Abstract
Background: Neuropeptide Y is a key peptide affecting adiposity and has been related to obesity risk. However, little is known about the role of NPY variations in diet-induced change in adiposity.
Objective: The objective was to examine the effects of NPY variant rs16147 on central obesity and abdominal fat distribution in response to dietary interventions.
Design: We genotyped a functional NPY variant rs16147 among 723 participants in the Preventing Overweight Using Novel Dietary Strategies trial. Changes in waist circumference (WC), total abdominal adipose tissue, visceral adipose tissue, and subcutaneous adipose tissue (SAT) from baseline to 6 and 24 mo were evaluated with respect to the rs16147 genotypes. Genotype–dietary fat interaction was also examined.
Results: The rs16147 C allele was associated with a greater reduction in WC at 6 mo (P < 0.001). In addition, the genotypes showed a statistically significant interaction with dietary fat in relation to WC and SAT (P-interaction = 0.01 and 0.04): the association was stronger in individuals with high-fat intake than in those with low-fat intake. At 24 mo, the association remained statistically significant for WC in the high-fat diet group (P = 0.02), although the gene–dietary fat interaction became nonsignificant (P = 0.30). In addition, we found statistically significant genotype–dietary fat interaction on the change in total abdominal adipose tissue, visceral adipose tissue, and SAT at 24 mo (P = 0.01, 0.05, and 0.04): the rs16147 T allele appeared to associate with more adverse change in the abdominal fat deposition in the high-fat diet group than in the low-fat diet group.
Conclusion: Our data indicate that the NPY rs16147 genotypes affect the change in abdominal adiposity in response to dietary interventions, and the effects of the rs16147 single-nucleotide polymorphism on central obesity and abdominal fat distribution were modified by dietary fat. This trial was registered at clinicaltrials.gov as NCT00072995.
Keywords: dietary fat, fat distribution, gene-nutrient interaction, neuropeptide Y, SNP
INTRODUCTION
Neuropeptide Y (NPY)16 is a 36–amino acid peptide mainly secreted by neurons (1) and plays an important role in food intake (2), obesity (3, 4), immune function (5), and cardiovascular regulation (6, 7). The adipogenic and antilipolytic effects of NPY have been widely reported (4, 8, 9). Evidence has indicated that the NPY gene is associated with obesity through stimulating food intake, decreasing energy expenditure (10), and increasing energy stored as adiposity (11). Previously, we found that genetic variants in the NPY gene affected long-term weight change in prospective cohorts (12). A functional single-nucleotide polymorphism (SNP) in the promoter region of NPY, rs16147 (C-399T), was found to show allele-specific effects on NPY gene expression and NPY peptide concentrations (12–16).
Central obesity and distribution of body fat have been related to a variety of metabolic disorders such as diabetes and cardiovascular disease, independent of overall obesity (17, 18). Previous studies have also indicated a pivotal role of NPY in determining body fat distribution. In brief, it has been postulated that NPY and adipokines form a feedback loop: NPY regulates the white adipose tissue (WAT) metabolism via the nerve endings housed in WAT (19); adipokines, secreted by WAT, then exert as a sensory input and inform the brain of body fat levels (20, 21).
To date, no studies have assessed the effect of NPY variants on long-term change in comprehensive measures of central obesity and abdominal fat distribution in clinical trials. In addition, the interaction between diet and NPY variants on central obesity and abdominal fat distribution has not been investigated in humans. Therefore, in this study, we aimed to investigate whether the rs16147 genotypes modulated the effects of weight loss diets varying in macronutrients on changes of waist circumference and abdominal fat distribution in a 2-y diet intervention study, the Preventing Overweight Using Novel Dietary Strategies (POUNDS LOST) trial.
METHODS
Study population
The POUNDS LOST trial is a 2-y randomized clinical trial to compare the effects on body weight of energy-reduced diets with different dietary intakes of macronutrients (22). In total, 811 (Caucasian: n = 643, African American: n = 127, Hispanic: n = 29, other: n = 12) overweight adults were randomly assigned to one of the 4 diets with targeted percentages of energy derived from fat, protein, and carbohydrates as 20%, 15%, and 65%; 20%, 25%, and 55%; 40%, 15%, and 45%; and 40%, 25%, and 35%. Of 811 participants who were assigned to a diet, 645 completed the study (white: n = 525, African American: n = 88, Hispanic: n = 23, other: n = 9). The baseline characteristics of participants who completed the study were similar to those who were assigned to one of the 4 diets. Our analysis was restricted to 723 participants with available DNA samples (white: n = 575, African American: n = 112, Hispanic: n = 25, other: n = 11).
Dietary intervention
The dietary program consisted of 4 diet groups and the targeted percentages of energy derived from fat, protein, and carbohydrate in the 4 diets as described above. The energy deficit was 750 kcal from baseline, and low glycemic index food was used for the diets. All participants were instructed to consume >20 g dietary fiber, ≤8% saturated fat, and ≤150 mg cholesterol/100 kcal. Participants were screened for potential causes of noncompliance. A multidimensional construct was used to conceptualize the adherence to the intervention (23). A 5-d diet record at baseline and a 24-h recall on 3 nonconsecutive days at months 6 and 24 in a random sample of 50% of all participants were used to assess the adherence to the dietary program. Biomarkers of adherence, including HDL cholesterol, urinary nitrogen, and respiratory quotient, were used to confirm the self-reported adherence.
Assessment of anthropometric and other traits
Body weight and waist circumference (WC) were measured in the morning before breakfast on 2 nonconsecutive days at baseline, 6 mo, and 24 mo, as well as on a single day at 12 and 18 mo. Height was measured at baseline on the same day when body weight was measured. Dietary intakes were assessed in a random sample of 50% of the participants by a review of a 5-d diet record at baseline and a 24-h recall during a telephone interview on 3 nonconsecutive days at 6 mo and 24 mo. Abdominal fat tissue mass was measured by computed tomography (CT) scanning with a General Electric High-Light CT scanner (General Electric) or a GE LightSpeed volume CT scanner (General Electric). A random subset of 25% of all participants was selected to receive CT scans, and CT scans were done at baseline, 6 mo, and 24 mo.
Genotyping
The previously identified functional NPY rs16147 SNP (13, 14) was genotyped in the study population. DNA was extracted from buffy coat by using a QIAmp Blood Kit (Qiagen), and genotyping was performed by using the OpenArray SNP Genotyping System (BioTrove). Control samples typed in duplicate ensured the internal quality of the genotyping and resulted in a concordance of >99%. The total genotype success rate was 99%. The NPY rs16147 SNP was successfully genotyped in all 723 participants with available DNA samples.
Ethics
The study was approved by the human subjects committee at the Harvard School of Public Health and Brigham and Women’s Hospital, Boston, Massachusetts, Pennington Biomedical Research Center, and by a data and safety monitoring board appointed by the National Heart, Lung, and Blood Institute. All participants gave written informed consent. The study was registered at clinicaltrials.gov as NCT00072995.
Statistical analysis
We conducted the post hoc analyses of data from the POUNDS LOST trial to evaluate the effects of the NPY genotypes and the genotype–dietary fat interaction on measures of central obesity and abdominal fat distribution. The primary outcome was the change in WC. The secondary outcomes included total abdominal adipose tissue (TAT), visceral adipose tissue (VAT), and subcutaneous adipose tissue (SAT). Because previous studies have reported the interaction between NPY and dietary fat (7, 24–26), the comparison between the low-fat group (20%) and the high-fat group (40%) was of the main interest in this study. Differences in baseline characteristics by genotypes were tested by using the χ2 test for categorical variables and ANCOVA for continuous variables (ANOVA for age). Additive genetic model was used in the analysis for the rs16147 genotypes. Main effects of the rs16147 genotypes and dietary fat intake on the changes in the primary and the secondary outcomes at 6 mo and 24 mo were examined by using generalized linear regression models. Covariates adjusted for the effects of genotypes include age, sex, ethnicity, baseline value for the respective outcome, and baseline BMI. Genotype–dietary fat interactions were evaluated by further adding the interaction terms into the model used to examine the main effects. Subgroup analyses were performed for groups on diets with different percentages of energy from fat adjusting for age, sex, ethnicity, baseline value for the respective outcome, and baseline BMI. Similar analyses were conducted among Caucasians to evaluate the influence of potential population stratification.
All statistical computation was conducted by using SAS (version 9.3; SAS Institute). P < 0.05 was used as the significance level, and all reported P values were 2-sided (except for likelihood ratio tests). Quanto 1.2.4 (University of Southern California, Los Angeles; http://hydra.usc.edu.gxe) was used to estimate the detectable effect sizes of genotype-diet (2 diet groups with different percentages of energy from fat) interactions (27). For power and effect size calculation, we assumed that the population mean ± SD change in WC was −6.87 ± 6.18 and −5.70 ± 7.68 cm at 6 mo and 24 mo. At the significance level of 0.05, the study had 80% power to detect the gene-diet interaction effect sizes of 1.85 and 2.51 cm for the change in WC from baseline to 6 mo and 24 mo, respectively.
RESULTS
Baseline characteristics of the study population
The allele frequency in all participants was in Hardy-Weinberg equilibrium (P > 0.05). The ethnic composition and the age distribution were statistically significantly different across the NPY genotypes (P = 0.01 and 0.02). There was no statistically significant difference in baseline BMI, WC, and abdominal adiposity distribution across the genotypes, after adjusting for age, sex, and ethnicity (Table 1). The dietary intakes were modified in the direction of the assigned interventions, which is validated by the analysis of the biomarkers of adherence (Supplemental Table 1). Similar associations between the rs16147 frequencies and baseline characteristics were observed in Caucasians (∼80% of the participants) (22, 28).
TABLE 1.
Baseline characteristics of the study participants1
|
NPY rs16147 genotype |
||||
| CC (n = 203) | CT (n = 341) | TT (n = 179) | P value | |
| Age, y | 52.5 ± 8.72 | 50.6 ± 9.5 | 49.9 ± 9.3 | 0.02 |
| Sex, n (%) | 0.66 | |||
| Female | 126 (28.6) | 211 (47.8) | 104 (23.6) | |
| Male | 77 (27.3) | 130 (46.1) | 75 (26.6) | |
| Ethnicity, n (%) | 0.01 | |||
| Caucasian | 146 (25.4) | 274 (47.6) | 155 (27.0) | |
| African American | 44 (39.3) | 53 (47.3) | 15 (13.4) | |
| Hispanic | 10 (40.0) | 11 (44.0) | 4 (16.0) | |
| Others | 3 (27.3) | 3 (27.3) | 5 (45.4) | |
| Diet groups (% energy from fat/protein/carbohydrate), n (%) | 0.85 | |||
| Group 1 (20/25/55) | 48 (27.3) | 88 (50.0) | 40 (22.7) | |
| Group 2 (20/15/65) | 51 (28.0) | 79 (43.4) | 52 (28.6) | |
| Group 3 (40/25/35) | 52 (28.7) | 84 (46.4) | 45 (24.9) | |
| Group 4 (40/15/45) | 52 (28.3) | 90 (48.9) | 42 (22.8) | |
| Dietary fat (% energy from fat/other), n (%) | 0.84 | |||
| Low fat (20) | 99 (27.6) | 167 (46.7) | 92 (25.7) | |
| High fat (40) | 104 (28.5) | 174 (47.7) | 87 (23.8) | |
| BMI, kg/m2 | 32.8 ± 4.1 | 32.7 ± 3.7 | 32.4 ± 5.0 | 0.58 |
| Waist circumference, cm | 104.0 ± 13.4 | 103.7 ± 12.7 | 103.0 ± 13.2 | 0.49 |
| TAT mass, kg | 16.1 ± 4.6 | 16.8 ± 3.8 | 17.0 ± 4.0 | 0.70 |
| VAT mass, kg | 5.4 ± 2.7 | 5.5 ± 2.5 | 5.9 ± 2.4 | 0.75 |
| SAT mass, kg | 16.1 ± 4.8 | 17.3 ± 4.0 | 16.7 ± 3.9 | 0.50 |
P values were calculated by the χ2 test for categorical variables and ANCOVA for continuous variables after adjusting for age, sex, and ethnicity (ANOVA for age). SAT, subcutaneous adipose tissue; TAT, total abdominal adipose tissue; VAT, visceral adipose tissue.
Mean ± SD (all such values).
Effect of the NPY rs16147 genotypes on measures of central obesity and abdominal fat distribution
We first examined the main genotype effects on the primary outcome: the change in WC from baseline. After adjusting for age, sex, ethnicity, dietary interventions, and baseline values of the outcome variables, the C allele of rs16147 was associated with a statistically significantly greater reduction in WC in all the participants at 6 mo (P = 0.03) (Table 2). The change in WC from baseline to 6 mo had a mean increase of 0.7 cm with each additional copy of the T allele. We further assessed measures of abdominal fat distribution: TAT, VAT, and SAT. No statistically significant overall genotype effects were observed (all P > 0.05).
TABLE 2.
Analyses of the effect of the NPY rs16147 genotype on changes in waist circumference and abdominal fat distribution from baseline to 6 and 24 mo1
| Month 6 |
Month 24 |
|||||||
| n | β | SE | P value | n | β | SE | P value | |
| WC, cm | 643 | 0.70 | 0.32 | 0.03 | 548 | 0.70 | 0.43 | 0.11 |
| TAT, kg | 112 | 0.37 | 0.29 | 0.20 | 84 | 0.03 | 0.45 | 0.95 |
| VAT, kg | 132 | 0.14 | 0.13 | 0.27 | 102 | 0.05 | 0.18 | 0.77 |
| SAT, kg | 112 | 0.36 | 0.28 | 0.21 | 84 | 0.04 | 0.61 | 0.93 |
Estimates and standard errors for the subgroup analyses were reported after adjusting for age, sex, ethnicity, diet intervention, baseline value for respective variable, and baseline BMI. β represents change in outcomes for the increase in each T allele of NPY rs16147. SAT, subcutaneous adipose tissue; TAT, total abdominal adipose tissue; VAT, visceral adipose tissue; WC, waist circumference.
Interactions between the NPY rs16147 genotypes and dietary fat on the change in WC
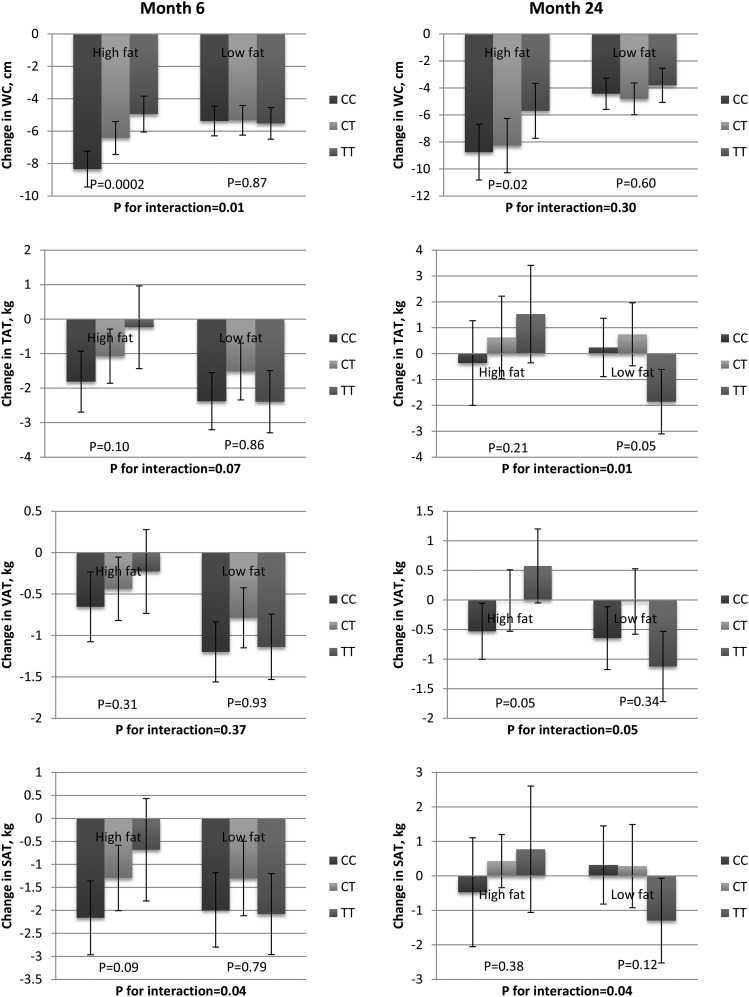
We next examined the interactions between the rs16147 genotypes and dietary interventions for WC at 6 mo and 24 mo. We found a statistically significant interaction between the NPY genotype and dietary fat on WC: the genotype effects were stronger in the high-fat diet group than in the low-fat diet group at 6 mo (P-interaction = 0.01) (Figure 1). Within the high-fat group, the T allele was statistically significantly associated with smaller loss in WC. At 24 mo, the association between the rs16147 variant and the reduction of WC remained statistically significant in the high-fat group (P = 0.02), although the gene–dietary fat interaction became not statistically significant. Analyses in Caucasians showed similar associations in terms of direction and magnitude. Similar genotype effects and genotype interaction effects were observed in Caucasians.
FIGURE 1.
Effect of NPY rs16147 and dietary fat on the change in WC, TAT, VAT, and SAT from baseline to 6 and 24 mo. Data are means ± SEs after adjustment for age, sex, ethnicity, baseline value for respective variables, and baseline BMI. P values for the interaction between genotype and dietary fat are presented in bold. P values for the genotype effects within each subgroup are presented for both high-fat and low-fat diet groups. The low-fat diet group consisted of 2 intervention groups with targeted percentages of energy from fat of 20%; the high-fat diet group consisted of 2 intervention groups with targeted percentages of energy from fat of 40%. The sample size for each dietary fat and genotype combination at 6 mo: WC, low fat, CC = 86, CT = 146, TT = 87; WC, high fat, CC = 90, CT = 160, TT = 74; TAT, low fat, CC = 15, CT = 25, TT = 18; TAT, high fat, CC = 18, CT = 29, TT = 7; VAT, low fat, CC = 17, CT = 30, TT = 19; VAT, high fat, CC = 20, CT = 35, TT = 11; SAT, low fat, CC = 15, CT = 25, TT = 18; and SAT, high fat, CC = 18, CT = 29, TT = 7. The sample size for each dietary fat and genotype combination at 24 mo: WC, low fat, CC = 76, CT = 127, TT = 74; WC, high fat, CC = 82, CT = 127, TT = 62; TAT, low fat, CC = 13, CT = 18, TT = 13; TAT, high fat, CC = 12, CT = 22, TT = 6; VAT, low fat, CC = 16, CT = 22, TT = 15; VAT, high fat, CC = 14, CT = 26, TT = 9; SAT, low fat, CC = 13, CT = 18, TT = 13; SAT, high fat, CC = 12, CT = 22, TT = 6. SAT, subcutaneous adipose tissue; TAT, total abdominal adipose tissue; VAT, visceral adipose tissue; WC, waist circumference.
Interactions between the NPY rs16147 genotypes and dietary fat on abdominal fat distribution
The interaction between the NPY rs16147 genotypes and dietary fat was also examined for the measures of abdominal fat distribution, including TAT, VAT, and SAT. At 6 mo, there was a statistically significant interaction between the NPY genotype and dietary fat on SAT (P-interaction = 0.04) (Figure 1). Similar trends were also found for TAT and VAT, but the interaction was not statistically significant at 6 mo (P-interaction = 0.07 and 0.37). Compared with the high-fat group, no clear trend was found within the low-fat group. We found a statistically significant genotype–dietary fat interaction for all 3 measures of abdominal fat deposition at 24 mo: among the high-fat diet consumers, the association between the rs16147 T allele and each measure of abdominal fat distribution was statistically significantly stronger than among the participants taking a low-fat diet (P-interaction = 0.01, 0.05, and 0.04). Similar results were also found among Caucasians.
DISCUSSION
In the POUNDS LOST trial, we found that the NPY rs16147 SNP was statistically significantly related to the change in WC at both 6 and 24 mo, especially among the participants assigned to the high-fat diet intervention. A statistically significant gene–dietary fat interaction was observed for WC and SAT at 6 mo. Also, a smaller reduction of WC was observed among the T allele carriers in the high-fat group compared with those who consumed the low-fat diet. At 24 mo, T allele carriers in the high-fat group were prone to regain abdominal fat. The associations of the rs16147 genotypes with the change in WC, TAT, VAT, and SAT from baseline to 24 mo were statistically significantly different between the high-fat group and the low-fat group.
The NPY variant rs16147 has been related to the expression of the NPY gene and the peptide concentration in some studies (12–16). Variation in the NPY expression can be largely explained by this SNP (13, 15). The rs16147 genotypes have also been reported to associate with adiposity measures (12). In children, the C allele was associated with lower body weight (29, 30), although the results obtained from adults are conflicting (12). However, our data showed that carriers of the rs16147 C allele were more responsive to weight loss diets in the reduction of central body fat, lending support to a role of the NPY genotypes in the regulation of abdominal fat distribution.
It has been postulated that NPY can regulate WAT metabolism via the nerve endings housed in WAT (19). Leptin, generated from WAT, then forms a feedback loop with NPY and informs the brain of body fat levels (20, 21). Anatomic evidence also suggests that reduced NPY expression leads to improved insulin and leptin sensitivity via afferent nerve signals from abdominal fat tissues to the hypothalamus (31). To our knowledge, the present study is the first one that comprehensively examined the NPY genetic effects and the interaction between the rs16147 genotypes and dietary fat with respect to central obesity and abdominal fat distribution.
Intriguingly, we found that dietary fat intake statistically significantly modified the genetic effect on change in WC, TAT, VAT, and SAT; individuals on a high-fat weight loss diet showed more effectiveness in reduction of central adiposity compared with those on a low-fat diet. Evidence from animal models shows that knockdown of NPY receptor 2, a major mediator of the NPY effects, in the peripheral tissues of mice prevented high-fat diet–induced obesity, whereas no such preventative effects were discovered among normal chow-fed mice (32). In addition, there is evidence that dietary fat might regulate NPY gene expression (33), supporting the potential interactions between NPY genotypes and dietary fat in determining body adiposity. Those findings, combined with the evidence supporting the adipose tissue–hypothalamus feedback loop, suggest that dietary fat may affect rs16147-regulated NPY expression and therefore modify the NPY effects on fat distribution via the leptin feedback. Unfortunately, the precise molecular mechanisms underlying the observed gene-diet interaction remain unclear, and further studies are warranted to investigate the functional basis for the interaction.
Our previous study found no genetic effects or interaction with dietary fat in relation to weight loss, which is confirmed by our analysis of BMI (7). Interestingly, we found a statistically significant genetic effect of rs16147 on WC, which is a measure of central obesity. Also, the rs16147 T allele appeared to associate with worse responses to a high-fat weight loss diet in terms of WC and abdominal fat deposition. It has been well known that WC, as a reliable indicator of abdominal fat, predicts various chronic diseases such as cardiovascular diseases, diabetes, and cancers, independent of BMI (17, 18, 34–36). More important, abdominal fat is a particularly strong risk factor of these metabolic disorders among overweight and obese people (36). The initial findings from the POUNDS LOST trial did not finding statistically significant dietary effects on WC (22). Our findings indicate that various diet interventions may differentially affect changes in adiposity when individuals’ genotype is not taken into consideration. Therefore, findings from our study provide important information for future research on the NPY genotype–dietary fat interaction in relation to other health outcomes. More important, our study has great clinical implications for the design of well-targeted dietary interventions.
Despite the statistically significant implications, there are several potential limitations in our study. First, the results should be interpreted and generalized with caution because the diet intervention was calorically restricted and all participants were overweight or obese. Second, we did not include low-frequency SNPs such as rs16139, which has also been reported (12). Because of the limited power to test gene-diet interactions for rare variants, we restricted our study to rs16147 with a frequency of 0.48. However, as discussed above, most of the variation in NPY expression can be explained by rs16147 (13, 15). Third, the analyses for the measures of abdominal fat distribution might be underpowered, although we observed statistically significant genotype–dietary fat interactions. This may explain why we did not observe statistically significant associations between rs16147 and measures of abdominal fat distribution in the high-fat group. To confirm our findings and to better understand the effects of the NPY genetic variants on abdominal fat distribution in response to dietary intervention, more studies with larger sample sizes are required.
In conclusion, we found that the NPY rs16147 genotypes affected the reduction of central adiposity and the deposition of abdominal fat in response to weight loss diets in overweight or obese participants. Dietary fat intake statistically significantly modified the genetic effects such that individuals carrying the C allele of the NPY rs16147 SNP might benefit more by taking a high-fat weight loss diet.
Acknowledgments
The authors’ responsibilities were as follows—GAB, FMS, and LQ: designed the research; XL, GAB, FMS, LL, and LQ: conducted the research; XL, QQ, LL, and LQ: analyzed the data or performed statistical analysis; XL and LQ: wrote the manuscript; XL, QQ, YZ, TH, ML, DZ, GAB, FMS, LL, and LQ: critically reviewed the manuscript and accepted it to be submitted; and LQ: had primary responsibility for the final content. The authors declared that they had no competing interests or conflicts of interest related to this study.
Footnotes
Abbreviations used: NPY, neuropeptide Y; POUNDS LOST, Preventing Overweight Using Novel Dietary Strategies; SAT, subcutaneous adipose tissue; SNP, single-nucleotide polymorphism; TAT, abdominal adipose tissue; VAT, visceral adipose tissue; WC, waist circumference.
REFERENCES
- 1.Ding B, Kull B, Liu Z, Mottagui-Tabar S, Thonberg H, Gu HF, Brookes AJ, Grundemar L, Karlsson C, Hamsten A, et al. . Human neuropeptide Y signal peptide gain-of-function polymorphism is associated with increased body mass index: possible mode of function. Regul Pept 2005;127:45–53. [DOI] [PubMed] [Google Scholar]
- 2.Lin S, Boey D, Herzog H. NPY and Y receptors: lessons from transgenic and knockout models. Neuropeptides 2004;38:189–200. [DOI] [PubMed] [Google Scholar]
- 3.Dryden S, Pickavance L, Frankish HM, Williams G. Increased neuropeptide Y secretion in the hypothalamic paraventricular nucleus of obese (fa/fa) Zucker rats. Brain Res 1995;690:185–8. [DOI] [PubMed] [Google Scholar]
- 4.Kuo LE, Kitlinska JB, Tilan JU, Li L, Baker SB, Johnson MD, Lee EW, Burnett MS, Fricke ST, Kvetnansky R, et al. . Neuropeptide Y acts directly in the periphery on fat tissue and mediates stress-induced obesity and metabolic syndrome. Nat Med 2007;13:803–11. [DOI] [PubMed] [Google Scholar]
- 5.Wheway J, Mackay CR, Newton RA, Sainsbury A, Boey D, Herzog H, Mackay F. A fundamental bimodal role for neuropeptide Y1 receptor in the immune system. J Exp Med 2005;202:1527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedrazzini T, Brunner HR, Waeber B. Neuropeptide Y and cardiovascular regulation. Curr Opin Nephrol Hypertens 1993;2:106–13. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Qi Q, Liang J, Hu FB, Sacks FM, Qi L. Neuropeptide y promoter polymorphism modifies effects of a weight-loss diet on 2-year changes of blood pressure: the preventing overweight using novel dietary strategies trial. Hypertension 2012;60:1169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosmaninho-Salgado J, Marques AP, Estrada M, Santana M, Cortez V, Grouzmann E, Cavadas C. Dipeptidyl-peptidase-IV by cleaving neuropeptide Y induces lipid accumulation and PPAR-gamma expression. Peptides 2012;37:49–54. [DOI] [PubMed] [Google Scholar]
- 9.Baker SB, Cohen M, Kuo L, Johnson M, Al-Attar A, Zukowska Z. The role of the neuropeptide Y2 receptor in liporemodeling: neuropeptide Y-mediated adipogenesis and adipose graft maintenance. Plast Reconstr Surg 2009;123:486–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flier JS. Obesity wars: molecular progress confronts an expanding epidemic. Cell 2004;116:337–50. [DOI] [PubMed] [Google Scholar]
- 11.Tatemoto K. Neuropeptide Y: history and overview. In: Michel MC, editor. Neuropeptide Y and related peptides; Springer (Berlin Heidelberg) 2004. p. 1–21. [Google Scholar]
- 12.Yeung EH, Zhang C, Chen J, Bowers K, Hu FB, Kang G, Qi L. Polymorphisms in the neuropeptide Y gene and the risk of obesity: findings from two prospective cohorts. J Clin Endocrinol Metab 2011;96:E2055–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Z, Zhu G, Hariri AR, Enoch MA, Scott D, Sinha R, Virkkunen M, Mash DC, Lipsky RH, Hu XZ, et al. . Genetic variation in human NPY expression affects stress response and emotion. Nature 2008;452:997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah SH, Freedman NJ, Zhang L, Crosslin DR, Stone DH, Haynes C, Johnson J, Nelson S, Wang L, Connelly JJ, et al. . Neuropeptide Y gene polymorphisms confer risk of early-onset atherosclerosis. PLoS Genet 2009;5:e1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buckland PR, Hoogendoorn B, Guy CA, Coleman SL, Smith SK, Buxbaum JD, Haroutunian V, O'Donovan MC. A high proportion of polymorphisms in the promoters of brain expressed genes influences transcriptional activity. Biochim Biophys Acta 2004;1690:238–49. [DOI] [PubMed] [Google Scholar]
- 16.Kim NS, Oh SM, Ko MM, Cha MH, Kang BK, Bang OS. Association of the C-399T promoter polymorphism of neuropeptide Y with susceptibility to ischemic stroke. Clin Biochem 2009;42:1699–704. [DOI] [PubMed] [Google Scholar]
- 17.Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, McQueen M, Budaj A, Pais P, Varigos J, et al. . Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet 2004;364:937–52. [DOI] [PubMed] [Google Scholar]
- 18.Gabriely I, Ma XH, Yang XM, Atzmon G, Rajala MW, Berg AH, Scherer P, Rossetti L, Barzilai N. Removal of visceral fat prevents insulin resistance and glucose intolerance of aging: an adipokine-mediated process? Diabetes 2002;51:2951–8. [DOI] [PubMed] [Google Scholar]
- 19.Kirkland JL, Dobson DE. Preadipocyte function and aging: links between age-related changes in cell dynamics and altered fat tissue function. J Am Geriatr Soc 1997;45:959–67. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Cline MA, Gilbert ER. Hypothalamus-adipose tissue crosstalk: neuropeptide Y and the regulation of energy metabolism. Nutr Metab 2014;11:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kos K, Harte AL, James S, Snead DR, O'Hare JP, McTernan PG, Kumar S. Secretion of neuropeptide Y in human adipose tissue and its role in maintenance of adipose tissue mass. Am J Physiol Endocrinol Metab 2007;293:E1335–40. [DOI] [PubMed] [Google Scholar]
- 22.Sacks FM, Bray GA, Carey VJ, Smith SR, Ryan DH, Anton SD, McManus K, Champagne CM, Bishop LM, Laranjo N, et al. . Comparison of weight-loss diets with different compositions of fat, protein, and carbohydrates. N Engl J Med 2009;360:859–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williamson DA, Anton SD, Han H, Champagne CM, Allen R, LeBlanc E, Ryan DH, McManus K, Laranjo N, Carey VJ, et al. . Adherence is a multi-dimensional construct in the POUNDS LOST trial. J Behav Med 2010;33:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stricker-Krongrad A, Cumin F, Burlet C, Beck B. Hypothalamic neuropeptide Y and plasma leptin after long-term high-fat feeding in the rat. Neurosci Lett 1998;254:157–60. [DOI] [PubMed] [Google Scholar]
- 25.Velkoska E, Cole TJ, Morris MJ. Early dietary intervention: long-term effects on blood pressure, brain neuropeptide Y, and adiposity markers. Am J Physiol Endocrinol Metab 2005;288:E1236–43. [DOI] [PubMed] [Google Scholar]
- 26.Belsky J, Pluess M. Beyond diathesis stress: differential susceptibility to environmental influences. Psychol Bull 2009;135:885–908. [DOI] [PubMed] [Google Scholar]
- 27.Gauderman W, Morrison J. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies [Internet]. 2006. [cited 2015 Jun 26]. Available from: http://biostats.usc.edu/software.
- 28.Qi Q, Bray GA, Smith SR, Hu FB, Sacks FM, Qi L. Insulin receptor substrate 1 gene variation modifies insulin resistance response to weight-loss diets in a 2-year randomized trial: the Preventing Overweight Using Novel Dietary Strategies (POUNDS LOST) trial. Circulation 2011;124:563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olza J, Gil-Campos M, Leis R, Ruperez AI, Tojo R, Canete R, Gil A, Aguilera CM. Influence of variants in the NPY gene on obesity and metabolic syndrome features in Spanish children. Peptides 2013;45:22–7. [DOI] [PubMed] [Google Scholar]
- 30.Hohmann S, Buchmann AF, Witt SH, Rietschel M, Jennen-Steinmetz C, Schmidt MH, Esser G, Banaschewski T, Laucht M. Increasing association between a neuropeptide Y promoter polymorphism and body mass index during the course of development. Pediatr Obes 2012;7:453–60. [DOI] [PubMed]
- 31.Yamada T, Katagiri H, Ishigaki Y, Ogihara T, Imai J, Uno K, Hasegawa Y, Gao J, Ishihara H, Niijima A, et al. . Signals from intra-abdominal fat modulate insulin and leptin sensitivity through different mechanisms: neuronal involvement in food-intake regulation. Cell Metab 2006;3:223–9. [DOI] [PubMed] [Google Scholar]
- 32.Shi YC, Lin S, Castillo L, Aljanova A, Enriquez RF, Nguyen AD, Baldock PA, Zhang L, Bijker MS, Macia L, et al. . Peripheral-specific y2 receptor knockdown protects mice from high-fat diet-induced obesity. Obesity (Silver Spring) 2011;19:2137–48. [DOI] [PubMed] [Google Scholar]
- 33.Ruipan Z, Xiangzhi M, Li L, Ying Z, Mingliang Q, Peng J, Jingwei L, Zijun Z, Yan G. Differential expression and localization of neuropeptide Y peptide in pancreatic islet of diabetic and high fat fed rats. Peptides 2014;54:33–8. [DOI] [PubMed] [Google Scholar]
- 34.Pischon T, Boeing H, Hoffmann K, Bergmann M, Schulze MB, Overvad K, van der Schouw YT, Spencer E, Moons KG, Tjonneland A, et al. . General and abdominal adiposity and risk of death in Europe. N Engl J Med 2008;359:2105–20. [DOI] [PubMed] [Google Scholar]
- 35.Bastien M, Poirier P, Lemieux I, Despres JP. Overview of epidemiology and contribution of obesity to cardiovascular disease. Prog Cardiovasc Dis 2014;56:369–81. [DOI] [PubMed] [Google Scholar]
- 36.Clinical guidelines on the identification, evaluation, and treatment of overweight and obesity in adults: executive summary. Expert Panel on the Identification, Evaluation, and Treatment of Overweight in Adults. Am J Clin Nutr 1998;68:899–917. [DOI] [PubMed] [Google Scholar]