Abstract
AIM: To investigate the value of chaperonin containing TCP1, subunit 3 (CCT3) to predict the prognosis of patients with hepatocellular carcinoma (HCC) and determine its function in HCC progression.
METHODS: CCT3 expression levels were examined in human non-cancerous liver tissues and a variety of HCC cell lines by quantitative real-time PCR and immunoblotting. CCT3 expression was suppressed by small interfering RNA. The effects of reducing CCT3 expression in HCC cells were tested. The 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyl tetrazolium bromide (MTT) assay, cell counting experiment, cell cycle assay, apoptosis assay and invasion assay were employed to evaluate cell functions in vitro. Immunohistochemistry was performed on HCC specimens. In addition, CCT3 expression in HCC specimens was also assessed at the protein and mRNA level. Associations between clinicopathological characteristics and prognosis were analyzed, along with the possible mechanisms involved in CCT3’s function in HCC progression.
RESULTS: The expression levels of CCT3 mRNA and protein were upregulated in HCC cell lines in contrast to adjacent non-cancerous tissues. Reducing CCT3 expression not only suppressed cell proliferation in cell counts, MTT assay, cell cycle assay and induced cell apoptosis (P < 0.05 vs negative control), but also inhibited the tumor cell invasion capacity in vitro (P < 0.01 vs negative control). Overexpression of CCT3 in the nuclei of cancer cells in HCC specimens (58 of 104 patients, 55.8%) was associated with poor prognosis in HCC patients (3-year survival rate, 55.5% vs 84.2%, P = 0.020) after hepatectomy. Mechanistic analyses showed that signal transducer and activator of transcription 3 (STAT3) activation was decreased even when stimulated by interleukin-6 after knocking down CCT3 in the HepG2 cell line.
CONCLUSION: Overexpression of CCT3 in the nuclei of cancerous cells is associated with HCC progression. CCT3 may be a target that affects the activation of STAT3 in HCC.
Keywords: Hepatocellular carcinoma; Chaperonin Containing TCP1, Subunit 3; Cell growth; Invasion; Prognosis
Core tip: Hepatocellular carcinoma (HCC) is a lethal disease and it is difficult to evaluate prognosis and manage the disease. This study showed that overexpression of chaperonin containing TCP1, subunit 3 (CCT3) in the nuclei of cancerous cells was an independent risk factor for the prognosis of HCC patients and was associated with tumor histological type and microvascular invasion. CCT3 affects activation of the interleukin-6/signal transducer and activator of transcription 3 signal pathway in vitro. CCT3 could play an essential role in the progression of HCC and might represent a prognostic biomarker in patients with HCC after hepatectomy.
INTRODUCTION
As the most common primary liver cancer, hepatocellular carcinoma (HCC) is the third most common cause of cancer death globally[1]. Approximately 750000 new cases of liver cancer are reported each year[2]. Eighty percent of HCC cases develop from fibrosis caused by viral hepatitis or alcohol[3]. In China, most HCC patients develop cirrhosis that is caused by hepatitis B virus hepatitis. The natural course of HCC is likely to be invasive and unpredictable. The prognosis of HCC has been assessed using different clinical classification criteria, the Barcelona Classification (BCLC), for instance, which enrolled tumor number, size, vascular invasion, Child-Pugh score and performance status[4]. However, patients with HCC may have a distinctly different disease course and outcome, even with the same BCLC score or classification. Molecular profiling is likely to be promising to assess prognosis and to target treatment in individuals with HCC[5].
Chaperonin containing TCP-1 [TCP-1 ring complex (TRiC), CCT/TRiC], a large molecular weight complex presents in eukaryotic cells, comprises eight homologous subunits (CCT1, 2, 3, 4, 5, 6, 7, 8). These subunits are assembled in two stacked octameric rings, which form a cage[6]. The subunits possess identical ATPase domains, whose polypeptide-binding regions have radically diverged while evolving to enhance binding specificity[7]. All of these subunits form a cage, which plays a critical role in protein folding and refolding. Being folded into specific three-dimensional structures is necessary for proteins to reach their native states and become functionally active. If incorrectly folded or misfolded, proteins can lose biological functions, and even gain harmful functions[8]. CCT/TRiC generally contributes to the folding of 10% to 15% of newly synthesized proteins into their native states in eukaryotic cells, in addition to refolding some proteins that have lost their native states under stress conditions[9,10]. CCT/TRiC is involved with a variety of proteins concerned with cell growth, proliferation, and apoptosis in normal and tumor cells, such as cyclin B, cyclin E, actins, tubulins, the von Hippel-Lindau (VHL) tumor suppressor protein, P53 and STAT3[11-16].
CCT3 (60 kDa) is a critical subunit in CCT/TRiC complexes, which plays a significant role in specifically binding these factors during protein folding or refolding. CCT3 is overexpressed in HCC tissues[17-19], ovarian cancer and cholangiocarcinoma[20,21]. Although the functions of CCT/TRiC involved in cell proliferation and the tumorigenesis of different tumors in vitro have been studied by targeting CCT1, CCT2, CCT4, and CCT8[22-24], few studies have been conducted on CCT3. Thus, it is unclear what effect CCT3 has on HCC.
In this study, the expression of CCT3 in HCC patients was evaluated. In addition, we investigated its effects on HCC cell proliferation, apoptosis, invasion and its potential mechanisms in vitro by targeted silencing of the CCT3 gene. These findings will contribute to the clarification of its function in HCC, and assess its value in clinical prognosis and targeted therapy in HCC.
MATERIALS AND METHODS
Sample collection and cell culture
Before this study, the protocol and sample collection were approved by the Institutional Review Board of Peking University People’s Hospital, Beijing, China. The human HCC cell lines SMMC-7721, HepG2, Huh-7 and Hep3B were purchased from Shanghai Institute for Biological Sciences, Shanghai, China. The cells were maintained in 5% CO2 at 37 °C in RPMI 1640 (Hyclone, Logan, UT, United States) supplemented with 10% fetal bovine serum (GIBCO, Carlsbad, CA, United States).
Patients and tissue specimens
Tumor tissues and adjacent non-cancerous tissues (n = 20) were collected from patients who had undergone resection for primary HCC in the Department of Hepatobiliary Surgery, Peking University People’s Hospital between 2013 and 2014. Neither radiofrequency ablation therapy nor transcatheter arterial chemoembolization (TACE) was performed in these patients preoperatively. Non-cancerous liver tissues were collected from patients with hepatic hemangiomas who underwent hepatectomy. After resection, the tissues were washed with 0.9% sodium chloride solution, then immersed in liquid nitrogen and stored in -80 °C. One hundred and four paraffin-embedded HCC samples were collected from primary HCC patients who had undergone hepatectomy in the Department of Hepatobiliary Surgery, Peking University People’s Hospital between 2008 and 2012. Patients with extrahepatic metastasis confirmed by computed tomography (CT), magnetic resonance imaging (MRI), or positron emission tomography were excluded. Tumor stages were determined according to the TNM system of the American Joint Committee on Cancer[25]. The histological grade of each tumor was determined based on the Edmondson-Steiner grading system[26]. Macrovascular invasion was defined by the thrombus adjacent to the tumor, with a blurred boundary presented in the portal vein, which was confirmed by at least one imaging modality, CT or MRI[27]. Microvascular invasion (MVI) was defined by a thrombus that was formed by cancerous cells presented in the vascular space encircled by vascular endothelial cells, located in the tumor capsule or in the surrounding liver parenchyma, either in the portal vein or hepatic vein branches. Clusters of cancerous cells or a few cancerous cells, which floated in isolated vessels, but were not covered by endothelium, were not diagnosed as MVI[28]. Patients enrolled in this study had detailed medical records of the etiology of hepatitis, gender, age, pre-operative alpha-fetoprotein (AFP) level, cirrhosis description, number and size of tumor nodules, Child-Pugh scores, radical resection or palliative resection and post-operative TACE. Follow-up was undertaken at outpatient visits, which included liver function tests, AFP and CT or MRI at 3 mo post-operatively, and every 3 mo for two years. Thereafter, laboratory tests and imaging were repeated at 6-month intervals. The study endpoint was February 28, 2015, and the median follow-up period was 36.5 mo. All patients who did not survive, died of HCC or related complications. Three-year survival (time from date of surgery to date of death or last follow-up) rate was used to evaluate prognosis.
RNA and protein extraction
Extraction of RNA was conducted using Trizol solution (Invitrogen, Shanghai, China). Total and nuclear proteins from cells and tissue were extracted with a total protein extraction kit or a nuclear-cytosol extraction kit (Applygen Technologies Inc., Beijing, China). The samples were then stored at -80 °C.
RNA reverse transcription, and quantitative real-time PCR
Quantitative real-time PCR (qPCR) amplification was undertaken using a Takara TP800 Real-time PCR system (Takara, Shiga, Japan). RNA was converted into cDNA. The amplification reactions were conducted with specific primers as follows: CCT3 forward primer 5’-CCTCCAGGTATCTTTTCCACTCT-3’, reverse primer: 5’-TCAGTCGGTGGTCATCTTTGG-3’. GAPDH forward primer 5’-TGACTTCAACAGCGACACCCA-3’, reverse primer 5’-CACCCTGTTGCTGTAGCCAAA-3’. The PCR conditions were performed as follows: 95 °C for 15 s to activate DNA polymerase; followed by 45 cycles of 95 °C for 5 s, 60 °C for 30 s; 1 cycle of 95 °C for 60 s, 55 °C for 60 s; and 81 cycles of heating from 55 °C to 95 °C for 4 s. The set point temperature was increased after cycle 2 by 0.5 °C. Specificities of the reaction products were evaluated by melting curve analyses. All experiments were repeated three times for the genes.
Western blotting
Western blotting was carried out on lysates of HCC tissues and cell lines as described previously[15] using rabbit polyclonal anti-CCT3 antibodies (1:1000, Abcam, Cambridge, MA, United States), rabbit monoclonal anti-GAPDH (1:2000, Cwbiotech, Beijing, China), rabbit monoclonal anti-(p)STAT3, rabbit monoclonal anti-STAT3 (1:2000, Abcam) and rabbit polyclonal anti-Histone 3 antibodies (1:2000, Abcam). The secondary antibody was an HRP-conjugated anti-rabbit IgG antibody (1:4000; Zhongshan, Beijing, China). The enhanced chemiluminescence reagent (Thermo, South Logan, UT, United States) was used for signal detection.
Immunohistochemistry
Paraffin-embedded blocks were sectioned at 3-μm thickness. The Polink-1 HRP DAB Detection System One-step polymer detection method was used as follows: tissue sections were deparaffinized in xylene, and then rehydrated through ethanol to water. Endogenous peroxidase activity blocking was performed with 3% H2O2 for 15 min. Antigen retrieval was performed as follows: sections were heated in a high pressure cooker with 10 mmol/L citrate buffer (pH 6.0) at 1000 kW power heated by an induction cooker for 2.5 min, and then left to cool naturally. Non-specific binding was abolished using 5% goat serum (Beyotime Biotechnology, Beijing, China) for 30 min. Prediluted primary polyclonal rabbit anti-CCT3 antibodies (1:80, Abcam Co., Hong Kong, China) were used. Negative control slides without antibody were included in this step. The slides were incubated for 16 h at 4 °C. After washing with phosphate-buffered saline (PBS), secondary antibody at working dilutions (rat anti-rabbit IgG, Beijing XiYa Jinqiao Biology Technology Company, Beijing, China) was added to the section, which was incubated for 1 h at room temperature. After washing, diaminobenzene (DAB) was added to develop the color reaction. After ending the reaction, the sections were counterstained with hematoxylin, cleared and mounted.
Immunocytochemistry
Cells were plated on coverslips at a density of 2 × 104/mL, then washed with PBS three times after incubating at 37 °C for 6 h. Cells on the coverslips were fixed with 4% paraformaldehyde for 15 min, then air dried, incubated with 0.5% Triton X-100 for 20 min, washed with PBS three times and incubated with 3% H2O2 for 15 min. The follow-up procedures were all the same as the immunohistochemistry (IHC) procedures, except that the antigen retrieval step was omitted.
Evaluation of staining
The IHC slides were reviewed and scored by two pathologists (Qian LH and Song JQ), who were blinded to the clinical parameters, from the Pathology Department of Peking University People’s Hospital, independently. CCT3 staining in the nuclei and the cytoplasm was evaluated separately. The score was calculated based on the sum of staining intensity and the percentage of positive staining of cancerous cells. The percentage of positively stained areas in the cytoplasm of cells was defined using a scale of 0-3 (0: < 10%, 1: 10%-25%, 2: 26%-75%, and 3: > 76%). Staining intensity was scored as “0” (no staining), “1” (weak staining), “2” (moderate staining), or “3” (strong staining). The percentage of positively stained nuclei were scored as 0: < 20%, 1: 20%-49%, 2: 50%-79%, and 3: > 80%) in cancerous cells. The total immunostaining score of nuclei or cytoplasm ranged from 0 to 6. CCT3 expression levels was classified as: “-” (0-1), “+” (2-3), “++” (3-4) and “+++” (4-6). Patients were then divided into low CCT3 expression (“-” and “+”) and high CCT3 expression (“++” and “+++”) groups[29,30].
Lentivirus vector construction and cell transfection
A lentiviral shRNA vector was used to target the expression of CCT3 in the SMMC-7721 and HepG2 cell lines. CCT3 knockdown recombined lentivirus vector, control vector pGCSIL-GFP and the small interfering ribonucleic acid (siRNA) were constructed by Gene Chem (Gene Chem, Shanghai, China). siRNA molecules used for CCT3 siRNA were as follows: CCT3 sense: 5'-CGGGCCAAGTCCATGATCGAAATTCTC-GAGAATTTCGATCATGGACTTGGCTTTTTG-3', anti-sense: 5’-AATTCAAAAAGCCAAGTCCATGATCGAAA TTCTCGAGAATTTCGATCATGGACTTGGC-3’. For the negative control sense: 5'-CCGGTTCTCCGAACGTGTCA-CGTTTCAAGAGAACGTGACACGTTCGGGAATTTTTG-3', anti-sense: 5’-AATTCAAAAATTCTCCGAACGTGTCACG TTCTCTTGAAACGTGACACG TTCGGAGAA-3’.
Transient transfection was performed when cells reached 30% confluence. After the infected cells with green fluorescent protein (GFP) exceeded 80% of the total in 72 h, the cells were harvested for further assays. Knockdown efficiency was evaluated by western blotting and qPCR.
Cell proliferation assay
The 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay and cell counting were used to assess cell proliferation and viability. Cells were plated in 96-well plates at approximately 2 × 103/well, and then incubated for 24, 48, 60, 72, 96 or 120 h. Twenty μL of MTT (5 mg/mL) (Sigma, St. Louis, MO, United States) was added to the wells at each time point. Supernatants in the wells were removed, and 100 μL dimethyl sulfoxide (DMSO) (Sigma) was used to end the reaction. The optical density in each well was detected by the Biotek Elx800 microplate reader (Biotek, Winooski, VT, United States). Cells infected with GFP-CCT3-siRNA (CCT3-siRNA) and control vector pGCSIL-GFP (negative control, NC) were plated in the 96-well plates at approximate 1 × 103/well, and then incubated for 120 h. The cells with GFP were counted using a Thermo Cellomics ArrayScan VT1 (Thermo) each day during the experiment. For each experimental group, five wells were used. All experiments were repeated in triplicate.
Cell cycle assay
Cells were collected after reaching 80% confluence, washed once in ice cold PBS, and fixed in 70% ice ethanol overnight. The fixed cells were then resuspended in 25 μL (2 mg/mL) propidium iodide (Sigma), 10 μL (10 mg/mL) RNaseA (Fermentas, MBI, Lithuania), and 1000 μL PBS, and then incubated in the dark at 4 °C for 1 h. They were subjected to flow cytometry analysis using a FACSCalibur device (BD, Franklin Lakes, NJ, United States). Triplicate experiments were conducted in each group.
Apoptosis assay
Transfected cells were collected after reaching 85% confluence. After washing twice with ice PBS solution, they were resuspended using 400 μL 1 × binding buffer followed by 1 mL 1 × staining buffer. After incubating with 5 μL Annexin V-APC (Ebioscience, San Diego, CA, United States), the cells were subjected to flow cytometry using a FACSCalibur device (BD). Each experiment was performed three times.
Matrigel invasion assay
The assays were conducted with the Chemicon Cell Invasion Assay Kit ECM550 (Chemicon, Temecula, CA, United States). The CCT3-siRNA group and NC group were collected after reaching 80% confluence. The cells (2 × 104/well) in 200 μL RPMI 1640 were added to the upper chamber. Five hundred μL RPMI 1640 containing 10% FBS was then added to the bottom chamber. After incubating for 48 h, non-migrating cells were removed from the top surface of the membrane. The membranes were fixed with 4% paraformaldehyde at room temperature for 30 min, then stained with 0.5% crystal violet, washed three times with PBS and counted under a microscope. Ten random fields were counted to calculate the number of stained cells. Triplicate experiments were performed in the groups.
IL-6 stimulation of cells
CCT3-siRNA and NC HepG2 cells were incubated overnight in 5% CO2 at 37 °C. IL-6 (Sigma) at 0, 0.1, 0.3 and 1 ng/mL was administered to the cells. After culturing at 37 °C for 30 min, the lysates were harvested.
Statistical analysis
SPSS 17.0 software (SPSS Inc., Chicago, IL, United States) and Graph Pad Prism 5.0 (GraphPad Software Inc., La Jolla, CA, United States) software were used for statistical analyses and plotting the data. All continuous variables were expressed as the mean ± SD and evaluated with the Mann-Whitney U nonparametric test or two-tailed paired t test. The χ2 test (Pearson’s test or continuity correction test) or Fisher’s exact test were employed to evaluate qualitative variables. The Kaplan-Meier method and log-rank test were used for plotting survival curves and for comparisons. The Cox proportional hazards regression model was used to evaluate the independent risk factors associated with prognosis. Statistical significance was set at P < 0.05.
RESULTS
CCT3 expression level in cell lines and liver tissues
Western blotting analyses and qPCR confirmed that the mRNA and protein transcription levels of CCT3 in HCC cell lines were increased compared with those in the non-cancer liver homogenate (HNCL) (Figure 1A and B).
Figure 1.
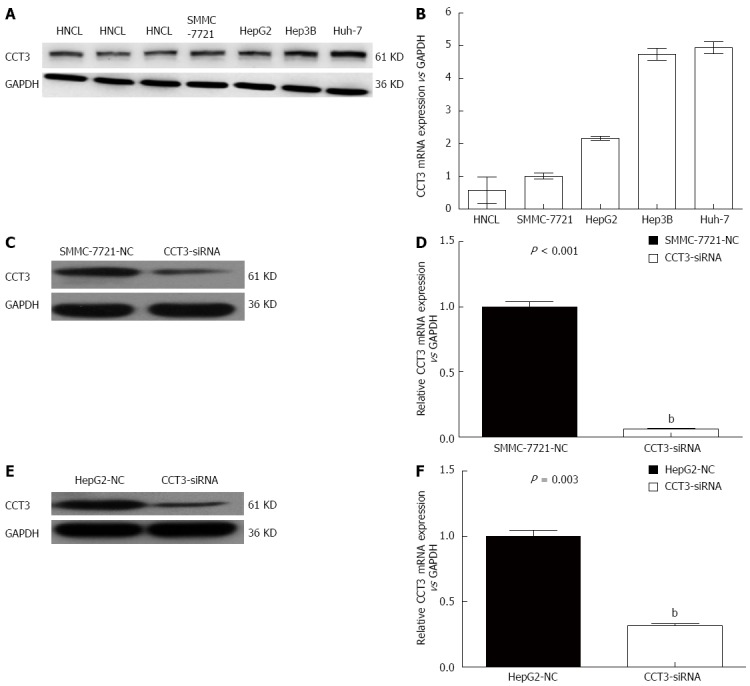
Expression of subunit 3 in tissues and cell lines. A: Expression of the subunit 3 (CCT3) protein in human non-cancerous liver tissues (HNCL) obtained from patients with hepatic hemangioma and in hepatocellular carcinoma (HCC) cells by western blotting; B: Expression of CCT3 mRNA in HNCL and HCC cells; C-F: Effect of knocking down the expression of CCT3 by siRNA in the HCC cell lines SMMC-7721 and HepG2, as shown by western blotting and qPCR (bP < 0.01 vs negative control). NC: Negative control.
Downregulation of CCT3 in vitro
To determine the function of CCT3 in HCC progression, SMMC-7721 and HepG2 cells were transfected with siRNA oligonucleotides, and found that CCT3 protein and mRNA expressions were downregulated (P < 0.01), which was verified by western blotting (Figure 1C and E) and qPCR (Figure 1D and F).
Role of CCT3 in HCC cell proliferation and cycle progression
We calculated the proliferation rates of the negative control (NC) and siRNA-CCT3 groups in SMMC-7721 and HepG2 cells, using the cellomics assay. A decrease in the proliferation rate was observed in the CCT3-siRNA group in contrast to the NC group at days 3-5 (P < 0.05, P < 0.01) (Figure 2A). The MTT assay revealed that inhibition of CCT3 expression significantly inhibited the proliferation of HCC cells (P < 0.01, Figure 2B). The DNA content of CCT3-siRNA and NC cells was determined by flow cytometry. The percentage of cells in the S stage in the CCT3-siRNA group increased compared with the NC group (P < 0.05, P < 0.01) (Figure 2C).
Figure 2.
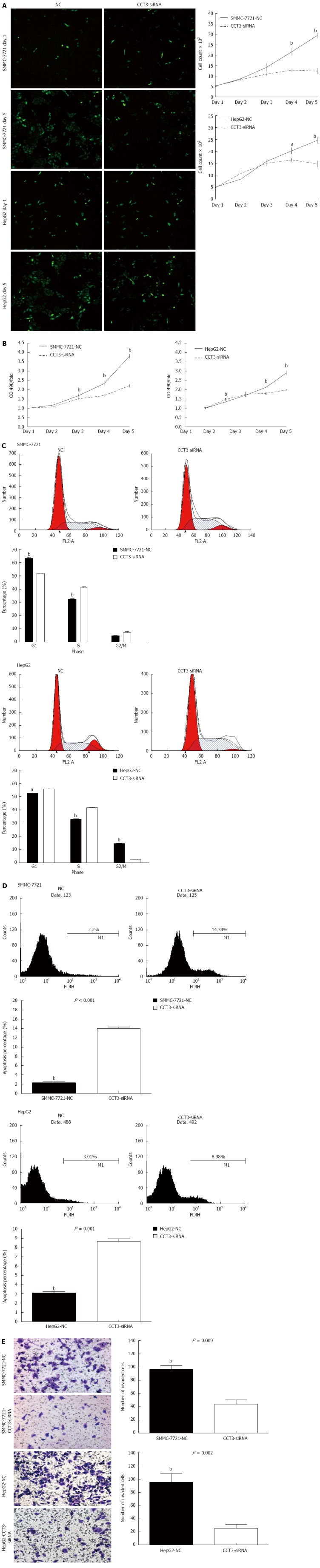
Knockdown of subunit 3 expression suppressed proliferation, apoptosis and invasion in hepatocellular carcinoma cell lines. A: Knockdown of subunit 3 (CCT3) gene expression suppressed proliferation in hepatocellular carcinoma (HCC) cells, as assessed by the cellomics assay [aP < 0.05, bP < 0.01 vs negative control (NC)]; B: Proliferation was inhibited in the CCT3-siRNA group, as assessed by the MTT assay, absorbance was recorded at 490 nm (bP < 0.01 vs negative control); C: Suppression of CCT3 expression changed the G1, S, or G2/M level in HCC cells, as assessed by flow cytometry (aP < 0.05, bP < 0.01 vs negative control); D: Inhibiting CCT3 expression in HCC cells induced more apoptosis compared with the negative control (bP < 0.01 vs negative control); E: Invasion was markedly reduced in the CCT3-siRNA group (bP < 0.01 vs negative control).
Role of CCT3 in HCC cell apoptosis
To determine whether inhibition of HCC cell growth was attributed to induction of apoptosis, an Annexin V-APC binding assay was performed. This revealed that reducing CCT3 expression induced increased apoptosis (P < 0.01) (Figure 2D).
Knock-down of CCT3 suppresses HCC cell invasion in vitro
An invasion assay showed that suppression of CCT3 expression could reduce cell invasion. The number of cells in the CCT3-siRNA group that invaded the lower compartment of the migration chamber was less than that in the NC group (P < 0.01) (Figure 2E).
Clinical and demographic data
In 20 paired fresh frozen HCC tissue samples, increased mRNA and protein levels of CCT3 were observed in 16 tumor samples compared with adjacent non-cancerous tissues, as shown by semi-quantitative western blotting analyses (sum of ranks 314.4 vs 505.5, Mann-Whitney U = 104.5, P = 0.01) (Figure 3A and B) and qPCR analyses (sum of ranks 315 vs 505, Mann-Whitney U = 105, P = 0.01) (Figure 3C). We performed immunohistochemical staining in 104 primary HCC patients who had undergone hepatectomy. The characteristics of the selected patients are shown in Table 1. The patients’ ages ranged from 35 to 84 years, with a mean age of 58.7 years. Eighty-three patients were male and 21 were female. Eighty-one patients (81.7%) had hepatitis B virus surface antigen, six patients (5.8%) had hepatitis C virus antibodies (none of these patients were positive for both types of hepatitis) and 13 patients (12.5%) had neither hepatitis C virus antibodies nor hepatitis B virus surface antigen.
Figure 3.
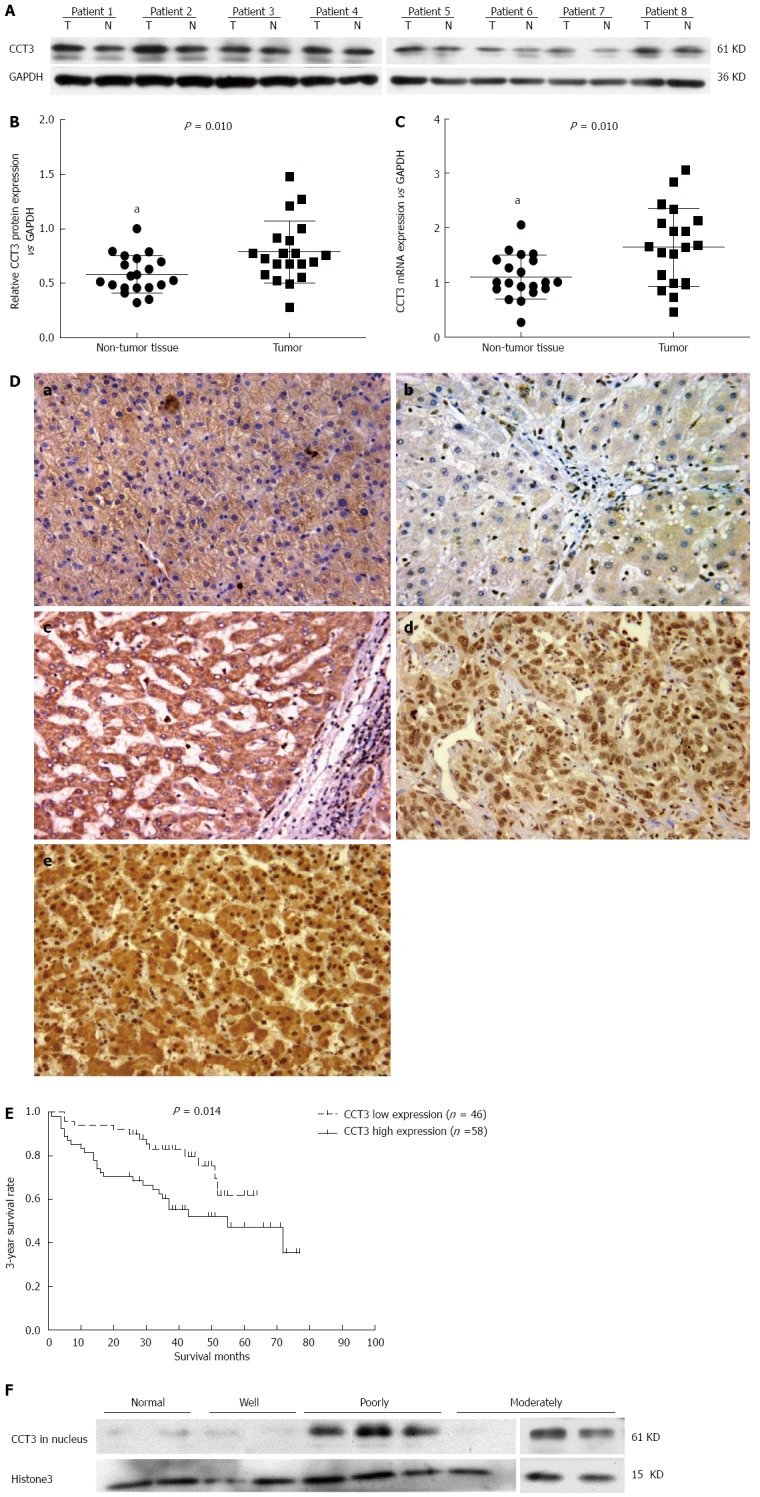
Subunit 3 expression in hepatocellular carcinoma specimens and its association with 3-year survival rate. A-C: Expression of subunit 3 (CCT3) protein in hepatocellular carcinoma (HCC) tissues was higher than that in adjacent non-cancerous tissue, as shown by western blotting and qPCR (aP < 0.05 vs control); D: Expression of CCT3 in various differentiated HCC specimens and non-cancerous liver tissues from patients with hepatic hemangioma by immunohistochemistry (Original magnification × 200). a: Non-cancerous liver tissue from hepatic hemangioma patients (CCT3 positive expression in the cytoplasm and negative in the nuclei); b: Highly-differentiated HCC (negative in both the cytoplasm and nuclei); c: Moderately-differentiated HCC (positive in the cytoplasm and negative in nuclei); d: Poorly-differentiated HCC (negative in the cytoplasm and positive in nuclei); e: Poorly-differentiated HCC (positive in the cytoplasm and nuclei); E: Survival curves according to CCT3 expression. Patients with low expression of CCT3 in the nuclei of cancerous cells had higher cumulative survival rates than patients with high expression; F: Expression of CCT3 in the nuclei of well-, moderately-, and poorly-differentiated HCC specimens, and in non-cancerous liver tissues.
Table 1.
Clinicopathological features of 104 patients with hepatocellular carcinoma
| Characteristics | n (%) |
| Gender (female/male) | 21 (20.2)/83 (79.8) |
| Age, mean (yr) | 58.7 (range 35-84, SD 11.2) |
| Hepatitis type | |
| HBV | 85 (81.7) |
| HCV | 6 (5.8) |
| None | 13 (12.5) |
| pT category (UICC) | |
| pT1 | 78 (75) |
| pT2 | 14 (13.5) |
| pT3a | 8 (7.7) |
| pT3b | 4 (3.8) |
| PT4 | 0 (0.0) |
| pN category | |
| pN0 | 104 (100.0) |
| pN1 | 0 (0.0) |
| pM category | |
| pM0 | 104 (100.0) |
| pM1 | 0 (0.0) |
| UICC stage | |
| I | 64 (61.5) |
| II | 12 (11.5) |
| IIIA | 13 (12.5) |
| IIIB | 7 (6.7) |
| IIIC | 8 (7.7) |
| IV | 0 (0.0) |
| Histological type | |
| Well-differentiated | 26 (25.0) |
| Moderately-differentiated | 60 (57.7) |
| Poorly-differentiated | 18 (17.3) |
| Macrovascular invasion | |
| None | 99 (95.2) |
| Present | 5 (4.8) |
| Microvascular invasion | |
| None | 78 (75.0) |
| Present | 26 (25.0) |
| Tumor size (cm) | |
| ≥ 5 | 58 (55.8) |
| < 5 | 46 (44.2) |
| Serum AFP level (ng/mL) | |
| < 400 | 79 (76.0) |
| ≥ 400 | 25 (24.0) |
| Liver encapsulation invasion | |
| None | 93 (89.4) |
| Present | 11 (10.6) |
| Tumor number | |
| Solitary | 83 (79.8) |
| Multiple | 21 (20.2) |
| Surgical intervention | |
| Radical resection | 85 (81.7) |
| Palliative resection | 19 (18.3) |
| Post-operative TACE | |
| No | 22 (21.2) |
| Yes | 82 (78.8) |
| Child-Pugh classification | |
| A | 92 (88.5) |
| B | 12 (11.5) |
| C | 0 (0.0) |
HBV: Hepatitis B virus; HCV: Hepatitis C virus; TACE: Transcatheter arterial chemoembolization.
Association between clinicopathological parameters and CCT3 expression in HCC patients
Immunohistochemistry analysis revealed that the CCT3 protein accumulated in the cytoplasm and nuclei of cancerous cells in HCC specimens. In non-cancerous liver cells, CCT3 accumulated mainly in the cytoplasm (Figure 3D). Overexpression of CCT3 was commonly found in the nuclei of tumor cells in moderately and poorly-differentiated HCC specimens. However, reduced overexpression of CCT3 was present in well-differentiated HCC and non-cancerous liver cells. We analyzed the associations between CCT3 expression and clinicopathological parameters. Seventy-five samples were found to have high expression of CCT3 in the cytoplasm of cancerous cells (immunostaining levels of “++” and “+++”) (Table 2), and 58 samples had high expression of CCT3 in the nuclei of cancerous cells. We observed relationships between the clinicopathological characteristics of HCC patients and CCT3 expression in the cytoplasm and nuclei of cancerous cells, respectively. No association was observed between CCT3 expression in the cytoplasm and other clinicopathological parameters.
Table 2.
Associations between subunit 3 expression in different subcellular localizations in cancerous cells and clinicopathological variables of patients with hepatocellular carcinoma n (%)
| Feature | n |
CCT3 expression in cytoplasm |
P value |
CCT3 expression in nucleus |
P value | ||
| (+) | (-) | (+) | (-) | ||||
| Gender (F/M) | |||||||
| Female | 21 | 14 (18.7) | 7 (24.1) | 11 (19.0) | 10 (21.7) | ||
| Male | 83 | 61 (81.3) | 22 (75.9) | 0.533 | 47 (81.0) | 36 (78.3) | 0.726 |
| Age (yr) | |||||||
| < 60 | 56 | 39 (52.0) | 17 (58.6) | 31 (53.4) | 25 (54.3) | ||
| ≥ 60 | 48 | 36 (48.0) | 12 (41.4) | 0.544 | 27 (46.6) | 21 (45.7) | 0.927 |
| Hepatitis type | |||||||
| HBV | 85 | 59 (78.7) | 26 (89.7) | 49 (84.5) | 36 (78.3) | ||
| HCV | 6 | 4 (5.3) | 2 (6.9) | 2 (3.4) | 4 (8.7) | ||
| None | 13 | 12 (16.0) | 1 (7.7) | 0.190 | 7 (12.1) | 6 (13.0) | 0.581 |
| Cirrhosis | |||||||
| Present | 78 | 54 (72.0) | 24 (82.8) | 43 (74.1) | 35 (76.1) | ||
| None | 26 | 21 (28.0) | 5 (17.2) | 0.2561 | 5 (25.9) | 11 (23.9) | 0.820 |
| UICC stage | |||||||
| I | 64 | 46 (61.3) | 18 (62.1) | 31 (53.4) | 33 (71.7) | ||
| II | 12 | 11 (14.7) | 1 (3.4) | 0.208 | 10 (17.2) | 2 (4.3) | 0.068 |
| IIIA | 28 | 18 (24.0) | 10 (34.5) | 17 (29.3) | 11 (23.9) | ||
| Histological type | |||||||
| Well-differentiated | 26 | 16 (21.3) | 10 (34.5) | 5 (8.6) | 21 (45.7) | ||
| Moderately-differentiated | 60 | 45 (60.0) | 15 (51.7) | 36 (62.1) | 24 (52.2) | ||
| Poorly-differentiated | 18 | 14 (18.7) | 4 (13.8) | 0.371 | 17 (29.3) | 1 (2.2) | < 0.001 |
| Macrovascular invasion | |||||||
| None | 99 | 71 (94.7) | 28 (96.6) | 54 (93.1) | 45 (97.8) | ||
| Present | 5 | 4 (5.3) | 1 (3.4) | 1.000 | 4 (6.9) | 1 (2.2) | 0.380 |
| Microvascular invasion | |||||||
| None | 78 | 55 (73.3) | 23 (79.3) | 39 (67.2) | 39 (84.8) | ||
| Present | 26 | 20 (26.7) | 6 (20.7) | 0.528 | 19 (32.8) | 7 (15.2) | 0.040 |
| Tumor size (cm) | |||||||
| ≥ 5 | 46 | 35 (46.7) | 11 (37.9) | 28 (48.1) | 18 (62.9) | ||
| < 5 | 58 | 40 (53.3) | 18 (62.1) | 0.421 | 30 (51.9) | 28 (37.1) | 0.351 |
| Serum AFP level (ng/mL) | |||||||
| < 400 | 79 | 56 (74.7) | 23 (79.3) | 43 (74.1) | 36 (78.3) | ||
| ≥ 400 | 25 | 19 (25.3) | 6 (20.7) | 0. 619 | 15 (25.9) | 10 (21.7) | 0.625 |
| Tumor number | |||||||
| Solitary | 83 | 62 (82.7) | 21 (72.4) | 46 (79.3) | 37 (80.4) | ||
| Multiple | 21 | 13 (17.3) | 8 (27.6) | 0.243 | 12 (20.7) | 9 (19.6) | 0.887 |
| Liver encapsulation invasion | |||||||
| None | 93 | 66 (88.0) | 27 (93.1) | 52 (89.7) | 41 (89.1) | ||
| Present | 11 | 9 (12.0) | 2 (6.9) | 0.687 | 6 (10.3) | 5 (10.9) | 1.000 |
HBV: Hepatitis B virus; HCV: Hepatitis C virus.
However, CCT3 expression in the nuclei was associated with histological type (P < 0.001) and microvascular invasion (P = 0.040). To confirm that the accumulation of CCT3 in the nuclei was associated with the differentiation level of HCC shown by immunohistochemistry-paraffin (IHC-P), we extracted nuclear protein from fresh frozen cancerous tissue taken from three patients with poorly-differentiated HCC, three patients with moderately-differentiated HCC and two patients with well-differentiated HCC and non-cancerous liver tissues from patients with hepatic hemangioma, and analyzed the level of CCT3 protein in the nuclei by western blotting. CCT3 expression in poorly-differentiated and moderately-differentiated cancerous tissues was higher than that in well-differentiated and non-cancerous liver tissues (Figure 3E).
To determine whether CCT3 could be a prognostic biomarker in HCC patients, we used Kaplan-Meier survival analysis to compare the associations between clinicopathological characteristics and the 3-year survival rate. The level of CCT3 expression in the nuclei correlated with the 3-year survival rate (P = 0.015) combined with postoperative TACE (P < 0.001), type of surgical intervention (P < 0.001), tumor encapsulation (P < 0.001), preoperative serum AFP (P = 0.013), tumor size (P = 0.005), microvascular invasion (P = 0.011), macrovascular invasion (P = 0.008) and TNM stage (P < 0.001). Patients with low CCT3 expression in the nuclei of cancerous cells had a better overall survival (OS) rate than patients with high expression (P = 0.014, Figure 3F). However, the level of CCT3 in the cytoplasm was not associated with these parameters. In the Cox proportional hazards regression model, high expression of CCT3 in the nuclei (HR = 2.387, 95%Cl: 1.144-4.981; P = 0.020) and no postoperative TACE (HR = 5.218, 95%Cl: 2.452-11.106; P < 0.001) were independent risk predictors for prognosis (Table 3).
Table 3.
Prognostic factors for the 3-year survival rate by univariate and Cox proportional multivariate analysis
| Factor |
Univariate analysis |
Multivariate analysis |
|||
| 3-year survival rate(%) | P value | HR (95%CI) | P value | ||
| Gender | Female/male | 70.7/71.2 | 0.921 | ||
| Age (yr) | < 60/≥ 60 | 69.5/72.6 | 0.518 | ||
| Hepatitis type | HBV/HCV/none | 73.0/83.3/52.7 | 0.265 | ||
| Cirrhosis | Present/none | 74.8/59.9 | 0.051 | ||
| TNM stage | I/II/III | 83.6/48.6/52.7 | < 0.001 | 0.254 (0.097-0.660) | 0.005 |
| 1.238 (0.465-3.295) | 0.670 | ||||
| Histological type | Well/moderately/poorlydifferentiated | 83.2/67.1/66.2 | 0.059 | ||
| Macrovascular invasion | Present/none | 60.0/71.8 | 0.008 | 1.623 (0.203-13.002) | 0.648 |
| Microvascular invasion | Present/none | 47.6/78.9 | 0.011 | 0.834 (0.358-1.939) | 0.673 |
| Tumor size (cm) | ≥ 5/< 5 | 58.6/80.2 | 0.005 | 1.488 (0.680-3.255) | 0.319 |
| Serum AFP level (ng/mL) | ≥ 400/< 400 | 59.5/77.6 | 0.013 | 1.181 (0.494-2.2820) | 0.708 |
| LiverEncapsulation invasion | Present/none | 18.2/76.5 | < 0.001 | 2.282 (0.340-15.307) | 0.395 |
| Tumor number | Solitary/multiple | 74.4/57.1 | 0.125 | ||
| Surgical intervention | Radical resection/palliative resection | 78.0/39.5 | < 0.001 | 0.811 (0.138-4.754) | 0.817 |
| Postoperative TACE | Yes/no | 77.0/48.5 | < 0.001 | 0.192 (2.157-9.159) | <0.001 |
| CCT3 expression in cytoplasm | Positive/negative | 66.8/82.2 | 0.185 | ||
| CCT3 expression in nucleus | Positive/negative | 55.5/84.2 | 0.015 | 2.387 (1.144-4.981) | 0.020 |
| Child-Pugh classification | A/B | 71.7/66.7 | 0.496 | ||
TACE: Transcatheter arterial chemoembolization.
CCT3 affected activation of STAT3
In immunocytochemistry sections, we also observed that CCT3 expression was mainly accumulated in the nuclei of NC cells. In the CCT3-siRNA group, CCT3 staining in the nuclei was reduced compared with the NC group (Figure 4A and B). To study the underlying mechanisms, we examined the effects of suppressing CCT3 expression on the level of STAT3 and (p)STAT3, which has been reported to bind with CCT3[15]. These experiments were conducted in HepG2 cells in which STAT3 had been constitutively activated[31]. In the nuclei, we observed that STAT3 and (p)STAT3 expression was downregulated in the CCT3-siRNA group (Figure 4C). To determine if suppression of CCT3 could inhibit the activation of IL-6/STAT3 signaling, we added IL-6 to the CCT3-siRNA group and NC group of HepG2 cells. The amount of (p)STAT3 and STAT3 protein in the nuclei decreased in the CCT3-siRNA group compared with the NC group (Figure 4D).
Figure 4.
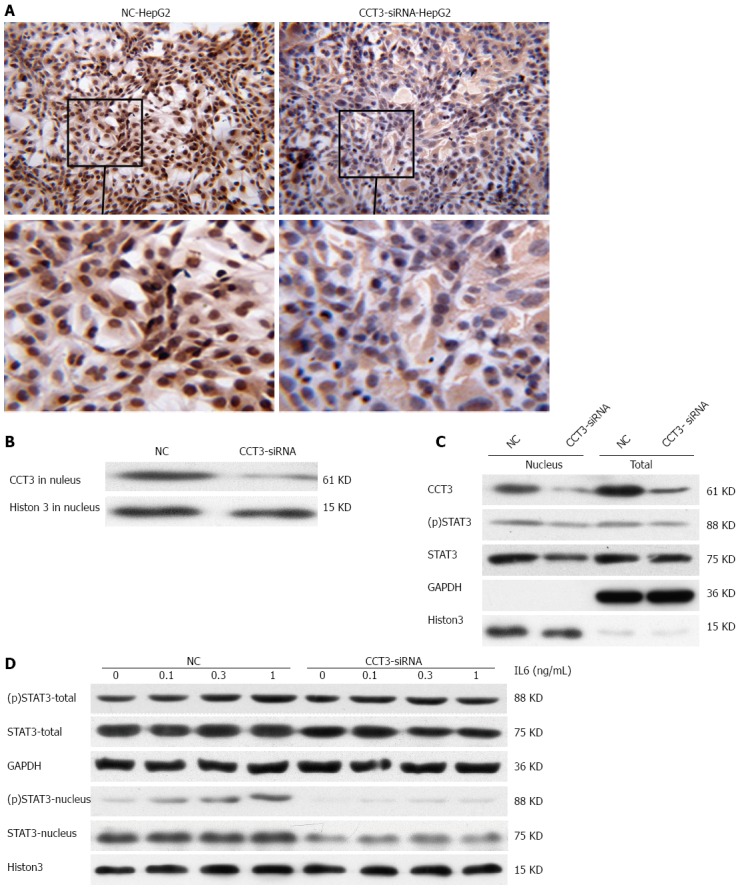
Expression of subunit 3 in HepG2 cells had an impact on the IL6/STAT3 signal pathway. A, B: Staining of subunit 3 (CCT3) in nuclei of the CCT3-siRNA group was reduced compared with the negative control (NC) group, as shown by immunocytochemistry (ICC, original magnification × 200) and western blotting; C: Knockdown of CCT3 expression reduced the level of total STAT3 in HepG2 cells and (p)STAT3 in the nucleus; D: Stimulation with IL-6 which could decreased the level of STAT3 and (p)STAT3 proteins in the nuclei of cells in the CCT3-siRNA compared with the NC group.
DISCUSSION
CCT/TRiC is an important chaperonin identified in eukaryotic cells[32]. Significant upregulation was observed in certain periods of the cell cycle, between the G1/S phase transition to the early S phase[33]. CCT/TRiC plays a critical role in cell proliferation via its effects on the folding or refolding of a variety of proteins such as: cytoskeletal proteins, which are involved in cell growth and division, for example: the actin and tubulin families; oncoproteins, such as p53, p65, cyclin E, cyclin B and VHL tumor suppressor protein[32,34].
CCT3 provides a binding site for proteins, such as tubulin and STAT3, in the folding and refolding process, which is mediated by the CCT/TRiC subunit complex[15,35]. These processes help newly translated proteins reach and maintain their native or biological state. Upregulated expression of CCT3 was observed in the log phase in eukaryotic cells[36]. To evaluate the biological functions of CCT3 in HCC, we suppressed the expression of CCT3 in vitro. The proliferation capability of cells in the CCT3-siRNA group was significantly inhibited, while apoptosis was promoted compared with the NC group. Knocking down CCT3 expression resulted in cell cycle arrest at the S phase, which seemed to prevent further downregulation of CCT3, as the expression of the CCT/TRiC subunit complex was at a low level during the G2/M phase[34]. The effects of reducing CCT3 expression on cell biological behavior were similar to knockdown of CCT1, 5 and 8[16,23,25]. This might reflect the fact that TRiC/CCT complexes would be degraded if CCT subunits are not combined with them[37]. In addition, we observed that CCT3 contributed to the invasion capacity of cells. This showed that CCT3 expression may be associated with metastasis in HCC.
Previous studies demonstrated that CCT3 was significantly elevated in many malignancies. To assess whether CCT3 could be used as a biomarker for HCC, we analyzed tumors and corresponding non-cancerous tissues from 20 HCC patients using qPCR and western blotting. Most tumor tissues showed upregulated expression of CCT3 compared with adjacent non-cancerous tissue.
To further examine the possible use of CCT3 in clinical studies, CCT3 expression was analyzed by IHC in archived formalin-fixed paraffin-embedded specimens. CCT3 staining was mainly present in the cytoplasm of normal liver cells, adjacent non-cancerous cells and cancerous cells. An interesting finding in the present study was that CCT3 staining was also present in the nuclei of some cancerous cells, especially in the poorly-differentiated and moderately-differentiated HCC specimens. This was also shown by western blotting in some fresh frozen specimens. Few studies have reported this. We decided to divide patients into the cytoplasm staining group and the nuclei staining group, according to the various subcellular locations of CCT3 staining in cancerous cells, and to determine their associations with clinicopathological variables. In the cytoplasm staining group, differences in clinical parameters were not obvious between the positive CCT3 staining group and the negative group. In the nuclei staining group, CCT3 expression was associated with microvascular invasion and histological type. This indicated that CCT3 expression in the nuclei of HCC cancerous cells was associated with tumor proliferation and metastasis. To determine the prognostic value of CCT3 in HCC patients after hepatectomy, Kaplan-Meier survival analysis was performed in these groups. There was no significant difference in the 3-year survival rate between the groups with high CCT3 expression and low expression in the cytoplasm, while patients with high CCT3 expression in the nuclei of cancerous cells had lower OS rates compared with patients with low CCT3 expression in the nuclei. In Cox proportional hazards regression, CCT3 expression in the nuclei, together with no post-operative TACE, and high TNM stage were independent risk factors in predicting poor prognosis of HCC patients. These findings indicated that CCT3 overexpression in the nuclei could be an oncogenic factor for HCC tumorigenesis.
CCT3 is mainly accumulated in the cytoplasm of cells. The reasons why CCT3 is translocated into nuclei, resulting in different prognoses, are unknown. As a critical component of CCT/TRiC, more CCT3 enters into the nuclei of cells, which may mean more CCT/TRiC is needed for biological processes in the nuclei. CCT/TRiC may participate in the following biological processes in the nuclei: (1) CCT/TRiC could interact with complexes involved in chromatin remodeling; (2) CCT/TRiC is able to interact with core nuclear pore complex (NPC) subunits and some proteins related to nuclear transport[38]; and (3) CCT/TRiC is capable of taking part in RNA processing/RNA splicing, which contributes to growth regulation[39]. However, these mechanisms require further clarification.
Recently, it was reported that STAT3 is involved with CCT3. CCT3 can enter into the nuclei with (p)STAT3[15]. STAT3 is the key transcription factor in the IL-6/STAT3 signaling pathway. Its activated form, (p)STAT3 (phosphorylation of STAT3 at Tyr705), enters the nuclei of cancerous cells and is a common phenomenon in HCC[40]. Continuous activation of STAT3 is positively correlated with proliferation and invasion in various tumor cells[41-43] and predicts poor prognosis in malignancies[26,27,43]. Whether CCT3 is a target for inhibition of STAT3 activation has not been extensively studied. To determine these effects, the expression of total STAT3 and (p)STAT3 in the nuclei was evaluated in the CCT3-siRNA group and NC group. The expressions of (p)STAT3 and STAT3 were reduced in the nuclei of the CCT3-siRNA group; however, the total decrease in STAT3 and (p)STAT3 expression was not significant. To further assess whether suppressing CCT3 expression inhibits activation of the IL-6/STAT3 pathway, various doses of IL-6 were added to the cell groups. (p)STAT3 and STAT3 in the nuclei were markedly reduced in the CCT3-siRNA group when stimulated with IL-6. CCT3 may play a key role in the translocation of (p)STAT3 and STAT3 from the cytoplasm into the nuclei. Knockdown of CCT3 may negatively regulate activation of the IL6/STAT3 signaling pathway. This could provide an explanation as to why overexpression of CCT3 in the nuclei of cancerous cells is associated with progression of HCC.
In summary, CCT3 expression in the nuclei may have significant value as an indicator of poor prognosis in HCC patients after hepatectomy. CCT3 may affect the progression of HCC partly by having an impact on the transport of (p)STAT3/STAT3 into the nuclei of HCC cells. These findings might provide a novel insight to target activated STAT3 in the treatment of HCC.
ACKNOWLEDGMENTS
We would like to thank Dr. Li-Hua Qian and Jin-Qiu Song, pathologists in the Pathology Department of Peking University People’s Hospital, for their help with the evaluation of the immunohistochemical staining. We would also like to thank Chun-Fang Zhang, an biostatistian in Peking University People’s Hospital, for her help in statistical analysis.
COMMENTS
Background
As the most common primary liver cancer, hepatocellular carcinoma (HCC) is the third most common cause of cancer death worldwide. The natural course of HCC seems to be invasive and unpredictable, and the prognosis of HCC has been assessed using different clinical classification criteria. Biomarker detection may provide important clinical evidence to predict prognosis and to design individual treatment following surgical intervention.
Research frontiers
Chaperonin Containing TCP1, Subunit 3 (CCT3) is a critical subunit of the Chaperonin containing TCP-1 (CCT/TRiC), which is essential for the folding of certain proteins into their native structure for biological function. CCT3 is associated with various proteins involved in growth, proliferation, apoptosis in normal cells and tumorigenesis. CCT3 was previously found to be upregulated in some types of malignancies, including HCC; however, its function in the progression of HCC and predicting the prognosis of patients after hepatectomy have not been reported extensively.
Innovations and breakthroughs
In this study, the authors attempted to assess the association between the expression of CCT3 and prognosis of HCC patients after hepatectomy and determine the mechanism in vitro. Overexpression of CCT3 in specific subcellular locations in cancerous cells was identified as a biomarker for poor prognosis in these patients. The mechanism may be partly attributed to its effect on the activation of IL-6/STAT3.
Applications
This study provides evidence that CCT3 may serve as a promising biomarker to evaluate the prognosis in HCC patients after hepatectomy. CCT3 may be used for targeted therapy of HCC following surgical intervention.
Peer-review
The authors assessed the value of CCT3 in different subcellular locations in cancerous cells for predicting prognosis of HCC patients and explored the mechanisms of its function in HCC progression. The study is well designed; great work has been done on a challenging research subject. The results are interesting and suggest that CCT3 could be a target for HCC treatment and to evaluate prognosis of HCC patients.
Footnotes
Supported by Beijing Key Laboratory Special Fund, No. Z141107004414042.
Institutional review board statement: This study was reviewed and approved by Institutional Review Board of Peking University People’s Hospital, Beijing, China.
Conflict-of-interest statement: The authors declare no conflict of interest in this study.
Data sharing statement: No additional data was available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: April 8, 2015
First decision: May 18, 2015
Article in press: July 3, 2015
P- Reviewer: Eghtesadi-Araghi P, Rukavina M, Varona MA S- Editor: Yu J L- Editor: Stewart G E- Editor: Zhang DN
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM, Di Bisceglie AM, Bruix J, Kramer BS, Lencioni R, Zhu AX, Sherman M, Schwartz M, Lotze M, Talwalkar J, et al. Design and endpoints of clinical trials in hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:698–711. doi: 10.1093/jnci/djn134. [DOI] [PubMed] [Google Scholar]
- 5.van Malenstein H, van Pelt J, Verslype C. Molecular classification of hepatocellular carcinoma anno 2011. Eur J Cancer. 2011;47:1789–1797. doi: 10.1016/j.ejca.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 6.Ditzel L, Löwe J, Stock D, Stetter KO, Huber H, Huber R, Steinbacher S. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell. 1998;93:125–138. doi: 10.1016/s0092-8674(00)81152-6. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, Willison KR, Horwich AL. Cystosolic chaperonin subunits have a conserved ATPase domain but diverged polypeptide-binding domains. Trends Biochem Sci. 1994;19:543–548. doi: 10.1016/0968-0004(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 8.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Yam AY, Xia Y, Lin HT, Burlingame A, Gerstein M, Frydman J. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol. 2008;15:1255–1262. doi: 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valpuesta JM, Martín-Benito J, Gómez-Puertas P, Carrascosa JL, Willison KR. Structure and function of a protein folding machine: the eukaryotic cytosolic chaperonin CCT. FEBS Lett. 2002;529:11–16. doi: 10.1016/s0014-5793(02)03180-0. [DOI] [PubMed] [Google Scholar]
- 11.Sternlicht H, Farr GW, Sternlicht ML, Driscoll JK, Willison K, Yaffe MB. The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proc Natl Acad Sci USA. 1993;90:9422–9426. doi: 10.1073/pnas.90.20.9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen WJ, Ohh M, Moslehi J, Kondo K, Kaelin WG, Welch WJ. Diverse effects of mutations in exon II of the von Hippel-Lindau (VHL) tumor suppressor gene on the interaction of pVHL with the cytosolic chaperonin and pVHL-dependent ubiquitin ligase activity. Mol Cell Biol. 2002;22:1947–1960. doi: 10.1128/MCB.22.6.1947-1960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melki R, Batelier G, Soulié S, Williams RC. Cytoplasmic chaperonin containing TCP-1: structural and functional characterization. Biochemistry. 1997;36:5817–5826. doi: 10.1021/bi962830o. [DOI] [PubMed] [Google Scholar]
- 14.Won KA, Schumacher RJ, Farr GW, Horwich AL, Reed SI. Maturation of human cyclin E requires the function of eukaryotic chaperonin CCT. Mol Cell Biol. 1998;18:7584–7589. doi: 10.1128/mcb.18.12.7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasembeli M, Lau WC, Roh SH, Eckols TK, Frydman J, Chiu W, Tweardy DJ. Modulation of STAT3 folding and function by TRiC/CCT chaperonin. PLoS Biol. 2014;12:e1001844. doi: 10.1371/journal.pbio.1001844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trinidad AG, Muller PA, Cuellar J, Klejnot M, Nobis M, Valpuesta JM, Vousden KH. Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol Cell. 2013;50:805–817. doi: 10.1016/j.molcel.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong N, Chan A, Lee SW, Lam E, To KF, Lai PB, Li XN, Liew CT, Johnson PJ. Positional mapping for amplified DNA sequences on 1q21-q22 in hepatocellular carcinoma indicates candidate genes over-expression. J Hepatol. 2003;38:298–306. doi: 10.1016/s0168-8278(02)00412-9. [DOI] [PubMed] [Google Scholar]
- 18.Skawran B, Steinemann D, Weigmann A, Flemming P, Becker T, Flik J, Kreipe H, Schlegelberger B, Wilkens L. Gene expression profiling in hepatocellular carcinoma: upregulation of genes in amplified chromosome regions. Mod Pathol. 2008;21:505–516. doi: 10.1038/modpathol.3800998. [DOI] [PubMed] [Google Scholar]
- 19.Peters DG, Kudla DM, Deloia JA, Chu TJ, Fairfull L, Edwards RP, Ferrell RE. Comparative gene expression analysis of ovarian carcinoma and normal ovarian epithelium by serial analysis of gene expression. Cancer Epidemiol Biomarkers Prev. 2005;14:1717–1723. doi: 10.1158/1055-9965.EPI-04-0704. [DOI] [PubMed] [Google Scholar]
- 20.Midorikawa Y, Tsutsumi S, Taniguchi H, Ishii M, Kobune Y, Kodama T, Makuuchi M, Aburatani H. Identification of genes associated with dedifferentiation of hepatocellular carcinoma with expression profiling analysis. Jpn J Cancer Res. 2002;93:636–643. doi: 10.1111/j.1349-7006.2002.tb01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Y, Deng X, Zhan Q, Shen B, Jin X, Zhu Z, Chen H, Li H, Peng C. A prospective proteomic-based study for identifying potential biomarkers for the diagnosis of cholangiocarcinoma. J Gastrointest Surg. 2013;17:1584–1591. doi: 10.1007/s11605-013-2182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Lin CY, Lei M, Yan S, Zhou T, Erikson RL. CCT chaperonin complex is required for the biogenesis of functional Plk1. Mol Cell Biol. 2005;25:4993–5010. doi: 10.1128/MCB.25.12.4993-5010.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boudiaf-Benmammar C, Cresteil T, Melki R. The cytosolic chaperonin CCT/TRiC and cancer cell proliferation. PLoS One. 2013;8:e60895. doi: 10.1371/journal.pone.0060895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang X, Wang X, Cheng C, Cai J, He S, Wang H, Liu F, Zhu C, Ding Z, Huang X, et al. Chaperonin containing TCP1, subunit 8 (CCT8) is upregulated in hepatocellular carcinoma and promotes HCC proliferation. APMIS. 2014;122:1070–1079. doi: 10.1111/apm.12258. [DOI] [PubMed] [Google Scholar]
- 25.He G, Yu GY, Temkin V, Ogata H, Kuntzen C, Sakurai T, Sieghart W, Peck-Radosavljevic M, Leffert HL, Karin M. Hepatocyte IKKbeta/NF-kappaB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell. 2010;17:286–297. doi: 10.1016/j.ccr.2009.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calvisi DF, Ladu S, Gorden A, Farina M, Conner EA, Lee JS, Factor VM, Thorgeirsson SS. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117–1128. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Kikuchi LO, Paranaguá-Vezozzo DC, Chagas AL, Mello ES, Alves VA, Farias AQ, Pietrobon R, Carrilho FJ. Nodules less than 20 mm and vascular invasion are predictors of survival in small hepatocellular carcinoma. J Clin Gastroenterol. 2009;43:191–195. doi: 10.1097/MCG.0b013e31817ff199. [DOI] [PubMed] [Google Scholar]
- 28.Poté N, Cauchy F, Albuquerque M, Voitot H, Belghiti J, Castera L, Puy H, Bedossa P, Paradis V. Performance of PIVKA-II for early hepatocellular carcinoma diagnosis and prediction of microvascular invasion. J Hepatol. 2015;62:848–854. doi: 10.1016/j.jhep.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z, Li L, Yang Z, Luo W, Li X, Yang H, Yao K, Wu B, Fang W. Increased expression of MMP9 is correlated with poor prognosis of nasopharyngeal carcinoma. BMC Cancer. 2010;10:270. doi: 10.1186/1471-2407-10-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Y, Luo Q, Long H, Hu Z, Que T, Zhang X, Li Z, Wang G, Yi L, Liu Z, et al. Alpha-enolase as a potential cancer prognostic marker promotes cell growth, migration, and invasion in glioma. Mol Cancer. 2014;13:65. doi: 10.1186/1476-4598-13-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun X, Zhang J, Wang L, Tian Z. Growth inhibition of human hepatocellular carcinoma cells by blocking STAT3 activation with decoy-ODN. Cancer Lett. 2008;262:201–213. doi: 10.1016/j.canlet.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Archibald JM, Logsdon JM, Doolittle WF. Origin and evolution of eukaryotic chaperonins: phylogenetic evidence for ancient duplications in CCT genes. Mol Biol Evol. 2000;17:1456–1466. doi: 10.1093/oxfordjournals.molbev.a026246. [DOI] [PubMed] [Google Scholar]
- 33.Yokota S, Yanagi H, Yura T, Kubota H. Cytosolic chaperonin is up-regulated during cell growth. Preferential expression and binding to tubulin at G(1)/S transition through early S phase. J Biol Chem. 1999;274:37070–37078. doi: 10.1074/jbc.274.52.37070. [DOI] [PubMed] [Google Scholar]
- 34.Pejanovic N, Hochrainer K, Liu T, Aerne BL, Soares MP, Anrather J. Regulation of nuclear factor κB (NF-κB) transcriptional activity via p65 acetylation by the chaperonin containing TCP1 (CCT) PLoS One. 2012;7:e42020. doi: 10.1371/journal.pone.0042020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alcolea PJ, Alonso A, Larraga V. Proteome profiling of Leishmania infantum promastigotes. J Eukaryot Microbiol. 2011;58:352–358. doi: 10.1111/j.1550-7408.2011.00549.x. [DOI] [PubMed] [Google Scholar]
- 36.Dekker C, Stirling PC, McCormack EA, Filmore H, Paul A, Brost RL, Costanzo M, Boone C, Leroux MR, Willison KR. The interaction network of the chaperonin CCT. EMBO J. 2008;27:1827–1839. doi: 10.1038/emboj.2008.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunisawa J, Shastri N. The group II chaperonin TRiC protects proteolytic intermediates from degradation in the MHC class I antigen processing pathway. Mol Cell. 2003;12:565–576. doi: 10.1016/j.molcel.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Fofaria NM, Srivastava SK. STAT3 induces anoikis resistance, promotes cell invasion and metastatic potential in pancreatic cancer cells. Carcinogenesis. 2015;36:142–150. doi: 10.1093/carcin/bgu233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang R, Yu M, Li CY, Zhan YQ, Xu WX, Xu F, Ge CH, Li W, Yang XM. New insights into the functions and localization of nuclear CCT protein complex in K562 leukemia cells. Proteomics Clin Appl. 2012;6:467–475. doi: 10.1002/prca.201200009. [DOI] [PubMed] [Google Scholar]
- 40.Li WC, Ye SL, Sun RX, Liu YK, Tang ZY, Kim Y, Karras JG, Zhang H. Inhibition of growth and metastasis of human hepatocellular carcinoma by antisense oligonucleotide targeting signal transducer and activator of transcription 3. Clin Cancer Res. 2006;12:7140–7148. doi: 10.1158/1078-0432.CCR-06-0484. [DOI] [PubMed] [Google Scholar]
- 41.Xuan X, Li S, Lou X, Zheng X, Li Y, Wang F, Gao Y, Zhang H, He H, Zeng Q. Stat3 promotes invasion of esophageal squamous cell carcinoma through up-regulation of MMP2. Mol Biol Rep. 2015;42:907–915. doi: 10.1007/s11033-014-3828-8. [DOI] [PubMed] [Google Scholar]
- 42.Fofaria NM, Srivastava SK. Critical role of STAT3 in melanoma metastasis through anoikis resistance. Oncotarget. 2014;5:7051–7064. doi: 10.18632/oncotarget.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rebouissou S, Amessou M, Couchy G, Poussin K, Imbeaud S, Pilati C, Izard T, Balabaud C, Bioulac-Sage P, Zucman-Rossi J. Frequent in-frame somatic deletions activate gp130 in inflammatory hepatocellular tumours. Nature. 2009;457:200–204. doi: 10.1038/nature07475. [DOI] [PMC free article] [PubMed] [Google Scholar]


