Abstract
Matrix metalloproteinases (MMP) play an important role in extracellular matrix remodeling. Excessive activity of these enzymes can be induced by UV light and leads to skin damage, a process known as photoaging. In this study we investigated the collagenase inhibition potential of mycosporine-like amino acids (MAA), compounds that have been isolated from marine organisms and are known photoprotectants against UV-A and UV-B. For this purpose the commonly used collagenase assay was optimized and for the first time validated in terms of relationships between enzyme-substrate concentrations, temperature, incubation time and enzyme stability. Three compounds were isolated from the marine red algae Porphyra sp. and Palmaria palmata, and evaluated for their inhibitory properties against Chlostridium histolyticum collagenase (Chc). A dose-dependent, but very moderate inhibition was observed for all substances and IC50 values of 104.0 μM for shinorine, 105.9 μM for porphyra and 158.9 μM for palythine were determined. Additionally, computer-aided docking models suggested that the MAA binding to the active site of the enzyme is a competitive inhibition.
Keywords: collagenase, mycosporine-like amino acids, algae, skin aging
Introduction
Collagen is the major structural protein in human skin. Consisting of a triple helix scaffold, the tensile strength of its fibers provide structural support for bones, skin, tendons, ligaments, blood vessels and are responsible for a dynamic strength of the skin. Mature dermal collagen is formed from precursor molecules called procollagens [1]. Collagenases, which belong to the family of matrix metalloproteinases (MMPs), are transmembrane zinc endopeptidase enzymes. By digesting collagen and elastin fibers they play important roles in many processes including tissue remodeling during development, tissue homeostasis and repair after wounding [2]. However, an over activation due to photoaging and chronical aging leads to alterations in the collagen and elastin composition of the extracellular matrix (ECM) and results in wrinkles, laxity, sagging and a coarse appearance of the human skin [3,4]. Up regulation of certain MMPs have shown to promote cancer progression and to induce angiogenesis by two modes. First, by activating growth factors such as the transforming growth factor beta (TGF-ß) and the vascular endothelial growth factor (VEGF) and second, by degrading the ECM and E-cadherin molecules, which are important to maintain intercellular interactions [5]. Activity of these enzymes can also be pathologically upregulated in response to UV light exposure (photodamage) or release of reactive oxygen species (ROS). In addition, in vivo studies prove that UV irradiation results in a reduction of procollagen synthesis [3-5]. Several authors have reported that upregulated MMPs and diminished procollagen synthesis are mechanisms involved in natural skin aging [6,7]. On the contrary, Chung et al. [8] considered a reduced collagen production and a rise in MMP levels typical for naturally aged skin, while in photoaged skin both parameters tend to be enhanced. The observed effect results in collagen depletion due to the much higher amount of MMP.
The discovery of new substances that can prevent connective tissue damage is of great interest for pharmaceutical and cosmetic industry, and many studies about matrix-metalloproteinase inhibitors from higher plants can be found in literature [9-11]. Marine organisms are interesting sources for new active biomolecules as they are known to produce potent photoprotective metabolites for their defense. To date, only few studies support the collagenase inhibition potential of natural products isolated from marine algae. Among the compounds tested so far, oligosaccharides, marine polyphenols and fatty acids have been found to inhibit MMP-2 and MMP-9 [12]. However, up to our knowledge, this is the first study that investigates mycosporine-like amino acids (MAA) for their collagenase inhibition potential. In addition, no marine substance has been tested on bacterial collagenase (Chlostridiopeptidase A). The investigated compounds were isolated from marine red algae and assayed for their inhibitory potential against microbial collagenase from Clostridium histolyticum. Molecular modelling was used to depict potential interactions between the isolated compounds and the enzyme. Furthermore, we validated and optimized the anti-collagenase assay for high-throughput screening (HTS) purposes.
Results
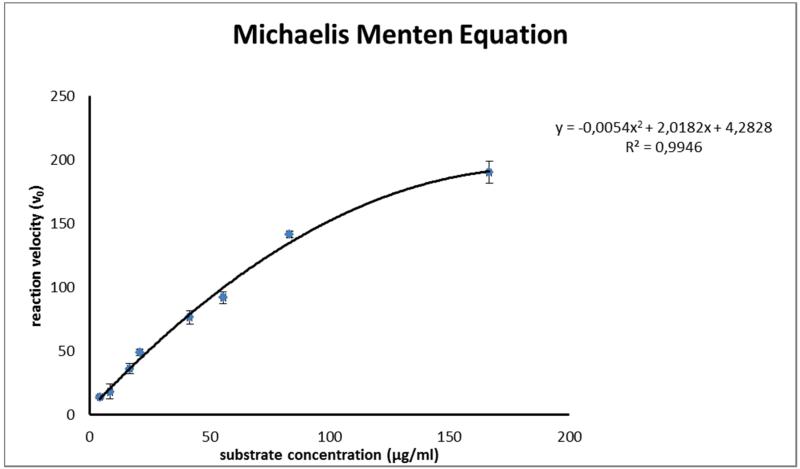
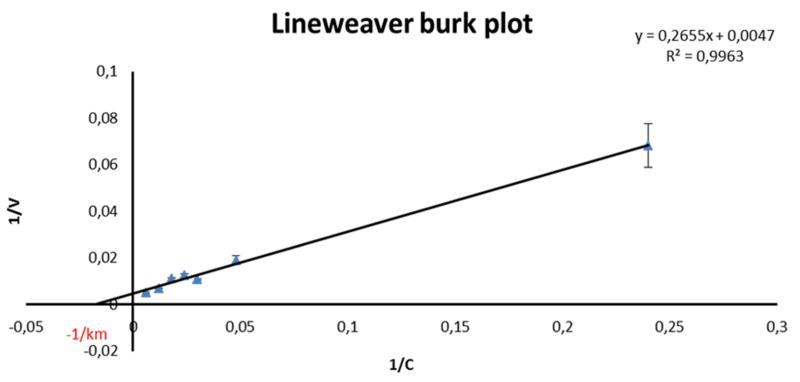
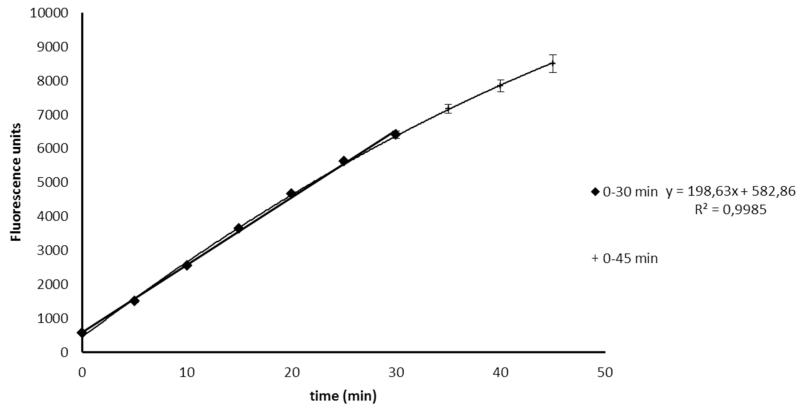
Assay optimization was initiated by the evaluation of the optimal enzyme to substrate ratio. This was necessary, as a broad range of different substrate and enzyme concentrations is described in literature. Moreover those assays vary in the types of substrates and enzymes and none of these previously conducted assays has been validated, which makes a comparison of generated data rather difficult. Therefore optimization was set in by testing different enzyme concentrations, ranging from 400 μg/mL to 40 μg/mL (initial concentration) resulting in an optimum concentration of 100 μg/mL (initial), which showed a suitable ratio of enzyme activity. To optimize the substrate concentration, the Michaelis Menten constant (Km) and the maximum velocity (Vmax) were determined by measuring the velocity of MMP-2 fluorogenic substrate conversion in a concentration range from 4.17 μg/mL to 166.67 μg/mL (initial concentrations; Fig. 1). The values for Km (51.67 μM) and Vmax 212.76 μmol/min were obtained by linear regression of the Michaelis-Menten curve (Lineweaver Burk plot) as shown in Fig. 2. As the enzyme constantly cleaves the substrate, the concentration of the latter is diminished during the assay, resulting in increasing non linearity of the reaction when [S] << Km [13]. Therefore, the optimum substrate concentration was chosen around the determined value of Km. A final substrate concentration of 13.89 μg/mL was used in all experiments. The established ratio of approximately 1 part of enzyme (100 μg/mL) to 2 parts of substrate (55.55 μg/mL) showed a stable reaction performance, as indicated by a linear curve over more than 30 minutes reaction time (Fig. 3). Longer reaction times resulted in trending slightly to a maximum. In a next step, the optimal reaction temperature was assessed by performing the assay at room temperature (RT), 30°C and 37°C and by using the standard inhibitor phosphoramidon at an initial concentration of 50 μg/mL. For 30°C and 37°C reaction temperature the same inhibition rate of 53.4% was obtained, while at RT inhibition was only slightly decreased (49.1%). This indicates that in case thermo-labile substances are tested, the assay could also be carried out at temperatures around 25°C. For the MAAs this was not required, thus the incubation time was evaluated at 37°C. Optimum incubation time of inhibitor-enzyme reactions was also investigated. No measurable differences were observed in the inhibition rate of phosphoramidon, whether incubation time was set to 5, 10 or 15 minutes. Longer incubation times however, resulted in lower rates of enzyme inhibition; for example an incubation time of 20 minutes resulted in a 10% lower inhibitory capacity compared to that of 15 min. The utilized collagenase was stable in a concentration of 1 mg/mL when kept on ice for up to 6 hours. Finally, assay validation was carried out with phosphoramidon and 1,10-phenanthroline, two standard inhibitors reported in literature [19,20]. Incubation was carried out at 37°C for 10 minutes, and the reaction was monitored over 30 minutes. The tested concentration range for phosphoramidon was from 0.78 to 62.5 μg/mL (final concentration). An IC50 of 18.8 μM ± 1.6 (11.07 μg/mL ± 1.30) was determined for phosphoramidon (Mr 587.47). The concentration range for the second standard inhibitor 1,10-phenanthroline (Mr 180.21) ranged from 6.25 to 250 μg/mL, and the IC50 value was calculated to be 238.1 μM ± 3.4 (42.86 μg/mL ± 1.35). This result does not indicate significant or physiologically relevant inhibition, but the value is according to literature [16].
Fig. 1).
Michaelis Menten equation performed with 8 different MMP-2 fluorogenic substrate concentrations ranging from 4.17 to 166.67 μg/mL (initial conc.); enzyme: 100 μg/mL (initial conc.).
Fig. 2).
Lineweaver burk plot resulting in a Km of 51.7 μM (12.9 μM final), in order to determine the initial substrate concentration required for a consistent reaction.
Fig. 3).
Confirmation of a linear and time dependent cleavage of the substrate under optimized assay conditions; 0-30 min: linear conditions; 0-45 min: longer assay duration leads to non-linearity.
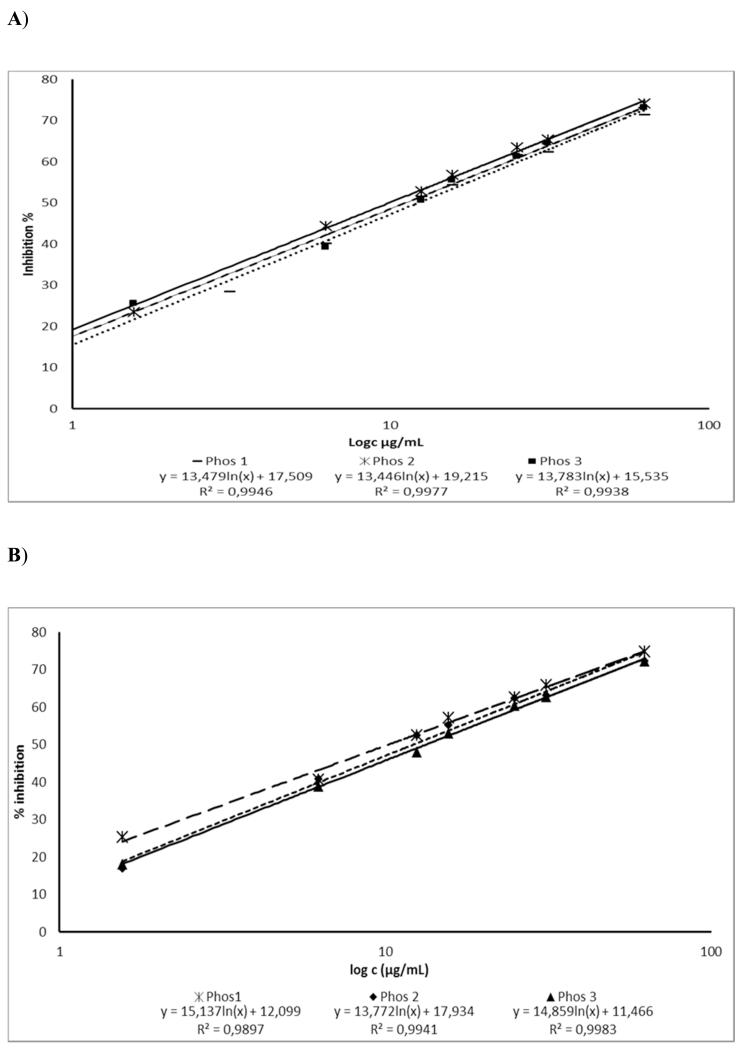
Method validation was carried out via endpoint reading mode and kinetic mode. For the latter, fluorescence was measured every 5 minutes over a total assay period of 30 minutes. The area under the curve was calculated for 7 different concentrations for the standard inhibitor phosphoramidon (serial dilution steps). For each approach, the IC50 of phosphoramidon was determined (Fig. 4). In addition, limit of quantification (LOQ) and limit of detection (LOD) were determined. Table 1 compares the two different approaches of data analysis. As can be seen both ways of data evaluation are comparable to each other. Validation results are slightly better in the endpoint-reading mode and more useful for high throughput screening purposes. However, the kinetic reading mode shows a consistent evaluation of inhibition based on a given time frame and not only in terms of a value of fluorescent units at a given point in time.
Fig. 4).
IC50 value of the standard inhibitor phosphoramidon as determined by endpoint reading (A) or calculated based on AUC from 0 to 30 min (B); n = 3.
Table 1.
Comparison of endpoint reading mode at 30 min versus kinetic reading mode (0-30 min) using the standard inhibitor phosphoramidon as an example (n=3).
| Parameter | Endpoint reading mode | Kinetic reading mode (AUC) |
|---|---|---|
|
| ||
| Linearity | 1) Y= −1400 ln x + 8932.2 R2=0.9936 |
1) Y= −657.3 ln x + 3883.0 R2=0.9906 |
| 2) Y= −1512 ln x + 9152.2 R2=0.9957 |
2) Y= −639 ln x + 3812.6 R2=0.9991 |
|
| 3) Y= −1183 ln x + 8503.3 R2=0.9913 |
3) Y= −923.5 ln x + 5066.1 R2=0.9869 |
|
|
| ||
| IC 50 | 11.07 μg/mL ± 0.95 | 11.95 μg/mL ± 1.28 |
|
| ||
| Linear range | 62.5 - 0.78 μg/mL | 62.5 - 0.78 μg/mL |
|
| ||
| LOD / LOQ | LOD: 1.14 μg/mL | LOD: 2.45 μg/mL |
| LOQ: 3.47 μg/mL | LOQ: 7.43 μg/mL | |
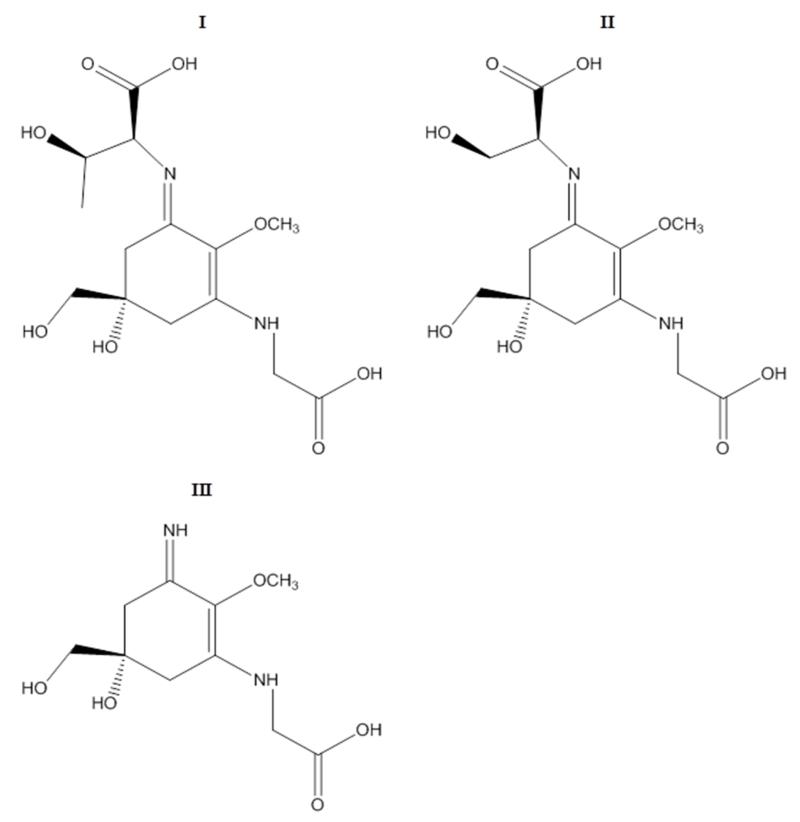
NMR data of the isolated compounds and their stereochemistry were in agreement with literature [14,15]. Palythine, porphyra and shinorine are based on the same cyclohexenimine structure, and the amino acid, which is conjugated to the ring system in position 3 is always glycine (Fig. 5).
Fig. 5).
Chemical structures of porphyra (I), shinorine (II) and palythine (III).
The isolated substances were tested using the fully validated procedure described above. The IC50 values for palythine and shinorine were evaluated using 8 serial dilutions in a concentration range from 12.5 to 125 μg/mL final concentration. The IC50 value of palythine was found to be 158.9 μM ± 3.2 (38.82 μg/mL ± 0.78), and that of shinorine 104.0 μM ± 3.7 (34.42 μg/mL ± 1.23). Porphyra was diluted in a concentration range from 1.87 to 150 μg/mL, and an IC50 value of 105.9 μM ± 2.3 (37.73 μg/mL ± 1.95) was calculated. The three MAA showed a moderate inhibition of collagenase, even the determined values were lower than for 1,10-phenanthroline (238.1 μM ± 3.4; 42.86 μg/mL ± 1.35), a standard inhibitor for collagenase described in literature (70.4 μg/mL) [16]. The results are comparable to inhibitors found in higher plants such as ecdysteroids with IC50 values from 28-50 μg/mL [16]. Others like (+) catechin-aldehyde polycondensates (IC50 values from 200 to 350 μM; [17]) and epigallocatechingallate (IC50 = 114 μg/mL; [15]) are less active than the investigated MAA. No marine species or substances have been tested on bacterial collagenase (clostridiopeptidase A) so far. Yet, a small number of substances, like marine saccharides (chitooligosaccharides), glycosaminoglycan from L. vannamei, flavonoids and fatty acids from marine source have been investigated for their MMP-2 (gelatinase A, collagenase type IV) and MMP-9 (gelatinase B; type IV precursor collagenase) inhibition in cell lines [16].
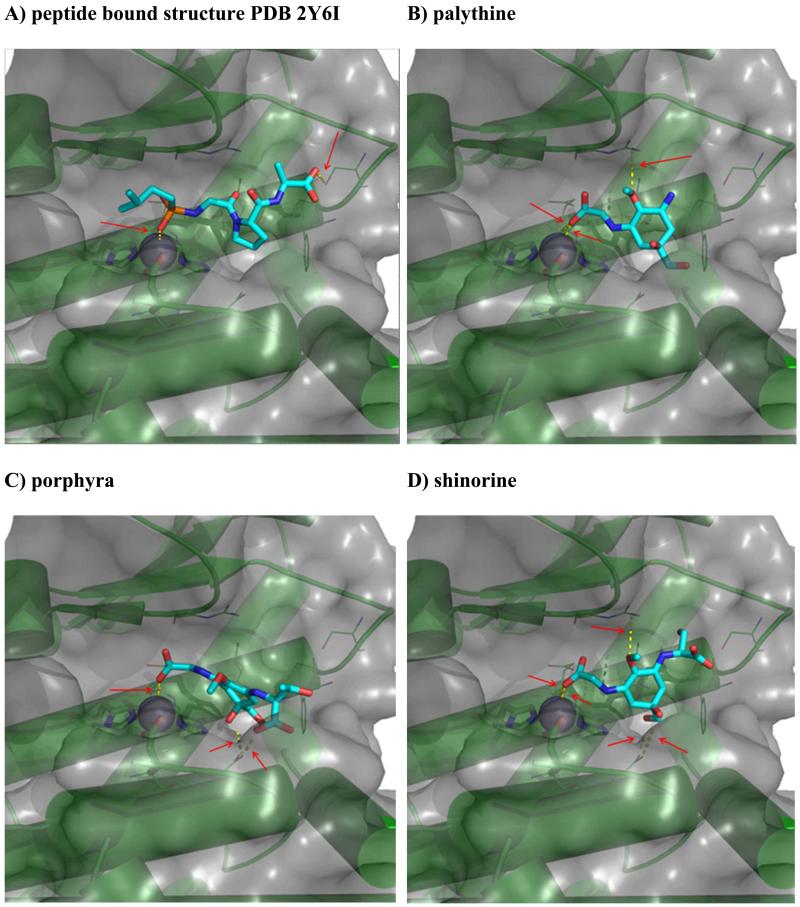
To rationalize collagenase inhibition on a structural level, we performed in silico docking studies for porphyra, palythine and shinorine to the crystal structure of collagenase G from Clostridium histolyticum (PDB: 2Y6I) [20]. We generated the protein structure using protonate3d [21] and ionized the MAAs for neutral pH in MOE (Molecular Operating Environment MOE 2014.09; Chemical Computing Group Inc., Montreal, Canada). Water molecules coordinating the active site Zn2+ ion were preserved to mimic the peptide-bound state resolved in the crystal structure with observed docking results. We used MOE’s default settings for flexible docking including placement by Triangle Matcher, scoring by London dG, a forcefield refinement of 30 intermediate poses and rescoring by GBVI/WSA dG. We introduced additional pharmacophore constraints during docking enforcing an acidic group coordinating to the Zn2+ ion as well as a hydrophobic group in the S2 pocket, thus replacing the proline side-chain in the native peptide position P2′. Resulting complexes were visualized using the software package Pymol (version 1.5.0.2). [22]. Poses are shown in Figure 6, and docking scores are given as supplementary information.
Fig. 6).
Predicted binding modes of MAA to collagenase: Protein is shown as green cartoon and semi-transparent Van der Waals surface with the active site Zn2+ represented as grey sphere. Side-chain heavy atoms of metal coordinating amino acids are shown as sticks, pocket residues as thin lines. Hydrogen bonding and metal coordination are indicated as yellow dotted lines and marked with arrows. A) Experimental binding mode of the peptide-derived inhibitor isoamylphosphonyl-Gly-Pro-Ala (PDB: 2Y6I). Ligand heavy atoms are shown in stick representation in atomic coloring with carbon in cyan. B-D) Predicted binding modes of the MAA porphyra, shinorine, and palythine.
The three investigated MAA are predicted to occupy a similar region within the binding site of the collagenase that overlaps with the native peptide’s P1′-P3′ region. The Zn2+ ion is consistently bound to the carboxylate group of the glycine substructure in the MAA replacing the native P1′ glycine residue. This nicely matches the strong preference of collagenases for small amino acids at the P1′ position [23,24]. The MAA’s hydrophobic rings occupy the position of the P2′ proline, again matching collagenases’ preferences for this particular amino acid. The larger MAA shinorine and porphyra stretch further towards the S3′ pocket. As all MAA are predicted to occupy the catalytic center of collagenase, a competitive binding mode would be expected, whereby the compounds are expected to inhibit the enzyme due to direct coordination to its active site.
Discussion
A number of collagenase inhibitors isolated from higher plants are described in literature, belonging to different compound classes like flavonoids [25,26], catechines [27], polyphenols (e.g. resveratrol) [28,29], or steroids [16]. Also, several multicomponent extracts were reported to inhibit collagenase activity [18]. Only few attempts were made to utilize marine algae as source for potential collagenase inhibitors, although these organisms are enormously rich in bioactive compounds that are produced in response to UV-light. This includes MAA, which derive from the shikimic acid pathway with 4-desoxygadusol as direct precursor molecule. The latter is known to be a strong antioxidant [30]. MAA also act as ROS scavengers after UV-radiation exposure [31,32]. Being known as sunscreen compounds, MAA could also play a role in the collagenase metabolism, a fact that never has been investigated before. Several different assay protocols are described in literature for the identification of collagenase inhibitors from natural sources [29,33-35]. In many cases, bacterial collagenase from Clostridium histolyticum was used because of its unique and extensive degradation ability on collagen compared to vertebrate collagenases [36]. Therefore, this type of enzyme was selected in our experiments. Substrates reported in literature varied a lot, also largely depending on the applied detection mode (fluorescence vs absorbance) [29,33]. However, none of the described assay procedures was validated, and thus the results of different studies are hardly comparable. By presenting a, for the first time fully validated method for high throughput screening purposes of collagenase inhibitors, we established a protocol that ensures reproducible results.
To our best knowledge, this is the first study on mycosporine-like amino acids serving as collagenase inhibitors. Even the observed effects on collagenase are moderate, MAA, which are already known as sunscreen compounds, could possibly serve as anti-skin aging molecules. It is possible that they protect from wrinkled skin as a result of chronically aging or photoaging. Additionally, they may have beneficial effects in the prevention of skin cancer. All these aspects can serve as an interesting starting point for the development of new cosmetics or skin protective products. Till date over 30 different MAA mainly from marine sources are known, which shows that possible applications are not limited to the here tested compounds. Our results indicate many options for the use of marine MAA in cosmetic or pharmaceutical industry.
Materials and Methods
Reagents and Chemicals
Collagenase type V from Clostridium histolyticum (EC 3.4.24.3), fluorogenic substrate peptide MMP-2 (MCA-Pro-Leu-Ala-Nva-DNP-Dap-Ala-Arg-NH2), phosphoramidon disodium salt (purity > 97%) and the protease inhibitor 1,10-phenanthroline (purity > 99%) were purchased from Sigma Aldrich. All other solvents used for isolation and extraction (methanol, butanol, hydrochloric acid and acetonitrile) were of analytical grade and purchased from Merck; black 96-well plates with flat bottom came from BD Biosciences.
Biological material and extraction
For the isolation of MAA, two commercially available algae, Palmaria palmata (Irish Seaweeds, LOT-Nr. 5391513420184, Belfast, UK) and Porphyra sp. (Asia Express Food, LOT-Nr. 120516, Kampen, NL) were used. The material was identified by microscopic means by one of the authors (A. Hartmann), and respective vouchers (2014-MAA-1 and 2014-MAA-2) are deposited at the Institute of Pharmacy, University of Innsbruck, Austria. Dried algal material was crushed to powder in a grinding mill prior to extraction using methanol/water (25:75) in an ultrasonic bath (Bandelin Sonorex 35 KHz,) at 45°C for 2 hours [37]. After centrifugation at 3000 rpm for 10 minutes, the supernatant was collected and evaporated at 45°C in a vacuum evaporator (Büchi). To obtain completely dried extracts, the resulting pasty liquid was transferred into a beaker and lyophilized (Heto power dry PL 6000, Thermo Fisher).
Isolation of MAAs
Dried extracts were dissolved in water and partitioned three times with 1-butanol to remove less polar components. The water fractions were combined, evaporated and separated on an ion exchange resin (Dowex 50WX H+ form,100-200 mesh, Sigma Aldrich) according to the protocol of Carignan et al. [38]. Briefly, the resin was placed in a glass column, the sample (aqueous solution) applied and the column first rinsed with water. Elution of MAA containing fractions was possible with 0.25 M HCl. To remove high salt concentrations, the obtained fractions were then applied on activated carbon cartridges (Supelco Envi-Carb, Sigma), they were washed with water and finally MAA-enriched fractions were eluted with pure methanol. Individual MAA were isolated from these pre-purified extracts by semi-preparative HPLC on a Dionex UltiMate 3000 preparative HPLC system (Thermo Fisher). The optimum separation was carried out on a Lichrosorb C18 100 Å column (200 mm × 10.00 mm; 7 μm) from Merck by using a mobile phase comprising 0.1% acetic acid in water (A) and acetonitrile (B). The method was run isocratic for 30 min with 2% mobile phase B. Then the column was washed for 10 min with 90% B, before being re-equilibrated for 15 minutes prior to the next injection. Detection was performed at 320 nm, the column was maintained at 20°C and the flow rate was set to 0.8 mL/min. The injected sample volume was 70 μL with a sample concentration of 25 mg/mL. After approx. 40 – 50 injections 13.0 mg of porphyra and 8.5 mg shinorine (both with a purity of ≥ 95%) were obtained from the pre-purified extract of Porphyra sp. Palythine (7.8 mg, purity ≥ 94%) could be isolated from Palmaria palmata using the same strategy. Detailed information on the isolation steps including HPLC-chromatograms are provided in the supplementary information.
During separation and purification, performance of each step was monitored by repeated HPLC-MS analysis, using a HP 1100 HPLC system from Agilent, coupled to an Esquire 3000 plus iontrap mass spectrometer (Bruker). For analysis in analytical scale a Luna C-18 column (250 × 3.00 mm, 5 μm, Phenomenex) was used under the same conditions as described above; only the flow was set to 0.5 mL/min. MS-Spectra were obtained in alternating ESI mode by setting the temperature to 350°C, the nebulizer gas (nitrogen) to 40 psi, and a nebulizer flow (nitrogen) of 8 L/min.
Structural analysis of MAA
NMR spectra were recorded at 25°C on an Ultra-Shield 600 MHz instrument (Bruker) using the following experiments: 1H- and 13 C- NMR, two dimensional correlation spectroscopy (2D COSY), heteronuclear multiple quantum coherence (HMQC) and heteronuclear multiple bond coherence (HMBC). All samples were dissolved in deuterated water and tetramethylsilan (both from Euriso-Top) was used as internal standard.
Collagenase Inhibition Assay
The fluorimetric assay was carried out in 96-well microplate formate on an Infinite F 200 pro microplate reader equipped with filter based technology (Tecan). The general framework of the assay was taken from literature [29]. In brief, reaction was monitored at an excitation wavelength of 320 nm and 400 nm emission wavelength, and the positive controls phosphoramidon and 1,10-phenanthroline were used as described previously [16]. However, the assay was optimized in terms of enzyme and substrate concentration, temperature and incubation time. A peptide with an amino acid sequence of MCA-Pro-Leu-Ala-Nva-DNP-Dap-Ala-Arg-NH2 was chosen as substrate, which contains a fluorogenic residue (7-methoxycoumarin-4-yl acetic acid; MCA). If no inhibitor is present the latter is released by the enzyme and a constant increase in fluorescence can be measured. Using the bacterial collagenase from Clostridium histolyticum (Chc) has several advantages. It cleaves not only the x-gly bond in collagen but also synthetic peptides and ECM, where it hydrolyses triple-helical collagen under both physiological and in vitro conditions [17,18]. In favor of a HTS screening protocol, the volumes were kept to a minimum, obtaining a total final reaction volume of 100 μL, composed of 25 μL substrate, 25 μL enzyme, 25 μL buffer solution and 25 μL sample / positive controls. Enzyme and substrate were dissolved in the reagent buffer (10 mM Tris-HCl; pH 7.3) in suitable concentrations. Both enzyme and substrate were diluted from stock solutions to 100 μg/mL (initial) enzyme concentration and 55.55 μg/mL (initial) substrate concentration directly before use. Test samples were dissolved in water/DMSO, always maintaining a final DMSO concentration of 1% in each well. Buffer and sample solutions were added first, followed by the enzyme. This mixture was incubated for 10 min. Finally, the readout reaction was started by adding the fluorogenic substrate. A negative control was evaluated in the same manner by adding 25 μL 10 mM Tris-HCl buffer to the reaction instead of the test substance. For the determination the IC50 values (MAA and standard inhibitors), seven different concentrations were prepared in a serial dilution step. Each substance was tested in triplicate, with three replicates on each plate. In addition, a blank was taken for each sample concentration to avoid false positive results due to a possible native fluorescence of the tested substances. The half maximal inhibitory concentration (IC50) was calculated using the formula: Enzyme inhibition activity (%) = [FU control-FU sample)/FU control]*100.
Supplementary Material
Table 2.
Collagenase inhibitory activity of the tested compounds.
| Compound | IC50 (μM) |
|---|---|
| shinorine | 104.0 ± 3.7 |
| porphyra | 105.9 ± 2.3 |
| palythine | 158.9 ± 3.2 |
|
| |
| phosphoramidon | 18.8 ± 1.6 |
| 1,10-phenanthroline | 238.1 ± 3.4 |
Values represent mean ± SD (n=3).
Acknowledgements
This work was financially supported by FWF (Project nr P241680). J.E. Fuchs acknowledges funding from the UK Medical Research Council (grant MR/K020919/1).
Footnotes
NMR shift values of the isolated substances porphyra, shinorine and palythine, as well as the isolation protocol for Porphyra sp. and docking scores for mycosporine-like amino acids and phosphoramidon are shown as supporting information.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Smith LT, Holbrook KA, Madri JA. Collagen type-I, type-Iii and type-V in human-embryonic and fetal skin. Am J Anat. 1986;175:507–521. doi: 10.1002/aja.1001750409. [DOI] [PubMed] [Google Scholar]
- 2.Woessner JF. The family of matrix metalloproteinases. Inhibition of matrix metalloproteinases: Ann NY Acad. 1994;732:11–21. doi: 10.1111/j.1749-6632.1994.tb24720.x. [DOI] [PubMed] [Google Scholar]
- 3.Philips N, Conte J, Chen YJ, Natrajan P, Taw M, Keller T, Givant J, Tuason M, Dulaj L, Leonardi D, Gonzalez S. Beneficial regulation of matrixmetalloproteinases and their inhibitors, fibrillar collagens and transforming growth factor-beta by Polypodium leucotomos, directly or in dermal fibroblasts, ultraviolet radiated fibroblasts, and melanoma cells. Arch Dermatol Res. 2009;301:487–495. doi: 10.1007/s00403-009-0950-x. [DOI] [PubMed] [Google Scholar]
- 4.Philips N, Keller T, Hendrix C, Hamilton S, Arena R, Tuason M, Gonzalez S. Regulation of the extracellular matrix remodeling by lutein in dermal fibroblasts, melanoma cells, and ultraviolet radiation exposed fibroblasts. Arch Dermatol Research. 2007;299:373–379. doi: 10.1007/s00403-007-0779-0. [DOI] [PubMed] [Google Scholar]
- 5.Philips N, Keller T, Holmes C. Reciprocal effects of ascorbate on cancer cell growth and the expression of matrix metalloproteinases and transforming growth factor-beta. Cancer Lett. 2007;256:49–55. doi: 10.1016/j.canlet.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Fisher GJ, Datta SC, Talwar HS, Wang ZQ, Varani J, Kang S, Voorhees JJ. Molecular basis of sun-induced premature skin ageing and retinoid antagonism. Nature. 1996;379:335–339. doi: 10.1038/379335a0. [DOI] [PubMed] [Google Scholar]
- 7.Fisher GJ, Wang ZQ, Datta SC, Varani J, Kang S, Voorhees JJ. Pathophysiology of premature skin aging induced by ultraviolet light. New Engl J Med. 1997;337:1419–1428. doi: 10.1056/NEJM199711133372003. [DOI] [PubMed] [Google Scholar]
- 8.Chung JH, Seo JY, Choi HR, Lee MK, Youn CS, Rhie G, Cho KH, Kim KH, Park KC, Eun HC. Modulation of skin collagen metabolism in aged and photoaged human skin in vivo. J Invest Dermatol. 2001;117:1218–1224. doi: 10.1046/j.0022-202x.2001.01544.x. [DOI] [PubMed] [Google Scholar]
- 9.Wierzchacz C, Enis S, Kolander J, Gebhardt R. Differential inhibition of matrix metalloproteinases-2, -9, and -13 activities by selected anthraquinones. Planta Med. 2009;75:327–329. doi: 10.1055/s-0028-1112205. [DOI] [PubMed] [Google Scholar]
- 10.Park HY, Lim H, Kim HP, Kwon YS. Downregulation of matrix metalloproteinase-13 by the root extract of Cyathula officinalis Kuan and its constituents in IL-1ß-treated chrondrocytes. Planta Med. 2011;77:1528–1530. doi: 10.1055/s-0030-1270834. [DOI] [PubMed] [Google Scholar]
- 11.Lee SJ, Hong S, Yoo SH, Kim GY. Cyanidin-3-o-sambubioside from Acanthopanax sessiliflorus fruit inhibits metastasis by downregulating MMP-9 in breast cancer cells MDA-MB-231. Planta Med. 2013;79:1636–1640. doi: 10.1055/s-0033-1350954. [DOI] [PubMed] [Google Scholar]
- 12.Pallela R, Na-Young Y, Kim SK. Anti-photoaging and photoprotective compounds derived from marine organisms. Mar Drugs. 2010;8:1189–1202. doi: 10.3390/md8041189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ß-Glucuronidase Assay. DyNA Quant™ Application note 3 Amersham Biosciences. Available at: https://www.gelifesciences.com/gehcls_images/GELS/Related%20Content/Files/1314716762536/litdoc80623675_20110830181743.pdf.
- 14.La Barre JMK. Outstanding marine molecules. Wiley-Blackwell; 2014. pp. 387–430. [Google Scholar]
- 15.Klisch M, Richter P, Puchta R, Häder DP, Bauer W. The stereostructure of porphyra-334: an experimental and calculational NMR investigation. Evidence for an efficient proton sponge. Helv Chim Acta. 2007;90:488–511. [Google Scholar]
- 16.Nsimba RY, Kikuzaki H, Konishi Y. Ecdysteroids act as inhibitors of calf skin collagenase and oxidative stress. J Biochem Mol Toxic. 2008;22:240–250. doi: 10.1002/jbt.20234. [DOI] [PubMed] [Google Scholar]
- 17.Kim YJ, Uyama H, Kobayashi S. Inhibition effects of (+)-catechin-aldehyde polycondensates on proteinases causing proteolytic degradation of extracellular matrix. Biochem Biophys Res Co. 2004;320:256–261. doi: 10.1016/j.bbrc.2004.05.163. [DOI] [PubMed] [Google Scholar]
- 18.Thring TSA, Hili P, Naughton DP. Anti-collagenase, anti-elastase and anti-oxidant activities of extracts from 21 plants. BMC Complement Alt Med. 2009;9:27. doi: 10.1186/1472-6882-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C, Kim SK. Matrix metalloproteinase inhibitors (MMPIs) from marine natural products: the current situation and future prospects. Mar Drugs. 2009;7:71–84. doi: 10.3390/md7020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckhard U, Schonauer E, Nuss D, Brandstetter H. Structure of collagenase G reveals a chew-and-digest mechanism of bacterial collagenolysis. Nat Struct Mol Biol. 2011;18:1109–1139. doi: 10.1038/nsmb.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labute P. Protonate3D: Assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins. 2009;75:187–205. doi: 10.1002/prot.22234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Lano W. The Pymol Molecular Graphics System. 2008 Version1.5.0.2. [Google Scholar]
- 23.Fuchs JE, Von Grafensteiner S, Huber RG, Margreiter MA, Spitzer GM. Cleavage entropy as quantitative measure of protease specifity. PLoS Comput Biol. 2013;9:e1003007. doi: 10.1371/journal.pcbi.1003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu YB, Webb E, Singh J, Morgan B, Gainor A, Gordon JA, Siahaan TD. Rapid determination of substrate specificity of Clostridium histolyticum beta-collagenase using an immobilized peptide library. J Biol Chem. 2002;277:8366–8371. doi: 10.1074/jbc.M111042200. [DOI] [PubMed] [Google Scholar]
- 25.Huang XK, Chen S, Xu L, Liu YQ, Deb DK, Platanias LC, Bergan RC. Genistein inhibits p38 map kinase activation, matrix metalloproteinase type 2, and cell invasion in human prostate epithelial cells. Cancer Res. 2005;65:3470–3478. doi: 10.1158/0008-5472.CAN-04-2807. [DOI] [PubMed] [Google Scholar]
- 26.Rooprai HK, Kandanearatchi A, Maidment SL, Christidou M, Trillo-Pazos G, Dexter DT, Rucklidge GJ, Widmer W, Pilkington GJ. Evaluation of the effects of swainsonine, captopril, tangeretin and nobiletin on the biological behaviour of brain tumour cells in vitro. Neuropath Appl Neurobiol. 2001;27:29–39. doi: 10.1046/j.0305-1846.2000.00298.x. [DOI] [PubMed] [Google Scholar]
- 27.Kirszberg C, Esquenazi D, Alviano CS, Rumjanek VM. The effect of a catechin-rich extract of Cocos nucifera on lymphocytes proliferation. Phytother Res. 2003;17:1054–1058. doi: 10.1002/ptr.1297. [DOI] [PubMed] [Google Scholar]
- 28.Tang HJ, Martel K, Stribley J, Christman G. The effect of resveratrol on collagen expression in human uterine leiomyoma cells. J Soc Gynecol Invest. 2006;13:70a–71a. [Google Scholar]
- 29.Moon HI, Kim TI, Cho HS, Kim EK. Identification of potential and selective collagenase, gelatinase inhibitors from Crataegus pinnatifida. Bioorg Med Chem Lett. 2010;20:991–993. doi: 10.1016/j.bmcl.2009.12.059. [DOI] [PubMed] [Google Scholar]
- 30.Shick JM, Dunlap WC. Mycosporine-like amino acids and related gadusols: Biosynthesis, accumulation, and UV-protective functions in aquatic organisms. Annu Rev Physiol. 2002;64:223–262. doi: 10.1146/annurev.physiol.64.081501.155802. [DOI] [PubMed] [Google Scholar]
- 31.Aguilera J, Bishof K, Karsten U, Hanelt D, Wiencke C. Seasonal variation in ecophysiological patterns in macroalgae from an Arctic fjord. II. Pigment accumulation and biochemical defence systems against high light stress Mar Biol. 2002;141:603–604. [Google Scholar]
- 32.Llewellyn CA, Airs RL. Distribution and abundance of MAAs in 33 species of microalgae across 13 classes. Mar Drugs. 2010;8:1273–1291. doi: 10.3390/md8041273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrantes E, Guinea M. Inhibition of collagenase and metalloproteinases by aloins and aloe gel. Life Sci. 2003;72:843–850. doi: 10.1016/s0024-3205(02)02308-1. [DOI] [PubMed] [Google Scholar]
- 34.Rennert B, Melzig MF. Free fatty acids inhibit the activity of Clostridium histolyticum collagenase and human neutrophil elastase. Planta Med. 2002;68:767–769. doi: 10.1055/s-2002-34411. [DOI] [PubMed] [Google Scholar]
- 35.Teramachi F, Koyano T, Kowithayakorn T, Hayashi M, Komiyama K, Ishibashi M. Collagenase inhibitory quinic acid esters from Ipomoea pes-caprae. J Nat Prod. 2005;68:794–796. doi: 10.1021/np0500631. [DOI] [PubMed] [Google Scholar]
- 36.Seifter SEH. The Collagenases. In: Boyer PD, editor. The Enzymes. Vol. 3. Academic Press; New York: 1971. pp. 649–697. [Google Scholar]
- 37.Tartarotti B, Sommaruga R. The effect of different methanol concentrations and temperatures on the extraction of mycosporine-like amino acids (MAAs) in algae and zooplankton. Arch Hydrobiol. 2002;154:691–703. [Google Scholar]
- 38.Carignan MO, Cardozo KHM, Oliveira-Silva D, Colepicolo P, Carreto JI. Palythine-threonine, a major novel mycosporine-like amino acid (MAA) isolated from the hermatypic coral Pocillopora capitata. J Photoch Photobio B. 2009;94:191–200. doi: 10.1016/j.jphotobiol.2008.12.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.