Abstract
Mycobacterium tuberculosis enters the host by inhalation of an infectious aerosol and replicates in the alveolar macrophages until the host's immune defense causes bacteriostasis, which leads the pathogen to go into nonreplicative drug-resistant dormancy. The dormant pathogen can survive for decades till the host's immune system is weakened and active tuberculosis develops. Even though fatty acids are thought to be the major energy source required for the persistence phase, the source of fatty acids used is not known. We postulate that the pathogen uses triacylglycerol (TG) as a storage form of fatty acids. Little is known about the biosynthesis of TG in M. tuberculosis. We show that 15 mycobacterial genes that we identified as putative triacylglycerol synthase (tgs) when expressed in Escherichia coli showed TGS activity, and we report some basic catalytic characteristics of the most active enzymes. We show that several tgs genes are induced when the pathogen goes into the nonreplicative drug-resistant state caused by slow withdrawal of O2 and also by NO treatment, which is known to induce dormancy-associated genes. The gene (Rv3130c) that shows the highest TGS activity when expressed in E. coli shows the highest induction by hypoxia and NO treatment. Biochemical evidence shows that TG synthesis and accumulation occur under both conditions. We conclude that TG may be a form of energy storage for use during long-term dormancy. Therefore, TG synthesis may be an appropriate target for novel antilatency drugs that can prevent the organism from surviving dormancy and thus assist in the control of tuberculosis.
Virulent Mycobacterium tuberculosis enters the host by inhalation of an infectious aerosol. The pathogen replicates in the alveolar macrophages, but in a great majority of cases, the host's immune defense causes bacteriostasis that leads the pathogen to go into a state of nonreplicative, drug-resistant dormancy (7, 14, 36, 40). One-third of the world population is estimated to be latently infected (9, 41). When the host's immune system is weakened, the pathogen replicates, leading to active tuberculosis. Most cases of active tuberculosis arise from the small fraction of people who have had the dormant organism for years or decades (10, 15, 18, 19, 22, 37). Since the pathogen under dormancy is resistant to antimicrobial drugs, the ability of the organism to survive long periods in such a state creates great difficulty in the control of tuberculosis. Molecular mechanisms that allow the pathogen to go into dormancy, survive in the host for decades under such conditions, and resume replication upon weakening of the immune system of the host are poorly understood.
Since the pathogen in the latent lesions is likely to be under hypoxic conditions, oxygen depletion has been tested as a means to induce dormancy in in vitro cultures. A gradual depletion of O2 in M. tuberculosis caused the pathogen to reach a nonreplicating persistent state that manifested drug sensitivity and structural changes suggestive of a dormant state (32, 39). Analysis of the changes in the gene expression patterns induced by hypoxia reveals a putative transcription factor, DosR (Rv3133c), that is required for transcriptional activation of most of the genes known to be strongly regulated by hypoxia (5, 26, 33). More recently, inhibition of respiration by NO, which is normally produced by activated macrophages, was found to induce a gene expression pattern that was quite similar to that found under a hypoxia-induced nonreplicating state (31, 38). Thus, both hypoxia and inhibition of respiration by NO may induce the pathogen to go into latency.
The efforts to explore metabolic events that might allow the pathogen to go into the persistence phase suggested that fatty acids may be the key source of energy needed for persistence (4, 27). Thus, genes that encode enzymes required to live on fatty acids as the chief carbon source, such as isocitrate lyase, were found to be essential for persistence (21). However, little is known about the source of the fatty acid substrates. For long-term survival with very low metabolic rates, such as that encountered in hibernating animals, triacylglycerol (TG) is the commonly used storage form of energy (1). Similarly, oil seeds store TG before they go into a very low metabolic state at which dry seeds remain until germination, when fatty acids are catabolized via the glyoxylate cycle (23). We postulate that M. tuberculosis may also use TG as a storage form of energy for its long-term survival under dormancy. Fatty acids that become available from the degenerating host tissue around the pathogen in the granuloma may be converted into TG for storage. This hypothesis is supported by the finding of intracellular TG inclusion bodies in M. tuberculosis organisms obtained from organ lesions (16). Under nutrient deprivation such as low nitrogen, TG-containing inclusion bodies appear upon availability of fatty acids (16). If this hypothesis has validity, dormancy-inducing conditions should induce TG synthesis. Such a possibility has not been explored, and little is known about the enzymes and genes involved in TG synthesis in M. tuberculosis.
The M. tuberculosis genome does not contain any classical triacylglycerol synthase (tgs) genes, but it contains nonannotated genes whose products have significant amino acid identity to a dual-function wax synthase-TGS from Acinetobacter calcoaceticus that have homologues with no known function also in other mycobacteria, streptomyces, and Arabidopsis thaliana (17). We designated 15 such genes that we have identified in the mycobacterial genome as tgs, but they do not show significant homology to any other reported tgs. Eleven of these genes have the conserved active-site motif HHxxxDG, three have modified versions of this motif, and one has no recognizable motif. We report that these gene products, when expressed in Escherichia coli, show TGS activity. We also report that in vitro induction of a persistent state by hypoxia upregulates some of the tgs genes whose products show the highest TGS activity. The same genes are also upregulated upon NO induction of the dormancy genetic program. Furthermore, the induction of this gene expression pattern is associated with elevated TGS activity and TG accumulation in M. tuberculosis H37Rv. These results suggest that M. tuberculosis may adopt the same energy storage and metabolic strategy as other hibernating organisms for long-term survival in the dormant state.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
For the different experiments, M. tuberculosis H37Rv (ATCC 25618) was grown in Middlebrook 7H9 (supplemented with 0.05% Tween 80, 10% oleic acid-albumin-dextrose-catalase enrichment, and 0.2% glycerol), in Dubos-Tween-albumin medium (prepared from Dubos broth base and Dubos medium albumin as per the manufacturer's instructions) and Sauton's medium (4 g of asparagine, 2 g of sodium citrate, 0.5 g of K2HPO4 · 3H2O, 0.5 g of MgSO4 · 7H2O, 0.05 g of ferric ammonium citrate, 60 g of glycerol, and 0.5 g of Tween in 1 liter of H2O; pH adjusted to 7.2). All media were purchased from Difco. E. coli DH5α and BL21 Star (DE3) (Invitrogen) used as host strains for cloning and expression experiments were grown on Luria-Bertani broth or agar and, when required, antibiotics were added to the culture media at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml. The NO donor spermine NONOate [(Z)-1-{N-[3-aminopropyl]-N-[4-(3-aminopropylammonio)butyl]-amino}-diazen-1-ium-1,2-diolate; SPER/NO]and its reference compound, spermine tetrahydrochloride (N,N′-bis[3-aminopropyl]-1,4-butanediamine tetrahydrochloride; SPER), were purchased from Alexis Corporation. Other chemicals and antibiotics were from Sigma Chemical Co. and Fisher Scientific.
Slow withdrawal of O2.
M. tuberculosis H37Rv cultures were subjected to hypoxia essentially as described by Wayne and Hayes (39). Seed cultures of M. tuberculosis H37Rv, grown in Middlebrook 7H9 aerobically at 37°C in roller bottles to an optical density at 600 nm (OD600) of 0.6 were used to inoculate Dubos-Tween-albumin medium to an OD600 of 0.006 in screw-cap tubes (with 0.5 headspace ratio) that were tightly sealed with solid caps having a latex rubber lining inside or with septum caps with plug-seal rubber septum, which were used to add antibiotic during the course of the experiment. To monitor gradual depletion of oxygen, the medium contained methylene blue (1.5 μg/ml). The culture was gently stirred using a magnetic stirring bar (120 rpm), and growth was monitored by measuring the OD600. Cultures from a set of tubes were pooled and divided into three parts for (i) RNA isolation, (ii) an in vivo radioactive tracer experiment to assess TG synthesis, and (iii) TGS activity measurement in cell extracts. In separate experiments, aliquots of the culture undergoing hypoxia were incubated with 0.64 μM oleic acid-0.5% bovine serum albumin (BSA) for 6 h, and the lipids were extracted and analyzed for TG by thin-layer chromatography (TLC).
Antibiotic resistance and sensitivity of hypoxic cultures were tested by determining the percent survival in medium containing isoniazid (4 μg/ml) or metronidazole (12 μg/ml) by determination of CFU after serial dilution and plating (39).
NO treatment.
The NO treatment was done essentially as previously described (25). M. tuberculosis H37Rv was grown in Middlebrook 7H9-Tween to an OD600 of 0.6 to 0.8, and this seed culture was used for growth in Sauton medium to an OD600 of 0.6. The culture was centrifuged, and the cells were washed twice and resuspended in the original volume of Sauton medium. The NO donor (SPER/NO) was added to a final concentration of 100 μM. The control set of cultures received 100 μM SPER. These cultures were incubated on a roller bottle incubator (120 rpm) for various periods of time at 37°C. Sixteen hours after the initial NO treatment, additional 100 μM NO donor was added in some cultures. At different intervals, a desired volume of the cultures was collected for RNA isolation, reverse transcription-PCR (RT-PCR), in vivo radioactive tracer assay for TG synthesis, total lipid extraction, and TGS enzyme assay in cell extracts.
General DNA techniques and data search.
All recombinant DNA techniques were performed according to standard procedures (28). DNA restriction and modifying enzymes were obtained from Invitrogen. We selected Rv3740c, which showed the highest degree of identity to the wax synthase-TGS of A. calcoaceticus (17), and used it to screen the genome of M. tuberculosis for related gene products using the Protein-Protein BLAST search program, available at http://www.ncbi.nlm.nih.gov/BLAST, yielding a total of 15 genes. We did pairwise alignment of 14 TGS proteins with the Rv3130c product using ALIGN, from http://xylian.igh.cnrs.fr/bin/align-guess.cgi, to determine the percent identity.
RNA isolation, RT-PCR, and quantitative real-time RT-PCR analysis.
M. tuberculosis H37Rv cultures were mixed with 2 volumes of RNA Protect bacteria reagent (QIAGEN), incubated for 5 min at room temperature, and centrifuged at 3,000 × g for 12 min at 4°C, and the cells were kept frozen at −80°C. Frozen bacterial pellets were thawed and resuspended in RNeasy lysis buffer (QIAGEN), transferred to a 2-ml tube containing silica beads (FastRNA Blue), and disrupted using the FastPrep F120 instrument (QBIOgene). The extract collected by centrifugation was used to isolate total RNA with an RNeasy kit (QIAGEN) according to the manufacturer's protocol. Equal amounts of DNase-treated total RNA were reverse transcribed using random primers and SuperScript RNase H− reverse transcriptase following the manufacturer's instructions (Invitrogen). RT-PCR amplification conditions comprised an initial cycle of denaturation at 94°C for 4 min, 29 cycles of 94°C for 55 s, 64°C for 50 s, and 72°C for 1 min, and a final incubation for 7 min at 72°C. The different primers used in RT-PCRs were selected to amplify fragments ranging between 496 and 812 bp (Table 1) for semiquantitative RT-PCR and between 206 and 246 bp for real-time PCR. 23S rRNA gene sequence amplification from each cDNA sample using different dilutions of cDNA stock was performed to quantify the level of expression of each gene. A control without reverse transcriptase verified the absence of DNA contamination. Different dilutions of cDNA for 23S rRNA were used as templates, and values obtained at a cDNA range that gave amplification product levels that showed linear dependence on template level were used for normalization. cDNA samples for each tgs gene product were also subjected to dilution before PCR to assure linear amplification.
TABLE 1.
Primers used for RT-PCR and real-time RT-PCR analyses of transcripts of tgs, dosR, and 23S rRNA genes
| Primer for tgs, dosR, or 23S rRNA gene | Primer sequence (5′-3′)a | Product length (bp) |
|---|---|---|
| RT-PCR primers | ||
| Rv3130c | F: CGTGCTAAGTCCCGCCGCGTCGTC | 737 |
| R: CTCCGCGCCTGCGAGTCACCTTGC | ||
| Rv3734c | F: GGTGGAATCGCCCGTGTGGCATGG | 765 |
| R: TGGGTCGTCGACATGGGTGGCGAG | ||
| Rv3234c | F: CTGGCCAGGCCGGTGTGGATCGAC | 595 |
| R: AGTCGTAGCGAGCCCGCACCGTGC | ||
| Rv3088 | F: CCGCGCCTGTTCGATGCCTACCGG | 780 |
| R: CATGAACGCCACCAGCGGCCGGTC | ||
| Rv1760 | F: ACCTCGACGATGCCGGGCGGCTAC | 750 |
| R: GCCGTGCTCGAGTAGGAAGCGCCG | ||
| Rv2285 | F: CGACCTGCCTAAGGGAGCACCGCG | 722 |
| R: CCTGCAGGTAGCGTCGGCAAGCCC | ||
| Rv0221 | F: GGGGGATGGATCTGCTGCCGGGAC | 715 |
| R: AGATCTCGGTGACCAGCGCGGCCC | ||
| Rv3740c | F: GAGCATCCGCTGCATGTCGGCGCG | 792 |
| R: CGTGTCCGGCAGCGCGTCGTTGTC | ||
| Rv3087 | F: CCGGTCTTCCGGCTGCGGTACCTG | 721 |
| R: TCGTGGCCACCATCGCGTCGGTGG | ||
| Rv3371 | F: CGACCTCACCCAGCACGTGCGACG | 703 |
| R: GGGTTGACGACGGCCGCTCTGCTG | ||
| Rv3480c | F: AGCCTGTCCACCGACCCGCACGAC | 812 |
| R: CGCAGGTAGAGCACGTCCTCGGGC | ||
| Rv1425 | F: GACTGGACCGGCCGTGGTTCGTCG | 802 |
| R: GTGCTCTCGTGGATGGCCGCCAGG | ||
| Rv0895 | F: GGTCGCTCACCTCGCTCGGGCAGT | 751 |
| R: CCTGGACCCTGGCCGAACACGTCG | ||
| Rv2484c | F: GGCGAGTCCCCCAGGTTGTCCGAC | 762 |
| R: CTCGGTACGGGTGGTCAGGCTGCG | ||
| dosR (Rv3133c) | F: GGTCGATGACCACGAGGTGGTGCGT | 496 |
| R: GGCGATCTGCTTGTTGGTCAGGCCC | ||
| 23S rRNA | F: GTGGCGTGTTCTGGACCCGAAGCG | 730 |
| R: GTCCATCGACTACGCCTGTCGGCC | ||
| Real-time RT-PCR primers | ||
| Rv3130c | F: AACGAAGACCAGTTATTCGAGC | 206 |
| R: CTCATACTTTCATCGGAGAGCC | ||
| Rv3734c | F: TACTACCTGATAGAGCGGAATGC | 215 |
| R: TCGGAGAGCACTTTCTTGTTG | ||
| Rv3234c | F: GTATGTCGGGTTGCTGTTGAT | 241 |
| R: GTGCAGTTGCTCGTCACTACC | ||
| Rv3088 | F: TGTGGTGATATCAAACATGAAGG | 241 |
| R: GGTTTGGAGCTCGGTGAAT | ||
| dosR (Rv3133c) | F: GTCGTCAAAGACATCAAGGGAAT | 246 |
| R: CGTCTTTTCGGCTAGGAACATT | ||
| 23S rRNA | F: ACGACACTTTCTGACTGCCTCTC | 224 |
| R: TCTGAATATATAGGGTGCGGGAG |
F, forward primer; R, reverse primer.
Real-time RT-PCR was performed using the iCycler iQ real-time detection system and iQ supermix according to the manufacturer's instructions (Bio-Rad Laboratories, Inc.). Amplification reactions consisted of 95°C for 3 min followed by 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Primers used for amplification reactions are listed in Table 1.
Expression of tgs genes in E. coli and determination of the TGS and WES activities of the expressed proteins.
DNA corresponding to the tgs open reading frames was amplified using Pfu Turbo Hotstart DNA polymerase (Stratagene), and expression was performed using the pET directional TOPO expression vector (Invitrogen). Rv3233c, Rv3234c, Rv3734c, Rv3740c, Rv3087, and Rv3088 were expressed in pET100/D-Topo. Rv2484c, Rv1760, Rv1425, and Rv0895 were expressed in pET102/D-Topo. Rv0221, Rv3371, Rv2285, Rv3480c, and Rv3130c were expressed in pET200/D-Topo. In these vectors, the open reading frames were directionally cloned and expressed as His-fusion proteins in E. coli strain BL21 Star (DE3) according to protocols provided by manufacturers. Total cell lysates were used for TGS and wax ester synthase (WES) activity measurements. Untransformed BL21 strain extracts showed extremely low TGS and WES activities.
TGS activity in the extracts was measured by the incorporation of 14C from [1-14C]oleoyl-coenzyme A (CoA) (specific activity, 55 Ci/mole; American Radiolabeled Chemicals Inc.) into triolein in the presence of diolein. In the absence of information about the substrate specificity of TGSs, we used oleoyl-CoA as a model substrate. Each reaction mixture containing total cell lysates (100 to 200 μg of protein), 14.5 μM (or the specified concentration) [1-14C]oleoyl-CoA, 1 mM (or the specified concentration) diolein, 10 mM MgCl2, and 1 mg of BSA in 250 μl of 0.1 M potassium phosphate buffer (pH 7.2) was incubated for 2 h at 37°C. The reaction products were extracted with chloroform-methanol (2:1 [vol/vol]), and 14C in the TG fraction was assayed after TLC in silica gel G using n-hexane-ethyl ether-formic acid (65:35:2 [vol/vol/vol]). Assays of pH dependence of activity were done in 50 mM citrate-phosphate buffer.
WES activity was determined by measuring the incorporation of [1-14C]palmityl alcohol (synthesized from [1-14C]palmitic acid; specific activity, 57 Ci/mole) into wax esters in the presence of palmitoyl-CoA. Assays were identical to the TGS assays with the exception that 20 μM (or the specified concentration) [1-14C]palmityl alcohol and 50 μM palmitoyl-CoA were used as substrates. 14C in the wax ester fraction was measured after TLC on silica gel with n-hexane-ethyl ether-acetic acid (90:10:1 [vol/vol/vol]). Silica gel from areas of the TLC that matched with the internal triolein or hexadecyl palmitate standards was assayed for 14C by liquid scintillation counting.
Incorporation of 14C-labeled precursors into lipids by M. tuberculosis.
M. tuberculosis cultures (40 ml) withdrawn after different treatments were incubated with 2 μCi of [1-14C]oleic acid (specific activity, 54 Ci/mole; Amersham Bioscience Corp.) for 6 h in the case of hypoxia and 1 h in the case of NO treatment. After incubation, the cells were collected by centrifugation and autoclaved, and total lipids were extracted with chloroform-methanol (2:1 [vol/vol]) as previously described (35). Radioactivity in the total extracted cellular lipids and the growth medium was measured. Lipids were analyzed by TLC using n-hexane-ethyl ether-formic acid (45:5:1 [vol/vol/vol]), and the radioactivity in the silica gel corresponding to the TG band was measured using a liquid scintillation counter (Packard). An autoradiogram of the TLC was prepared. The amount of TG was visualized by sulfuric acid-dichromate charring of the TLC plates as described before (35). The charred TLC plate was also scanned for quantification of TG accumulation by using the AlphaImager 2200 Gel Doc system (AlphaInnotech). At different time intervals after the initial NO treatment, the cells were incubated with 2 μCi of [1-14C]oleic acid for 1 h and the lipids were extracted and analyzed as described above. Similar procedures were used for incorporation of [1-14C]acetate (4-h incubation with 10 μCi; specific activity, 56.7 Ci/mole; American Radiolabeled Chemicals Inc.) and [1-14C]palmitic acid (1-h incubation with 2 μCi; specific activity, 60 Ci/mole; Amersham Bioscience Corp.). Fatty acid composition of labeled TG was determined by radio-gas chromatography (radio-GC) of the total methyl esters or after argentation TLC (11).
TGS activity in cell extracts from M. tuberculosis subjected to hypoxia and NO treatment.
At each time point, cells were collected by centrifugation and washed in lysis buffer consisting of 50 mM HEPES, pH 7.5, containing 150 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol, and 10 μg of phenylmethylsulfonyl fluoride/ml and resuspended in 1 ml of the same lysis buffer, and cells were disrupted using a FastPrep F120 instrument (QBIOgene). The extract was centrifuged, and the supernatant was filter sterilized (0.2-μm-pore-size filter). The protein concentration in the supernatant was measured by the Bio-Rad method and used for measuring TGS activity. The reaction mixture consisted of [1-14C]oleic acid (0.2 μCi), 5 mM ATP, 5 mM MgCl2, 100 μM CoA, 100 μM diolein, and enzyme extract (200 μg of protein) in a total volume of 400 μl at 37°C for 2 h. The reaction products were analyzed as indicated above for TGS expressed in E. coli, except that n-hexane-ethyl ether-formic acid (45:5:1 [vol/vol/vol]) was used as the solvent system. All experiments were repeated at least three times, and typical results are shown in all cases. Since details such as cell density were not absolutely identical in all repetitions we did not average the values, but the results from a typical experiment are shown.
RESULTS
Expression of M. tuberculosis tgs genes in E. coli and characterization of the expressed proteins.
The source of energy used by dormant and reactivating M. tuberculosis within the host remains unclear. We postulate that TG may be stored by the organism as it enters into the dormant phase for utilization during and after dormancy. Since the enzymes involved in the biosynthesis of TG in M. tuberculosis have not been identified, we examined the M. tuberculosis genome for putative tgs genes. No classical tgs genes have been identified in the genome. Homology to a bifunctional wax synthase-TGS gene in A. calcoaceticus revealed 13 conserved hypothetical protein genes in the mycobacterial genome (17). We used the mycobacterial gene Rv3740c that showed the highest degree of identity to the A. calcoaceticus gene to screen the M. tuberculosis H37Rv genome for related genes. A total of 15 M. tuberculosis H37Rv genes were identified, and we designated these genes as tgs; these genes showed little homology to other tgs genes and, thus, belong to the novel family of bacterial tgs's (Table 2). We postulate that some of these genes may be involved in TG synthesis as the organism adapts to dormancy. Eleven of these genes contain the HHxxxDG active-site motif that is thought to catalytically participate in the acyl-CoA acyltransferase reactions involved in TG synthesis. The others have modified active-site motifs. Rv3371 has a 16-amino-acid insertion in the active-site motif, while in Rv2484c the first histidine of the motif is replaced by serine, and in Rv3234c the second histidine is replaced by glutamine. Rv3233c does not have any recognizable motif. All tgs genes would encode products with calculated molecular masses of 47 to 54 kDa except Rv3234c and Rv3233c, which would yield 30- and 21-kDa proteins, respectively (Table 2). The theoretical pI values range from 4.69 to 10.38. Eight of the tgs gene products are predicted to be membrane-bound proteins, and six are predicted to be cytoplasmic.
TABLE 2.
Characteristics of putative tgs genes in the genome of M. tuberculosis
| tgs gene | Identitya (%) | Active-site motif | Theoretical molecular mass (kDa) | Theoretical pI |
|---|---|---|---|---|
| Rv3130c | 100.0 | HHCMADG | 50.7 | 10.11 |
| Rv3371 | 42.8 | HHCMAGAMSAAHLLARLCDDADG | 48.9 | 9.48 |
| Rv3740c | 29.5 | HHALVDG | 48.4 | 9.35 |
| Rv0221 | 28.8 | HHALADG | 51.9 | 7.90 |
| Rv3480c | 28.0 | HHSLIDG | 53.3 | 6.10 |
| Rv3734c | 27.7 | HHALIDG | 49.3 | 6.36 |
| Rv1425 | 26.9 | HHAIVDG | 50.1 | 5.79 |
| Rv0895 | 26.9 | HHALADG | 53.9 | 10.38 |
| Rv3087 | 26.7 | HHAYSDG | 52.6 | 6.93 |
| Rv1760 | 26.3 | HHAVVDG | 54.1 | 6.27 |
| Rv3088 | 25.9 | HHALIDG | 50.9 | 9.58 |
| Rv2285 | 25.1 | HHCAVDG | 47.7 | 7.70 |
| Rv2484c | 22.5 | SHAVTDG | 52.3 | 7.78 |
| Rv3234c | 22.0 | HQALING | 30.4 | 9.83 |
| Rv3233c | 14.5 | No motif | 20.9 | 4.69 |
Since the Rv3130c-encoded enzyme showed the highest TGS activity, amino acid sequence identities of the other tgs gene products were compared to it by pairwise alignment with ALIGN, available at http://xylian.igh.cnrs.fr/bin/align-guess.cgi.
When the tgs genes from M. tuberculosis were expressed in E. coli and the total cell lysates were fractionated by centrifugation at 100,000 × g, it was found that most of the TGS activity was localized in the pellet. Since many tgs genes were predicted to be hydrophobic and membrane bound, purification of each of the 15 TGS proteins was not attempted at this stage, and total cell lysates were used for all the initial characterization studies reported in this paper. Extracts of the wild-type E. coli cells showed little TGS activity. For all expressed proteins, TGS activity linearly increased with protein levels up to 200 μg and 180 min of incubation time (data not shown). The pH dependence of TGS activity was determined for each expressed enzyme. Most of the enzymes showed a broad pH optimum near neutral pH. Among the most active enzymes, the Rv3130c product that we designated TGS1 showed maximal activity at pH 4.5, and 85% of this activity was retained up to pH 6.5; subsequent increases in pH caused sharp decreases in activity. The Rv3734c product (TGS2) showed maximal activity above pH 7.0 but retained 85% of activity at pH 6.5. For each enzyme, the range at which at least 85% of the maximal activity was observed is shown in Table 3. The Rv3234c product (TGS3) and the Rv3371 product showed more sharp pH optima at 6.5 and 4.5, respectively. Activity of each recombinant enzyme was normalized for expression level in total cell lysate. In most cases, expressed protein levels ranged from 15 to 25% of total protein. All 15 tgs genes displayed TGS activity when tested with diolein and oleoyl-CoA as substrates (Table 3). TGS activity was high for products of certain tgs genes, such as TGS1, TGS2, TGS3, and TGS4 (Rv3088), but very low for others, such as the products of Rv3233c, Rv1425, Rv8895, and Rv2484c (Table 3). The dependence of TGS activity on the concentration of diolein and oleoyl-CoA was investigated, and Km and Vmax values for the more active enzymes are shown in Table 4. The tgs gene products were also assayed for WES activity, which was found to be at much lower levels than the corresponding TGS activity for most of the enzymes. As can be seen from the data in Table 3, TGS and WES activities were not directly correlated. The highest WES activity was displayed by TGS2 and the products of Rv3740c and Rv3480c (Table 3), whereas the highest TGS activity was displayed by TGS1.
TABLE 3.
TGS and WES activities of M. tuberculosis genes expressed in E. coli
| tgs gene | TGS activitya (pmol/min/mg) | Optimal pHb | WES activitya (pmol/min/mg) |
|---|---|---|---|
| Rv3130c (TGS1) | 226 | 4.5-6.5 | 0.9 |
| Rv3734c (TGS2) | 119 | 6.5-7.2 | 9.5 |
| Rv3234c (TGS3) | 93 | 6.5 | 0 |
| Rv3088 (TGS4) | 79 | 6.5-7.2 | 1.8 |
| Rv1760 | 38 | 5.5-6.5 | 2.4 |
| Rv2285 | 37 | 5.5-7.2 | 3.5 |
| Rv0221 | 24 | 5.5-6.5 | 1.7 |
| Rv3740c | 17 | 5.5-7.2 | 5.3 |
| Rv3087 | 14 | 5.5-6.5 | 0 |
| Rv3371 | 14 | 4.5 | 0.2 |
| Rv3480c | 12 | 4.5-6.5 | 4 |
| Rv3233c | 4.4 | 5.5-6.5 | 1.1 |
| Rv1425 | 4.1 | 6.5-7.2 | ND |
| Rv0895 | 3.5 | 4.5-6.5 | 0.5 |
| Rv2484c | 3.3 | 5.5-7.7 | 0.06 |
The enzymatic activities were normalized for expression level as indicated in Materials and Methods. ND, not determined.
A range of optimal pH is given in cases where at least 85% of activity was retained within that range. Single pH values indicate a more narrow optimal pH range.
TABLE 4.
Kinetic parameters for TGS of M. tuberculosis expressed in E. colia
| TGS |
Km (μM)
|
Vmax (pmol/min) | |
|---|---|---|---|
| Diolein | Oleoyl-CoA | ||
| Rv3130c | 28 | 161 | 476 |
| Rv3734c | 370 | 200 | 217 |
| Rv3234c | 63 | 200 | 19 |
| Rv3088 | 714 | 26 | 4 |
| Rv1760 | 56 | 667 | 80 |
| Rv2285 | 152 | 30 | 11 |
Experimental conditions are indicated under Materials and Methods.
Induction of TG synthesis by hypoxia.
Our hypothesis is that TG constitutes the long-term storage form of energy that allows the pathogen to survive through long dormancy periods. If this were true, conditions that induce dormancy should induce TG synthesis. We postulate that the tgs genes we identified may be involved in the dormancy-associated TG synthesis. Since hypoxia is thought to induce a nonreplicating persistent state resembling dormancy (40), we tested whether the tgs genes are upregulated under such a condition. Cells were grown in an in vitro culture model in which gradual oxygen depletion was achieved, leading to a hypoxic condition as previously seen (39). The growth pattern we observed was very similar to that previously described (39). The hypoxic conditions we used, in fact, caused the pathogen to go into a nonreplicative state that showed isoniazid resistance (0.4 to 10% survival up to 6 days and 93 to 95% survival at 8 to 17 days) and metronidazole sensitivity (100% survival up to 4 days and 33% survival at 17 days) characteristic of dormant bacilli.
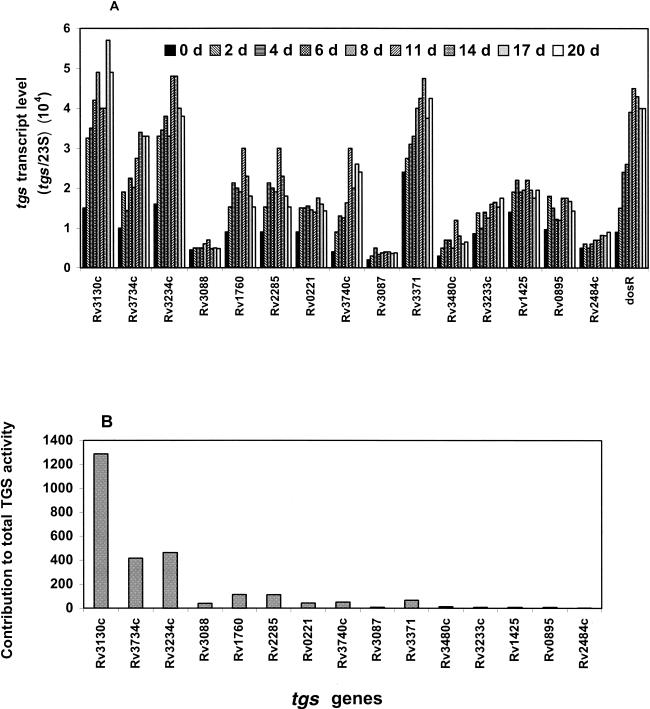
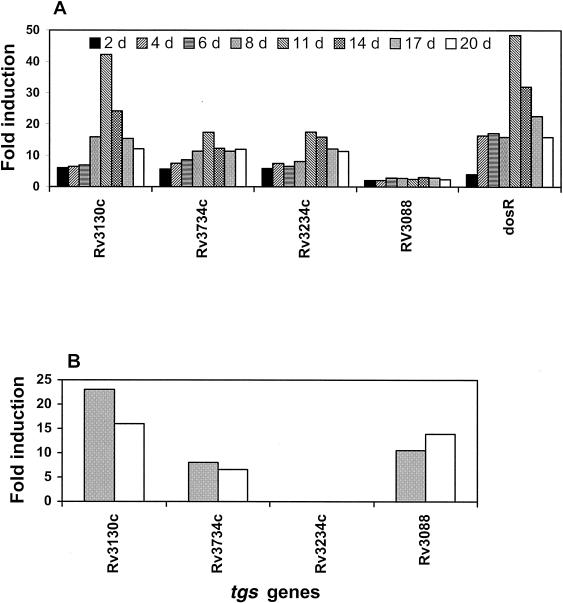
Induction levels of the 15 tgs genes were assessed by RT-PCR analyses of mRNAs isolated from the cells grown under hypoxic conditions. The tgs transcript levels are expressed as the fraction of 23S rRNA transcript (Fig. 1A). All 15 tgs genes were found to be expressed in the cells before subjecting them to hypoxia. Several of these tgs genes were significantly upregulated following the gradual depletion of oxygen. We also measured by real-time PCR the level of induction of the tgs genes whose products showed the highest levels of enzymatic activity to confirm the results obtained with the semiquantitative RT-PCR (Fig. 2A).
FIG. 1.
(A) RT-PCR assessment of induction of tgs genes in M. tuberculosis H37Rv during the gradual depletion of O2. Transcript levels measured by RT-PCR are shown as a fraction of 23S rRNA transcripts. The method used for quantitation and experimental details are given in Materials and Methods. Each bar represents the induction level at a different sampling day as shown on the top of the graph. The induction level of dosR (Rv3133c) is shown for comparison. Since in the different experiments the initial cell density was slightly different, we did not average the values; instead, we represent a typical experiment. The same pattern was observed in the individual experiments. (B) Estimated potential relative contribution of the tgs gene products to the total TGS activity. The maximal level of each tgs transcript achieved during hypoxia was multiplied by the TGS activity of each expressed enzyme.
FIG. 2.
(A) Real-time PCR measurement of the most highly induced tgs genes in M. tuberculosis H37Rv during the gradual depletion of O2. Transcript levels were measured by real-time PCR, and data were analyzed by comparative CT method (ΔΔCT) for relative quantitation of gene expression. The induction level of dosR (Rv3133c) is shown for comparison. (B) Real-time PCR measurement of the most highly induced tgs genes in M. tuberculosis H37Rv by NO treatment. Quantitation of transcript levels was done by real-time PCR, and data were analyzed as for panel A but using the spermine control as the reference. The maximal level was reached within 4 h of the first NO treatment (gray bars) and within 4 h of the second NO treatment 16 hours after the initial NO treatment (open bar).
The tgs (Rv3130c) gene whose expressed product showed the highest TGS activity also showed the highest induction as the cells entered into the nonreplicative state, consistent with a possible role for this gene product in the establishment of dormancy. Both semiquantitative RT-PCR and real-time RT-PCR showed that the induction level of this gene was similar to that of dosR. Both methods showed similar relative levels of induction of the other tgs genes which encode the most active enzymes, although the values for induction revealed by real-time PCR were higher than those indicated by the semiquantitative RT-PCR. In most cases, the induction level increased as the hypoxia developed and the cells reached the isoniazid-resistant, metronidazole-sensitive nonreplicative state. The highest tgs transcript levels were reached between the 11th and 17th days, and the level remained high till the end of the experiment (20 days). To assess the possible relative contributions of the various tgs products to the level of TGS activity that cells may contain, we multiplied the maximal transcript level of each tgs gene with the TGS activity level of each expressed gene product (Fig. 1B). The Rv3130c product (TGS1) showed the highest potential for participating in TG synthesis. With the exception of the Rv3234c and Rv3734c products, the other tgs gene products probably would not have the ability to make significant contributions to TG synthesis under these conditions.
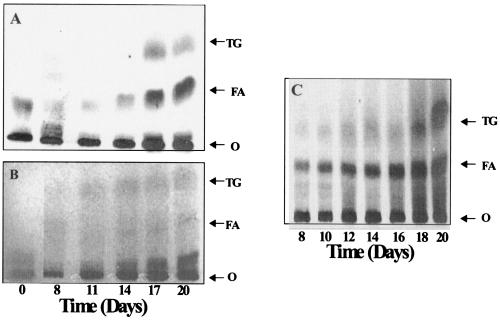
To test whether the induction of the tgs genes leads to TG synthesis, we examined whether the bacilli acquire increased TGS activity during the development of the hypoxia-induced nonreplicating state. As the bacilli acquired isoniazid resistance and metronidazole sensitivity, incorporation of exogenous [1-14C]oleic acid into TG increased (Fig. 3A). The amount of oleic acid incorporated into TG was very low at day zero; from 8 to 20 days, as the antibiotic sensitivity changes developed, incorporation of oleic acid into TG increased. Incorporation into polar lipids decreased from 90% of the recovered 14C at 8, 11, and 14 days to 70% at 17 and 20 days. To test whether the bacilli store TG, the total lipids extracted from the cells were subjected to TLC and lipids were visualized by charring (Fig. 3B). The chemical level of TG in the cells increased as the bacilli reached the nonreplicating state. In an attempt to mimic the possible availability of fatty acids released from the degrading host tissue in the developing lesions, we provided exogenous oleic acid to the pathogen as it entered hypoxic conditions. TLC analysis clearly showed accumulation of TG as the pathogen went into the nonreplicative state (Fig. 3C).
FIG. 3.
Induction of TG synthesis in M. tuberculosis during gradual depletion of O2. (A) Autoradiogram showing [1-14C]oleic acid incorporation into TG. (B and C) Dichromate-sulfuric acid charring of lipids showing TG accumulation in M. tuberculosis cells going into the nonreplicative state without exogenous oleic acid (B) and after 6 h of incubation with 0.64 μM oleic acid-0.5% BSA (C). Lipids were separated by TLC using n-hexane-ethyl ether-formic acid (45:5:1 [vol/vol/vol]) as the solvent system. O, origin; FA, fatty acids. Time after the initiation of O2 depletion is shown in days.
Induction of TG synthesis by NO treatment.
Since NO treatment was recently shown to induce the same set of genes as those induced under the hypoxia-induced nonreplicative state (26, 38), we tested whether NO treatment would also induce the tgs genes. We treated M. tuberculosis cells with the NO donor SPER/NO, with controls being treated with only spermine. NO treatment caused detectable suppression of growth, as noted by others (25). Induction levels of the 15 tgs genes were tested by RT-PCR analyses of mRNA isolated from M. tuberculosis H37Rv cells treated with the NO donor. The tgs transcript levels are expressed as the fraction of 23S rRNA transcript (Fig. 4A). Total duration of the experiment was for 20 h, but we have represented the data as the maximum induction level achieved within 4 h of initial NO treatment and the maximum induction level obtained within 4 h of the second NO treatment, administered 16 h after the initial NO treatment. Among the15 tgs genes tested, 11 were found to be induced (Fig. 4A). The maximum level of induction was detected for Rv3130c (tgs1) after 2 and 4 h of initial NO treatment. We tested the induction level of dosR (Rv3133c), as a control, and it also showed a similar extent of induction as Rv3130c (Fig. 4A). Levels of most tgs transcripts reached a maximum level 2 to 4 h after NO treatment and subsequently decreased. To test whether repeated NO treatment would induce tgs transcript levels, we treated the cells with NO 16 h after the initial treatment, when tgs transcript levels had returned to basal levels. This second treatment caused induction of the same tgs genes as those induced by the first NO treatment and in each case to about the same levels as those reached by the first treatment (Fig. 4A). The tgs levels induced by the second treatment also reached maximal levels by 4 h after the second treatment. Real-time PCR analysis of induction of the tgs genes whose products showed the highest enzymatic activities confirmed the results obtained with the semiquantitative RT-PCR (Fig. 2B). The level of induction of dosR revealed by the real-time PCR was considerably higher than that indicated by the semiquantitative RT-PCR; according to real-time PCR, the dosR level showed a 60-fold induction, compared to the 15-fold induction for Rv3130c, whereas semiquantitative RT-PCR showed similar levels of induction for both genes.
FIG. 4.
(A) Induction of tgs genes in M. tuberculosis H37Rv by NO treatment. Transcript levels were measured by RT-PCR and expressed as a fraction of the 23S rRNA transcript level. In each case the values obtained with the spermine control were subtracted, and the maximal level reached within 4 h after NO treatment is shown (gray bars). Sixteen hours after the initial NO treatment additional treatment with NO was done, and the maximal transcript levels reached within the next 4 h are shown (open bars). Induction level of dosR (Rv3133c) is shown for comparison. Since in the different experiments the initial cell density was slightly different, we did not average the values; instead, we represent a typical experiment. The same pattern was observed in the individual experiments. (B) Estimated potential relative contribution of the tgs gene products to the total TGS activity in M. tuberculosis cells. The maximal level of each tgs transcript achieved during the first 4 h of initial NO treatment was multiplied by the TGS activity of each expressed enzyme.
To assess the probable relative contributions of the various tgs products to the total level of TGS activity in the NO-treated cells, we multiplied the maximal transcript level achieved within 4 h of the first NO treatment of each tgs gene with the TGS activity level measured for each product expressed in E. coli (Fig. 4B). The Rv3130c product (TGS1) showed the highest potential for contributing to TG synthesis (Fig. 4B), as we have also observed in the hypoxia experiments.
To test whether induction of tgs genes by NO treatment of M. tuberculosis cells results in actual TG synthesis, we examined the ability of the cells to synthesize TG both in vivo and in vitro. [1-14C]oleic acid incorporation into TG by the intact cells was significantly increased after the NO treatment, whereas the level of TG synthesized remained more or less constant in the control samples containing only spermine, or was slightly increased at the later time points, but never reached the TG level found in NO-treated samples (Fig. 5A). The induction of 14C incorporation into TG reached maximal levels by 8 h after NO treatment and subsequently started decreasing. After a second NO treatment, 16 h after the initial NO treatment, the 14C incorporation into TG again markedly increased and reached even a slightly higher level than the maximum level reached after the first NO treatment (Fig. 5A).
FIG. 5.
Induction of TG synthesis in M. tuberculosis by NO treatment. (A) Autoradiogram showing [1-14C]oleic acid incorporation into TG. (B) Dichromate-sulfuric acid charring of lipids showing TG accumulation. Lipids were separated by TLC using n-hexane-ethyl ether-formic acid (45:5:1 [vol/vol/vol]) as the solvent system. S, spermine control; N, NO treatment. Sixteen hours after the initial NO treatment additional treatment with NO was done, and samples were taken at 2 h (18B) and 4 h (20B) after the second NO treatment. In panel A, incorporation of 14C into TG is shown as a percentage of the total 14C administered. In panel B, the bar graph shows the intensity of the TG band determined in arbitrary units by the AlphaImager 2200 Gel Doc system. O, origin; FA, fatty acids.
Induction of TGS activity by NO treatment was also tested in cell extracts of M. tuberculosis. Incorporation of [1-14C]oleic acid into TG in the presence of diolein increased (Fig. 6). After reaching a maximum level 8 h after this NO treatment, TGS activity decreased to the initial level by 16 h. A second NO treatment 16 h after the first NO treatment showed an increase in TGS activity (Fig. 6, 20B). This increased level was comparable to that reached after the first treatment. At each time point, TGS activity in spermine-added control samples remained more or less constant.
FIG. 6.
Induction of TGS activity in cell extracts of M. tuberculosis cells after NO treatment. In each case, 200 μg of protein was assayed as indicated in Materials and Methods, and values obtained with spermine control cultures were subtracted. Sixteen hours after the initial NO treatment additional NO treatment was done, and samples were taken at 2 h (18B) and 4 h (20B) after the second NO treatment.
To test whether the increases in tgs transcript levels and in TGS activity levels, detected by oleic acid incorporation into TG in vivo and in cell-free preparations, cause actual TG accumulation in the cells, the lipids extracted from the cells at various time points after NO treatment were subjected to TLC and the chromatograms were charred after spraying them with dichromate-sulfuric acid. TG accumulation caused by NO treatment compared with that in spermine-treated controls was clearly seen (Fig. 5B); the pattern of TG accumulation was quite similar to the pattern of oleic acid incorporation into TG.
We also examined the fatty acid constituents of the labeled TG produced from [1-14C]oleic acid in M. tuberculosis cells (Fig. 5A). Analysis by radio-GC of the fatty acid constituents of the TG fraction isolated from 14C-labeled lipids extracted from M. tuberculosis cells isolated 4 h after the second NO treatment showed 14C in oleic acid and longer-chain saturated fatty acids, with C26 as the major component (Fig. 7). Argentation TLC confirmed the labeling of the saturated acids (data not shown). The chemical composition of the TG showed that n-C26 acid was a major component in the TG of M. tuberculosis H37Rv.
FIG. 7.
Radio-GC of fatty acids in TG derived from exogenous [1-14C]oleic acid in NO-treated M. tuberculosis. Methyl esters were prepared from [14C]TG from the 4-h sample after the second NO treatment 16 h after the initial NO treatment. The top panel shows the radioactivity detector response, and the lower panel shows the flame ionization detector (FID) response. Retention times of n-fatty acids are indicated above.
To further examine the induction of TG synthesis by NO treatment, we administered [1-14C]acetate and [1-14C]palmitate to the M. tuberculosis cultures 4 h after NO treatment. Stimulation of the incorporation of both of these substrates into TG by NO treatment was readily seen in the TLC of the lipids derived from these substrates (Fig. 8). Incorporation of acetate and palmitate into TG was stimulated almost 10- and 3-fold, respectively. Radio-GC analysis of the labeled fatty acid constituents of the TG showed that acetate was incorporated into C16 to C28 fatty acids, with C26 as the major labeled acid. Palmitate was incorporated directly into TG, and 14C was also found in the longer-chain acids up to C28 (Fig. 9).
FIG. 8.
Autoradiogram showing induction of TG synthesis from 14C-labeled precursors by NO treatment in M. tuberculosis. After 4 h of NO treatment, cells were incubated with [1-14C]palmitic acid (for 1 h) and [1-14C]acetate (for 4 h), and the lipids were separated by TLC using n-hexane-ethyl ether-formic acid (45:5:1 [vol/vol/vol]) as the solvent system. S, spermine control; N, NO treatment. The bar graph shows the percentage of total administered radioactivity incorporation into TG. O, origin.
FIG. 9.
Radio-GCs of fatty acids from TG derived from [1-14C]acetate and [1-14C]palmitic acid in NO-treated M. tuberculosis. Methyl esters were prepared from [14C]TG isolated after incubation with 14C-labeled precursors at 4 h after the NO treatment. Retention times of n-fatty acids are indicated above.
DISCUSSION
TG is a common and efficient form of energy storage in organisms for utilization during long-term survival. M. tuberculosis survives for decades within the host in a state of dormancy, and the use of fatty acids has been associated with the persistence of the pathogen in the murine host (21). However, the source of the fatty acids used during dormancy has not been identified. Since TG would be an ideal source of fatty acids for use under such conditions, we examined the genome for genes that might encode TGS. Based on homology to a dual function protein from the wax-producing A. calcoaceticus (17), we have found a total of 15 M. tuberculosis genes which could potentially encode TGS. RT-PCR analysis revealed that all 15 tgs genes are expressed in this pathogen in culture. All 15 tgs gene products showed TGS activity when expressed in E. coli. TGS1 (Rv3130c) showed the highest TGS activity, and three other tgs gene products (TGS2 [Rv3734c], TGS3 [Rv3234c], and TGS4 [Rv3088]) also showed considerable TGS activity, while others showed much lower activity. Among these weakly active enzymes, two (Rv3371 and Rv2484c) have a modified active-site motif. Even though Rv3234c has some modification in its active-site motif, its product showed fairly high TGS activity. Rv3371 showed the highest degree of identity to TGS1; however, it showed only a low TGS activity, presumably because its active-site motif contains a 16-amino-acid insertion. The relative activities we have reported were determined with diolein and oleoyl-CoA as substrates. In view of the fact that the TG in the pathogen also contains other fatty acids, especially very-long-chain fatty acids (2, 16), it is possible that some of the TGSs may have selectivity for such acids. An example could be the product of Rv3371 that has an insertion of a hydrophobic 16-amino-acid segment in its active site and showed a high level of induction under hypoxia and moderately high-level expression upon NO treatment. The substrate specificities of the TGSs have not been studied.
The potential contributions of the tgs products to the total TGS activity might be reflected by the multiplication product of the transcript level and the TGS activity level of the expressed tgs genes. Such an assessment shows that the Rv3130c product makes by far the highest contribution to TGS activity induced under hypoxia or NO treatment. Induction of Rv3130c in static cultures (versus shaking) has been detected by RT-PCR analysis (13). Microarray analyses showed that Rv3130c was induced under hypoxia and NO treatment (26, 38), although it was not recognized as a tgs gene. In fact, this is the only tgs gene that has been found to be upregulated under such dormancy-inducing conditions.
All TGS proteins would have a calculated molecular mass of around 50 kDa. However, the molecular masses of Rv3234c and Rv3233c together add up to 50 kDa, and Rv3233c does not possess the conserved active-site motif. Therefore, it is possible that both genes are transcribed as one open reading frame. In fact, RT-PCR with primers spanning the junction between the two annotated open reading frames gave a product with the size and sequence expected from a single transcript containing both Rv3234c and Rv3233c (data not shown). Thus, the two annotated genes are transcribed together. Consistent with this conclusion is the finding that both Rv3233c and Rv3234c were downregulated under nutrient starvation (3) and both were unaffected by NO treatment (38). We confirmed the presence of a termination codon followed by a 2-nucleotide gap before the translation initiation codon of the next gene, consistent with the assignment of two open reading frames. However, the proteins encoded by these genes in M. tuberculosis have not been studied.
An examination of the genomic neighborhoods of the tgs genes revealed that several of them are located near transcriptional regulatory genes, suggesting coregulation with a related set of genes. Interestingly, some of the tgs genes are located near two-component transcriptional regulatory proteins. The best example is the presence of devS/devR (dosR) near the TGS1-encoding gene (26). Disruption of the dosR gene has been demonstrated to abolish the induction of tgs1 (Rv3130c) when the organism was exposed to hypoxia (26), although this gene was not known to be a tgs. We also found upregulation of fas, and the acyl-CoA carboxylase components (accD4 and accD5), under hypoxia (data not shown), consistent with the previous reports of induction of fas (26). Rv3087 and Rv3088 are located in the mymA operon under the control of virS, which is a transcriptional regulator of the ARAC family (34). The other genes in this operon include lipR (Rv3084), alcohol dehydrogenases (Rv3085 and Rv3086), and fadD13 (Rv3089), which is an acyl-CoA synthase. Interestingly, this operon was shown to be preferentially induced at acidic pH and upon infection of macrophages and has been suggested to utilize the accumulated C24 and C26 fatty acids produced by the downregulation of FAS II under acidic conditions (12). The environmental stresses the organism may encounter within the granuloma in the human host are thought to include hypoxia, acid pH, cytokines, reactive nitrogen intermediates produced by the host nitric oxide synthase (NOS2), reactive oxygen species, and nutritional stress (6, 8, 22, 24, 30). Different sets of tgs genes may be turned on in response to different stress factors encountered in the host by the pathogen in order to enable the organism to synthesize TG with maximum efficiency.
Ten of the 15 tgs genes are located adjacent or proximal to 11 lip genes that are annotated as probable phospholipases or lipases-esterases-carboxylesterases. Some lip genes may be cotranscribed with neighboring tgs genes under unique environmental stresses and may possibly play important roles in making fatty acids from host lipids available for synthesizing TG stores. lip gene products may also function as TG hydrolase and function in releasing fatty acids from TG for utilization during dormancy and upon reactivation after dormancy. Alternatively, the lip gene product may release a newly synthesized fatty acid chain from a polyketide synthase for TG synthesis or transfer to appropriate acceptors. We have expressed many lip genes, and their hydrolytic activities have been detected (unpublished results). Rv0221 is located near lipC (Rv0220), lipW (Rv0217c), acyl-CoA synthetase (Rv0214), acyl-CoA dehydrogenase (Rv0215c), and an integral membrane acyltransferase (Rv0228), suggesting that these genes may be cotranscribed under specific stimuli and may be involved in the degradation of lipids. The tgs gene product (Rv2484c), which has a significant degree of identity (72%) to a Mycobacterium leprae gene product (ML1244), is located next to a carboxylesterase lipQ (Rv2485c), a probable glycerol-3-phosphate acyltransferase (Rv2482c), a lysophosphatidic acid acyltransferase-like protein (Rv2483c), and a probable enoyl-CoA hydratase (Rv2486), suggesting a possible involvement in synthesis of TG via the Kennedy pathway. A few tgs genes (Rv3234c, Rv3233c, Rv2285, and Rv1425) are located proximal to lipoproteins, which may serve as donors or acceptors of fatty acids.
To test for the validity of our hypothesis that M. tuberculosis stores fatty acids in the form of TG for use in dormancy, we subjected the pathogen to slow depletion of O2 and to NO treatment, the two conditions thought to induce a dormancy-like state in vitro. Both of these conditions caused induction of several tgs genes, particularly those that show the highest TGS activity when expressed in E. coli. The most striking observation is that the tgs1 (Rv3130c) gene whose product has the highest TGS activity is the one that is induced the most under both dormancy-inducing conditions. The level of induction of this tgs gene was similar to that of dosR, a transcription regulator of a two-component system that has been previously shown to be upregulated by hypoxia and NO treatment (26, 38). Real-time PCR confirmed the relative levels of induction of tgs genes indicated by the semiquantitative RT-PCR. The tgs2 gene (Rv3734c), whose product shows the next highest TGS activity, is also strongly induced under both stress conditions. tgs3 (Rv3234c) was upregulated under hypoxia but not induced by NO treatment. On the other hand, tgs4 (Rv3088) was induced by NO treatment but only weakly induced under hypoxia. Under nutrient starvation, tgs2 and tgs4 were reported to be induced (3). tgs4 has been reported to be induced also under acidic conditions (12). Two of the tgs genes, Rv3087 and Rv3371, were suggested to be required for survival in mice (29). It is possible that different tgs genes are induced under the influence of the different host factors that contribute to the dormancy of the pathogen in vivo.
The increase in tgs transcript levels caused by hypoxia and NO treatment was reflected in the ability of the pathogen to synthesize TG. Exogenous oleic acid incorporation into TG increased as the cultures became hypoxic. Incorporation of exogenous oleic acid into other cellular lipids did not show major changes. Accumulation of TG caused by hypoxia could be readily detected at a chemical level by charring TLC. NO treatment that caused induction of the tgs genes also caused an increase in TGS activity, incorporation of [1-14C]acetate and exogenous fatty acids into TG, and accumulation of chemically detected TG levels. The exogenous fatty acids were incorporated directly and after elongation into TG. Incorporation of β-oxidation products into fatty acids might happen, as indicated by the incorporation of 14C from [1-14C]oleic acid into saturated C16 to C28 fatty acids. These results are consistent with previous reports of induction of β-oxidation enzymes by hypoxia and NO (26, 31). The presence of very-long-chain acids in TG has been found in other mycobacterial species (2, 16, 20). The increased level of TG was maintained for up to 8 h in NO-treated M. tuberculosis cells and subsequently began to decrease. However, additional NO treatment restored the increased level of TG synthesis. In the host, the pathogen is probably continuously exposed to NO and, therefore, the NO-induced TG synthesis would be maintained for long periods, probably as long as the organism is in dormancy. Induction of TG synthesis in Mycobacterium smegmatis under nitrogen-limiting growth conditions has been observed (16), and we have confirmed these observations in M. tuberculosis H37Rv (unpublished results). The lipophilic inclusion bodies containing TG observed in the pathogen recovered from sputum (16) might represent TG stored during dormancy or TG produced in the expanding granuloma from the fatty acids released from the degrading host tissue. Experiments with tgs disruptants, which are in progress, will determine whether the induction of TG synthesis is required for dormancy and reactivation. If so, the TGS(s) involved in this process could offer targets for novel drugs that could prevent dormancy and thus help in the control of tuberculosis.
Acknowledgments
This work was supported in part by grants AI46582 and AI35272 from the National Institutes of Health.
We thank Alexander Steinbuchel for revealing to us information about wax synthase-TGSs before publication.
REFERENCES
- 1.Alvarez, H. M., and A. Steinbuchel. 2002. Triacylglycerols in prokaryotic microorganisms. Appl. Microbiol. Biotechnol. 60:367-376. [DOI] [PubMed] [Google Scholar]
- 2.Asselineau, C., J. Asselineau, G. Laneelle, and M. A. Laneelle. 2002. The biosynthesis of mycolic acids by mycobacteria: current and alternative hypotheses. Prog. Lipid Res. 41:501-523. [DOI] [PubMed] [Google Scholar]
- 3.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 4.Bishai, W. 2000. Lipid lunch for persistent pathogen. Nature 406:683-685. [DOI] [PubMed] [Google Scholar]
- 5.Boon, C., and T. Dick. 2002. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J. Bacteriol. 184:6760-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, E. D., J. Chan, and N. W. Schluger. 2001. What is the role of nitric oxide in murine and human host defense against tuberculosis? Current knowledge. Am. J. Respir. Cell Mol. Biol. 25:606-612. [DOI] [PubMed] [Google Scholar]
- 7.Clark-Curtiss, J. E., and S. E. Haydel. 2003. Molecular genetics of Mycobacterium tuberculosis pathogenesis. Annu. Rev. Microbiol. 57:517-549. [DOI] [PubMed] [Google Scholar]
- 8.Cooper, A. M., L. B. Adams, D. K. Dalton, R. Appelberg, and S. Ehlers. 2002. IFN-γ and NO in mycobacterial disease: new jobs for old hands. Trends Microbiol. 10:221-226. [DOI] [PubMed] [Google Scholar]
- 9.Corbett, E. L., C. J. Watt, N. Walker, D. Maher, B. G. Williams, M. C. Raviglione, and C. Dye. 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 163:1009-1021. [DOI] [PubMed] [Google Scholar]
- 10.Cosma, C. L., D. R. Sherman, and L. Ramakrishnan. 2003. The secret lives of the pathogenic mycobacteria. Annu. Rev. Microbiol. 57:641-676. [DOI] [PubMed] [Google Scholar]
- 11.Dubey, V. S., T. D. Sirakova, M. H. Cynamon, and P. E. Kolattukudy. 2003. Biochemical function of msl5 (pks8 plus pks17) in Mycobacterium tuberculosis H37Rv: biosynthesis of monomethyl branched unsaturated fatty acids. J. Bacteriol. 185:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher, M. A., B. B. Plikaytis, and T. M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Florczyk, M. A., L. A. McCue, A. Purkayastha, E. Currenti, M. J. Wolin, and K. A. McDonough. 2003. A family of acr-coregulated Mycobacterium tuberculosis genes shares a common DNA motif and requires Rv3133c (dosR or devR) for expression. Infect. Immun. 71:5332-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flynn, J. L., and J. Chan. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19:93-129. [DOI] [PubMed] [Google Scholar]
- 15.Flynn, J. L., and J. Chan. 2003. Immune evasion by Mycobacterium tuberculosis: living with the enemy. Curr. Opin. Immunol. 15:450-455. [DOI] [PubMed] [Google Scholar]
- 16.Garton, N. J., H. Christensen, D. E. Minnikin, R. A. Adegbola, and M. R. Barer. 2002. Intracellular lipophilic inclusions of mycobacteria in vitro and in sputum. Microbiology 148:2951-2958. [DOI] [PubMed] [Google Scholar]
- 17.Kalscheuer, R., and A. Steinbuchel. 2003. A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J. Biol. Chem. 278:8075-8082. [DOI] [PubMed] [Google Scholar]
- 18.Lillebaek, T., A. Dirksen, E. Vynnycky, I. Baess, V. O. Thomsen, and A. B. Andersen. 2003. Stability of DNA patterns and evidence of Mycobacterium tuberculosis reactivation occurring decades after the initial infection. J. Infect. Dis. 188:1032-1039. [DOI] [PubMed] [Google Scholar]
- 19.Manabe, Y. C., and W. R. Bishai. 2000. Latent Mycobacterium tuberculosis—persistence, patience, and winning by waiting. Nat. Med. 6:1327-1329. [DOI] [PubMed] [Google Scholar]
- 20.McCarthy, C. M. 1984. Free fatty acid and triglyceride content of Mycobacterium avium cultured under different growth conditions. Am. Rev. Respir. Dis. 129:96-100. [DOI] [PubMed] [Google Scholar]
- 21.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 22.McKinney, J. D., and J. E. Gomez. 2003. Life on the inside for Mycobacterium tuberculosis. Nat. Med. 9:1356-1357. [DOI] [PubMed] [Google Scholar]
- 23.Murphy, D. J. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40:325-438. [DOI] [PubMed] [Google Scholar]
- 24.Nathan, C. 2002. Inducible nitric oxide synthase in the tuberculous human lung. Am. J. Respir. Crit. Care Med. 166:130-131. [DOI] [PubMed] [Google Scholar]
- 25.Ohno, H., G. Zhu, V. P. Mohan, D. Chu, S. Kohno, W. R. Jacobs, Jr., and J. Chan. 2003. The effects of reactive nitrogen intermediates on gene expression in Mycobacterium tuberculosis. Cell. Microbiol. 5:637-648. [DOI] [PubMed] [Google Scholar]
- 26.Park, H. D., K. M. Guinn, M. I. Harrell, R. Liao, M. I. Voskuil, M. Tompa, G. K. Schoolnik, and D. R. Sherman. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell, D. G. 2003. Phagosomes, fatty acids and tuberculosis. Nat. Cell Biol. 5:776-778. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saviola, B., S. C. Woolwine, and W. R. Bishai. 2003. Isolation of acid-inducible genes of Mycobacterium tuberculosis with the use of recombinase-based in vivo expression technology. Infect. Immun. 71:1379-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seiler, P., T. Ulrichs, S. Bandermann, L. Pradl, S. Jorg, V. Krenn, L. Morawietz, S. H. Kaufmann, and P. Aichele. 2003. Cell-wall alterations as an attribute of Mycobacterium tuberculosis in latent infection. J. Infect. Dis. 188:1326-1331. [DOI] [PubMed] [Google Scholar]
- 33.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh, A., S. Jain, S. Gupta, T. Das, and A. K. Tyagi. 2003. mymA operon of Mycobacterium tuberculosis: its regulation and importance in the cell envelope. FEMS Microbiol. Lett. 227:53-63. [DOI] [PubMed] [Google Scholar]
- 35.Sirakova, T. D., A. K. Thirumala, V. S. Dubey, H. Sprecher, and P. E. Kolattukudy. 2001. The Mycobacterium tuberculosis pks2 gene encodes the synthase for the hepta- and octamethyl-branched fatty acids required for sulfolipid synthesis. J. Biol. Chem. 276:16833-16839. [DOI] [PubMed] [Google Scholar]
- 36.Smith, I. 2003. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 16:463-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tufariello, J. M., J. Chan, and J. L. Flynn. 2003. Latent tuberculosis: mechanisms of host and bacillus that contribute to persistent infection. Lancet Infect. Dis. 3:578-590. [DOI] [PubMed] [Google Scholar]
- 38.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
- 41.World Health Organization. 2003. Global tuberculosis control. [Online.] http://www.who.int/gtb/publications/globrep/index.html.