko E, Czimmerer Z, et al. Differentially expressed microRNAs in small cell lung cancer. Exp Lung Res. 2009;35(8):646–664. doi: 10.3109/01902140902822312. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno Y, Tokuzawa Y, et al. miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Lett. 2009;583(13):2263–2268. doi: 10.1016/j.febslet.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Mochel F, Knight MA, et al. Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am J Hum Genet. 2008;82(3):652–660. doi: 10.1016/j.ajhg.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Yasuda T, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25(17):2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutharasan RK, Nagpal V, et al. MicroRNA-210 is upregulated in hypoxic cardiomyocytes through Akt- and p53-dependent pathways and exerts cytoprotective effects. Am J Physiol Heart Circ Physiol. 2011;301(4):H1519–H1530. doi: 10.1152/ajpheart.01080.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada C, Tsukamoto Y, et al. Overexpression of miR-210, a downstream target of HIF1α, causes centrosome amplification in renal carcinoma cells. J Pathol. 2011;224(2):280–288. doi: 10.1002/path.2860. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Patrushev N, et al. Role of protein tyrosine phosphatase 1B in vascular endothelial growth factor signaling and cell–cell adhesions in endothelial cells. Circ Res. 2008;102(10):1182–1191. doi: 10.1161/CIRCRESAHA.107.167080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y, Han B-M, et al. Identification of MicroRNAs involved in hypoxia- and serum deprivation-induced apoptosis in mesenchymal stem cells. Int J Biol Sci. 2011;7:762–768. doi: 10.7150/ijbs.7.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pineles BL, Romero R, et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol. 2007;196(3):261 e261–266. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Place RF, Li L-C, et al. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porkka KP, Pfeiffer MJ, et al. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67(13):6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- Presti JC, Jr, Rao PH, et al. Histopathological, cytogenetic, and molecular characterization of renal cortical tumors. Cancer Res. 1991;51(5):1544–1552. [PubMed] [Google Scholar]
- Puissegur MP, Mazure NM, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18(3):465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulkkinen K, Malm T, et al. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 2008;582(16):2397–2401. doi: 10.1016/j.febslet.2008.05.048. [DOI] [PubMed] [Google Scholar]
- Qin L, Chen Y, et al. A deep investigation into the adipogenesis mechanism: profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/beta-catenin signaling pathway. BMC Genomics. 2010;11:320. doi: 10.1186/1471-2164-11-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raponi M, Dossey L, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69(14):5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- Redova M, Poprach A, et al. MiR-210 expression in tumor tissue and in vitro effects of its silencing in renal cell carcinoma. Tumor Biol. 2012;34(1):481–491. doi: 10.1007/s13277-012-0573-2. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rothe F, Ignatiadis M, et al. Global MicroRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS One. 2011;6(6):e20980. doi: 10.1371/journal.pone.0020980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan K, Song G, et al. Role of hypoxia in the hallmarks of human cancer. J Cell Biochem. 2009;107(6):1053–1062. doi: 10.1002/jcb.22214. [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, et al. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar J, Gou D, et al. MicroRNA-21 plays a role in hypoxia-mediated pulmonary artery smooth muscle cell proliferation and migration. Am J Physiol Lung Cell Mol Physiol. 2010;299(6):L861–L871. doi: 10.1152/ajplung.00201.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satzger I, Mattern A, et al. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int J Cancer. 2009;126(11):2553–2562. doi: 10.1002/ijc.24960. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Angiogenesis ischemic and neoplastic disorders. Annu Rev Med. 2003;54(1):17–28. doi: 10.1146/annurev.med.54.101601.152418. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010a;29(5):625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Vascular responses to hypoxia and ischemia. Arterioscler Thromb Vasc Biol. 2010b;30(4):648–652. doi: 10.1161/ATVBAHA.108.181644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391(6665):404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Simone NL, Soule BP, et al. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS One. 2009;4(7):e6377. doi: 10.1371/journal.pone.0006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvang J, Silahtaroglu AN, et al. The utility of LNA in microRNA-based cancer diagnostics and therapeutics. Semin Cancer Biol. 2008;18(2):89–102. doi: 10.1016/j.semcancer.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Tabernero J, Shapiro GI, et al. First-in-man trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Disco. 2013;3(4):406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Thum T, Galuppo P, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116(3):258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- Tong WH, Rouault TA. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 2006;3(3):199–210. doi: 10.1016/j.cmet.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Toyama T, Kondo N, et al. High expression of microRNA-210 is an independent factor indicating a poor prognosis in Japanese triple-negative breast cancer patients. Jpn J Clin Oncol. 2012;42(4):256–263. doi: 10.1093/jjco/hys001. [DOI] [PubMed] [Google Scholar]
- Tsuchiya S, Fujiwara T, et al. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1) J Biol Chem. 2011;286(1):420–428. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, et al. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103(48):18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, et al. Switching from repression to activation: MicroRNAs can upregulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26(2):225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136(4):586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, Mathur R, et al. MiRNA response to DNA damage. Trends Biochem Sci. 2011;36(9):478–484. doi: 10.1016/j.tibs.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Olson EN. AngiomiRs–key regulators of angiogenesis. Current Opin Genet Dev. 2009;19(3):205–211. doi: 10.1016/j.gde.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci U S A. 1993;90(9):4304–4308. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270(3):1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, et al. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen J, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res. 2009;2(9):807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JA, Baxter DH, et al. The MicroRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NMA, Bao TT, et al. MiRNA profiling for clear cell renal cell carcinoma: biomarker discovery and identification of potential controls and consequences of miRNA dysregulation. J Urol. 2011;186(3):1077–1083. doi: 10.1016/j.juro.2011.04.110. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, et al. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. Elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11(6):393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- Wu F, Yang Z, et al. Role of specific microRNAs for endothelial function and angiogenesis. Biochem Biophys Res Commun. 2009;386(4):549–553. doi: 10.1016/j.bbrc.2009.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Yang W, Sun T, et al. Downregulation of miR-210 expression inhibits proliferation, induces apoptosis and enhances radiosensitivity in hypoxic human hepatoma cells in vitro. Exp Cell Res. 2012;318(8):944–954. doi: 10.1016/j.yexcr.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Zhang H, Gao P, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11(5):407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Sun H, et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8(17):2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wan G, et al. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell. 2011;41(4):371–383. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao A, Li G, et al. Serum miR-210 as a novel biomarker for molecular diagnosis of clear cell renal cell carcinoma. Exp Mol Pathol. 2013;94(1):115–120. doi: 10.1016/j.yexmp.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Zhu XM, Han T, et al. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol. 2009;200(6):661 e661–667. doi: 10.1016/j.ajog.2008.12.045. [DOI] [PubMed] [Google Scholar]
- Zundel W, Schindler C, et al. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 2000;14(4):391–396. [PMC free article] [PubMed] [Google Scholar]


