Abstract
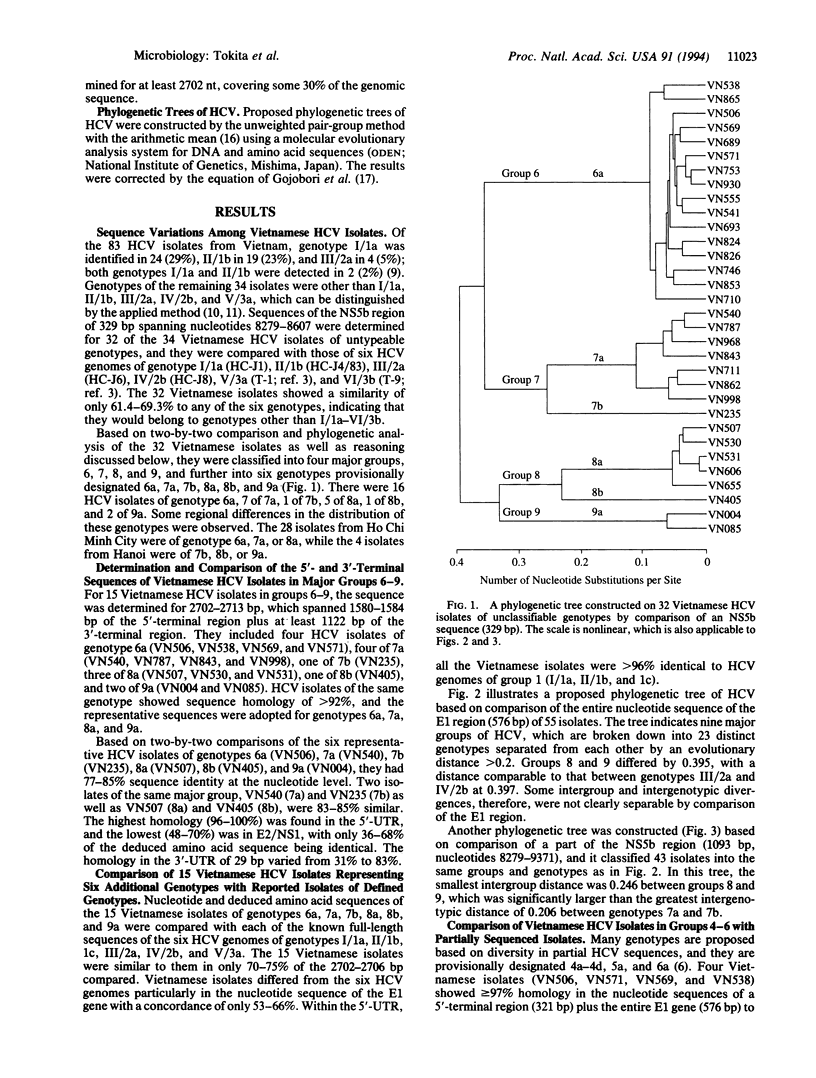
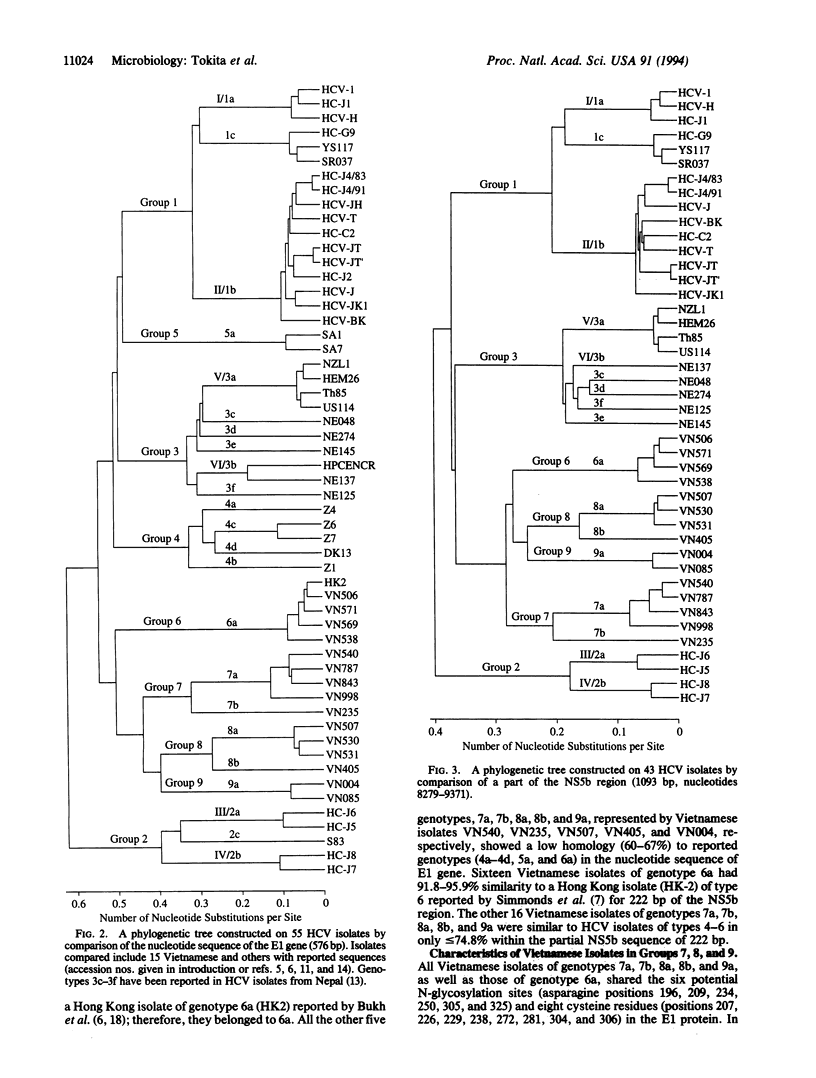
Thirty-four (41%) of 83 hepatitis C virus (HCV) isolates from commercial blood donors in Vietnam were not classifiable into genotype I/1a, II/1b, III/2a, IV/2b, or V/3a; for 15 of them, the sequence was determined for 1.6 kb in the 5'-terminal region and 1.1 kb in the 3'-terminal region. Comparison of the 15 Vietnamese isolates among themselves and with reported full or partial HCV genomic sequences indicated that they were classifiable into four major groups (groups 6-9) divided into six genotypes (6a, 7a, 7b, 8a, 8b, and 9a). Vietnamese HCV isolates of genotypes 7a, 7b, 8a, 8b, and 9a were significantly different from those classified into groups 4, 5, and 6 based on divergence within partial sequences; those of genotype 6a were homologous to a Hong Kong isolate (HK2) of genotype 6a. Phylogenetic trees based on the envelope 1 (E1) gene (576 bp) of 55 isolates and a part of the nonstructural 5 (NS5) region (1093 bp) of 43 isolates revealed at least nine major groups, three of which (groups 7, 8, and 9) were identified only in Vietnamese blood donors. With a prospect that many more HCV isolates with significant sequence divergence will be reported from all over the world, the domain of the HCV genome to be compared and criteria for grouping/typing and genotyping/subtyping will have to be determined, so that they may be correlated with virological, epidemiological, and clinical characteristics.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bukh J., Purcell R. H., Miller R. H. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):8234–8238. doi: 10.1073/pnas.90.17.8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh J., Purcell R. H., Miller R. H. Sequence analysis of the 5' noncoding region of hepatitis C virus. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):4942–4946. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha T. A., Beall E., Irvine B., Kolberg J., Chien D., Kuo G., Urdea M. S. At least five related, but distinct, hepatitis C viral genotypes exist. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7144–7148. doi: 10.1073/pnas.89.15.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojobori T., Ishii K., Nei M. Estimation of average number of nucleotide substitutions when the rate of substitution varies with nucleotide. J Mol Evol. 1982;18(6):414–423. doi: 10.1007/BF01840889. [DOI] [PubMed] [Google Scholar]
- Hijikata M., Kato N., Ootsuyama Y., Nakagawa M., Ohkoshi S., Shimotohno K. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem Biophys Res Commun. 1991 Feb 28;175(1):220–228. doi: 10.1016/s0006-291x(05)81223-9. [DOI] [PubMed] [Google Scholar]
- Houghton M., Weiner A., Han J., Kuo G., Choo Q. L. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991 Aug;14(2):381–388. [PubMed] [Google Scholar]
- Mori S., Kato N., Yagyu A., Tanaka T., Ikeda Y., Petchclai B., Chiewsilp P., Kurimura T., Shimotohno K. A new type of hepatitis C virus in patients in Thailand. Biochem Biophys Res Commun. 1992 Feb 28;183(1):334–342. doi: 10.1016/0006-291x(92)91648-a. [DOI] [PubMed] [Google Scholar]
- Nagayama R., Tsuda F., Okamoto H., Wang Y., Mitsui T., Tanaka T., Miyakawa Y., Mayumi M. Genotype dependence of hepatitis C virus antibodies detectable by the first-generation enzyme-linked immunosorbent assay with C100-3 protein. J Clin Invest. 1993 Sep;92(3):1529–1533. doi: 10.1172/JCI116731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata N., Alter H. J., Miller R. H., Purcell R. H. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3392–3396. doi: 10.1073/pnas.88.8.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H., Kojima M., Okada S., Yoshizawa H., Iizuka H., Tanaka T., Muchmore E. E., Peterson D. A., Ito Y., Mishiro S. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology. 1992 Oct;190(2):894–899. doi: 10.1016/0042-6822(92)90933-g. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Kojima M., Sakamoto M., Iizuka H., Hadiwandowo S., Suwignyo S., Miyakawa Y., Mayumi M. The entire nucleotide sequence and classification of a hepatitis C virus isolate of a novel genotype from an Indonesian patient with chronic liver disease. J Gen Virol. 1994 Mar;75(Pt 3):629–635. doi: 10.1099/0022-1317-75-3-629. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Kurai K., Okada S., Yamamoto K., Lizuka H., Tanaka T., Fukuda S., Tsuda F., Mishiro S. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology. 1992 May;188(1):331–341. doi: 10.1016/0042-6822(92)90762-e. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Sugiyama Y., Okada S., Kurai K., Akahane Y., Sugai Y., Tanaka T., Sato K., Tsuda F., Miyakawa Y. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol. 1992 Mar;73(Pt 3):673–679. doi: 10.1099/0022-1317-73-3-673. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Tokita H., Sakamoto M., Horikita M., Kojima M., Iizuka H., Mishiro S. Characterization of the genomic sequence of type V (or 3a) hepatitis C virus isolates and PCR primers for specific detection. J Gen Virol. 1993 Nov;74(Pt 11):2385–2390. doi: 10.1099/0022-1317-74-11-2385. [DOI] [PubMed] [Google Scholar]
- Sakamoto M., Akahane Y., Tsuda F., Tanaka T., Woodfield D. G., Okamoto H. Entire nucleotide sequence and characterization of a hepatitis C virus of genotype V/3a. J Gen Virol. 1994 Jul;75(Pt 7):1761–1768. doi: 10.1099/0022-1317-75-7-1761. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Holmes E. C., Cha T. A., Chan S. W., McOmish F., Irvine B., Beall E., Yap P. L., Kolberg J., Urdea M. S. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993 Nov;74(Pt 11):2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- Simmonds P., McOmish F., Yap P. L., Chan S. W., Lin C. K., Dusheiko G., Saeed A. A., Holmes E. C. Sequence variability in the 5' non-coding region of hepatitis C virus: identification of a new virus type and restrictions on sequence diversity. J Gen Virol. 1993 Apr;74(Pt 4):661–668. doi: 10.1099/0022-1317-74-4-661. [DOI] [PubMed] [Google Scholar]
- Song P., Duc D. D., Hien B., Nakata S., Chosa T., Watanabe J., Tsuda F., Murata K., Okamoto H. Markers of hepatitis C and B virus infections among blood donors in Ho Chi Minh City and Hanoi, Vietnam. Clin Diagn Lab Immunol. 1994 Jul;1(4):413–418. doi: 10.1128/cdli.1.4.413-418.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuyver L., Rossau R., Wyseur A., Duhamel M., Vanderborght B., Van Heuverswyn H., Maertens G. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J Gen Virol. 1993 Jun;74(Pt 6):1093–1102. doi: 10.1099/0022-1317-74-6-1093. [DOI] [PubMed] [Google Scholar]
- Takada N., Takase S., Enomoto N., Takada A., Date T. Clinical backgrounds of the patients having different types of hepatitis C virus genomes. J Hepatol. 1992 Jan;14(1):35–40. doi: 10.1016/0168-8278(92)90128-c. [DOI] [PubMed] [Google Scholar]
- Takamizawa A., Mori C., Fuke I., Manabe S., Murakami S., Fujita J., Onishi E., Andoh T., Yoshida I., Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991 Mar;65(3):1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokita H., Shrestha S. M., Okamoto H., Sakamoto M., Horikita M., Iizuka H., Shrestha S., Miyakawa Y., Mayumi M. Hepatitis C virus variants from Nepal with novel genotypes and their classification into the third major group. J Gen Virol. 1994 Apr;75(Pt 4):931–936. doi: 10.1099/0022-1317-75-4-931. [DOI] [PubMed] [Google Scholar]
- Weiner A. J., Brauer M. J., Rosenblatt J., Richman K. H., Tung J., Crawford K., Bonino F., Saracco G., Choo Q. L., Houghton M. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991 Feb;180(2):842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- Yoshioka K., Kakumu S., Wakita T., Ishikawa T., Itoh Y., Takayanagi M., Higashi Y., Shibata M., Morishima T. Detection of hepatitis C virus by polymerase chain reaction and response to interferon-alpha therapy: relationship to genotypes of hepatitis C virus. Hepatology. 1992 Aug;16(2):293–299. doi: 10.1002/hep.1840160203. [DOI] [PubMed] [Google Scholar]




