Abstract
Arsenic (As) tops the ATSDR list of hazardous environmental chemicals and is known to cause liver injury. Although the concentrations of As found in the US water supply are generally too low to directly damage the liver, subhepatotoxic doses of As sensitize the liver to experimental NAFLD. It is now suspected that GI microbiome dysbiosis plays an important role in development of NALFD. Importantly, arsenic has also been shown to alter the microbiome. The purpose of the current study was to test the hypothesis that the prebiotic oligofructose (OFC) protects against enhanced liver injury caused by As in experimental NAFLD. Male C57Bl6/J mice were fed low fat diet (LFD), high fat diet (HFD), or HFD containing oligofructose (OFC) during concomitant exposure to either tap water or As-containing water (4.9 ppm as sodium arsenite) for 10 wks. HFD significantly increased body mass and caused fatty liver injury, as characterized by an increased liver weight-to-body weight ratio, histologic changes and transaminases. As observed previously, As enhanced HFD-induced liver damage, which was characterized by enhanced inflammation. OFC supplementation protected against the enhanced liver damage caused by As in the presence of HFD. Interestingly, arsenic, HFD and OFC all caused unique changes to the gut flora. These data support previous findings that low concentrations of As enhance liver damage caused by high fat diet. Furthermore, these results indicate that these effects of arsenic may be mediated, at least in part, by GI tract dysbiosis and that prebiotic supplementation may confer significant protective effects.
Keywords: liver injury, gut:liver axis, metagenomics
Introduction
Exposure to high concentrations of arsenic is associated with a myriad of health effects including skin lesions, hypertension, cardiovascular disease, respiratory disease, and malignancies of the skin and internal organs (Waalkes et al., 2004). The liver, a major site of arsenic metabolism, is a known target of arsenic toxicity (Santra et al., 1999). Indeed, chronic arsenic exposure causes hepatomegaly, non-cirrhotic portal fibrosis and portal hypertension (Mazumder, 2005; Santra et al., 1999; 2000). The concentrations of arsenic found in the US water supply are generally considered lower than those necessary to cause overt liver damage, however lower concentrations of arsenic can cause more subtle changes in the liver (Straub et al., 2007) and can sensitize the liver to injury by another insult (Arteel et al., 2008). Therefore, although arsenic levels in the US may not be high enough to directly injure the liver, there is evidence that low levels of arsenic may interact with other risk-modifying factors to cause hepatic injury.
Obesity is another major health concern for the US and globally. More than one third of American adults are obese (Ogden et al., 2012), and it is expected that the number of obese individuals in the US will double by 2050 (Fakhouri et al., 2013). Obesity is associated with a myriad of health effects including insulin resistance, diabetes, and non-alcoholic fatty liver disease (NAFLD). NAFLD is a spectrum of disease states including simple steatosis, non-alcoholic steatohepatitis, and fibrosis and cirrhosis. Studies have shown that about 20–30% of the general population has NAFLD (Browning et al., 2004) however the prevalence of NAFLD can more than double in obese cohorts (Bellentani et al., 2000). Despite the high incidence of NALFD in obese populations, only minority of obese individuals ever progress to more severe stages of liver disease, such as non-alcoholic steatohepatitis. Risk factors for primary NAFLD include obesity, type II diabetes and dyslipidemia (Clark, 2006); however it is likely that other unidentified risk factors contribute to progression to more severe stages of liver disease (i.e. NASH).
Interestingly, there is potential overlap in areas of risk for arsenic exposure and obesity. For example, rural communities, which have both artesian water supplies that are unregulated by Clean Water Act, often have high arsenic concentrations; these areas also generally have a high prevalence of obesity. In a study investigating the possible interaction between obesity and arsenic, this group demonstrated that low concentrations of arsenic synergistically enhance liver damage caused by high fat diet (Tan et al., 2011). These data suggest that arsenic may be an underlying factor that modifies the risk of hepatic injury caused by obesity. However, the mechanism(s) underlying this enhancement of NAFLD remain unknown.
The role of the gut:liver axis in liver disease is well recognized. Indeed, liver injury is associated with dysbiosis, disruption of gut barrier integrity, and leakage of inflammatory bacterial-derived components (i.e., endotoxin) from the gut into the systemic circulation. Obesity can affect gut permeability by decreasing the barrier function of the epithelium (Cani et al., 2008). Additionally, both obesity and arsenic have been shown to alter gut flora (Choudhry et al., 2009). Interestingly, probiotics or prebiotics that promote commensal bacteria growth, can preserve intestinal barrier integrity and protect against liver injury in animal models of NAFLD (Li et al., 2003). Recently, arsenic exposure has been shown to also cause dysbiosis in mice (Lu et al., 2014), which could, in principle, exacerbate the effect of HFD. The purpose of the current study was to test this hypothesis and to determine if the prebiotic oligofructose, an oligosaccharide found in vegetables and other plants, can protect against enhanced liver injury caused by arsenic in a mouse model of NAFLD.
Methods
Animals and treatment
Four week old male C57Bl6/J mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were housed in a pathogen-free barrier facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care and procedures were approved by the local Institutional Animal Care and Use Committee. Food and tap water were allowed ad libitum. Mice were fed AIN-76 Purified diet (Harlan Laboratories) for one week to reduce potential confounding factors of arsenic present in standard laboratory chow (Kozul et al., 2008). Mice were exposed to sodium arsenite (4.9 ppm in drinking water) or tap water for one week prior to initiating feeding with either low fat diet (LFD; 13% fat in calories), high fat diet (HFD; 42% fat in calories), or HFD containing oligofructose (OFC; 5% w/w) (Harlan Laboratories, Madison WI) for 10 weeks (Figure 1A). This exposure concentration of arsenic was determined by previous range-finding experiments to cause no overt liver damage in mice fed low-fat diet (Tan et al., 2011). Food and water consumption were measured twice a week. Body weight was measured once a week. At termination of the experiment, mice were anesthetized with ketamine/xylazine (100/15 mg/kg i.m.). Blood was collected from the vena cava just prior to sacrifice by exsanguination and citrated plasma was stored at −80°C until further analysis. Contents of the cecum were removed at sacrifice and snap-frozen in liquid nitrogen. Some portions of liver tissue were snap frozen in liquid nitrogen, while other portions were either fixed in 10% neutral buffered formalin or embedded in frozen specimen medium (Tissue-Tek OCT compound, Sakura Finetek, Torrance, CA) for subsequent sectioning and mounting on microscope slides.
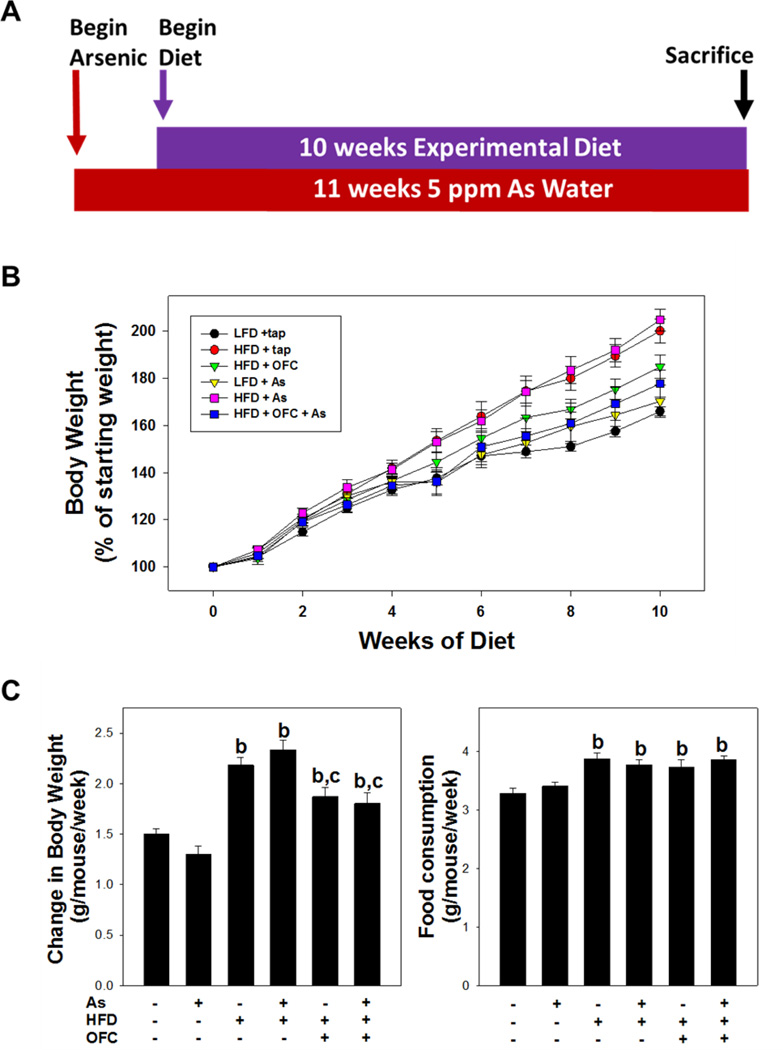
Figure 1. Model parameters.
As shown in panel A, animals were exposed to arsenic (or tap) water for 11 weeks. One week after the start of arsenic exposure experimental diet (e.g. LFD, HFD, or HFD +OFC) was provided as described in Methods. Body weight and food consumption were recorded once per week for the duration of the experiment. Panel B shows change in body weight as a % of starting body weight over the course of the experimental diet feeding. Panel C shows total change in body weight in each experimental group in grams of body weight/mouse/week (left) and food consumption (right). Data are shown as means ± SEM (n = 6–10). a, p < 0.05 compared to tap water control; b, p < 0.05 compared to LFD; c, p < 0.05 compared to no OFC.
Clinical Chemistry and Pathologic Evaluation
The plasma activities of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were determined spectrophotometrically using standard kits (ThermoScientific, Waltham, MA). Formalin fixed, paraffin embedded sections were cut at 5 µm and mounted on glass slides. Sections were deparaffinized and stained with hematoxylin and eosin (H&E).
Lipid determination
Total lipid was extracted from mouse livers as previously described (Tan et al., 2011). Briefly, mouse liver was first homogenized in 2× phosphate buffered saline. Hepatic lipids were extracted from pulverized liver tissue with methanol:chloroform (2:1), dried in an evaporating centrifuge, and resuspended in 5% fat-free bovine serum albumin. Hepatic triglyceride (Thermo Scientific, Middletown, VA) non-esterified fatty acids (Sigma Aldrich, St. Louis MO) were determined using standard kits. Values were normalized to initial liver tissue weight.
Immunostaining
Immunohistochemical staining for the marker F4/80 was performed as previously described (Tan et al., 2011). Immunofluorescent detection of fibrin deposition was performed as described in (Beier et al., 2008). Bright field staining and fluorescence for Alexa 488 (490ex, 520em nm) and Hoechst (350ex, 460em nm) were visualized using a Nikon Eclipse E600 microscope (Nikon Corporation, Tokyo, Japan). Metamorph software (Molecular Devices, Sunnyvale, CA) was used to acquire photomicrographs and to quantitate number of positive cells per 1000 hepatocytes (F4/80 staining) or total fluorescent signal per field (fibrin staining)(Tan et al., 2011).
RNA Isolation and Quantitative Reverse-Transcriptase Polymerase Chain Reaction
The hepatic expression of select genes was detected by quantitative reverse-transcriptase polymerase chain reaction (rt-PCR), which is routine for this group (Tan et al., 2011). PCR primers and probes for TNFα, PAI-1 and β-actin were used as described previously (Tan et al., 2011). Total RNA was extracted from liver tissue by a guanidinium thiocyanate-based method (RNA STAT 60 Tel-Test, Ambion, Austin, TX). RNA concentrations were determined spectrophotometrically and 1 µg of total RNA was reverse transcribed using a kit (Quanta Biosciences, Gaithersburg, MD). PerfeCta qPCR Fast Mix (Quanta Biosciences, Gaithersburg, MD) was used to prepare the PCR reaction mixture. Amplification reactions were carried out using the ABI StepOne Plus machine and software (Quanta Biosciences, Gaithersburg, MD). The comparative CT method (2−ΔΔCt) was used to determine fold changes in mRNA expression compared to an endogenous reference gene (β-actin).
Bacterial DNA extraction and deep sequencing of the cecal content
DNA was extracted from frozen cecal pellet using the QIAamp DNA Stool Mini Kit, according to the manufacturer’s directions. Deep sequencing of bacterial 16S rDNA regions was performed with 454 FLX Titanium sequencing using previously established protocols (Bull-Otterson et al., 2013). The V3–V5 16S rDNA gene variable regions were amplified by PCR using 454 adapter-linked bar-coded primers 375F and 926R as previously described (Jumpstart Consortium Human Microbiome Project Data Generation Working Group., 2012). Amplicons were purified using a SPRI bead clean-up step, quantitated by picogreen assay, normalized, pooled, and then sequenced on a 454 instrument using the FLX Titanium chemistry.
The raw sequencing dataset, consisting of an average of 28,590 reads per sample with an average read length of 253 nucleotides, was deconvoluted via the PyNAST alignment tool (Caporaso et al., 2010) using the Greengenes reference database as described by (DeSantis et al., 2006); the database employed was released in August 2013. Quality filtering was performed filtered with cutoff parameters of: less than 200 and greater than 1,000 bp, quality score below 30, number of mismatches in primer greater than 0, number of ambiguous bases greater than 6, and maximum homopolymer runs greater than 6. Following quality filtering, 514,622 sequences remained from the original 531,138 with an average read length of 253 nucleotides. Chimeric sequences were removed by reference and de novo chimeric filtering with UCHIME (Edgar et al., 2011) utilizing the Greengenes reference database released in August 2013 (DeSantis et al., 2006). Operational taxonomic unit (OTU) tables based on sequences within the filtered data set were then generated by taxonomic binning via the Quantitative Insights Into Microbial Ecology tool (Qiime) and its various components (Edgar et al., 2011). Open reference OTU picking was performed via Usearch 6.1 software using 97% similarity to the clustered Greengenes database as described in (McDonald et al., 2012); the database used was released in August 2013. Singleton OTUs were removed from the OTU table as further quality filtering to improve specificity. The taxonomic classifications of sequence reads were identified from the phylum to the species level by the Uclust based consensus taxonomy classifier with phylogenetic tree creation by Fast Tree (Price et al., 2010).
For diversity measures, an even sequence sampling depth of 16,000 sequences was used to avoid artifacts due to differing numbers of sequences per sample. Briefly, alpha (α) diversity was used as an index of diversity within a sample and was determined by counting the number of OTUs found in multiple samplings of bacterial DNA sequences. During the first sampling, all OTUs identified were counted towards the number of observed OTUs; for each additional sampling, only unique OTUs were counted and added to the number of observed OTUs for that sample. This process was repeated until a total of 16,000 sequences are sampled. Principle coordinate analyses (PCoA) were performed to compare microbial community structure between samples utilizing weighted UniFrac measures (Lozupone and Knight, 2005). Linear effect size Linear Discriminant Analysis Effect Size (LefSe) analyses were performed using the freely distributed Galaxy efSe analysis module hosted at huttenhower.sph.harvard.edu/lefse/ (Segata et al., 2011). Hypothesis testing using the Dirichlet-Multinomial distribution was performed via the HMP package for R (La Rosa et al., 2012) within the R Statistical Computing Environment (Dean and Nielsen, 2007).
Statistical Analysis
Results are reported as means +/− SEM (n = 6–10). ANOVA with Bonferroni post-hoc test (for parametric data) or Mann-Whitney Rank Sum (for nonparametric data) were used for the determination of statistical significance among treated groups, as appropriate. A p value of less than 0.05 was selected before the study as the level of significance.
Results
Body and organ weight
All animals gained weight and there was no mortality or morbidity in any group during the course of the study. As observed previously (Tan et al., 2011), arsenic did not significantly affect food consumption or body mass gain in animals fed either low fat diet (LFD) or high fat diet (HFD) (Figure 1A and B). HFD feeding significantly increased food consumption ~20% and body mass gain ~50% compared to LFD alone (Figure 1C). Whereas oligofructose (OFC) supplementation did not affect food consumption, it did significantly attenuate the increase in body mass gain caused by HFD; this effect of OFC was independent of the presence of arsenic in the diet.
Liver Injury
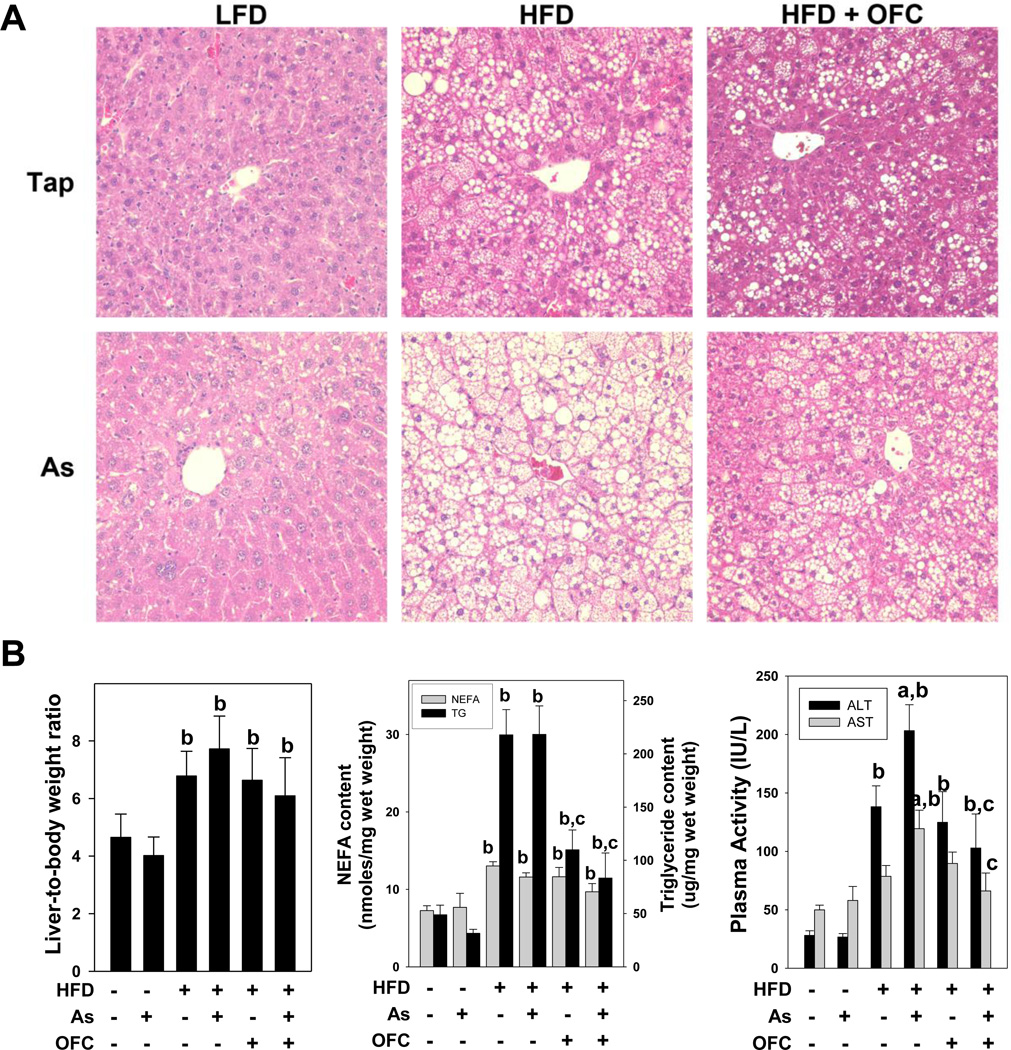
HFD significantly increased the liver-to-body weight ratio compared to low fat diet (Figure 2B). Liver weight to body weight percent was not significantly affected by concomitant arsenic exposure or by OFC feeding. As expected by the hepatomegaly observed, 10 weeks of HFD feeding dramatically increased lipid accumulation in the liver (Figure 2A); this pathologic change consisted of both macrovesicular and microvesicular steatosis. Changes in quantity of hepatic triglycerides and non-esterified fatty acids (NEFA) (Figure 2B) were also determined. HFD alone increased hepatic triglyceride and NEFA content compared to LFD alone by more than 4 fold. Arsenic administration did not affect steatosis or hepatic triglyceride or NEFA content in either LFD- or HFD-fed animals. OFC administration blunted fat accumulation and significantly decreased triglyceride content compared to animals that received either HFD or arsenic + HFD without OFC. Interestingly, OFC had no significant effect on NEFA content in the liver.
Figure 2. OFC protects against liver injury and steatosis.
Animals and exposures to high fat diet (HFD), arsenic (As), and the prebiotic oligofructose (OFC) were as described in the Methods. Panel A shows representative photomicrographs depicting hematoxylin & eosin staining (H&E, 200×; top left) of liver tissue. The left graph of panel B shows liver-to-body weight ratio (LW/BW%; left), which was determined as described in Methods. The middle graph shows hepatic non-esterified fatty acid (NEFA, gray bars) and triglyceride (TG, black bars) content which were determined using kits as described in Methods. The right graph shows plasma activity of alanine aminotransferase (ALT; black bars) and aspartate aminotransferase (AST; gray bars), which were used an index of liver injury and were measured as described in Methods. Quantitative data are shown as means ± SEM (n = 6–10). a, p < 0.05 compared to tap water control; b, p < 0.05 compared to LFD; c, p < 0.05 compared to no OFC.
HFD feeding also increased the appearance of necroinflammatory foci in the liver (Figure 2A). Circulating levels of the liver enzymes alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were determined as indices of liver damage (Figure 2B). HFD alone significantly increased ALT activity, but did not significantly affect AST activity. Whereas arsenic did not significantly affect plasma activities of either ALT or AST in the LFD group, it significantly enhanced the increase in these variables caused by HFD under these conditions by ~50%. The addition of OFC to animals receiving HFD did not affect the transaminase activities in the absence of arsenic. However, OFC administration completely blunted the increase in both ALT and AST caused by the combined exposure of arsenic and HFD.
Inflammation
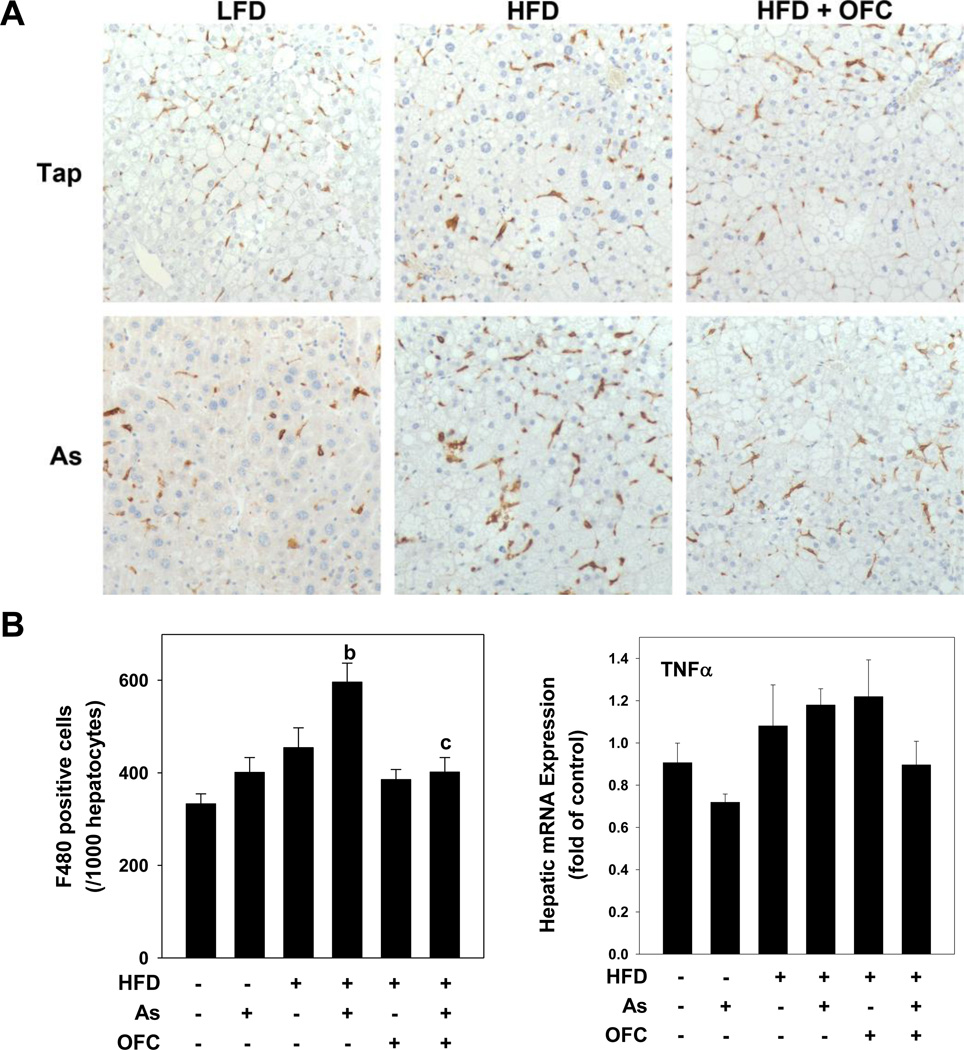
Immunohistochemistry for F4/80, a marker of macrophages, was performed on liver tissue as an index of monocytic inflammation (Figure 3A). Neither arsenic nor HFD alone altered the number of F4/80 positive macrophages; however the combination of arsenic + HFD significantly increased the number of hepatic macrophages compared to arsenic or HFD alone, as was observed previously (Tan et al., 2011). The administration of OFC significantly blunted the increase in number of activated macrophages caused by the combination of arsenic and HFD and values were similar to those of LFD controls. Hepatic mRNA expression of TNFα (Figure 3B) was not significantly changed by either arsenic or HFD alone.
Figure 3. OFC protects against hepatic inflammation.
Animals and exposures to high fat diet (HFD), arsenic (As), and the prebiotic oligofructose (OFC) were as described in the Methods. Panel A shows representative photomicrographs of immunohistochemical staining for F4/80 (200×) in which hepatic macrophages are stained brown. The left graph of panel B shows quantitation of F4/80 staining, which was determined by blinded counting of the number of F4/80 positive cells per 1000 hepatoctyes as described in Methods. The right graph of panel B shows real time RT-PCR for the pro-inflammatory gene TNFα, which was performed as described in Methods. Quantitative data are shown as means ± SEM (n = 6–10). a, p < 0.05 compared to tap water control; b, p < 0.05 compared to LFD; c, p < 0.05 compared to no OFC.
Fibrin/PAI-1
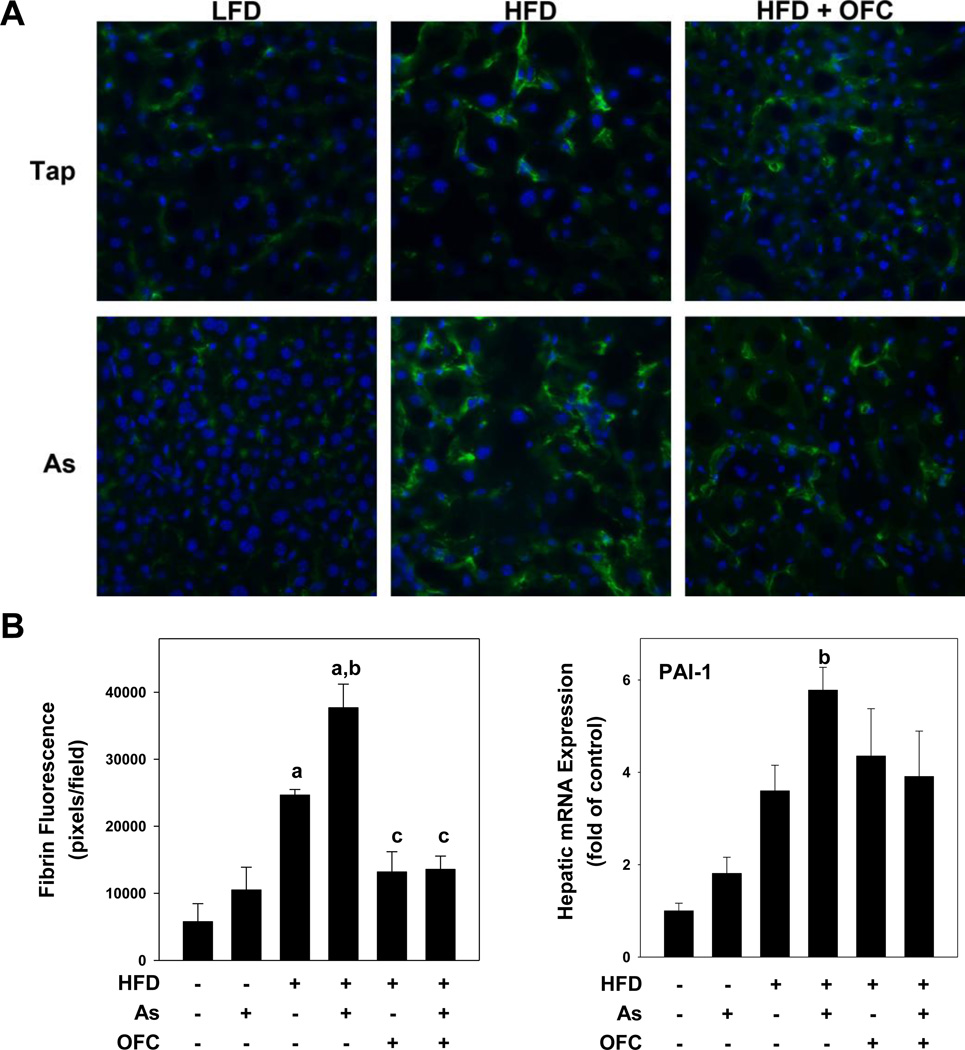
Previously, the enhancement of HFD-induced liver injury caused by arsenic correlated with a significant increase in hepatic expression of PAI-1 (Tan et al., 2011). PAI-1 is a major inhibitor of both tissue-type plasminogen activator (tPA) and urokinase-type plasminogen activator (uPA), and is therefore a key inhibitor of fibrin degradation (i.e., fibrinolysis) by plasmin. Fibrin deposition was determined by immunofluorescent staining and quantitation of fluorescent signal. LFD-fed animals given either tap or arsenic-containing water had minimal fibrin deposition in the liver (Figure 4A). HFD alone increased deposition of fibrin by ~2-fold. As observed previously (Tan et al., 2011), the combination of HFD + arsenic significantly increased the expression of PAI-1 and fibrin deposition in liver (Figure 4B). OFC administration almost completely prevented the changes caused by arsenic under these conditions.
Figure 4. OFC protects against sinusoidal fibrin accumulation.
Animals and exposures to high fat diet (HFD), arsenic (As), and the prebiotic oligofructose (OFC) were as described in the Methods. Panel A shows representative photomicrographs (400×) of immunofluorescent staining for fibrin in which fibrin is stained with FITC (green) and cell nuclei are stained with DAPI (blue). Panel B shows quantitation of fibrin immunofluorescence which was determined using Metamorph software as described in Methods. Hepatic PAI-1 mRNA expression was determined using real time RT-PCR as described in Methods. Quantitative data are shown as means ± SEM (n = 6–10). a, p < 0.05 compared to tap water control; b, p < 0.05 compared to LFD; c, p < 0.05 compared to no OFC.
Effects of HFD, arsenic, and OFC on the cecal microbiome
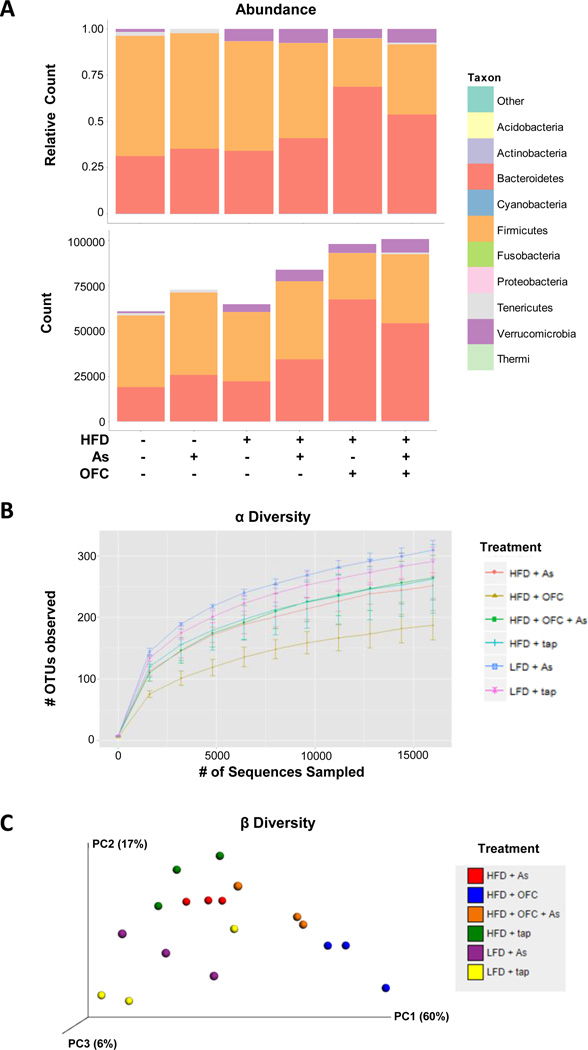
The effects of arsenic, HFD, and OFC on relative abundance and total abundance of bacteria was determined at the phylum level (Figure 5A). As expected, the most abundance bacteria phyla were Bacteroidetes and Firmicutes. Arsenic exposure alone caused a loss of the Verrucobiorobia in animals that were fed a LFD. Interestingly, this effect was lost in animals that were fed a HFD; however, the combination of arsenic and HFD did lead to an increase in the gram-negative phylum Bacteroidetes. Oligofructose feeding increased the relative abundance of Bacteroidetes with a concomitant decrease in Firmicutes compared to all other groups that did not receive the prebiotic. Additionally, OFC restored the growth of Tenericutes which was lost in animals that received a HFD. OFC also resulted in a robust increase in total bacteria compared to animals that did not receive OFC.
Figure 5. Effects of HFD, arsenic, and OFC on bacterial abundance and diversity.
Animals and exposures to high fat diet (HFD), arsenic (As), and the prebiotic oligofructose (OFC) were as described in the Methods. Bacterial DNA in the cecal content was isolated and sequenced as described in Methods. Panel A shows the relative abundance (top) and total count (bottom) of bacteria at the phyla level for each experimental group. Total count (bottom) was determined by averaging the total number of OTUs found in each bacterial phyla per group. Panel B shows alpha diversity, a measure of unique OTUs present in a given sample. Panel C shows beta diversity which serves as a measure of diversity between samples. Samples that are closer in proximity have microbiomes that are more similar in term of phylogenetic diversity and OTU abundance.
Alpha (α) diversity (Figure 5B) is shown as an index of diversity within a sample and was determined by counting the number of OTUs found in multiple samplings of bacterial DNA sequences. Interestingly, the greatest α diversity was observed in animals exposed to a LFD. HFD feeding reduced α diversity and the combination of HFD and OFC resulted in the lowest α diversity of all six groups. β diversity was used as a measure of diversity between samples (Figure 5C). As expected, samples that represent a single treatment group clustered more closely together, suggested a higher degree of similarity among the samples.
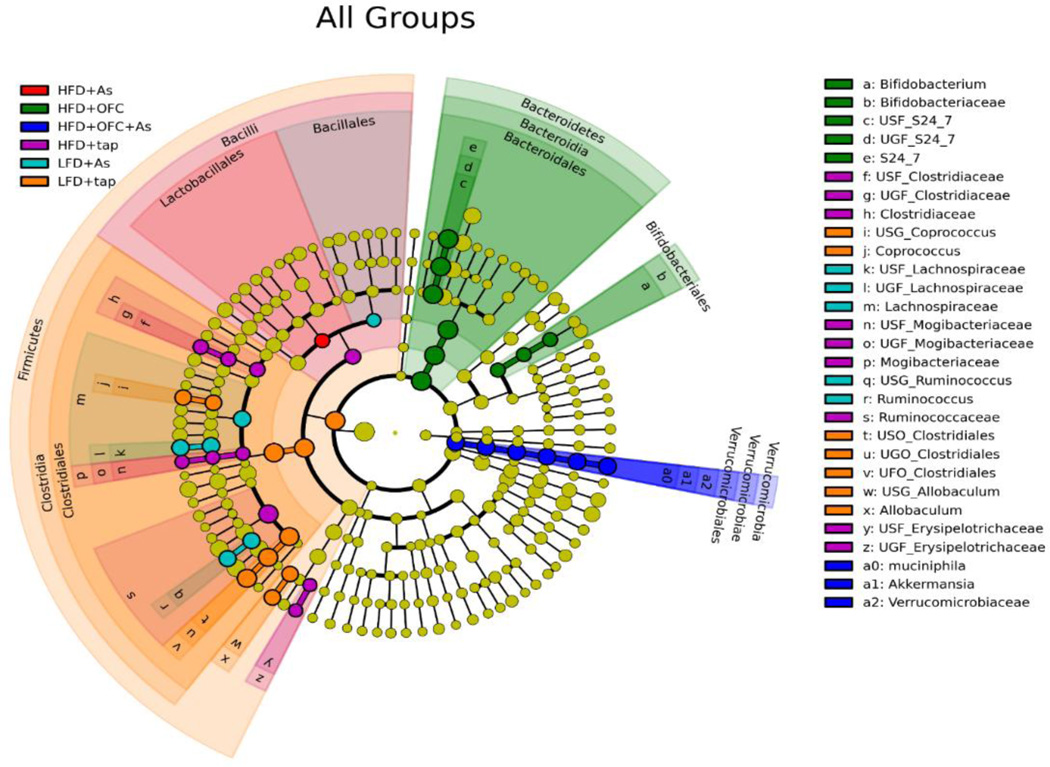
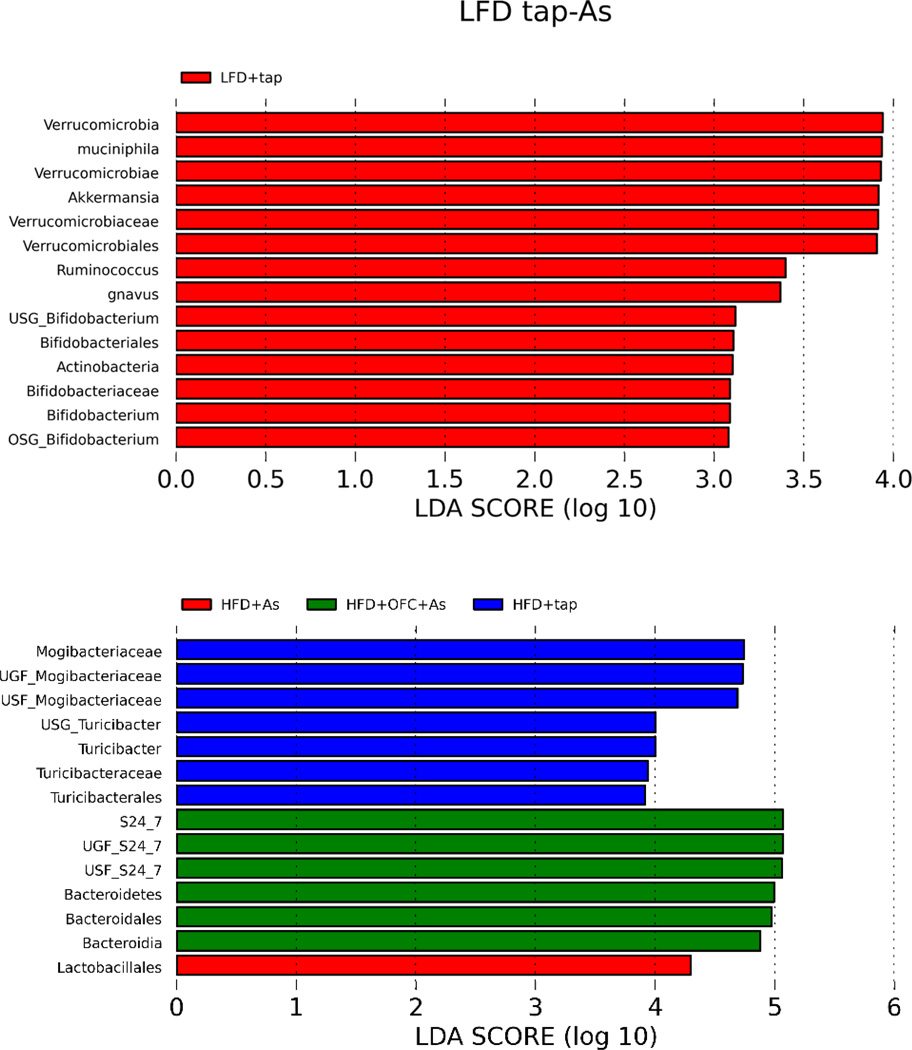
Linear discriminant analysis effect size (LefSe) was used to identify the enrichment of bacteria within treatment groups. When all six treatment groups were compared, unique enrichment profiles were observed for each group (Figure 6). In a LefSe comparison between the LFD and LFD + arsenic, enrichment was primarily seen in animals that received tap water, suggesting that arsenic exposure resulted in decreases in these bacteria including the commensal Bifidobacterium (Figure 7). Comparisons between animals fed HFD with or without arsenic and OFC showed that arsenic exposure was associated with a decrease in Mogibacteriacaceae and Turicibacter (Figure 7).
Figure 6. HFD, arsenic, and OFC exposure caused unique changes to the microbiome.
Animals and exposures to high fat diet (HFD), arsenic (As), and the prebiotic oligofructose (OFC) were as described in the Methods. Bacterial DNA in the cecal content was isolated and sequenced as described in Methods. Linear Discriminant Analysis Effect Size (LefSe) was used to determined enrichment of bacterial taxa between groups. Effect size was estimated using linear discriminant analysis (LDA). Panel A shows LefSe analysis of all six treatment groups, which is plotted on a cladogram. Each treatment group is represented by a unique color; highlighted taxa indicate enrichment of that taxa within the treatment group that corresponds to the highlighting color. Effect size is represented by the size of the circular intersection points between the taxa.
Figure 7. LefSe analysis of microbiome changes.
Panel A shows a bar plot of LefSe analysis comparing the gut microbiomes of animals fed either LFD or a LFD and arsenic. Each bar represents a bacterial taxa that was enriched by LFD. The length of the bar represents the effect size. Panel B shows a bar plot of LefSe analysis comparing the gut microbiomes of animals fed either HFD (blue), HFD and arsenic (red), or HFD, arsenic, and OFC (green). The length of the bar represents effect size.
Discussion
The GI tract is home to up to 100 trillion gut microbes including bacteria and viruses (Ley et al., 2005). The microbes residing in the gut influence many important physiological functions, including metabolic processing, energy production, and immune system development (Ley et al., 2005). Therefore, it is not surprising that dysbiosis of the gut flora is associated with a myriad of disease states locally in the GI tract, including diarrhea/malabsorption (Keeney et al., 2014) and irritable bowel disease (Sokol et al., 2006). Because of the critical importance of the microbiome in immune surveillance and tolerance, changes in the gut microbiome are also linked to autoimmune diseases and allergies (Ochoa-Reparaz et al., 2009; West, 2014). Additionally, the gut microbiome can contribute to diseases of distal organ systems, including the CNS which is negatively affected by gut dysbiosis (Bested et al., 2013).
A major mechanism by which gut dysbiosis may contribute to systemic disease is via changing/impairing the GI tract barrier function. For example, rodent models of obesity have been associated with altered gut microbiota that favor gut leakiness and endotoxemia (Brun et al., 2007; Cani et al., 2007; 2008). Furthermore, dysbiosis associated with obesity contributes to the development of inflammation (Cani et al., 2007; 2008) metabolic syndrome (Cani and Delzenne, 2010), and non-alcoholic fatty liver injury (Brun et al., 2007). The understanding that the GI tract contributes to liver injury (i.e., the gut-liver axis) has been understood for over a century and was first shown experimentally more than 60 years ago by Rutenberg et al, when they demonstrated that nonabsorbable antibiotics protected against diet-induced liver injury (Rutenburg et al., 1957). Since then, many studies using a myriad of models including antibiotics (Adachi et al., 1995), and germ-free mouse models (Luckey et al., 1954) have provided additional evidence implicating the microbiome in the susceptibility and development of liver disease. Today, the gut-liver axis and enteric dysbiosis, including that caused by a HFD, is a generally accepted component in the development of liver disease.
Although the role of arsenic in altering the microbiome is less well studied, arsenic is known to alter the microbial components of soil (Sheik et al., 2012). A recent study by Lu et al has shown that high concentrations of arsenic can cause dysbiosis in mice (Lu et al., 2014). In their study, metagenomic analysis of the gut flora of mice exposed to 50 ppm arsenic showed that arsenic significantly altered the abundance of some microbial families and also resulted in functional changes in the microbiome’s metabolic profile (Lu et al., 2014). The changes in the gut flora caused by arsenic in the current study were more subtle than those seen by Lu and colleagues; this difference may be attributed, in part, to the >10-fold lower arsenic concentrations used in the current study. Nevertheless, arsenic exposure here did change the mouse microbiome in the current study; importantly, arsenic further exacerbated the dysbiosis caused by HFD feeding in mice. Importantly, supplementation with oligofructose, a prebiotic that has been demonstrated to protect against dysbiosis and favor commensal bacteria, almost completely prevented the enhancement of HFD-induced liver injury caused by arsenic. These results therefore support the hypothesis that arsenic mediates liver injury under these conditions via mechanisms that involve the gut-liver axis.
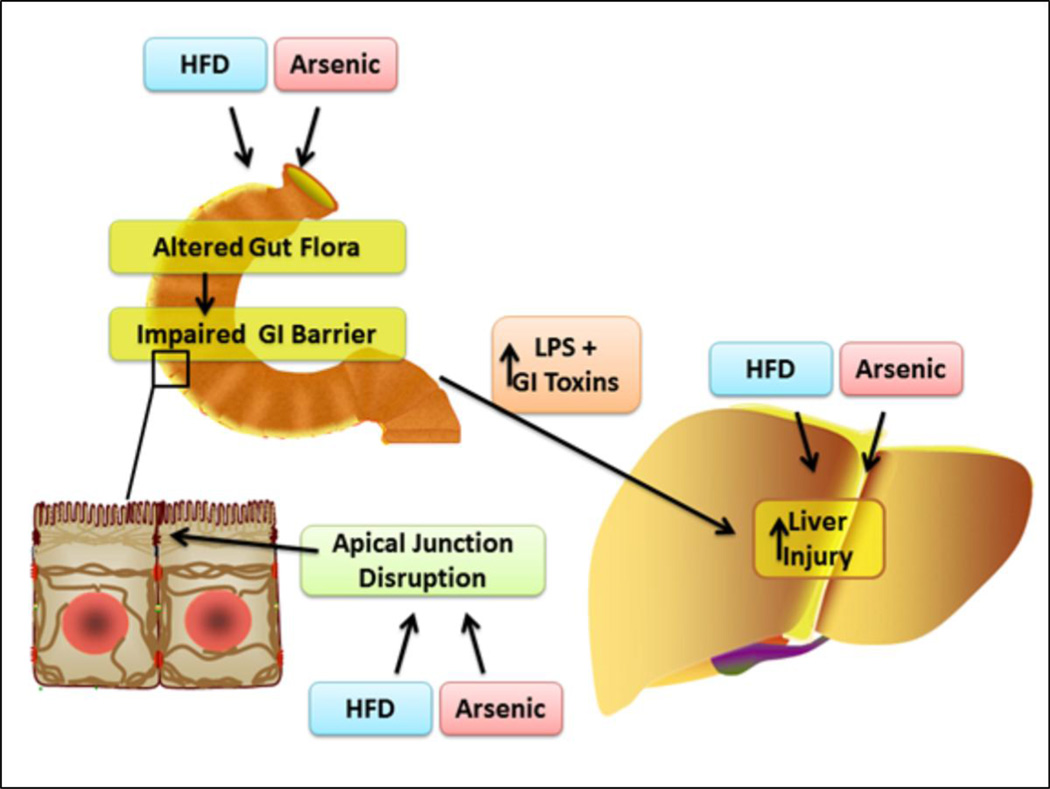
There are several potential mechanisms by which the microbiome may mediate arsenic-induced liver injury (Figure 8). First, the changes in the microbiome caused by diet (HFD and OFC) may affect arsenic metabolism and absorption. Indeed, interactions between gut microbes and arsenic are bidirectional. For instance, Pinyayev et al showed that bacteria from the mouse cecum can metabolize arsenic (Pinyayev et al., 2011) and others have shown that the human gut flora can metabolize arsenic into toxic metabolites (Rubin SS et al., 2014; Van de Wiele et al., 2010). Whereas this is distinctly possible, it should be noted that concentrations that are 10-fold higher than used here were not directly hepatotoxic (Arteel et al., 2008). Alternatively, the effects of arsenic on the microbiome may mediate the observed hepatotoxicity. Indeed, arsenic exposure expanded the population of Gram (−) bacteria that could produce endotoxins, which are well known for contributing to fatty liver injury. However, whether the effects are driven by microbiome changes on arsenic levels, or by arsenic changes on the microbiome, these changes may potentiate arsenic toxicity locally on the gut epithelium (Laparra et al., 2006) and contribute to enhanced arsenic toxicity distally in the liver.
Figure 8. Scheme of the working hypothesis.
Previous studies suggest that arsenic toxicity may be a hidden risk factor for developing severe fatty liver disease; indeed this group has shown that arsenic enhances liver injury in a model of NASH (6). Here we propose that the mechanisms underlying enhanced hepatic injury involve changes in both the gut and liver. We propose that HFD and arsenic favor the production of GI tract toxins (e.g. LPS) and increase gut leakiness by altering the GI tract flora. We further propose that HFD and arsenic sensitize the liver to toxic injury caused by gut-derived toxins. Prebiotics have been shown to protect against changes in gut flora and permeability caused by high fat diet (8). Therefore, the aim of this study was to test the hypothesis that the prebiotic oligofructose can protect against hepatic injury caused by HFD and arsenic.
The results of LEfSe analysis (Figures 6 and 7) may yield insight into the impact of arsenic and OFC supplementation on liver injury under these conditions (Segata et al., 2011). LEfSe combines statistical significance with additional tests for biological consistency and effect relevance, and determines the metagenomic features that most likely explain differences between the treatment groups. The output are “biomarkers” that drive the assumed differences between the experimental groups. First, the “biomarker” of an increase in Turibacterales in the HFD group validates previous findings (Serino et al., 2012); importantly, the changes in this population correlated with insulin resistance in that study. Furthermore, decreases in Verrucomicrobia (Akkermansia) and Actinobacteria (Bifidobacteria) are “biomarkers” for the effect of arsenic in LFD-fed mice. Both commensal bacterial populations are associated with protective effects against glucose intolerance, inflammation and/or metabolic endotoxemia (Shin et al., 2014; Everard et al., 2013; Cani et al., 2007) and their loss may contribute to effect of arsenic under these conditions. The main “biomarker” in the HFD As +OFC group was in the Bacteroidetes spp, a population which is known to be increased by prebiotic fibers (Serino et al., 2012) and correlates with protection against metabolic effects of HFD feeding in mice. It is likely that these changes in bacterial populations drives, at least in part, the interaction between As and HFD, as well as the protection by OFC under these conditions (Figures 1–4).
In addition to direct effects on the microbiome, the protective effect of OFC feeding under these conditions could also be ‘upstream’ of arsenic. Specifically, OFC feeding significantly attenuated the increase in hepatic lipids caused by HFD, both in the presence and absence of arsenic, which is in-line with previous findings [e.g., (Serino et al., 2012)]. The multi-hit hypothesis of fatty liver disease involves not only an increase in gut-derived products that damage the liver, but also an increased sensitivity to the damage caused by these gut-derived products. Indeed, fatty livers are more sensitive to a second toxic ‘hit,’ including gut-derived LPS (Harmon et al., 2011). Therefore, the favorable effects of OFC on hepatic steatosis may indirectly protect against the injury caused by HFD and/or arsenic by blunting this sensitization. In support of this hypothesis, enhanced liver injury caused by arsenic + HFD co-exposure was not due to an increase in steatosis per se, but rather due to an enhanced hepatic inflammatory response. Therefore, OFC may also be protecting against damage in this model by preventing hepatic sensitization caused by HFD-induced endotoxemia. Such a mechanism is supported by previous studies that arsenic-exposed livers are more sensitive to exogenous LPS (Arteel et al., 2008) and that liver injury here in OFC-fed mice was insensitive to arsenic administration.
In conclusion, protection against enhanced liver injury conferred by OFC is likely due to both direct effects on the microbiome, as well indirect effects on hepatic sensitization. These findings are in-line with previous work, in which OFC supplementation was shown to protect against direct effects of HFD on the gut microbiome and barrier function (Cani et al., 2007). Prebiotics favor an environment in which commensal bacteria can thrive, in lieu of direct delivery of commensal bacteria (i.e., probiotics). As such, this approach may yield more predictable and stable results. Nevertheless, these results serve proof-of-principle that protecting against microbiome changes caused by arsenic can prevent the enhancement of inflammatory liver injury caused by this environmental contaminant.
Highlights.
Arsenic (As) enhances liver damage caused by a high-fat (HFD) diet in mice.
Oligofructose protects against As-enhanced liver damage caused by HFD.
As causes dysbiosis in the GI tract and exacerbates the dysbiosis caused by HFD.
OFC prevents the dysbiosis caused by HFD and As, increasing commensal bacteria.
Acknowledgements
This work was supported, in part, by a grants from the National Institute of Environmental Health Sciences (ES016367). VLM and RHS were supported by a predoctoral fellowship (ES011564).
Abbreviation list
- ALT
alanine aminotransferase
- As
arsenic
- AST
aspartate aminotransferase
- GI
gastrointestinal tract
- HFD
high fat diet
- LefSe
linear discriminant analysis effect size
- LFD
low fat diet
- LPS
lipopolysaccharide
- NAFLD
non-alcoholic fatty liver disease
- NEFA
non-esterified fatty acids
- OFC
oligofructose
- OTU
operational taxonomic unit
- PAI-1
Plasminogen activator inhibitor 1
- PCoA
principle coordinate analysis
- QIIME
Quantitative Insights Into Microbial Ecology tool
- TG
triglycerides
- TNFα
tumor necrosis factor alpha
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Adachi Y, Moore LE, Bradford BU, Gao W, Thurman RG. Antibiotics prevent liver injury in rats following long-term exposure to ethanol. Gastroenterology. 1995;108:218–224. doi: 10.1016/0016-5085(95)90027-6. [DOI] [PubMed] [Google Scholar]
- Arteel GE, Guo L, Schlierf T, Beier JI, Kaiser JP, Chen TS, Liu M, Conklin DJ, Miller HL, von Montfort C, States JC. Subhepatotoxic exposure to arsenic enhances lipopolysaccharide-induced liver injury in mice. Toxicol. Appl. Pharmacol. 2008;226:128–139. doi: 10.1016/j.taap.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier JI, Guo L, von Montfort C, Kaiser JP, Joshi-Barve S, Arteel GE. New role of resistin in lipopolysaccharide-induced liver damage in mice. J. Pharmacol. Exp. Ther. 2008;325:801–808. doi: 10.1124/jpet.108.136721.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellentani S, Saccoccio G, Masutti F, Croce L, Brandi G, Sasso F, Cristanini G, Tiribelli C. Prevalence of and risk factors for hepatic steatosis in Northern Italy. Ann Intern Med. 2000;132:112–117. doi: 10.7326/0003-4819-132-2-200001180-00004. [DOI] [PubMed] [Google Scholar]
- Bested AC, Logan AC, Selhub EM. Intestinal microbiota, probiotics and mental health: from Metchnikoff to modern advances: Part I - autointoxication revisited. Gut Pathog. 2013;5:5. doi: 10.1186/1757-4749-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, Grundy SM, Hobbs HH. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- Brun P, Castagliuolo I, Di Leo V, Buda A, Pinzani M, Palu G, Martines D. Increased intestinal permeability in obese mice: new evidence in the pathogenesis of nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2007;292:G518–G525. doi: 10.1152/ajpgi.00024.2006. [DOI] [PubMed] [Google Scholar]
- Bull-Otterson L, Feng W, Kirpich I, Wang Y, Qin X, Liu Y, Gobejishvili L, Joshi-Barve S, Ayvaz T, Petrosino J, Kong M, Barker D, McClain C, Barve S. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One. 2013;8:e53028. doi: 10.1371/journal.pone.0053028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- Cani PD, Delzenne NM. Involvement of the gut microbiota in the development of low grade inflammation associated with obesity: focus on this neglected partner. Acta Gastroenterol. Belg. 2010;73:267–269. [PubMed] [Google Scholar]
- Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, Gibson GR, Delzenne NM. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry ZK, Misbahuddin M, Hosain AK, Saleh AA. Inhibitory effect of arsenic on aerobic gut flora in rat. Bangladesh Med. Res. Counc. Bull. 2009;35:79–83. [PubMed] [Google Scholar]
- Clark JM. The epidemiology of nonalcoholic fatty liver disease in adults. J. Clin. Gastroenterol. 2006;40:S5–S10. doi: 10.1097/01.mcg.0000168638.84840.ff. [DOI] [PubMed] [Google Scholar]
- Dean CB, Nielsen JD. Generalized linear mixed models: a review and some extensions. Lifetime. Data Anal. 2007;13:497–512. doi: 10.1007/s10985-007-9065-x. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhouri TH, Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of obesity among older adults in the United States, 2007–2010. NCHS Data Brief. 2013;106:1–8. [PubMed] [Google Scholar]
- Harmon RC, Tiniakos DG, Argo CK. Inflammation in nonalcoholic steatohepatitis. Expert. Rev. Gastroenterol. Hepatol. 2011;5:189–200. doi: 10.1586/egh.11.21. [DOI] [PubMed] [Google Scholar]
- Jumpstart Consortium Human Microbiome Project Data Generation Working Group. Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One. 2012;7:e39315. doi: 10.1371/journal.pone.0039315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney KM, Yurist-Doutsch S, Arrieta MC, Finlay BB. Effects of antibiotics on human microbiota and subsequent disease. Annu. Rev. Microbiol. 2014;68:217–235. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- Kozul CD, Nomikos AP, Hampton TH, Warnke LA, Gosse JA, Davey JC, Thorpe JE, Jackson BP, Ihnat MA, Hamilton JW. Laboratory diet profoundly alters gene expression and confounds genomic analysis in mouse liver and lung. Chem. Biol. Interact. 2008;173:129–140. doi: 10.1016/j.cbi.2008.02.008. [DOI] [PubMed] [Google Scholar]
- La Rosa PS, Brooks JP, Deych E, Boone EL, Edwards DJ, Wang Q, Sodergren E, Weinstock G, Shannon WD. Hypothesis testing and power calculations for taxonomic-based human microbiome data. PLoS One. 2012;7:e52078. doi: 10.1371/journal.pone.0052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laparra JM, Velez D, Barbera R, Granero L, Polache A, Montoro R, Farre R. Cytotoxic effect of As(III) in Caco-2 cells and evaluation of its human intestinal permeability. Toxicol. In Vitro. 2006;20:658–663. doi: 10.1016/j.tiv.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yang S, Lin H, Huang J, Watkins PA, Moser AB, Desimone C, Song XY, Diehl AM. Probiotics and antibodies to TNF inhibit inflammatory activity and improve nonalcoholic fatty liver disease. Hepatology. 2003;37(2):343–350. doi: 10.1053/jhep.2003.50048. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Abo RP, Schlieper KA, Graffam ME, Levine S, Wishnok JS, Swenberg JA, Tannenbaum SR, Fox JG. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environmental Health Perspectives. 2014;122:284–291. doi: 10.1289/ehp.1307429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckey TD, Reyneirs JA, Gyorgy P, Forbes M. Germfree animals and liver necrosis. Ann N. Y. Acad Sci. 1954;57:932–935. doi: 10.1111/j.1749-6632.1954.tb36472.x. [DOI] [PubMed] [Google Scholar]
- Mazumder DN. Effect of chronic intake of arsenic-contaminated water on liver. Toxicol. Appl. Pharmacol. 2005;206:169–175. doi: 10.1016/j.taap.2004.08.025. [DOI] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME. J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Foureau DM, Haque-Begum S, Kasper LH. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J Immunol. 2009;183:6041–6050. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, Kit BK, Flegal KM. NCHS data brief. Vol. 82. National Center for Health Statistics; 2012. Prevalence of obesity in the United States, 2009–2010. [PubMed] [Google Scholar]
- Pinyayev TS, Kohan MJ, Herbin-Davis K, Creed JT, Thomas DJ. Preabsorptive metabolism of sodium arsenate by anaerobic microbiota of mouse cecum forms a variety of methylated and thiolated arsenicals. Chem. Res Toxicol. 2011;24:475–477. doi: 10.1021/tx200040w. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin SS, D C, Alava P, Zekker I, Du LG, Van de Wiele T. Arsenic thiolation and the role of sulfate-reducing bacteria from the human intestinal tract. Environmental Health Perspectives. 2014;122:817–822. doi: 10.1289/ehp.1307759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutenburg AM, Sonnenblick E, Koven I, Aprahamian HA, Reiner L, Fine J. The role of intestinal bacteria in the development of dietary cirrhosis in rats. Journal of Experimental Medicine. 1957;106:1–14. doi: 10.1084/jem.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra A, Das GJ, De BK, Roy B, Guha Mazumder DN. Hepatic manifestations in chronic arsenic toxicity. Indian J. Gastroenterol. 1999;18:152–155. [PubMed] [Google Scholar]
- Santra A, Maiti A, Das S, Lahiri S, Charkaborty SK, Mazumder DN. Hepatic damage caused by chronic arsenic toxicity in experimental animals. J. Toxicol. Clin. Toxicol. 2000;38:395–405. doi: 10.1081/clt-100100949. [DOI] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino M, Luche E, Gres S, Baylac A, Berge M, Cenac C, Waget A, Klopp P, Iacovoni J, Klopp C, Mariette J, Bouchez O, Lluch J, Ouarne F, Monsan P, Valet P, Roques C, Amar J, Bouloumie A, Theodorou V, Burcelin R. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut. 2012;61:543–553. doi: 10.1136/gutjnl-2011-301012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheik CS, Mitchell TW, Rizvi FZ, Rehman Y, Faisal M, Hasnain S, McInerney MJ, Krumholz LR. Exposure of soil microbial communities to chromium and arsenic alters their diversity and structure. PLoS One. 2012;7:e40059. doi: 10.1371/journal.pone.0040059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, Bae JW. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63:727–735. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- Sokol H, Seksik P, Rigottier-Gois L, Lay C, Lepage P, Podglajen I, Marteau P, Dore J. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm. Bowel. Dis. 2006;12:106–111. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- Straub AC, Stolz DB, Ross MA, Hernandez-Zavala A, Soucy NV, Klei LR, Barchowsky A. Arsenic stimulates sinusoidal endothelial cell capillarization and vessel remodeling in mouse liver. Hepatology. 2007;45:205–212. doi: 10.1002/hep.21444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Schmidt RH, Beier JI, Watson WH, Zhong H, States JC, Arteel GE. Chronic subhepatotoxic exposure to arsenic enhances hepatic injury caused by high fat diet in mice. Toxicol. Appl. Pharmacol. 2011;257:356–364. doi: 10.1016/j.taap.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Wiele T, Gallawa CM, Kubachka KM, Creed JT, Basta N, Dayton EA, Whitacre S, Du LG, Bradham K. Arsenic metabolism by human gut microbiota upon in vitro digestion of contaminated soils. Environmental Health Perspectives. 2010;118:1004–1009. doi: 10.1289/ehp.0901794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalkes MP, Liu J, Chen H, Xie Y, Achanzar WE, Zhou YS, Cheng ML, Diwan BA. Estrogen signaling in livers of male mice with hepatocellular carcinoma induced by exposure to arsenic in utero. J. Natl. Cancer Inst. 2004;96:466–474. doi: 10.1093/jnci/djh070. [DOI] [PubMed] [Google Scholar]
- West CE. Gut microbiota and allergic disease: new findings. Curr. Opin. Clin Nutr Metab Care. 2014;17:261–266. doi: 10.1097/MCO.0000000000000044. [DOI] [PubMed] [Google Scholar]