Abstract
The 22q11.2 deletion syndrome is one of the most common deletion syndromes in newborns. Some affected newborns may be diagnosed shortly after birth because of the presence of heart defects, palatal defects, or severe immune deficiencies. However, diagnosis is often delayed in patients presenting with other associated conditions that would benefit from early recognition and treatment, such as speech delays, learning difficulties, and schizophrenia. Fluorescence in situ hybridization (FISH) is the gold standard for deletion detection, but it is costly and time consuming and requires a whole blood specimen. Our goal was to develop a suitable assay for population-based screening of easily collectible specimens, such as buccal swabs and dried blood spots (DBS). We designed a pyrosequencing assay and validated it using DNA from FISH–confirmed 22q11 deletion syndrome patients and normal controls. We tested DBS from nine patients and paired buccal cell and venous blood specimens from 20 patients. Results were 100% concordant with FISH assay results. DNA samples from normal controls (n = 180 cell lines, n = 15 DBS, and n = 88 buccal specimens) were negative for the deletion. Limiting dilution experiments demonstrated that accurate results could be obtained from as little as 1 ng of DNA. This method represents a reliable and low-cost alternative for detection of the common 22q11.2 microdeletions and can be adapted to high-throughput population screening.
The 22q11.2 deletion syndrome (22q11DS), alias DiGeorge syndrome (DGS), shprintzen syndrome, and velocardiofacial syndrome, is estimated to be the most common inheritable genetic deletion syndrome, with a reported prevalence ranging from approximately 1:9700 to as high as 1:3900 live births.1,2 This syndrome is the result of hemizygous deletions of chromosome 22q11.2, with a 3-Mb deletion representing most (approximately 90%) of all deletions.3 Approximately 7% of cases are accounted for by the smaller 1.5-Mb deletion nested within the 3-Mb region, with the remaining 3% consisting of various other microdeletions.3 Clinical presentation is highly variable, and there is often poor correlation between genotype and phenotype. In addition, lesser-known microduplications, which are presumed to be reciprocal rearrangements to the microdeletions characterized in the 22q11.2 region, generally result in milder, yet highly variable, phenotypes.4 Well-designed, population-based studies are needed for a more comprehensive evaluation of the impact, distribution, and clinical presentation of this highly dynamic 22q11.2 region.
The current gold standard clinical laboratory assay for 22q11DS uses the TUPLE fluorescence in situ hybridization (FISH) probe, which is located in the common 3-Mb deletion and in the less common 1.5-Mb nested deletion. FISH is costly, requires a whole blood sample, and takes at least 48 hours to perform. This makes the test feasible as a diagnostic test only for those patients for whom it is clinically warranted. Alternative molecular technologies, such as multiplex ligation-dependent probe amplification (MLPA),5–8 microsatellite marker analysis,6 and real-time quantitative PCR,9–11 have been developed for possible use as population screening tests in the diagnosis of 22q11DS. Although economically more cost-effective and less labor intensive than FISH, some methods are still technologically challenging, data analysis is not always straightforward, and the high costs of commercial kits or expensive fluorescent dye-labeled probes may make higher-throughput screening cost prohibitive. In addition, many of these methods typically require a minimum of 10 to 100 ng of high-quality DNA obtained from whole blood specimens.
Although large quantities of high-quality DNA can be obtained from whole blood collected by venipuncture, the costs of collection, transport, and storage are important considerations. Use of alternative specimens, such as dried blood spots (DBS) and buccal cells that are less invasive than whole blood, has become widely accepted. Buccal cell collection is a cost-effective method that has become increasingly popular in large-scale studies and is particularly suitable for infants, young children, and widely dispersed populations.12 DBSs are easily transported and stored and are obtained on all newborns in the United States through newborn screening programs. If properly stored, DBSs are stable for many years.13
We used pyrosequencing, a solution-based, fluorescent, real-time DNA sequencing method, to determine the relative copy number of the 22q11.2 region. During the reaction, pyrophosphate is stoichiometrically split off from the deoxynucleoside triphosphate as it is incorporated into the growing strand. This initiates a reaction cascade leading to quantifiable light emission. The resulting peak heights are directly proportional to the number of individual nucleotides incorporated, making it a quantitative technology with the added benefit of providing confirmatory context sequence data.14 We designed an assay that targets the ubiquitin fusion degradation (UFDL1) gene located within both the commonly deleted 3-Mb region and the nested 1.5-Mb deletion on chromosome 22 (Chr22). The assay also targets a reference sequence on chromosome 18 (Chr18) that serves as an undeleted control. The high degree of homology of the reference sequence to the Chr22 region allows for co-amplification of both loci using a single PCR primer pair and one pyrosequencing primer. After the two primary amplicons are generated, the sequencing primer is used to extend the PCR products by addition of nucleotides in a specific order, according to the known sequence. The amplicons differ from each other at several nucleotide positions, and the relative contribution of nucleotides specific to Chr18 and Chr22 is then used to determine the copy number. This approach is based on the paralogue ratio test,15–17 and proved to be reliable even for DNA from buccal and DBS specimens, which typically yield lower quantity and quality DNA compared with whole blood.13,18
Materials and Methods
Assay Design
To identify a homologous reference sequence, we compared the genomic DNA sequence of genes located within the 3-Mb and the nested 1.5-Mb deletion (Figure 1A) against the entire human genome sequence using the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi, last accessed April 30, 2014). A 780-bp intergenic region on Chr18 was identified, which shares 93% identity to an exonic region in the UFD1L gene on Chr22. A search of the Database of Genomic Variants (http://dgv.tcag.ca/dgv/app/home, last accessed April 30, 2014) showed no known deletions in the Chr18 region. This is a curated database of structural variation in >14,000 unique samples in approximately 44 different populations. In recent years, most data have been generated by high-resolution microarrays and next-generation sequencing technologies, which contributed to significant improvements in accuracy.19 Primer location and sequences used for the pyrosequencing assay are shown in Figure 1B. The PCR products contained several nucleotide sequence differences between the two loci. The forward sequencing primer was positioned upstream of two key nucleotide site differences. A selective nucleotide dispensation order was used to generate Chr22- and Chr18-specific peaks that allow for clear distinction between the two loci in the resulting pyrograms. Negative nucleotide dispensations were inserted to serve as internal controls for nucleotide misincorporation. All peaks in the pyrograms were used to ensure the specificity of the assay. The pyrosequencing assay was designed using PSQ Assay Design software version 1.0 (Biotage AB, Uppsala, Sweden). The specificity of the PCR primer pairs was checked using University of California, Santa Cruz, Genome Bioinformatics in silico PCR (http://genome.ucsc.edu/cgi-bin/hgPcr?command=start, last accessed April 30, 2014) to confirm that only the targeted Chr22 and Chr18 regions would be amplified.
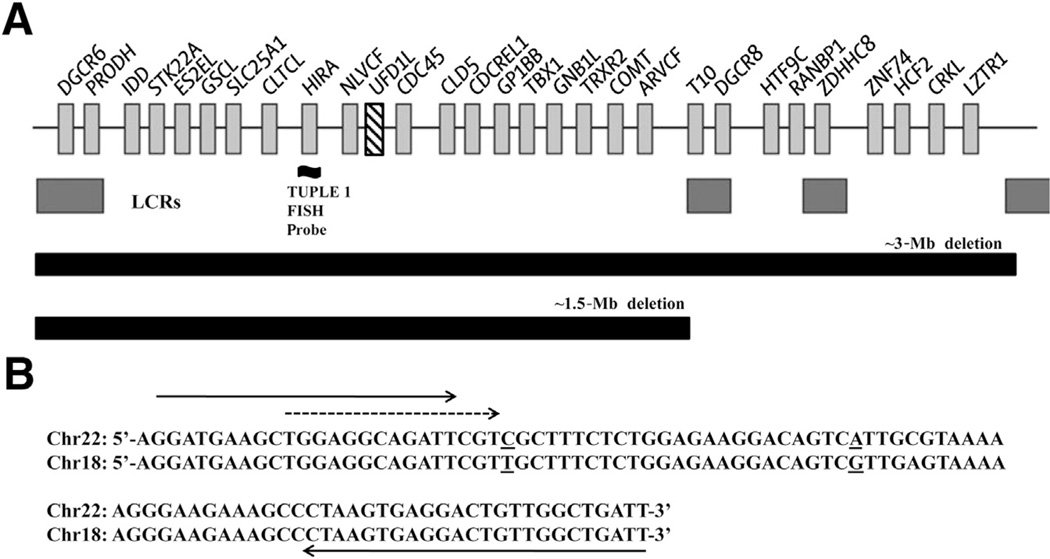
Figure 1.
A: Map of the 22q11.2 deleted region showing the location of the TUPLE1 FISH probe and UFD1L gene (hatch marked). Light gray boxes with associated Human Genome Organization (HUGO) gene names denote surrounding genes in this region. Dark gray boxes indicate locations of the four low copy repeats (LCRs) that mediate deletions of 22q11.2.10 B: Alignment of Chr22 (UFD1L) and Chr18 target sequences. Solid arrows indicate forward (5′-GGATGAAGCTGGAGGCAGATT-3′) and 3′-biotinylated reverse (5′-AATCAGCCAACAGTCCTCACTTAG-3′) PCR primers. The broken arrow indicates the forward sequencing primer (5′-TGGAGGCAGATTCGT-3′). Underlined nucleotides represent sequence differences between the two chromosomes that are detected in the pyrosequencing assay.
Data Analysis for Pyrosequencing Assay
Peak height values for Chr22- and Chr18-specific peaks were exported from the PSQHS 96A Software version 1.2 (Biotage AB) into a text file, which was imported into Microsoft Excel (Redmond, WA) for peak height ratio calculations. Two peak height ratios were determined: the C/T ratio was obtained by dividing the Chr22-specific peak height at dispense 3 (C) by the Chr18-specific peak height at dispense 2 (T), and the A/G peak height ratio was obtained by dividing the Chr22-specific peak height at dispense 24 (A) by the Chr18-specific peak height at dispense 23 (G). Theoretical ratios of 0.5, 1.0, and 2.0 are expected for deleted, nondeleted, and duplicated samples, respectively. All graphs and calculations for mean, median, coefficient of variation (CV), and SD were performed within Microsoft Excel.
Samples
We performed initial assay development and validation using cell line–derived DNA from Coriell Cell Repositories (Camden, NJ). Samples included FISH-confirmed 22q11.2 deleted DNA from patients (n = 3) with clinically diagnosed DGS (NA07215 or NA13325) and velocardiofacial syndrome (NA07939). DNA from the Human Variation Collection (n = 180) served as nondeleted controls and included samples from the SNP500 Cancer Resource panel (n = 108; SNP500V), African American panel (n = 24; HD24AA), European Caucasian panel (n = 24; HD24EC), and Han Chinese panel (n = 24 HD24CHI). We also evaluated whole blood, DBS, and buccal cell specimens obtained from patients attending the Southeastern Regional Center for 22q (Emory University, Atlanta, GA). This set of specimens consisted of paired whole blood and buccal swabs obtained from 19 patients and one nonpaired patient buccal swab. DBSs were obtained from an additional nine patients. The Emory Cytogenetics laboratory confirmed the presence of the 22q11.2 deletion in all patients by FISH. We also included buccal samples (n = 88) and DBS (n = 15) from subjects who lack the deletion. Last, two DNA samples with verified 22q11.2 microduplications were obtained from the MGL Cytogenetic and Microarray Laboratories at Baylor College of Medicine (Houston, TX). The duplication was confirmed by two-color FISH and array-comparative genomic hybridization. All samples were de-identified before receipt. Samples were obtained with informed consent under study protocols with Institutional Review Board approval at Emory University and Baylor College of Medicine. The Centers for Disease Control and Prevention’s roles in laboratory analysis of de-identified samples did not constitute engagement in human subject research.
DNA Extraction
DNA was isolated from 4-mL whole blood specimens using Gentra Puregene reagents (Qiagen, Valencia, CA), according to the manufacturer’s instructions. DNA from one 3.2-mm DBS punch was isolated using the Qiagen Investigator kit, according to the manufacturer’s protocol, with a 60-µL elution volume. Buccal swabs were collected using cytobrushes, and DNA was extracted by phenol-chloroform.20 Genomic DNA yields were assessed by quantitative real-time PCR using the human RNase P gene. Primer sequences are available on request. DNA concentrations were determined, in duplicate, using the ABI 7500 Sequence Detection System and established manufacturer’s protocols (Applied Biosystems, Foster City, CA).
MLPA of Nondeleted Controls
The copy number of the 22q11.2 region in non–FISH-confirmed Coriell, buccal, and DBS samples that served as nondeleted controls was analyzed by MLPA. The SALSA P250-A1 MLPA-DGS test kit (MRC-Holland, Amsterdam, the Netherlands) was used following the manufacturer’s instructions. The P250 probe mix is a high-density kit that contains 48 different probes and is designed to detect deletions and duplications in the 22q11.2 region. All runs included DNA from three normal controls to calibrate unknown samples. The reaction products were detected with an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). The ABI GeneMapper software version 3.5 (Applied Biosystems) was used to size the PCR products and to obtain peak areas. These data were imported into Coffalyser Software version 9.4 (MRC-Holland) for MLPA analysis.
PCR Optimization
PCR conditions were evaluated for each specimen type using 5-ng input DNA from deleted, nondeleted, and duplicated samples. All reactions were performed in a final volume of 20 µL containing 0.5 µmol/L of each primer (Integrated DNA Technologies, Coralville, IA) and a commercial PCR master mix (described below) at 1× concentration. The optimum annealing temperature was 60.5°C for DNA from blood, cell lines, and buccal specimens and 63.5°C for DBS, as determined by gradient cycling (Eppendorf Mastercycler Gradient; Eppendorf, Hauppauge, NY), ranging from 55.0°C to 64.0°C. We chose the highest annealing temperature that maintained good separation between deleted and nondeleted C/T ratios, yet remained close to theoretical values of 1.0 and 0.5 for nondeleted and deleted, respectively.
Thermal cycling conditions were as follows: 95°C for 10 minutes, 40 cycles at 94°C for 10 seconds, followed by either 60.5°C or 63.5°C for 20 seconds, 72°C for 1 minute, and a final extension at 72°C for 2 minutes. All PCR runs included a no template control, a deleted DNA control, and a nondeleted DNA control derived from the same specimen type as the test samples.
Three PCR master mixes were evaluated: i) AmpliTaq Gold PCR Master Mix (Applied Biosystems), ii) Amplitaq Gold PCR Master Mix with the addition of PCRboost (Biomatrica, San Diego, CA), and iii) PyroMark PCR Kit (Qiagen). A subset of blood and cell line–derived DNA (n = 19 deleted samples, and n = 147 nondeleted samples) was amplified using all three master mixes. On the basis of these results, buccal DNA samples (n = 6 deleted, and n = 16 nondeleted) were amplified using two of the master mixes: Amplitaq Gold PCR Master Mix with PCRboost and Pyro-Mark PCR Kit.
Pyrosequencing
PCR products (5 µL) were processed in a 96-well format for pyrosequencing analysis following the standard manufacturer’s protocol (Qiagen). Single-stranded DNA was prepared by immobilization of the biotinylated PCR product onto streptavidin-coated Sepharose high-performance beads (GE Healthcare, Piscataway, NJ) with subsequent removal of nonbiotinylated single strands using the Vacuum Prep workstation (Biotage AB). The pyrosequencing reaction was conducted in a PSQ 96HSA System (Biotage AB) using PyroMark Gold reagents (Qiagen).
Analytic Sensitivity and Reproducibility
Sensitivity of the optimized PCR system was investigated by limiting dilution experiments using DNA from whole blood (n = 2 deleted), cell lines (n = 2 nondeleted, n = 1 deleted), buccal cells (n = 3 nondeleted, n = 3 deleted), and DBS (n = 3 nondeleted, n = 2 deleted). Serially diluted DNA ranging from 0.125 to 2.0 ng was amplified in quadruplicate using a PyroMark PCR kit followed by pyrosequencing. Mean C/T ratios (Chr22/Chr18 ratio) were determined for each quantity of input DNA. We evaluated assay reproducibility by amplifying replica sets of DNA from 88 nondeleted and 20 deleted buccal samples on 2 different days.
Results
MLPA Confirmation of Nondeleted Controls
MLPA analysis of Coriell and buccal DNA samples used as nondeleted controls confirmed the absence of the 22q11.2 deletion in all samples. Analysis of DNA from DBS was unsuccessful by MLPA, most likely because of the limited quantity and quality of the DNA necessary for this method. However, 4 of 15 nondeleted control DBS samples were FISH confirmed. Pyrosequencing analyses between these four samples and the remaining 11 non–FISH-confirmed controls were concordant for the absence of the 22q11.2 deletion.
Assay Design and Interpretation
A quantitative pyrosequencing assay was designed for the relative quantitation of the 22q11.2 microdeletion region using a homologous region of Chr18 as a reference. The choice of a reference sequence with high homology allowed us to amplify both targets in a single tube using the same PCR and sequencing primers. The relative quantities of amplification products were determined by pyrosequencing through short regions that differ between the 22q11.2 deleted region and the Chr18 reference sequence. The resulting signal peak pattern (pyrogram) is a product of the two sequencing reactions that are performed simultaneously. The three types of peaks that were generated were peaks specific for the 22q11.2 region, peaks specific for the Chr18 region, and peaks that represent a combined signal from both regions. We determined the relative copy number of the 22q11.2 region by calculating the ratio between Chr22- and Chr18-specific peaks.
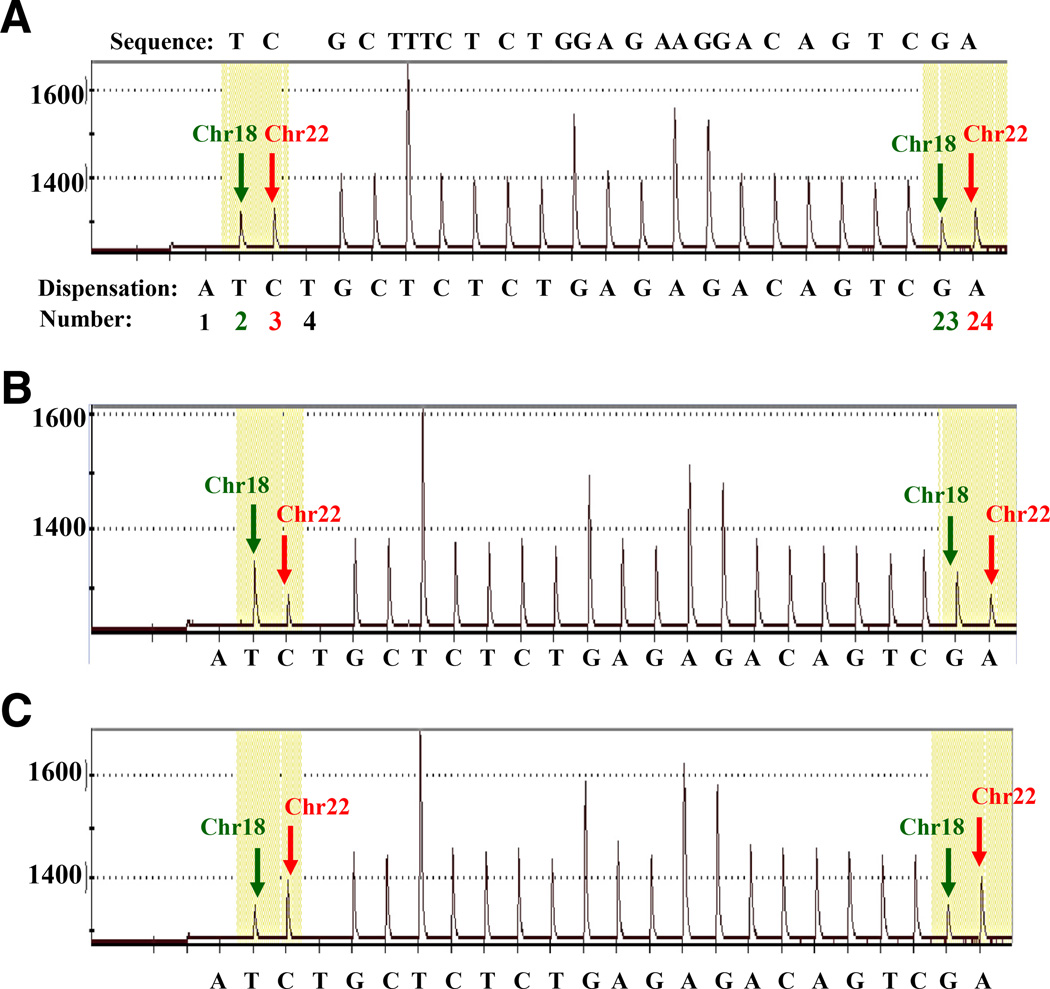
Representative pyrograms from selected nondeleted, deleted, and duplicated samples are shown in Figure 2. The nondeleted (2N) sample (Figure 2A) produced Chr22- and Chr18-specific peak heights that were similar in magnitude to each other. The deleted (hemizygous or 1N) sample (Figure 2B) showed Chr22-specific peaks that were approximately half the height of Chr18-specific peaks, whereas the duplicated (3N) sample (Figure 2C) showed Chr22-specific peaks that were approximately twice the height of Chr18-specific peaks. These results are consistent with the expected peak height ratios for each 22q11.2 copy number status. The consistent peak heights for the Chr18 region in all samples showed no evidence of copy number variability.
Figure 2.
Representative pyrograms generated by pyrosequencing. A: The expected sequence (forward direction) appears above the pyrogram, and the dispensation order is shown under each pyrogram. Dispensations 1 and 4 are negative and serve as internal controls for nucleotide misincorporation. The Chr18-specific nucleotide peaks (dispensations 2 and 23, green arrows) and the Chr22-specific nucleotide peaks (dispensations 3 and 24, red arrows). Pyrogram for a nondeleted DNA sample from buccal cells shows Chr18- and Chr22-specific peaks of approximate equivalent height (A), for a 22q11.2 deletion DNA sample from buccal cells shows Chr22-specific peak heights decreased by approximately half of Chr18-specific peak heights (B), and for a 22q11.2 duplication DNA sample from whole blood shows Chr22-specific peak heights increased approximately twofold higher than that of Chr18-specific peak heights (C).
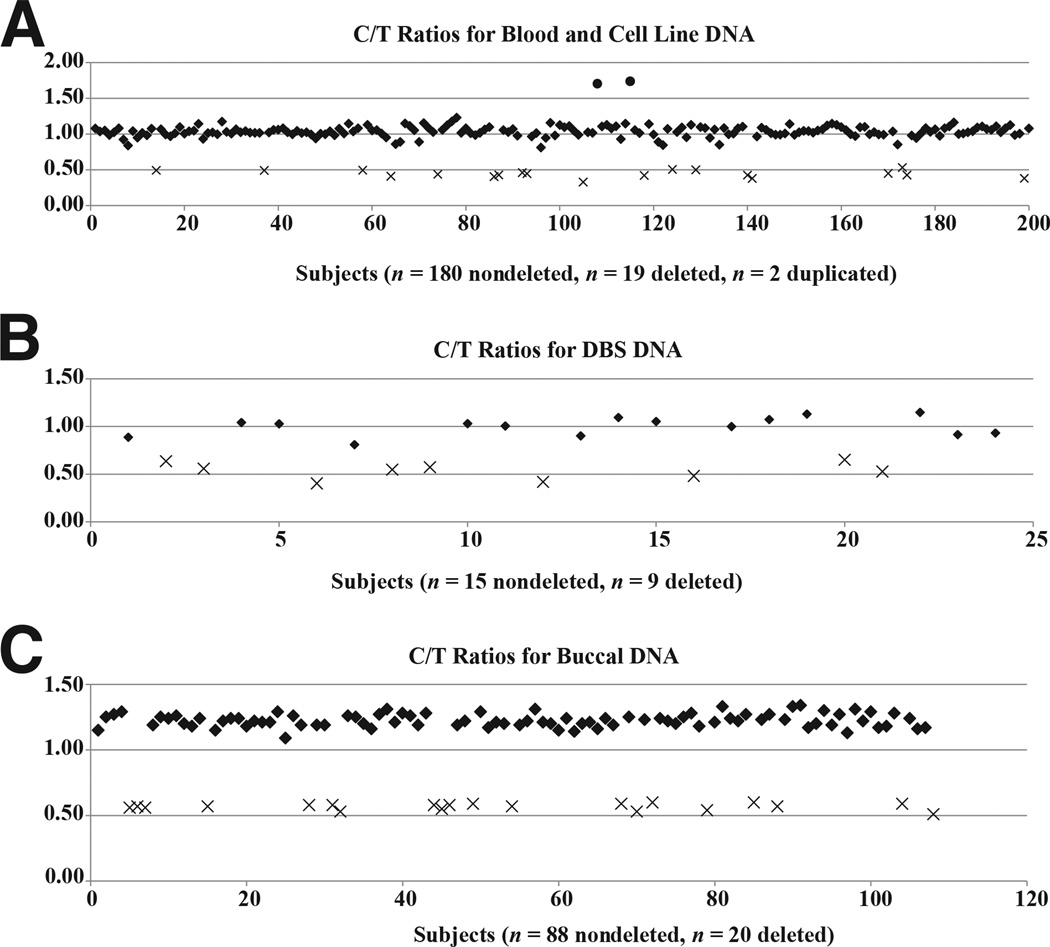
The distribution of C/T peak height ratios is shown in Figure 3 scatter plots. Cell line and whole blood–derived DNA segregated into two groups (Figure 3A). The deleted samples clustered into one group with a median (mean ± SD) peak height ratio of 0.44 (0.44 ± 0.05), and the nondeleted samples clustered into a second group with a median (mean ± SD) peak height ratio of 1.03 (1.04 ± 0.07). The two duplication samples gave a mean peak height ratio value of 1.72. Likewise, DBS and buccal DNA samples correctly segregated into two nonoverlapping groups according to their deletion status (Figure 3, B and C). DBS DNA gave a median (mean ± SD) peak height ratio of 0.54 (0.53 ± 0.08) for deleted samples and 1.03 (1.0 ± 0.1) for nondeleted samples. Median (mean ± SD) peak height ratios in buccal DNA were 0.57 (0.57 ± 0.02) and 1.22 (1.23 ± 0.05) for deleted and nondeleted samples, respectively. All pyrosequencing results from deleted samples were 100% concordant with FISH assay results, and all nondeleted controls were correctly called.
Figure 3.
Scatter plots show C/T ratios for nondeleted, deleted, and duplicated (whole blood only) DNA samples from whole blood and cell lines (A), DBS (B), and buccal cells (C). Nondeleted (two copies), deleted (one copy), and duplicated (three copies) subjects are indicated by theoretical C/T ratios of 1.0, 0.5, and 1.5, respectively. Duplication samples (circles), nondeletion samples (diamonds), and deletion samples (X marks) are shown.
In most cases, the deletion status calls on the basis of A/G peak height ratios were concordant with those on the basis of C/T peak height ratios; however, we observed that A/G ratios were less robust in discriminating between deleted and nondeleted samples. Receiver operating characteristic curve analysis showed 100% sensitivity and 100% specificity at C/T ratio cutoff values for deletion samples of 0.53, 0.65, and 0.6 for blood, DBS, and buccal specimens, respectively. Although receiver operating characteristic curve analysis also showed 100% sensitivity and 100% specificity at A/G ratio cutoff values of 0.75 for DBS and 0.67 for buccal DNA samples, the cutoff value for blood DNA samples of 0.6 represents 100% sensitivity and 98.9% specificity because of two observations that generated an overlap between deletion and nondeletion samples. The adenosine (A) nucleotide used in pyrosequencing is an ATP analog; therefore, peak heights generated from adenosine dispensations are consistently higher than those generated from addition of other bases.21 In addition, the pyrosequencing reaction tends to become less robust as peaks are generated further downstream in the sequence where the A and G peaks reside. Because of the aforementioned issues, the A/G site alone is not reliable for detection of the 22q11.2 deletion. A search in the University of California, Santa Cruz, genome browser yielded several low-frequency single-nucleotide polymorphisms (SNPs) that reside just upstream or downstream of the A/G site on either Chr22 or Chr18. One SNP on Chr22 (rs199813245) occurred in 1 (0.046%) of 2179 samples tested and may be a private mutation. Two other SNPs on Chr18, rs138982743 and rs78664154, have a prevalence of 3 (0.138%) of 2179 and 15 (0.653%) of 2296, respectively. Because these SNPs do not interfere with the upstream diagnostic C/T site, there would be no effect on final assay outcomes.
Assay Optimization
Gradient cycler experiments showed minimal influence of PCR annealing temperature on peak height ratios in DNA from whole blood, cell line, DBS, and buccal cell specimens. We selected annealing temperatures that provided the greatest separation between nondeleted and deleted C/T ratios, which were also closest to the theoretical values of 1.0 and 0.5 for nondeleted and deleted, respectively. This was slightly different for DNA from blood, cell lines, and buccal specimens (60.5°C) compared with DNA from DBS (63.5°C).
A summary of the PCR master mix evaluation is shown in Table 1. The SD and CV of C/T ratios obtained from nondeleted blood, cell line, and buccal DNA samples were significantly smaller (P < 0.0001, Student’s t-test) when the PyroMark PCR kit was used compared with both Amplitaq Gold and Amplitaq Gold plus PCRboost master mixes. Likewise, the SD and CV of C/T ratios for deleted buccal DNA samples were significantly smaller (P < 0.0023, Student’s t-test) when the PyroMark PCR kit was used compared with the Amplitaq Gold plus PCRboost master mix. Although results from deleted blood and cell line DNA samples had lower variability using the PyroMark PCR kit, they did not reach statistical significance. This underlines the importance of optimizing assays for each specimen type to achieve the most reliable results.
Table 1.
Effect of PCR Master Mixes on C/T Ratio Values
| DNA source | Master mix | Mean | Median | SD | % CV |
|---|---|---|---|---|---|
| Nondeleted | |||||
| Blood/cell line (n = 147) | Amplitaq Gold | 0.97 | 0.99 | 0.14 | 14.26 |
| Amplitaq Gold + PCRboost | 1.09 | 1.07 | 0.13 | 12.14 | |
| PyroMark PCR | 1.02 | 1.03 | 0.06 | 6.12 | |
| Buccal (n = 16) | Amplitaq Gold + PCRboost | 1.37 | 1.36 | 0.15 | 10.77 |
| PyroMark PCR | 1.12 | 1.13 | 0.05 | 4.02 | |
| Deleted | |||||
| Blood/cell line (n = 19) | Amplitaq Gold | 0.48 | 0.48 | 0.09 | 18.48 |
| Amplitaq Gold + PCRboost | 0.42 | 0.43 | 0.08 | 19.53 | |
| PyroMark PCR | 0.44 | 0.44 | 0.05 | 11.6 | |
| Buccal (n = 6) | Amplitaq Gold + PCRboost | 0.67 | 0.68 | 0.05 | 7.11 |
| PyroMark PCR | 0.58 | 0.59 | 0.03 | 4.35 |
Analytic Sensitivity and Interassay Variation
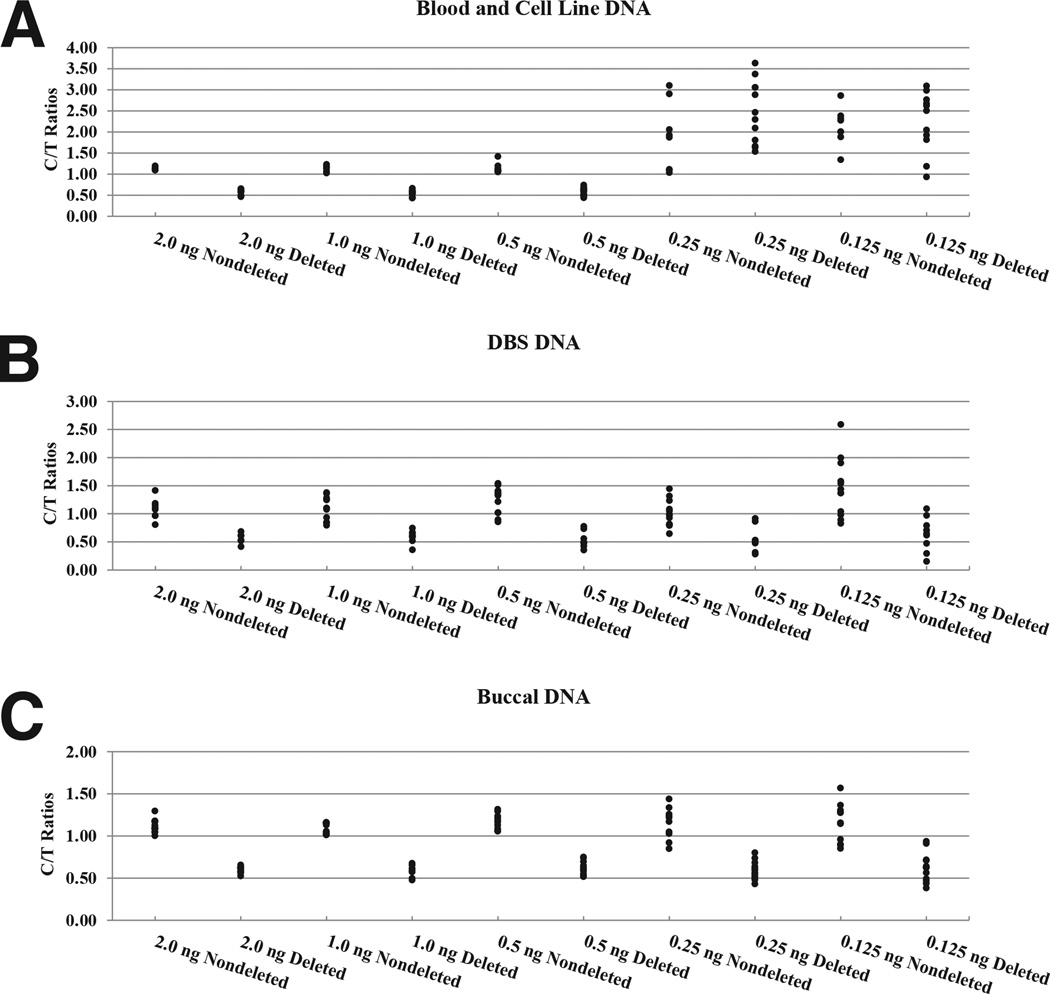
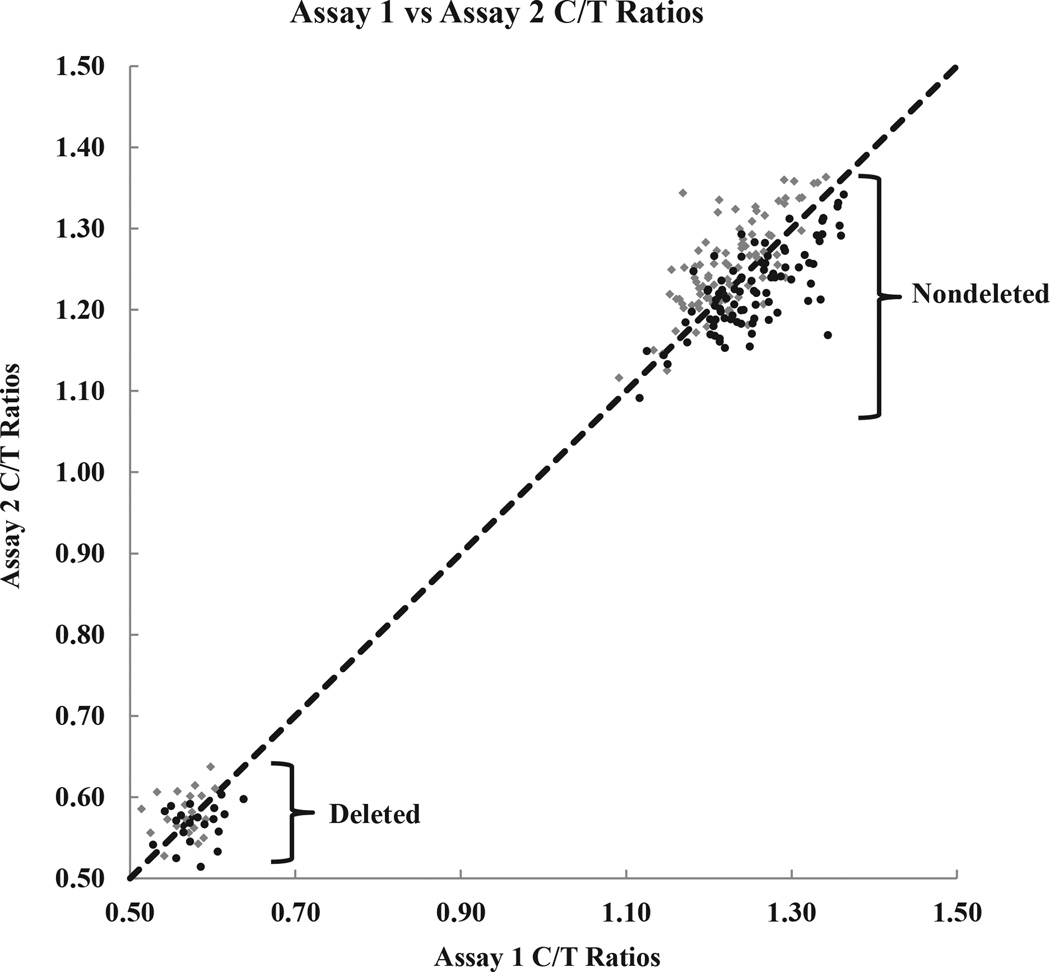
The sensitivity of the assay was evaluated by performing limiting dilution experiments (Figure 4). The deletion status could accurately be determined using as little as 0.5 ng of DNA from whole blood, cell lines, and buccal cells. At lower amounts of input DNA, the C/T ratios of deleted samples overlapped with those of nondeleted samples, which prevented an unambiguous determination of the deletion status. Although the C/T ratios between deleted and nondeleted samples were nonoverlapping for 1.0- and 0.5-ng input DNA from DBS, the cutoff values were close (0.74/0.79 and 0.77/0.85, respectively). This amount of input DNA should be used with caution; otherwise, 2.0-ng input DNA is most reliable in this case. Reproducibility was assessed by performing replica runs on different days using DNA from deleted and nondeleted buccal specimens. A pairwise comparison of the two assays is shown graphically in Figure 5. The scatter plot shows that the data for the two assays lie on the line y = x, which indicates good agreement. Both assays were able to accurately distinguish the deleted from the nondeleted samples.
Figure 4.
C/T peak height ratios for nondeleted and deleted samples as a function of input DNA (ng) extracted from blood and cell lines (A), DBS (B), and buccal cells (C).
Figure 5.
Pairwise graph shows reproducibility between duplicate assay runs on different days using DNA extracted from buccal cells. Assay 1 data (diamonds) and assay 2 data (circles). C/T ratio data points for assay 1 versus assay 2 fall along the line y = x, which shows strong agreement between the two. There is a clear separation between nondeleted and deleted C/T ratios.
Discussion
The goal of this study was to develop a robust and accurate screening tool for the detection of the 22q11DS that could reliably be used in population-based studies that collect alternative specimens, such as dried blood spots and buccal swabs. We designed a pyrosequencing assay that coamplifies a region of the ubiquitin fusion degradation (UFDL1) gene located within the commonly deleted regions on Chr22 and a highly homologous reference sequence on Chr18. Only a single PCR primer pair and one pyrosequencing primer are needed, and two nucleotide sequence differences between the two amplicons are used to determine copy number of the 22q11.2 region. Pyrosequencing has been widely used for the analysis of genetic variations, such as SNPs, insertions/deletions, short repeats, and gene copy number. It has several advantages over traditional methods for genotyping. The technique offers high accuracy, flexibility, automation, and parallel processing (96 samples in a single plate). Once a PCR product is obtained, the simple purification steps can be completed in 5 to 10 minutes and subsequent sequencing data can be obtained in 15 minutes. For increased throughput, a plate-stacking hotel can feed up to 10 plates in succession into the sample sequencing chamber. This method is cost-effective in that it obviates the need for electrophoresis, size separation, labeled nucleotides, or labeled primers (except one biotinylated PCR primer). Jansson et al22 and Söderbäck et al23 have also successfully applied pyrosequencing technology for gene copy analysis of the GSTM1 gene and the CY2PD6 gene, respectively. A related approach has been described by Deutsch et al15 for the diagnosis of trisomy 21.
In our assay, we used primers that perfectly match both Chr22 and Chr18 targets to increase the likelihood that amplification efficiencies would be nearly identical.23 Neither deletions nor duplications have been reported for the Chr18 reference region, which shares approximately 93% homology to the UFD1L region at the nucleotide level. This design simplifies method validation, assay setup, and data analysis. To our knowledge, this is the first report describing the detection of the 22q11.2 deletion that uses a highly homologous region on a separate gene as the 2N reference as opposed to an unrelated (nonhomologous) gene, which requires separate PCR and sequencing primers and may lead to variations in amplification efficiencies between the different genetic regions.
In conclusion, we developed a new assay for the reliable and accurate detection of 22q11.2 deletions and duplications in DNA samples of lower quality and quantity from such sources as buccal cells and DBS. This assay is an appealing alternative to existing methods that often require high-quality DNA and expensive fluorescent probes. The assay offers a cost-effective and high-throughput way in which to perform population-based screening to obtain more accurate prevalence and phenotypic data. This novel technique could be a useful method for newborn screening programs and for retrospective and prospective studies on the basis of a single small punch from a DBS or a single cytobrush. This assay design approach might also be considered for the development of assays that target atypical 22q11.2 deletions outside the common deletion regions.
Acknowledgments
We thank Dr. Mary Jenkins (National Center on Birth Defects and Developmental Disabilities) and Karlene Coleman (Children’s Healthcare of Atlanta) for the coordination and receipt of FISH-confirmed paired blood-buccal samples and Dr. Maya Sternberg (National Center for Environmental Health) for her input and statistical analysis of the data.
Supported by intramural funding from the Centers for Disease Control and Prevention.
The findings and conclusions herein are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry.
Footnotes
Disclosures: None declared.
References
- 1.Kobrynski LJ, Sullivan KE. Velocardiofacial syndrome, DiGeorge syndrome: the chromosome 22q11.2 deletion syndromes. Lancet. 2007;370:1443–1452. doi: 10.1016/S0140-6736(07)61601-8. [DOI] [PubMed] [Google Scholar]
- 2.Bales AM, Zaleski CA, McPherson EW. Newborn screening programs: should 22q11 deletion syndrome be added? Genet Med. 2010;12:135–144. doi: 10.1097/GIM.0b013e3181cdeb9a. [DOI] [PubMed] [Google Scholar]
- 3.Carlson C, Sirotkin H, Pandita R, Goldberg R, McKie J, Wadey R, Patanjali SR, Weissman SM, Anyane-Yeboa K, Warburton D, Scambler P, Shprintzen R, Kucherlapati R, Morrow BE. Molecular definition of 22q11 deletions in 151 velo-cardio-facial syndrome patients. Am J Hum Genet. 1997;61:620–629. doi: 10.1086/515508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ou Z, Berg JS, Yonath H, Enciso VB, Miller DT, Picker J, Lenzi T, Keegan CE, Sutton VR, Belmont J, Chinault AC, Lupski JR, Cheung SW, Roeder E, Patel A. Microduplications of 22q11.2 are frequently inherited and are associated with variable phenotypes. Genet Med. 2008;10:267–277. doi: 10.1097/GIM.0b013e31816b64c2. [DOI] [PubMed] [Google Scholar]
- 5.Stachon AC, Baskin B, Smith AC, Shugar A, Cytrynbaum C, Fishman l, Mendoza-Londono R, Klatt R, Teebi A, Ray PN, Weksberg R. Molecular diagnosis of 22q11.2 deletion and duplication by multiplex ligation dependent probe amplification. Am J Med Genet A. 2007;143A:2924–2930. doi: 10.1002/ajmg.a.32101. [DOI] [PubMed] [Google Scholar]
- 6.Fernández L, Lapunzina P, Arjona D, López Pajares I, García-Guereta L, Elorza D, Burgueros M, De Torres ML, Mori MA, Palomares M, García-Alix A, Delicado A. Comparative study of three diagnostic approaches (FISH, STRs and MLPA) in 30 patients with 22q11.2 deletion syndrome. Clin Genet. 2005;68:373–378. doi: 10.1111/j.1399-0004.2005.00493.x. [DOI] [PubMed] [Google Scholar]
- 7.Sørensen KM, Agergaard P, Olesen C, Andersen PS, Larsen LA, Østergaard JR, Schouten JP, Christiansen M. Detecting 22q11.2 deletions by use of multiplex ligation-dependent probe amplification on DNA from neonatal dried blood spot samples. J Mol Diagn. 2010;12:147–151. doi: 10.2353/jmoldx.2010.090099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vorstman JAS, Jalali GR, Rappaport EF, Hacker AM, Scott C, Emanuel BS. MLPA: a rapid, reliable, and sensitive method for detection and analysis of abnormalities of 22q. Hum Mutat. 2006;27:814–821. doi: 10.1002/humu.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kariyazono H, Ohno T, Ihara K, Igarashi H, Joh-o K, Ishikawa S, Hara T. Rapid detection of the 22q11.2 deletion with quantitative real-time PCR. Mol Cell Probes. 2001;15:71–73. doi: 10.1006/mcpr.2000.0340. [DOI] [PubMed] [Google Scholar]
- 10.Chen YF, Kou PL, Tsai SJ, Chen KF, Chan HH, Chen CM, Sun HS. Computational analysis and refinement of sequence structure on chromosome 22q11.2 region: application to the development of quantitative real-time PCR assay for clinical diagnosis. Genomics. 2006;87:290–297. doi: 10.1016/j.ygeno.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Weksberg R, Hughes S, Moldovan L, Bassett AS, Chow EWC, Squire JA. A method for accurate detection of genomic microdeletions using real-time quantitative PCR. BMC Genomics. 2005;6:180. doi: 10.1186/1471-2164-6-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freeman B, Powell J, Ball D, Hill L, Craig I, Plomin R. DNA by mail: an inexpensive and noninvasive method for collecting DNA samples from widely dispersed populations. Behav Genet. 1997;27:251–257. doi: 10.1023/a:1025614231190. [DOI] [PubMed] [Google Scholar]
- 13.Sjöholm MIL, Dillner J, Carlson J. Assessing quality and functionality of DNA from fresh and archival dried blood spots and recommendations for quality control guidelines. Clin Chem. 2007;53:1401–1407. doi: 10.1373/clinchem.2007.087510. [DOI] [PubMed] [Google Scholar]
- 14.Berg LM, Sanders R, Alderborn A. Pyrosequencing™ technology and the need for versatile solutions in molecular clinical research. Expert Rev Mol Diagn. 2002;2:361–369. doi: 10.1586/14737159.2.4.361. [DOI] [PubMed] [Google Scholar]
- 15.Deutsch S, Choudhury U, Merla G, Howald C, Sylvan A, Antonarakis SE. Detection of aneuploidies by paralogous sequence quantification. J Med Genet. 2004;41:908–915. doi: 10.1136/jmg.2004.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Jimenez N, Castellanos-Rubio A, Plaza-Izurieta L, Gutierrez G, Irastorza I, Castaneo L, Vitoria JC, Bilbao JR. Accuracy in copy number calling by qPCR and PRT: a matter of DNA. PLoS One. 2011;6:1–7. doi: 10.1371/journal.pone.0028910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armour JAL, Palla R, Zeeuwen PLJM, den Heijer M, Schalkwijk J, Hollox EJ. Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res. 2007;35:e19. doi: 10.1093/nar/gkl1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beckett SM, Laughton SJ, Pozza LD, McCowage GB, Marshall G, Cohn RJ, Milne E, Ashton LJ. Buccal swabs and treated cards: methodological considerations for molecular epidemiologic studies examining pediatric populations. Am J Epidemiol. 2008;167:1260–1267. doi: 10.1093/aje/kwn012. [DOI] [PubMed] [Google Scholar]
- 19.MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42(Database issue):D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallagher ML, Sturchio C, Smith A, Koontz D, Jenkins MM, Honein MA, Rasmussen SA. Evaluation of mailed pediatric buccal cytobrushes for use in a case-control study of birth defects. Birth Defects Res A Clin Mol Teratol. 2011;91:642–648. doi: 10.1002/bdra.20829. [DOI] [PubMed] [Google Scholar]
- 21.Chen G, Olson MT, O’Neill A, Norris A, Beierl K, Harada S, Debeljak M, Rivera-Roman K, Finley S, Stafford A, Gocke CD, Lin M-T, Eshleman JR. A virtual pyrogram generator to resolve complex pyrosequencing results. J Mol Diagn. 2012;14:149–159. doi: 10.1016/j.jmoldx.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jansson M, Rada A, Tomic L, Larsson LI, Wadelius C. Analysis of the glutathione S-transferase M1 gene using pyrosequencing and multiplex PCR: no evidence of association to glaucoma. Exp Eye Res. 2003;77:239–243. doi: 10.1016/s0014-4835(03)00109-x. [DOI] [PubMed] [Google Scholar]
- 23.Söderbäck E, Zackrisson AL, Lindblom B, Alderborn A. Determination of CYP2D6 gene copy number by pyrosequencing. Clin Chem. 2005;51:522–531. doi: 10.1373/clinchem.2004.043182. [DOI] [PubMed] [Google Scholar]