Abstract
HLA-DRB1*0101 is associated with susceptibility to human T lymphotropic virus type 1 (HTLV-1)–associated myelopathy/tropical spastic paraparesis (HAM/TSP). Here, we used a synthetic tetramer of DRB1*0101 and its epitope peptide to analyze HTLV-1–specific CD4+ T cells ex vivo. The frequency of tetramer+CD4+ T cells was significantly greater in patients with HAM/TSP than in healthy HTLV-1 carriers (HCs) at a given proviral load and correlated with HTLV-1 tax messenger RNA expression in HCs but not in patients with HAM/TSP. These cells displayed an early to intermediate effector memory phenotype and were preferentially infected by HTLV-1. T cell receptor gene analyses of 2 unrelated DRB1*0101-positive patients with HAM/TSP showed similar Vβ repertoires and amino acid motifs in complementarity-determining region 3. Our data suggest that efficient clonal expansion of virus-specific CD4+ T cells in patients with HAM/TSP does not simply reflect higher viral burden but rather reflects a rapid turnover caused by preferential infection and/or in vivo stimulation by major histocompatibility complex–peptide complexes.
Human T lymphotropic virus type 1 (HTLV-1)–associated myelopathy/tropical spastic paraparesis (HAM/TSP) [1–4] is a chronic, progressive myelopathy characterized by spastic paraparesis, sphincter dysfunction, and mild sensory disturbance in the lower extremities and is observed only in a minority of infected individuals. Most previous investigations of the specific cellular immune response during HTLV-1 infection have focused on CD8+ cytotoxic T lymphocytes (CTLs), which are typically abundant, chronically activated [5, 6], and mainly targeted to the viral transactivator protein Tax [7–9]. HTLV-1–specific CD8+ T cells have the potential to produce proinflammatory cytokines [10]. However, possession of the HLA-A2 allele, which efficiently presents epitopes of HTLV-1 Tax protein, has been associated with protection against HAM/TSP as well as with a lower proviral load [11]. Thus, there is debate concerning the role played by HTLV-1–specific CD8+ T cells—namely, whether these cells contribute to the inflammatory and demyelinating processes of HAM/TSP or whether the dominant effect of such cells in vivo is protective against disease (although these 2 mechanisms are not mutually exclusive).
Because CD4+ T cell help is necessary for optimal CTL and antibody responses, the CD4+ T cell response against HTLV-1 must also be important. We have previously reported that an HTLV-1 envelope (Env) gp21 immunodominant epitope was restricted by HLA-DRB1*0101 [12, 13] and that HLA-DRB1*0101 was associated with susceptibility to HAM/TSP in independent HTLV-1–infected populations in southern Japan [11, 14] and northeastern Iran [15], indicating the reproducibility of the effect at the population level. Also, CD4+ T cells are the main reservoir of HTLV-1 [16] in vivo and predominate in the mononuclear-cell infiltrate that is found in early active inflammatory spinal cord lesions in HAM/TSP [17] with spontaneous secretion of proinflammatory cytokines [18]. These data suggest that HLA-DRB1*0101 might be associated with susceptibility to HAM/TSP via an effect on CD4+ T cell activation and subsequent bystander damage in the central nervous system (CNS) [5, 19]. Major histocompatibility complex (MHC) class II tetramers have been used for direct ex vivo characterization studies of HIV-1–specific CD4+ T cell populations [20, 21], but such reagents have never been used for HTLV-1–specific CD4+ T cells.
In the present study, we have used an MHC class II tetramer formed between the disease susceptibility–associated allele HLA-DRB1*0101 and its immunodominant epitope to analyze the frequency, phenotype, and T cell receptor (TCR) specificities directly ex vivo in HTLV-1–infected individuals without in vitro cultivation.
PATIENTS, MATERIALS, AND METHODS
Patients and cells
This research was approved by the institutional review boards of the authors’ institutions, and written informed consent was obtained from all individuals. Peripheral blood was studied from 20 patients with HAM/TSP (diagnosed by World Health Organization criteria [22]) and from 19 healthy HTLV-1 carriers (HCs) from Kagoshima, an area in southern Japan where HTLV-1 infection is endemic. Characteristics of patients with HAM/TSP and HCs are shown in table 1. All individuals possessed HLA-DRB1*0101, determined by polymerase chain reaction (PCR) with sequence-specific primers, as described elsewhere [23]. Fresh peripheral blood mononuclear cells (PBMCs) were isolated by Histopaque-1077 (Sigma) density gradient centrifugation, washed, and stored in liquid nitrogen until use.
Table 1.
Characteristics of patients with human T lymphotropic virus type 1 (HTLV-1)–associated myelopathy/tropical spastic paraparesis (HAM/TSP) and of healthy HTLV-1 carriers (HCs).
| Characteristic | HAM/TSP (n = 20) | HCs (n = 19) |
|---|---|---|
| Age, mean ± SD, years | 59.1 ± 8.2 | 48.0 ± 15.2 |
| Sex, no. | ||
| Male | 10 | 7 |
| Female | 10 | 12 |
| Serum anti–HTLV-1 antibody titera | ||
| Mean ± SD | 43,300 ± 48,400 | 1770 ± 2390 |
| Median | 16,384 | 1024 |
| HTLV-1 proviral load in PBMCsb | ||
| Mean ± SD | 438 ± 322 | 245 ± 234 |
| Median | 354 | 151 |
Anti–HTLV-1 antibodies were titrated by the particle agglutination method.
HTLV-1 Tax copy no./1 × 104 peripheral blood mononuclear cells (PBMCs).
Quantification of HTLV-1 proviral load, tax mRNA expression, and anti–HTLV-1 antibody titers
We performed real-time quantitative PCR using an ABI Prism 7700 device (PE Applied Biosystems) to examine HTLV-1 proviral load and tax mRNA expression in PBMCs, as described elsewhere [24, 25]. The amount of HTLV-1 proviral DNA was calculated using β-actin as an internal control by the following formula: copy number of HTLV-1 (pX) per 1 × 104 PBMCs = [(copy number of pX)/(copy number of β-actin/2)] × 104. For mRNA quantification, serially diluted cDNA from HTLV-1–infected MT-2 cells was used for generating standard curves for the value of HTLV-1 tax mRNA and hypoxanthine ribosyl transferase (HPRT) mRNA, and the relative HTLV-1 tax mRNA load was calculated by the following formula: HTLV-1 tax mRNA load = (value of tax)/(value of HPRT) × 10,000. All assays were performed in triplicate. We used aliquots of the same standard MT-2 cDNA preparation for all assays, and the correlation values of standard curves were always >99%. Serum HTLV-1 antibody titers were determined by the particle agglutination method.
Monoclonal antibodies and DRB1*0101/HTLV-1 Env380 –394 tetramer
The following mouse anti–human monoclonal antibodies (MAbs) were used for flow cytometry: CD4–phycoerythrin (PE)–cyanine 5–succinimidylester (PC5), CD8–energy-coupled dye, CD28-PC5 (Beckman Coulter), CD45RA-PC5, CD27–fluorescein isothiocyanate (FITC), interferon (IFN)–γ–FITC (BD Pharmingen), and CCR7-FITC (R&D Systems). The PE-conjugated DRB1*0101/HTLV-1 Env380–394 (RGLDLLFWEQGGLCK) tetramer (DR1/Env tetramer) and control HLA-DRB1*0101/CLIP (PVSKMRMATPLLMQA) tetramer (DR1/CLIP tetramer) were generated by peptide exchange using DRB1*0101/CLIP complexes produced in CHO cells, as described elsewhere [26].
Flow cytometry
PBMCs first were surface stained with the DR1/Env tetramer or the control DR1/CLIP tetramer for 2 h at room temperature (RT). Various MAbs for costaining were added for the last 20 min of incubation. After cell-surface labeling, cells were washed with PBS and fixed in PBS containing 2% paraformaldehyde (Sigma) for 20 min and then resuspended in PBS. Isotype-matched mouse immunoglobulins were used as a control. Intracellular IFN-γ staining was done as described elsewhere [27], using freshly isolated PBMCs cultivated for 6 h at 37°C with 0.1 ng/mL phorbol myristate acetate (Sigma) and 0.5 μg/mL A23187 (Sigma), with brefeldin A (Sigma) at a final concentration of 10 μg/mL included for the last 5 h to inhibit secretion of cytokines from the cells. For intracellular Tax staining, PBMCs were incubated with PE-conjugated DR1/Env tetramer for 2 h at RT or with each MAb to cell-surface molecules for 15 min at RT. Then, cells were fixed and subjected to intracellular Tax staining with anti-Tax MAb (Lt-4; IgG3) [28] or with isotype control MAb, as described elsewhere [29]. Finally, the cells were washed and analyzed by use of an EPICS XL flow cytometer and EXPO32 analysis software (Beckman Coulter) in the lymphocyte gate, on the basis of forward versus side scatter. At least 500 DR1/Env tetramer+CD4+ cells were analyzed for each sample. The frequency of DR1/Env tetramer+CD4+ T cells denotes the frequency of tetramer+ cells among total CD4+ cells.
Enrichment of DR1/Env tetramer+ cells
Cells from 2 Japanese HLA-DRB1*0101–positive patients with HAM/TSP who had relatively high frequencies of DR1/Env tetramer+CD4+ T cells were subjected to enrichment. First, CD4+ T cells were negatively selected by use of the CD4+ T Cell Isolation Kit II (Miltenyi Biotec) in accordance with the manufacturer’s instructions, with purity determined to be >95% by flow cytometry (data not shown). Anti-PE MACS beads were incubated with purified CD4+ T cells for 15 min at 4°C following PE-conjugated DR1/Env tetramer staining for 2 h at RT. After incubation with MACS beads, the cells were passed over a separation column, in accordance with the manufacturer’s instructions.
Reverse-transcriptase PCR (RT-PCR) analysis and sequencing of TCR-Vβ transcripts
RT-PCR and sequencing analyses of TCR-Vβ transcripts were done as described elsewhere [30]. Briefly, cDNA was generated from 5 × 104 (for each) whole PBMCs and ex vivo–isolated DR1/Env tetramer+CD4+ T cells. Then, RT-PCR was performed with a panel of 26 TCR-Vβ–specific primers [31] and a reverse primer specific for the TCR-Vβ constant region (CB-R) that was end-labeled with 6-FAM (Applied Biosystems). The fluorescent intensity of each band was quantified using GeneScan software (version 3.1; Applied Biosystems). Finally, each purified PCR product was subcloned and sequenced [30].
Statistical analysis
The Mann-Whitney U test was used for comparing the differences in the frequencies of DR1/Env tetramer+CD4+ T cells and Tax-expressing cells between patients with HAM/TSP and HCs. Correlations between variables were examined by Spearman’s rank correlation analysis. The results represent mean ± SD values, where applicable. P < .05 was considered to indicate statistical significance.
RESULTS
Direct ex vivo detection and enrichment of HTLV-1 Env gp21–specific CD4+ T cells by an HLA class II tetramer
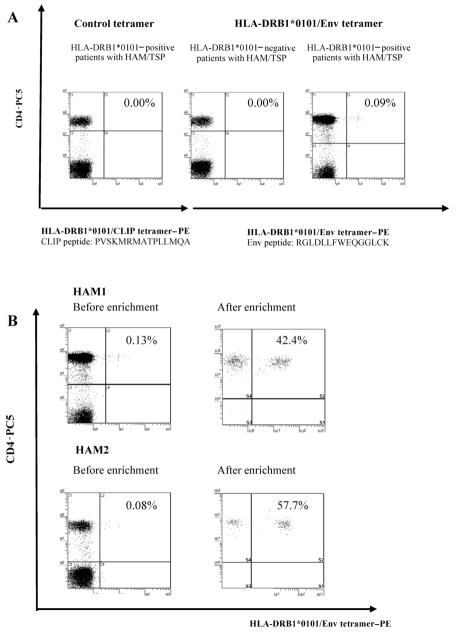
Specific binding of the DR1/Env tetramer is shown in figure 1. We confirmed the absence of detectable staining of PBMCs for 20 DRB1*0101-positive patients with HAM/TSP by use of the control DR1/CLIP tetramer (figure 1A left panel), for 10 HTLV-1–seronegative subjects (5 DRB1*0101 positive and 5 negative) (data not shown) by use of the DR1/Env tetramer, and for 5 DRB1*0101-negative patients with HAM/TSP by use of the DR1/Env tetramer (figure 1A, center panel). In contrast, clear and distinct staining was observed for DRB1*0101-positive patients with HAM/TSP and HCs by use of the DR1/Env tetramer (figure 1A, right panel). All DR1/Env tetramer+ cells were CD4+. Enrichment of PE-labeled DR1/Env tetramer+ cells from 2 patients with HAM/TSP (HAM1 and HAM2)—the 2 who had the highest frequencies of DR1/Env tetramer+CD4+ T cells (0.13% and 0.08% of PBMCs in HAM1 and HAM2, respectively) of 20 DRB1*0101-positive patients with HAM/TSP tested—was done by positive selection using anti-PE microbeads. After enrichment, we observed that 42.4% (HAM1) and 57.7% (HAM2) of CD4+ T cells were DR1/Env tetramer+(figure 1B).
Figure 1.
Direct ex vivo detection and enrichment of human T lymphotropic virus type 1 (HTLV-1) Env gp21–specific CD4+ T cells by the DRB1*0101/Env380-394 (DR1/Env) tetramer. A, No detectable staining of tetramer+ peripheral blood mononuclear cells in DRB1*0101-positive patients with HTLV-1–associated myelopathy/tropical spastic paraparesis (HAM/TSP) by the control HLA-DRB1*0101/CLIP tetramer (left panel) or in DRB1*0101-negative patients with HAM/TSP by the DR1/Env tetramer (center panel). In contrast, clear and distinct staining was observed in DRB1*0101-positive patients with HAM/TSP by the DR1/Env tetramer (right panel). All DR1/Env tetramer+ cells were CD4+. The percentages of cells in each quadrant are given in the upper right corner. B, Ex vivo major histocompatibility complex class II tetramer staining of HTLV-1 Env380-394 –specific CD4+ T cells (left panels). DR1/Env tetramer+ T cells were positively selected from negatively selected CD4+ T cells by use of anti-phycoerythrin (PE) MACS beads and a magnetic separation column (right panels). The purity was checked by flow cytometry with anti-CD4 monoclonal antibody staining. PC5, cyanine 5–succinimidylester.
Significantly greater in vivo frequency of HTLV-1 Env gp21–specific CD4+ T cells in patients with HAM/TSP than in HCs at a given proviral load
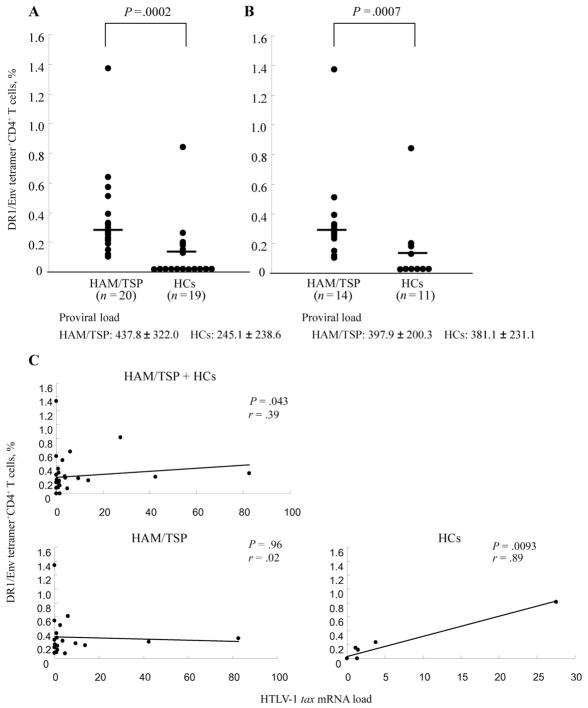
The mean frequency of DR1/Env tetramer+ cells among the CD4+ T cells from 20 patients with HAM/TSP (mean ± SD, 0.304% ± 0.287%) was significantly higher than that for 19 HCs (mean ± SD, 0.099% ± 0.189%; P = .0002, Mann–Whitney U test) (figure 2A). Because the mean HTLV-1 proviral load for the HAM/TSP group was significantly higher than that for the HCs (mean ± SD, 437.8 ± 322.0 for the HAM/TSP group vs. 245.1 ± 238.6 for the HC group; P = .043, Mann-Whitney U test), we compared the frequencies of DR1/Env tetramer+CD4+ T cells between the patients with HAM/TSP and the HCs who had a similar proviral load (mean ± SD, 397.9 ± 200.3 for patients with HAM/TSP [n = 14] and 381.1 ± 231.1 for HCs [n = 11]). The mean frequency of DR1/Env tetramer+CD4+ T cells was still significantly higher in patients with HAM/TSP (mean ±SD, 0.308% ± 0.318% [n = 14]) than in HCs (mean ± SD, 0.125% ± 0.238% [n = 11]) (P = .0007, Mann-Whitney U test) (figure 2B). In accordance with this observation, the frequency of DR1/Env tetramer+CD4+ T cells was not significantly correlated with HTLV-1 proviral load in patients with HAM/TSP alone, in HCs alone, or in both groups combined (data not shown). In contrast, the frequency of DR1/Env tetramer+CD4+ T cells was significantly correlated with HTLV-1 tax mRNA expression in the peripheral blood of HCs alone and in both groups combined but not in patients with HAM/TSP alone (figure 2C).
Figure 2.
Ex vivo frequencies of DRB1*0101/Env380-394 (DR1/Env) tetramer+ cells among peripheral blood mononuclear cells (PBMCs) from patients with human T lymphotropic virus type 1 (HTLV-1)–associated myelopathy/tropical spastic paraparesis (HAM/TSP) and from healthy HTLV-1 carriers (HCs). A, Frequency of DR1/Env tetramer+CD4+ T cells in 20 patients with HAM/TSP and in 19 HCs. The mean frequency of DR1/Env tetramer+CD4+ T cells among PBMCs from 20 patients with HAM/TSP was significantly higher than that of 19 HCs (mean ± SD, 0.304% ± 0.287% vs. 0.099% ± 0.189%; P = .0002, Mann-Whitney U test). HTLV-1 proviral load was also significantly higher in patients with HAM/TSP than in HCs (mean ± SD, 437.8 ± 322.0 vs. 245.1 ± 238.6; P = .043, Mann-Whitney U test). The percentage represents the frequency of the tetramer+ cells among total CD4+ T cells. Means are represented by horizontal lines. B, Frequency of DR1/Env tetramer+CD4+ T cells in 14 patients with HAM/TSP and in 11 HCs. Although there was no significant difference in HTLV-1 proviral load between these patients with HAM/TSP and HCs (mean ± SD, 397.9 ± 200.3 vs. 381.1 ± 231.1), the mean frequency of tetramer+CD4+ T cells was still significantly higher in the patients with HAM/TSP than in the HCs (mean ± SD, 0.308% ± 0.318% vs. 0.125% ± 0.238%; P = .0007, Mann-Whitney U test). The percentage represents the frequency of tetramer+ cells among total CD4+ cells. Means are represented by horizontal lines. C, Significant correlation between the frequency of tetramer+CD4+ T cells and HTLV-1 tax mRNA expression in the peripheral blood of HCs alone or both groups combined but not in patients with HAM/TSP alone. Data were analyzed by Spearman rank correlation. The solid line is the least-squares regression line. The relative HTLV-1 tax mRNA load was calculated by the following formula: HTLV-1 tax mRNA load = (value of tax)/(value of HPRT) × 10,000.
Preferential infection of HTLV-1 Env380 –399 –specific CD4+ T cells by HTLV-1 in patients with HAM/TSP
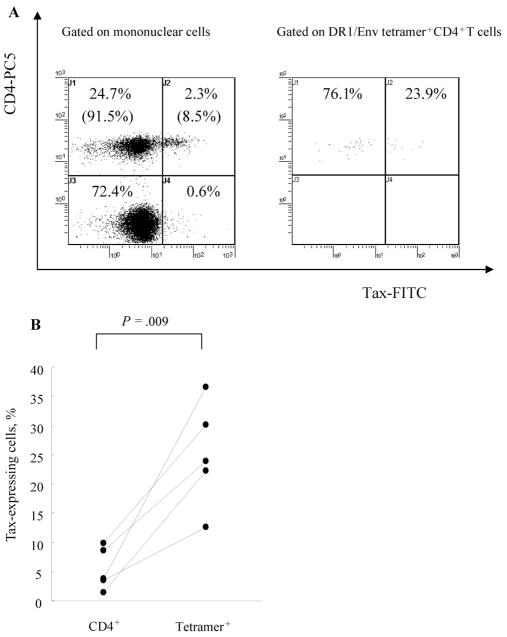
Intracellular detection of HTLV-1 Tax in DR1/Env tetramer+CD4+ T cells after 6 h of in vitro cultivation of PBMCs revealed that the frequency of HTLV-1 Tax–expressing cells among DR1/Env tetramer+CD4 T cells was always higher than that of Tax-expressing cells among CD4+ cells in 5 patients with HAM/TSP tested (for the number of Tax+ DR1/Env tetramer+CD4+ cells divided by total DR1/Env tetramer+CD4+ cells times 100, mean ± SD of 25.04% ± 8.99%; for the number of Tax+CD4+ cells divided by total CD4+ cells times 100, mean ± SD of 5.34% ± 3.61%; P = .009, Mann-Whitney U test), indicating that DR1/Env tetramer+CD4+ T cells were preferentially infected by HTLV-1 (figure 3). A significant correlation was observed between the percentage of Tax+CD4+ cells and proviral load but not between the percentage of Tax+DR1/Env tetramer+CD4+ cells and proviral load (data not shown).
Figure 3.
Increased frequency of Tax expression in human T lymphotropic virus type 1 (HTLV-1) Env380 –399 –specific CD4+ T cells compared with that in total CD4+ T cells, in patients with HTLV-1–associated myelopathy/tropical spastic paraparesis (HAM/TSP). A, Intracellular detection of HTLV-1 Tax in DRB1*0101/Env380-394 (DR1/Env) tetramer+CD4+ T cells after 6 h of in vitro cultivation of peripheral blood mononuclear cells. One representative result from those for 5 patients with HAM/TSP is shown. The frequency of HTLV-1 Tax– expressing cells among DR1/Env tetramer+CD4+ T cells (23.9%) was higher than that of Tax-expressing cells among CD4+ T cells (8.5%). B, Data on all 5 patients with HAM/TSP tested. The frequency of HTLV-1 Tax– expressing cells among DR1/Env tetramer+CD4+ T cells was always higher than that of Tax-expressing cells among CD4+ T cells (for the no. of Tax+DR1/Env tetramer+CD4+ cells divided by total DR1/Env tetramer+CD4+ cells times 100, mean ± SD of 25.04% ± 8.99%; for the no. of Tax+CD4+ cells divided by total CD4+ cells times 100, mean ± SD of 5.34% ± 3.61%; P = .009, Mann-Whitney U test). Lines link 2 data points for each patient. At least 500 DR1/Env tetramer+CD4+ cells were analyzed for each sample. FITC, fluorescein isothiocyanate; PC5, cyanine 5–succinimidylester.
Ex vivo phenotypic and functional analysis of Env380 –399 –specific CD4+ T cells
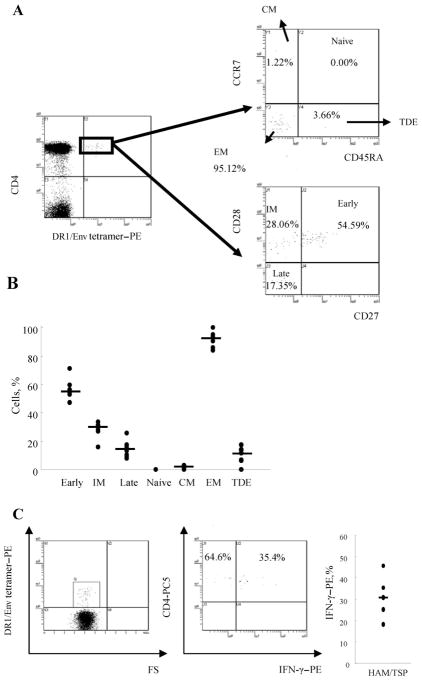
We could directly determine the phenotype of DR1/Env tetramer+CD4+ T cells in 7 patients with HAM/TSP by costaining for cell-surface markers of memory (CD45RA), lymph node homing (CCR7), and costimulation (CD27 and CD28) without in vitro cultivation (figure 4A). The expression of CCR7 and CD45RA in DR1/Env tetramer+CD4+ T cells showed that the vast majority of DR1/Env tetramer+CD4+ T cells were effector memory (CCR7−CD45RA−) cells (mean ± SD, 90.56% ± 6.06%; median, 90.54%) (figure 4B). The maturational phenotype of memory CD4+ T cells was determined by CD27 and CD28 staining, to separate antigen-specific CD4+ T cells into 3 distinct functional subsets—CD27+CD28+, CD27−CD28+, and CD27−CD28−, which correlate with early, intermediate, and late phenotype, respectively [32]. A majority of HTLV-1–specific CD4+ T cells were skewed to an early (mean ± SD, 56.43% ± 7.46%; median, 54.59%) to intermediate (mean ± SD, 28.27% ± 5.91%; median, 30.88%) memory phenotype in all 7 patients with HAM/TSP tested (figure 4B). Intracellular staining of IFN-γ in DR1/Env tetramer+CD4+ T cells after 6 h of in vitro stimulation with mitogen revealed that less than half of Env380–399–specific CD4+ T cells express IFN-γ (mean ± SD, 32.23% ± 11.43%; median, 31.03%; n = 6) (figure 4C). We could not obtain data on HCs because the frequencies of DR1/Env tetramer+CD4+ T cells were very low; therefore, sufficient amounts of blood specimens were not available.
Figure 4.
Phenotypic analysis of human T lymphotropic virus type 1 (HTLV-1) Env380 –399 –specific CD4+ T cells by HLA class II tetramer staining. A, Peripheral blood mononuclear cells from 7 patients with HTLV-1–associated myelopathy/tropical spastic paraparesis (HAM/TSP) were stained with cell-surface markers of memory (CD45RA), lymph node homing (CCR7), and costimulation (CD27 and CD28). One representative result is shown. The expression of CCR7 and CD45RA in DRB1*0101/Env380-394 (DR1/Env) tetramer+CD4+ T cells indicates that the vast majority of DR1/Env tetramer+CD4+ T cells were effector memory (EM; CCR7−CD45RA−) cells. The expression of CD27 and CD28 in DR1/Env tetramer+CD4+ T cells indicated that the majority of HTLV-1–specific CD4+ T cells were skewed to an early (54.59%) to intermediate (IM; 28.06%) memory phenotype. B, Ex vivo phenotypic profile of HTLV-1 Env380 –399 –specific CD4+ T cells in 7 patients with HAM/TSP. Each dot represents the percentage of DR1/Env tetramer+CD4+ T cells that showed the indicated phenotype (early, CD28+CD27+; IM, CD28+CD27−; late, CD28−CD27−; central memory [CM], CCR7+CD45RA−; EM, CCR7−CD45RA−; and terminary differentiated effector [TDE], CCR7−CD45RA+). Means are represented by horizontal lines. C, Interferon (IFN)–γ expression in DRB1*0101/Env380-394 tetramer+ cells. IFN-γ was detected in tetramer+ cells in all 5 patients with HAM/TSP tested. More than half of DR1/Env tetramer+CD4+ T cells did not express IFN-γ (for IFN-γ+tetramer+ cells, mean ± SD of 32.47% ± 12.76% and median of 31.03%). At least 500 DR1/Env tetramer+CD4+ cells were analyzed for each sample.
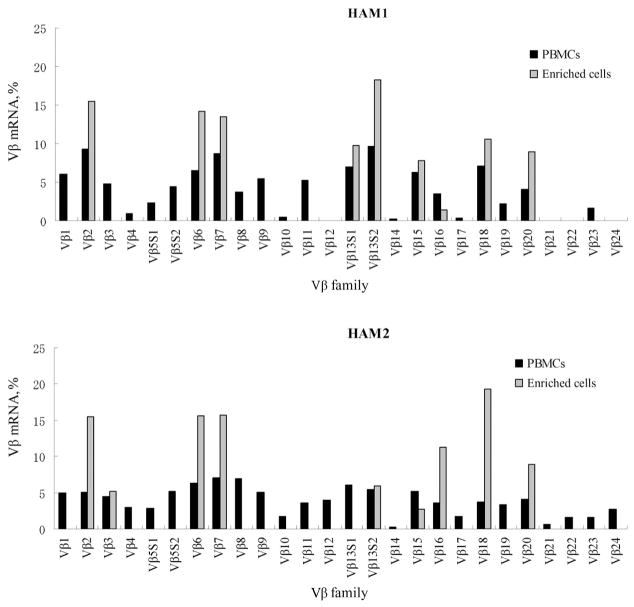
Expression of TCR-Vβ transcripts in enriched DR1/Env tetramer+CD4+ T cells and whole PBMCs from patients with HAM/TSP
TCR gene expression in enriched DR1/Env tetramer+CD4+ T cells and whole PBMCs is shown in figure 5. In contrast to the TCR-Vβ repertoire of whole PBMCs, which is composed of a diverse set of Vβ alleles, the Vβ repertoire of enriched DR1/Env tetramer+CD4+ T cells from 2 unrelated DRB1*0101-positive patients with HAM/TSP showed a similar and limited TCR-Vβ gene use. Namely, Vβ chains 2, 6, 7, 13S2, 15, 16, 18, and 20 were detected in enriched DR1/Env tetramer+CD4+ T cells derived from both HAM1 and HAM2, whereas Vβ13S1 was observed in only HAM1 and not in HAM2.
Figure 5.
Expression of T cell receptor (TCR)–Vβ transcripts in enriched DRB1*0101/Env380-394 tetramer+ cells and in whole peripheral blood mononuclear cells (PBMCs) from patients with human T lymphotropic virus type 1–associated myelopathy/tropical spastic paraparesis (HAM/TSP). Expression of TCR-Vβ transcripts in enriched DRB1*0101/Env380-394 (DR1/Env) tetramer+CD4+ T cells and whole PBMCs from 2 patients with HAM/TSP (HAM1 and HAM2) were analyzed by reverse-transcriptase polymerase chain reaction (RT-PCR). Two microliters of the final TCR-Vβ RT-PCR product mixture were subjected to electrophoresis on a 5% polyacrylamide sequencing gel, and the resulting bands were detected on an automated sequencer. The relative amounts of Vβ transcripts in enriched DR1/Env tetramer+CD4+ T cells (gray bars) and total PBMCs (black bars) were quantified by fluorescence with GeneScan software.
TCR junctional region sequences of enriched DR1/Env tetramer+CD4+ T cells
We have analyzed the nucleotide sequences of each different TCR-Vβ band. Conserved amino acid motifs were observed in the complementarity-determining region (CDR) 3 of enriched DR1/Env tetramer+CD4+ T cells derived from HAM1 and HAM2 (table 2). The T-X-G (where X indicates any amino acid) or S-X-T-G motifs were present in the CDR3 of Vβ2 PCR products of enriched DR1/Env tetramer+CD4+ cells. Similarly, the T-X-X-R, T-X-X-P, Q-E, P-G-X-G, and VG motifs were present in Vβ13S2, Vβ15, Vβ16, Vβ18, and Vβ20 CDR3 of the enriched DR1/Env tetramer+CD4+ cells derived from both HAM1 and HAM2, although we did not observe any amino acid motifs in Vβ7.
Table 2.
T cell receptor junctional region sequences of enriched DRB1*0101/Env380–394 tetramer+ T cells and Env380–394–specific T cell lines.
| Subject | Vβ | N-D-N | Jβ | Cβ | No. of clones/total |
|---|---|---|---|---|---|
| HAM1 | YICSA (Vβ2) | PTPGAS | EAFFGQGTRLTVV (Jβ1.1) | EDLNK | 13/19 |
| YICSA (Vβ2) | SKTGV | NTEAFFGQGTRLTVV (Jβ1.1) | EDLNK | 3/19 | |
| HAM2 | YICSA (Vβ2) | SHTGGR | QPQHFGDGTRLSIL (Jβ1.5) | EDLNK | 16/20 |
| YICSA (Vβ2) | GSGWP | NSPLHFGNGTRLTVT (Jβ1.6) | EDLNK | 3/20 | |
| HAM1 | YLCASS (Vβ6) | PRVPL | AKNIQYFGAGTRLSVL (Jβ2.4) | EDLKN | 16/20 |
| YLCASS (Vβ6) | PFTSGRD | SYEQYFGPGTRLTVT (Jβ2.7) | EDLKN | 2/20 | |
| HAM2 | YLCASS (Vβ6) | SRTGGS | SNQPQHFGDGTRLSIL (Jβ1.5) | EDLNK | 9/19 |
| YLCASS (Vβ6) | QTSGRPS | DTQYFGPGTRLTVL (Jβ2.3) | EDLKN | 5/19 | |
| YLCASS (Vβ6) | SDTLGS | YEQYFGPGTRLTVT (Jβ2.7) | EDLKN | 3/19 | |
| HAM1 | YLCASS (Vβ7) | LPWRT | YNSPLHFGNGTRLTVT (Jβ1.6) | EDLNK | 16/19 |
| YLCASS (Vβ7) | QSSGVQES | QYFGPGTRLLVL (Jβ2.5) | EDLKN | 2/19 | |
| HAM2 | YLCASS (Vβ7) | QDPNY | TGELFFGEGSRLTVL (Jβ2.2) | EDLKN | 20/20 |
| HAM1 | YFCASS (Vβ13S2) | TNKRTGDVA | QETQYFGPGTRLLVL (Jβ2.5) | EDLKN | 20/20 |
| HAM2 | YFCASS (Vβ13S2) | TTRRGTGKF | NQPQHFGDGTRLSIL (Jβ1.5) | EDLNK | 20/20 |
| HAM1 | YFCA (Vβ15) | TSAPETLK | QYFGPGTRLTVT (Jβ2.7) | EDLKN | 17/17 |
| HAM2 | YFCA (Vβ15) | TVKPYGNE | QYFGPGTRLTVT (Jβ2.7) | EDLKN | 20/20 |
| HAM1 | YFCASS (Vβ16) | QEAVSM | NYGYTFGSGTRLTVV (Jβ1.2) | EDLNK | 20/20 |
| HAM2 | YFCASS (Vβ16) | QEKDM | NTEAFFGQGTRLTVV(Jβ1.1) | EDLNK | 19/19 |
| HAM1 | YFCASS (Vβ18) | PGIGP | DTQYFGPGTRLTVL (Jβ2.3) | EDLKN | 20/20 |
| HAM2 | YFCASS (Vβ18) | RLPGQGAT | GNTIYFGEGSWLTVV (Jβ1.3) | EDLNK | 19/19 |
| HAM1 | YFCAW (Vβ20) | SVGGPGK | PQHFGDGTRLSIL (Jβ1.5) | EDLNK | 11/19 |
| YFCAW (Vβ20) | SH | QETQYFGPGTRLLVL (Jβ2.5) | EDLKN | 8/19 | |
| HAM2 | YFCAW (Vβ20) | KVGA | NTGELFFGEGSRLTVL (Jβ2.2) | EDLKN | 15/18 |
| YFCA (Vβ20) | GTYGI | YEQYFGPGTRLTVT (Jβ2.7) | EDLKN | 3/18 |
NOTE. The common amino acid motifs found in clonotypes from HAM1 and HAM2 are indicated in boldface.
DISCUSSION
Until recently, studies of the HTLV-1–specific cellular immune response have focused on the CD8+ CTL response, and the CD4+ T cell response to HTLV-1 has been little studied. This is mainly because HTLV-1 predominantly infects CD4+ T cells in vivo [29, 33] and because expression of HTLV-1 Tax in infected CD4+ T cells leads to cell activation and to IFN-γ production and proliferation, which are the basis of the standard assays used to study the antigen-specific CD4+ T cell response. To overcome this problem, we have used a synthetic tetramer of the disease susceptibility–associated allele DRB1*0101 and its immuno-dominant peptide to analyze HTLV-1–specific CD4+ T cells ex vivo. The advantage of this technique is that we could directly detect HTLV-1 Env gp21–specific CD4+ T cells among PBMCs from patients with HAM/TSP and HCs without in vitro cultivation, which may significantly alter the composition and functional properties of the T cells.
We found that there was no correlation between the frequency of DR1/Env tetramer+CD4+ T cells and HTLV-1 proviral load in either the patients with HAM/TSP alone, the HCs alone, or both groups combined, indicating that the higher frequency of HTLV-1–specific CD4+ T cells observed in patients with HAM/TSP did not simply reflect the higher proviral load (i.e., more HTLV-1–infected T cells). This is consistent with the findings of previous studies by Goon et al. [34, 35]. Goon et al. also showed that the total frequency of HTLV-1–specific CD4+ T cells was significantly higher in patients with HAM/TSP than in HCs with a similar proviral load, by use of an enzyme-linked immunospot assay to estimate the total frequency of HTLV-1–responsive CD4+ cells that produce IFN-γ. That the 2 independent experimental techniques give the same qualitative result increases confidence that this conclusion is correct. We observed a significantly greater frequency of HTLV-1–specific CD4+ T cells in patients with HAM/TSP than in HCs, although there was no significant difference between the groups with respect to mean proviral load. This observation suggests that more-efficient expansion of HTLV-1–specific CD4+ T cell clones irrespective of proviral load does occur in patients with HAM/TSP, probably because of the greater chronic antigenic stimulation in these patients than in HCs with a similar proviral load. This suggestion is consistent with the previous observation by our group [36] and others [25] that HTLV-1 proviral expression in patients with HAM/TSP is greater than that in HCs at a given proviral load. A second factor might also contribute to the efficient proliferation of HTLV-1–specific CD4+ T cells: these cells are preferentially infected with HTLV-1, which, when expressed, will drive proliferation of the host cells.
To further characterize the DR1/Env tetramer+CD4+ T cells, we analyzed (1) TCR-Vβ use and CDR3 sequence, which is critical to antigen recognition [37, 38]; (2) cell surface phenotypes; and (3) Tax and IFN-γ expression after 6 h of cultivation in the DR1/Env tetramer+CD4+ T cells from patients with HAM/TSP for whom sufficient blood was available for assay. The higher frequency of HTLV-1–specific CD4+ cells compared with the frequency of virus-specific CD4+ T cells determined using a class II tetramer in other viral infections—such as influenza A virus [39], HIV [20, 21], Eptein-Barr virus [40], and hepatitis C virus [26]—made these experiments possible. For example, in influenza A virus infection, the frequency of hemagglutinin-specific CD4+ T cells is typically only between 0.0012% and 0.0061% of circulating CD4+ T cells [39], whereas the mean frequency of the DR1/Env tetramer+CD4+ T cells is >10-fold higher (0.20%) in the present study. As for TCR analysis, although the limited TCR-Vβ repertoire of virus-specific CD4+ T cells has been reported in hepatitis C virus infection by use of a class II tetramer and Vβ monoclonal antibodies [26], the CDR3 sequences of directly isolated virus-specific CD4+ T cells have not been reported elsewhere. In spite of the lower purity of enriched DR1/Env tetramer+CD4+ T cells (42.4% for HAM1 and 57.7% for HAM2), we found similar patterns of TCR-Vβ use and conserved amino acid motifs in different CDR3 regions. These unexpected findings suggest the possibility that, although the cells “negative” for tetramer in the enriched population were polyclonal, the cells “positive” for tetramer in the enriched population were oligoclonal. These results are also closely analogous to our previous findings for HTLV-1 Tax 11–19–specific CD8+ T cells. Namely, an amino acid motif in the CDR3 of a particular TCR-Vβ chain was observed in freshly isolated HLA-A*0201/Tax11-19 tetramer+CD8+ T cells and muscle infiltrating cells from patients with HAM/TSP [30] and HTLV-1–infected patients with polymyositis [41]. It is therefore possible that the observed amino acid motifs are the result of in vivo selection by a complex of the particular MHC (DRB1*0101) and peptide (Env380–394) during the course of chronic infection. However, our data also suggest that DR1/Env tetramer+CD4+ T cells were preferentially infected by HTLV-1 in patients with HAM/TSP, again directly corroborating the findings of the previous study by Goon et al. [35], who showed that the frequency of CD4+ T cells infected with HTLV-1 was greater among HTLV-1–specific cells than among cytomegalovirus-specific cells by both Tax expression and proviral load quantification (i.e., preferential infection of HTLV-1 in HTLV-1–specific CD4+ T cells). Therefore, we cannot rule out the possibility that HTLV-1–specific CD4+ T cells efficiently proliferate in response to infection. Nevertheless, in either case, it is evident that particular HTLV-1–specific CD4+ T cell clones expand efficiently in vivo in patients with HAM/TSP.
Next, we ascertained surface phenotypes on the basis of CD27, CD28, CCR7, and CD45RA expression, which are considered to distinguish T cell differentiation phenotypes [42, 43] and to correlate with the cell’s capacity to migrate to secondary lymphoid tissues (i.e., the central vs. the effector memory T cell distribution) [44]. Our data indicate that the vast majority of DR1/Env tetramer+CD4+ cells in patients with HAM/TSP were effector memory (CCR7−CD45RA−) T cells, which are able to leave the circulation and migrate into the parenchyma of peripheral tissues, including the CNS [45]. Interestingly, this effector memory T cell phenotype is the same as that of previously reported HIV-specific tetramer+CD4+ T cells [20]. Moreover, the maturational phenotype of memory CD4+ T cells ascertained by CD27 and CD28 staining, which reflects the different phenotypic characteristics of CD4+ T cells specific for different pathogens [43], indicated that the majority of DR1/Env tetramer+CD4+ cells were skewed to an early to intermediate memory phenotype, again the same as during HIV-1 infection [32]. As for HIV-1 infection, the relatively immature phenotype of HTLV-1–specific CD4+ T cells may be due to a rapid turnover of HTLV-1–specific CD4+ T cells during the course of chronic infection. These immature virus–specific CD4+ T cells may produce a low frequency of IFN-γ production, leading to inefficient T cell surveillance and to the high proviral load seen in patients with HAM/TSP, given that we observed that more than half of DR1/Env tetramer+CD4+ T cells did not express IFN-γ after 6 h of in vitro stimulation with mitogen for all patients with HAM/TSP tested.
In conclusion, we have shown that HTLV-1–specific DR1/Env tetramer+CD4+ T cells that displayed a distinct early to intermediate effector memory phenotype expand more efficiently in vivo in patients with HAM/TSP than in HCs. These HTLV-1–specific CD4+ T cells may be associated with HAM/TSP via an inappropriate regulation of cellular immunity, leading to activation, more-efficient proliferation, migration into the CNS, and subsequent bystander damage in patients with HAM/TSP [5, 19]. Further longitudinal study combined with single-cell analysis may provide useful information on how HTLV-1–specific CD4+ T cells are involved in the initiation and pathogenesis of HAM/TSP.
Acknowledgments
Financial support: Ministry of Health, Labor, and Welfare, Japan (grant in aid for research on brain science to M.O. and M.S. and Neuroimmunological Disease Research Committee grant to M.O. and Y.O.); Ministry of Education, Science, Sports, and Culture, Japan (grant 17590886 to M.S.); Kanazawa Medical University (grant S2006-1 to M.S.).
We thank the staff and blood donors of Kagoshima University Hospital. We also thank Fumiko Inoue and Yoko Nishino of Kagoshima University and Sumie Saito of Kanazawa Medical University for their excellent technical assistance.
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–9. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc Natl Acad Sci USA. 1982;79:2031–5. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gessain A, Barin F, Vernant JC, et al. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet. 1985;2:407–10. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 4.Osame M, Usuku K, Izumo S, et al. HTLV-I associated myelopathy, a new clinical entity. Lancet. 1986;1:1031–2. doi: 10.1016/s0140-6736(86)91298-5. [DOI] [PubMed] [Google Scholar]
- 5.Bangham CR. The immune response to HTLV-I. Curr Opin Immunol. 2000;12:397–402. doi: 10.1016/s0952-7915(00)00107-2. [DOI] [PubMed] [Google Scholar]
- 6.Jacobson S. Immunopathogenesis of human T cell lymphotropic virus type I-associated neurologic disease. J Infect Dis. 2002;186(Suppl 2):S187–92. doi: 10.1086/344269. [DOI] [PubMed] [Google Scholar]
- 7.Jacobson S, Shida H, McFarlin DE, Fauci AS, Koenig S. Circulating CD8+ cytotoxic T lymphocytes specific for HTLV-I pX in patients with HTLV-I associated neurological disease. Nature. 1990;348:245–8. doi: 10.1038/348245a0. [DOI] [PubMed] [Google Scholar]
- 8.Kannagi M, Harada S, Maruyama I, et al. Predominant recognition of human T cell leukemia virus type I (HTLV-I) pX gene products by human CD8+ cytotoxic T cells directed against HTLV-I-infected cells. Int Immunol. 1991;3:761–7. doi: 10.1093/intimm/3.8.761. [DOI] [PubMed] [Google Scholar]
- 9.Parker CE, Daenke S, Nightingale S, Bangham CR. Activated, HTLV-1-specific cytotoxic T-lymphocytes are found in healthy seropositives as well as in patients with tropical spastic paraparesis. Virology. 1992;188:628–36. doi: 10.1016/0042-6822(92)90517-s. [DOI] [PubMed] [Google Scholar]
- 10.Kubota R, Kawanishi T, Matsubara H, Manns A, Jacobson S. Demonstration of human T lymphotropic virus type I (HTLV-I) tax-specific CD8+ lymphocytes directly in peripheral blood of HTLV-I-associated myelopathy/tropical spastic paraparesis patients by intracellular cytokine detection. J Immunol. 1998;161:482–8. [PubMed] [Google Scholar]
- 11.Jeffery KJ, Usuku K, Hall SE, et al. HLA alleles determine human T-lymphotropic virus-I (HTLV-I) proviral load and the risk of HTLV-I-associated myelopathy. Proc Natl Acad Sci USA. 1999;96:3848–53. doi: 10.1073/pnas.96.7.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamano Y, Kitze B, Yashiki S, et al. Preferential recognition of synthetic peptides from HTLV-I gp21 envelope protein by HLA-DRB1 alleles associated with HAM/TSP (HTLV-I-associated myelopathy/tropical spastic paraparesis) J Neuroimmunol. 1997;76:50–60. doi: 10.1016/s0165-5728(97)00041-6. [DOI] [PubMed] [Google Scholar]
- 13.Kitze B, Usuku K, Yamano Y, et al. Human CD4+ T lymphocytes recognize a highly conserved epitope of human T lymphotropic virus type 1 (HTLV-1) env gp21 restricted by HLA DRB1*0101. Clin Exp Immunol. 1998;111:278–85. doi: 10.1046/j.1365-2249.1998.00497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeffery KJ, Siddiqui AA, Bunce M, et al. The influence of HLA class I alleles and heterozygosity on the outcome of human T cell lymphotropic virus type I infection. J Immunol. 2000;165:7278–84. doi: 10.4049/jimmunol.165.12.7278. [DOI] [PubMed] [Google Scholar]
- 15.Sabouri AH, Saito M, Usuku K, et al. Differences in viral and host genetic risk factors for development of human T-cell lymphotropic virus type 1 (HTLV-1)-associated myelopathy/tropical spastic paraparesis between Iranian and Japanese HTLV-1-infected individuals. J Gen Virol. 2005;86:773–81. doi: 10.1099/vir.0.80509-0. [DOI] [PubMed] [Google Scholar]
- 16.Moritoyo T, Reinhart TA, Moritoyo H, et al. Human T-lymphotropic virus type I-associated myelopathy and tax gene expression in CD4+ T lymphocytes. Ann Neurol. 1996;40:84–90. doi: 10.1002/ana.410400114. [DOI] [PubMed] [Google Scholar]
- 17.Umehara F, Izumo S, Nakagawa M, et al. Immunocytochemical analysis of the cellular infiltrate in the spinal cord lesions in HTLV-I-associated myelopathy. J Neuropathol Exp Neurol. 1993;52:424–30. doi: 10.1097/00005072-199307000-00010. [DOI] [PubMed] [Google Scholar]
- 18.Umehara F, Izumo S, Ronquillo AT, Matsumuro K, Sato E, Osame M. Cytokine expression in the spinal cord lesions in HTLV-I-associated myelopathy. J Neuropathol Exp Neurol. 1994;53:72–7. doi: 10.1097/00005072-199401000-00009. [DOI] [PubMed] [Google Scholar]
- 19.Ijichi S, Izumo S, Eiraku N, et al. An autoaggressive process against bystander tissues in HTLV-I-infected individuals: a possible pathomechanism of HAM/TSP. Med Hypotheses. 1993;41:542–7. doi: 10.1016/0306-9877(93)90111-3. [DOI] [PubMed] [Google Scholar]
- 20.Scriba TJ, Zhang HT, Brown HL, et al. HIV-1-specific CD4+ T lymphocyte turnover and activation increase upon viral rebound. J Clin Invest. 2005;115:443–50. doi: 10.1172/JCI23084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seth N, Kaufmann D, Lahey T, Rosenberg ES, Wucherpfennig KW. Expansion and contraction of HIV-specific CD4 T cells with short bursts of viremia, but physical loss of the majority of these cells with sustained viral replication. J Immunol. 2005;175:6948–58. doi: 10.4049/jimmunol.175.10.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osame M. Review of WHO Kagoshima meeting and diagnostic guidelines for HAM/TSP. In: Blattner WA, editor. Human retrovirology: HTLV. New York: Raven Press; 1990. [Google Scholar]
- 23.Olerup O, Zetterquist H. HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours: an alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens. 1992;39:225–35. doi: 10.1111/j.1399-0039.1992.tb01940.x. [DOI] [PubMed] [Google Scholar]
- 24.Nagai M, Usuku K, Matsumoto W, et al. Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: high proviral load strongly predisposes to HAM/TSP. J Neurovirol. 1998;4:586–93. doi: 10.3109/13550289809114225. [DOI] [PubMed] [Google Scholar]
- 25.Yamano Y, Nagai M, Brennan M, et al. Correlation of human T-cell lymphotropic virus type 1 (HTLV-1) mRNA with proviral DNA load, virus-specific CD8+ T cells, and disease severity in HTLV-1-associated myelopathy (HAM/TSP) Blood. 2002;99:88–94. doi: 10.1182/blood.v99.1.88. [DOI] [PubMed] [Google Scholar]
- 26.Day CL, Seth NP, Lucas M, et al. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J Clin Invest. 2003;112:831–42. doi: 10.1172/JCI18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goon PK, Igakura T, Hanon E, et al. High circulating frequencies of tumor necrosis factor alpha- and interleukin-2-secreting human T-lymphotropic virus type 1 (HTLV-1)-specific CD4+ T cells in patients with HTLV-1-associated neurological disease. J Virol. 2003;77:9716–22. doi: 10.1128/JVI.77.17.9716-9722.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka Y, Yoshida A, Takayama Y, et al. Heterogeneity of antigen molecules recognized by anti-tax1 monoclonal antibody Lt-4 in cell lines bearing human T cell leukemia virus type I and related retroviruses. Jpn J Cancer Res. 1990;81:225–31. doi: 10.1111/j.1349-7006.1990.tb02554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanon E, Hall S, Taylor GP, et al. Abundant tax protein expression in CD4+ T cells infected with human T-cell lymphotropic virus type I (HTLV-I) is prevented by cytotoxic T lymphocytes. Blood. 2000;95:1386–92. [PubMed] [Google Scholar]
- 30.Saito M, Taylor GP, Saito A, et al. In vivo selection of T-cell receptor junctional region sequences by HLA-A2 human T-cell lymphotropic virus type 1 Tax11-19 peptide complexes. J Virol. 2001;75:1065–71. doi: 10.1128/JVI.75.2.1065-1071.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eiraku N, Hingorani R, Ijichi S, et al. Clonal expansion within CD4+ and CD8+ T cell subsets in human T lymphotropic virus type I-infected individuals. J Immunol. 1998;161:6674–80. [PubMed] [Google Scholar]
- 32.Yue FY, Kovacs CM, Dimayuga RC, Parks P, Ostrowski MA. HIV-1-specific memory CD4+ T cells are phenotypically less mature than cytomegalovirus-specific memory CD4+ T cells. J Immunol. 2004;172:2476–86. doi: 10.4049/jimmunol.172.4.2476. [DOI] [PubMed] [Google Scholar]
- 33.Richardson JH, Edwards AJ, Cruickshank JK, Rudge P, Dalgleish AG. In vivo cellular tropism of human T-cell leukemia virus type 1. J Virol. 1990;64:5682–7. doi: 10.1128/jvi.64.11.5682-5687.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goon PK, Hanon E, Igakura T, et al. High frequencies of Th1-type CD4+ T cells specific to HTLV-1 Env and Tax proteins in patients with HTLV-1-associated myelopathy/tropical spastic paraparesis. Blood. 2002;99:3335–41. doi: 10.1182/blood.v99.9.3335. [DOI] [PubMed] [Google Scholar]
- 35.Goon PK, Igakura T, Hanon E, et al. Human T cell lymphotropic virus type I (HTLV-I)-specific CD4+ T cells: immunodominance hierarchy and preferential infection with HTLV-I. J Immunol. 2004;172:1735–43. doi: 10.4049/jimmunol.172.3.1735. [DOI] [PubMed] [Google Scholar]
- 36.Asquith B, Mosley AJ, Heaps A, et al. Quantification of the virus-host interaction in human T lymphotropic virus I infection. Retrovirology. 2005;2:75. doi: 10.1186/1742-4690-2-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 38.Garcia KC, Degano M, Stanfield RL, et al. An alphabeta T cell receptor structure at 2. 5 A and its orientation in the TCR-MHC complex. Science. 1996;274:209–19. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- 39.Lucas M, Day CL, Wyer JR, et al. Ex vivo phenotype and frequency of influenza virus-specific CD4 memory T cells. J Virol. 2004;78:7284–7. doi: 10.1128/JVI.78.13.7284-7287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye M, Kasey S, Khurana S, et al. MHC class II tetramers containing influenza hemagglutinin and EBV EBNA1 epitopes detect reliably specific CD4+ T cells in healthy volunteers. Hum Immunol. 2004;65:507–13. doi: 10.1016/j.humimm.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 41.Saito M, Higuchi I, Saito A, et al. Molecular analysis of T cell clonotypes in muscle-infiltrating lymphocytes from patients with human T lymphotropic virus type 1 polymyositis. J Infect Dis. 2002;186:1231–41. doi: 10.1086/344315. [DOI] [PubMed] [Google Scholar]
- 42.Appay V, Dunbar PR, Callan M, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8:379–85. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 43.Amyes E, Hatton C, Montamat-Sicotte D, et al. Characterization of the CD4+ T cell response to Epstein-Barr virus during primary and persistent infection. J Exp Med. 2003;198:903–11. doi: 10.1084/jem.20022058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–63. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 45.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]