Abstract
The T cell receptor (TCR)–CD3 complex represents on of the most intricate membrane receptor structures since it is built from six distinct chains. This complexity led to a number of different proposals for the arrangement of the receptor subunits, its stoichiometry and the mechanisms responsible for receptor triggering. Early work had demonstrated that basic and acidic transmembrane (TM) residues were involved in the assembly but the molecular arrangement could not be deduced due to the complexity of the receptor. Using a novel method for the isolation of intact radiolabeled protein complexes, we demonstrated that the complex assembled in the ER contains only a single TCRαβ heterodimer and one copy of each of the CD3δε, CD3γε and ζ–ζ signaling dimers. Surprisingly, assembly of each of the three signaling dimers with TCR was dependent on one of the three basic TCR TM residues as well as both acidic residues located in the TM domains of the interacting signaling dimer. Each assembly step thus results in the formation of a three-helix interface in the membrane that involves one basic and two acidic TM residues, and this arrangement effectively shields these ionizable residues at protein–protein interfaces from the lipid. Since proteins whose TM domains have exposed ionizable residues are not stably integrated into the lipid bilayer, assembly based on shielding of ionizable residues permits full equilibration of the receptor into the lipid bilayer and prevents degradation. Assembly, export of intact receptor complexes and degradation of unassembled components thus rely on the same organizing principle.
Keywords: T cell receptor, Transmembrane, CD3
1. Introduction
Signaling through the T cell receptor (TCR) controls key events in the life of T cells: their development in the thymus from common lymphoid progenitors, the survival of naïve T cells following their exit from the thymus, and the differentiation of these cells into effector populations with discrete functional profiles. The basic components of the receptor have been known for a number of years, but it has been difficult to determine the subunit interactions and the mechanisms responsible for the assembly of this receptor. As a result, the mechanisms leading to the initial triggering of the receptor have not been possible to define with certainty. The TCR heterodimer is responsible for ligand recognition and the four associated signaling components, CD3γ, CD3δ, CD3ε and ζ bear ITAM motifs in their cytoplasmic domains that are phosphorylated following receptor triggering (Klausner et al., 1990; Exley et al., 1991; Dave et al., 1997; Marie-Cardine and Schraven, 1999; Kane et al., 2000). The signaling components are known to form three distinct dimers: CD3γε, CD3δε and ζ–ζ (Fig. 1). A remarkable feature of the transmembrane (TM) domains of these receptor components is the presence of a total of nine basic/acidic residues. The three basic residues are located in the TM domains of the TCR, while each of the three signaling dimers carries a pair of acidic residues in the center of the membrane-spanning segments. Mutation of some of these polar residues was shown to result in a loss of receptor expression at the cell surface (Alcover et al., 1990; Blumberg et al., 1990; Rutledge et al., 1992), but it was difficult to deduce the molecular arrangement given the number of subunits from which the receptor is built.
Fig. 1.
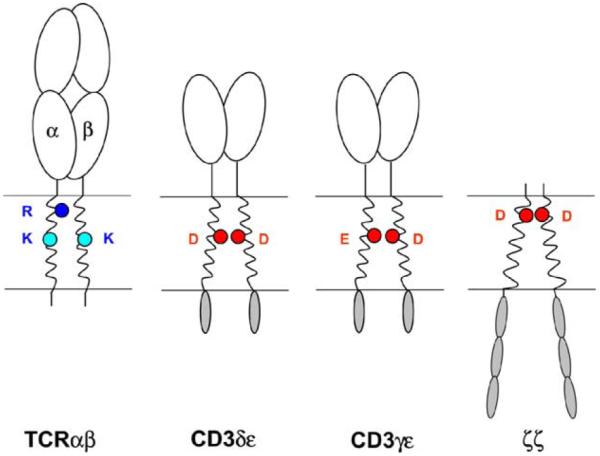
Components of the TCR–CD3 complex. The TCR heterodimer is responsible for ligand recognition, while the associated signaling dimers (CD3δε, CD3γε and ζ–ζ) induce the signaling events that result from receptor engagement. A remarkable property of the receptor is the presence of three basic amino acids in the TM domains of the TCR (R: arginine, K: lysine; blue circles) and of a pair of acidic TM residues in each of the three signaling dimers (D: aspartic acid, E: glutamic acid; red circles). Such ionizable amino acids are energetically highly unfavorable in the hydrophobic interior of the lipid bilayer and these polar groups are shielded from the lipid at protein–protein interfaces in a series of assembly events that lead to the formation of the intact receptor complex.
2. Can assembly be explained by pairwise charge–charge interactions among transmembrane domains?
Cosson et al. (1991) proposed that assembly is determined by pairwise interactions between basic and acidic transmembrane residues, based on the notion that polar residues would form interactions in the membrane that are similar to the well-studied ionic interactions in an aqueous environment. Experimental evidence for this hypothesis was provided by transfection experiments in COS cells with two chain combinations (Cosson et al., 1991; Manolios et al., 1991). An interaction was observed between CD3δ and a fusion protein carrying the TM and cytoplasmic domains of TCRα and the extracellular domain of CD25 (Tac) (Cosson et al., 1991). A major limitation of this approach was that higher-order assembly intermediates were not tested. Also, the interaction was maintained when either one of the two basic TCRα TM residues was mutated, raising questions regarding the specificity of these interactions. A number of other studies addressed this problem (John et al., 1989; Alcover et al., 1990; Bonifacino et al., 1990, 1991; Rutledge et al., 1992; Campbell et al., 1994), but it was not possible to assign a specific function to any of the basic or acidic TM residues due to limitations of the cellular systems that were available. In the Jurkat mutants that were used in transfection experiments only single chains could be experimentally manipulated. Expression of the entire receptor was accomplished in COS cells (Hall et al., 1991; Manolios et al., 1991), but expression levels were limiting since six different plasmids had to be simultaneously transfected.
3. Models of the TCR–CD3 complex: are one or multiple TCR present per complex?
These difficulties in explaining the subunit interactions led to a number of different models for the structural arrangement of the subunits and the mechanisms by which the receptor is triggered. These models fall into two groups: the first group of models proposes that there is a single TCR heterodimer and one copy of each of the three signaling dimers per complex (Manolios et al., 1991; Punt et al., 1994; Kearse et al., 1995; Call et al., 2002), while the second set of models proposes that two or more TCR heterodimers are present per complex (Exley et al., 1995; Jacobs, 1997; San Jose et al., 1998; Fernandez-Miguel et al., 1999). The intellectual driving force for the multivalent models has been to solve the perceived charge imbalance problem: if two TCR heterodimers were present in the complex, the number of basic transmembrane residues would exactly match the six acidic transmembrane residues of the associated signaling dimers. The models are thus distinct in a critical aspect, the valency of complex. If two or more TCR were to be present in a complex, receptor activation could result from a conformational change in a pre-assembled receptor dimer as reported for cytokine receptors like the erythropoetin receptor (Livnah et al., 1999; Remy et al., 1999) and other hormone receptors (Carr et al., 2001; He et al., 2001).
This question was experimentally addressed by different groups using T cells from TCR transgenic mice, and different conclusions were reached. Singer and colleagues (Punt et al., 1994) crossed two mouse strains that expressed distinct TCR heterodimers and found that the two different TCRα as well as TCRβ chains could be distinguished on 2D gels following deglycosylation. Immunoprecipitation experiments of surface receptors labeled with 125-iodine demonstrated that an antibody to one TCRα chain did not co-precipitate the second a chain; the same observation was made when antibodies to the different TCRβ chains were used. de La Hera and colleagues (Fernandez-Miguel et al., 1999) came to the opposite conclusion when they analyzed T cells from mice that expressed two transgenic TCRβ chains. Their experimental strategy was based on immunoprecipitation of one TCR Vβ, followed by Western blotting for the other Vβ and thus did not discriminate between mature complexes, assembly intermediates and potentially misfolded proteins. They observed that the other TCR Vβ was co-precipitated, but the major fraction of the co-precipitated second β chain was not part of a disulfide-linked TCR heterodimer.
4. Direct assessment of the stoichiometry of the TCR–CD3 complex
In order to examine the subunit interactions that drive assembly of the receptor, we developed a novel approach for the isolation of intact radiolabeled receptor complexes (Call et al., 2002). In conventional two-step immunoprecipitation experiments, denaturing conditions are used to elute the bound protein complex from the first antibody (a denaturing detergent or change in pH). The second IP step then only permits assessment of the presence of a particular component of the complex, but does not allow isolation of intact complexes in which all chains are represented in the same relative quantities as in the native receptor structure. We solved this problem by using specialized affinity tags that permit elution under non-denaturing conditions following the first IP: biotin elution for a streptavidin binding peptide [SBP] (Wilson et al., 2001; Call et al., 2002) and chelation of calcium for a calcium-dependent protein C antibody. We refer to this procedure as a sequential non-denaturing immunoprecipitation (snIP). Intact radiolabeled protein complexes can thus be isolated in a second IP step and the components can be quantitated using a phosphor imager following SDS-PAGE and transfer of radiolabeled proteins to a PVDF membrane. We chose to perform these experiments using an in vitro translation system in which radiolabeled proteins are synthesized from input RNAs and co-translationally inserted into ER membranes where they assemble into receptor structures. A number of studies had demonstrated that the same protein–protein interactions are observed in this system as in metabolic labeling experiments performed in cells (Ribaudo and Margulies, 1992; Bijlmakers et al., 1993, 1994; Ribaudo and Margulies, 1995; Huppa and Ploegh, 1997; Hebert et al., 1998). The in vitro translation system offers a key advantage over cell-based metabolic labeling experiments: only the input RNAs are translated into radiolabeled protein since endogenous RNAs have been removed by nuclease treatment. Such in vitro translation experiments had traditionally been performed with microsomes isolated from canine pancreas (Walter and Blobel, 1983) and we developed a procedure for the isolation of suitable membrane preparations from human and mouse cell lines of B cell origin (Call et al., 2002) since they are closely related to T cells, but do not synthesize any component of the TCR–CD3 complex.
We used this system to directly address the two major issues related to the composition of the receptor: the number of TCR heterodimers (the valency) and the relative quantities of the different chains (the stoichiometry). In order to address the valency question, we performed assembly experiments in which the TCRβ chain carried two distinct affinity tags at the C-terminus (SBP or HA tag). These affinity tags differ in their molecular weight and the resulting TCR heterodimers could thus be resolved by SDS-PAGE. These experiments demonstrated that only one TCRβ chain is present per complex since the heterodimer with the second affinity tag was not detected, regardless of whether the IP targeted the SBP or the HA tag. Complexes in which two differentially tagged TCRβ chains were incorporated could also not be detected in sequential non-denaturing IP experiments. The same results were obtained when two different TCRs (the MHC class I restricted A6 TCR and the MHC class II restricted HA1.7 TCR) were examined and when the tags were placed on either TCRα or β chains (Call et al., 2004, unpublished data). Since CD3ε is known to be present in two copies per complex we placed the same set of tags on this chain, and this positive control experiment demonstrated that the approach was suitable for addressing this question (Call et al., 2002). The results clearly demonstrated that the TCR–CD3 complex assembled in the ER contains only a single TCR heterodimer. These results are thus in agreement with the studies performed by Singer and colleagues (Punt et al., 1994), who assessed the valency of the TCR–CD3 complex expressed at the cell surface. How do we explain the opposite findings by de La Hera and colleagues (Fernandez-Miguel et al., 1999)? They detected complexes containing two TCRβ chains in lysates in which either 1% NP40 or Brij96 were used for solubilization. Since 1% NP40 is known to disrupt the TCR–CD3 complex (San Jose et al., 1998), the material detected in these immunoprecipitation reactions using whole cell lysates does not represent the intact receptor structure.
A direct assessment of the stoichiometry of all receptor components had not been feasible, due to a number of limitations of the cellular systems that were available. Direct stoichiometry measurements require homogenous labeling of all receptor components with the same tracer. Labeling of surface receptors with 125-iodine or biotin is not suitable since the number of labeled groups per chain is not known with certainty. Metabolic labeling is appropriate, provided that unlabeled receptor components synthesized prior to the initiation of labeling are not present in significant quantities. Since the half-life of the individual components and partial complexes varies greatly, such experiments are difficult to perform in cell lines of T cell origin. Again, the in vitro translation system provides an elegant solution to these experimental difficulties since homogenous metabolic labeling is achieved and membranes from closely related cell types can be utilized that do not contain components of the TCR–CD3 complex (Call et al., 2002). We thus isolated intact radiolabeled complexes by two-step non-denaturing IP and measured the radioactive signal for each component following separation by SDS-PAGE under non-reducing conditions (Call et al., 2004, unpublished data). Under these conditions, the TCR forms a disulfide-linked heterodimer and ζ a disulfide-linked homodimer. By taking the number of methionine residues present in each chain into consideration, we observed one copy of the ζ–ζ homodimer, one copy of CD3γ and CD3δ and two copies of CD3ε per TCR heterodimer. Since the valency experiments described above had demonstrated that a single TCR heterodimer is present per complex and CD3ε is known to pair with CD3γ and CD3δ (Berkhout et al., 1988; Bonifacino et al., 1988; Koning et al., 1990), we could conclude that the receptor is composed of one copy of each of the four dimers: αβ TCR, CD3δε, CD3γε and ζ–ζ. It was critical to exclude the possibility that the result was dependent on the choice of detergent used for solubilization of the complex. We thus examined a panel of different detergents for their ability to effectively solubilize the receptor without disrupting the interaction between the components, and performed the stoichiometry measurements with a second detergent belonging to a different structural class (Call et al., 2004, unpublished data). These experiments demonstrated the same relative quantities of the different components of the complex.
5. How are the three basic and six acidic transmembrane residues arranged in the TCR–CD3 complex?
The stoichiometry data indicate that a total of three basic and six acidic residues are present in an assembled TCR–CD3 complex and thus raise the question of how the TCR interacts with the other components of the complex given the apparent charge imbalance between basic and acidic transmembrane residues. The basic TCR TM residues are conserved among a variety of different of species and among different TCR forms (Fig. 2). The TCRδ chain that is expressed in γδ T cells is homologous to the TCRα chain, and both the TM arginine and the lysine are conserved among these chains. The two basic TM residues are also found in the pTα chain that assembles with TCRβ in developing thymocytes prior to rearrangement of the TCRα locus. Only three other residues are conserved among the TM domains of TCRα, TCRδ and pTα and two of these are leucine residues that are very common in TM domains due to their hydrophobic nature (see sequence alignments, Fig. 2). In TCRβ and the homologous TCRγ chain, the TM lysine residue is also conserved. The three basic TM residues are thus present in all three TCR forms (αβ TCR, γδ TCR and pre-TCR), suggesting that the principle mechanisms of receptor assembly are conserved among all TCR forms.
Fig. 2.
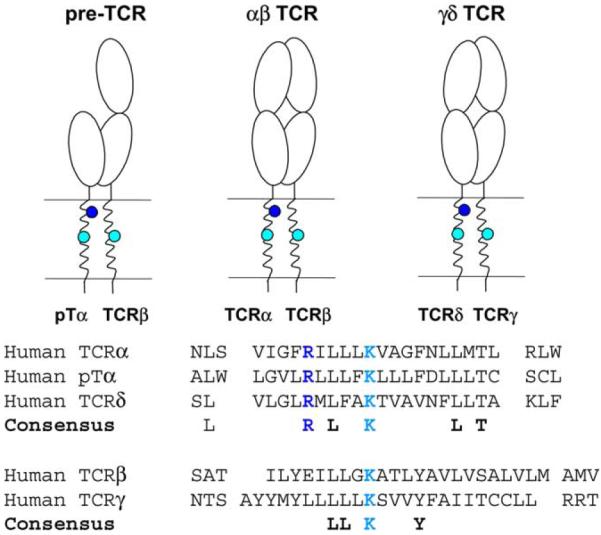
Conservation of basic TCR transmembrane residues. In the pre-TCR that is expressed in thymocytes prior to rearrangement of the TCRα locus, TCRβ forms a heterodimer with pTα. Both basic amino acids (R: arginine and K: lysine) are conserved between TCRα and pTα indicating that both αβ TCR and pre-TCR are assembled based on the same general mechanism. The three basic TCR TM residues are also conserved in the γδ TCR that is expressed by a second lineage of T cells. Only six other residues are conserved among these TM domains, and four of these represent leucine residues that are very common in TM domains due to their hydrophobic character.
Previous mutagenesis experiments in Jurkat cells had demonstrated that the basic TCR residues are required for surface expression of the receptor (Alcover et al., 1990; Blumberg et al., 1990), but given the complexity of the receptor it had not been possible to define the molecular arrangement among the chains. The two-chain transfection experiments in COS cells had suggested that either the lysine or the arginine in the TM domain of TCRα could interact with CD3δ (Cosson et al., 1991), and subsequent experiments had demonstrated many other potential two-chain interactions that could not be interpreted within the context of a single model of receptor assembly (Hall et al., 1991; Manolios et al., 1991). It was thus essential to determine whether each of the three basic TM residues served a particular function in the assembly process and to identify their interacting partners. A major advantage of the in vitro translation system was that mutations could be introduced in any chain and that the effect of mutations could be examined on defined chain combinations representing key assembly intermediates identified in cellular systems. We therefore examined mutations at the three basic TM residues in the context of all TCR–CD3 chains or relevant assembly intermediates (Call et al., 2002). These experiments demonstrated that each of the three basic TM residues serves a discrete role in the assembly process (Fig. 3). Mutation of the arginine in the TM segment of TCRα resulted in a selective loss of the ζ–ζ homodimer from the complex. The arginine of TCRα and the aspartic acid pair of the ζ–ζ homodimer are located in the upper third of their respective TM segments, while all other basic and acidic TM residues are located in the center of their TM domains. Studies in mutant cell lines that lack expression of the ζ chain have shown that all other components of the TCR–CD3 complex can assemble in the absence of the ζ chain (Sussman et al., 1988; Geisler et al., 1989; Weissman et al., 1989), indicating that it is the last component to join the complex.
Fig. 3.
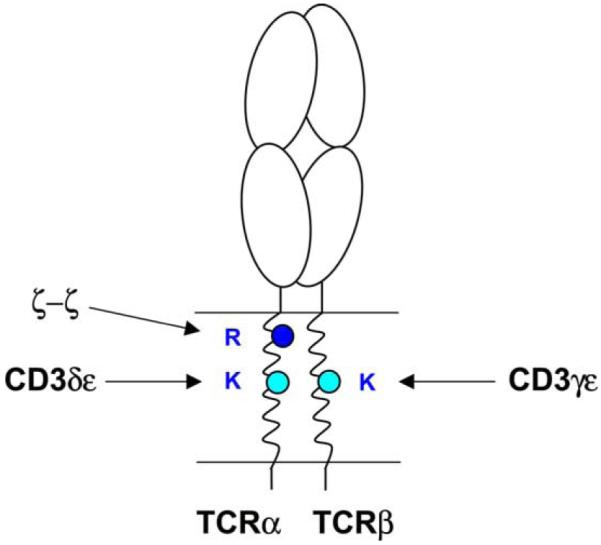
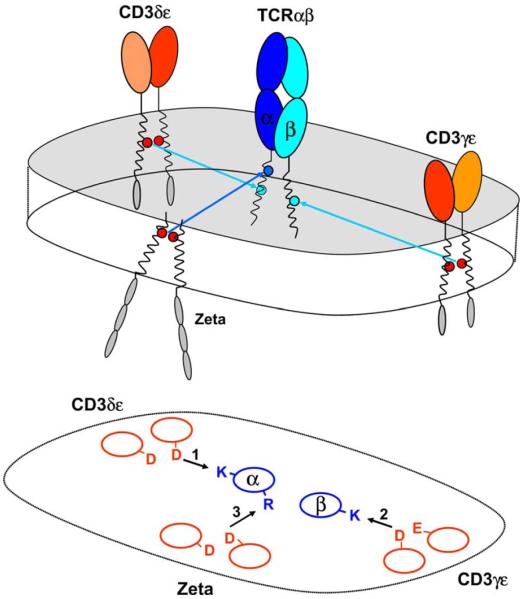
Each basic TCR transmembrane residue is required for assembly with a particular signaling dimer. Assembly experiments using an in vitro translation system permitted systematic mutagenesis of basic and acidic TCR–CD3 TM residues. Each basic TCR TM residue was found to be essential for TCR assembly with a particular signaling dimer. The assembly with the ζ–ζ dimer occurs as the last step, and mutation of the arginine in the TM of TCRα (R) resulted in a selective loss of the ζ–ζ dimer from the complex. The two lysine residues (K) are located in the center of the TM domains of TCRα and β and serve as critical contact points for assembly with the CD3δε and CD3γε dimers, respectively. The TCRαβ–CD3δε complex forms efficiently in the absence of CD3γ and represents an important initial assembly step. Association of CD3γε with TCR is more efficient in the presence of the CD3δε dimer indicating that the kinetically preferred sequence is assembly of TCR with CD3δε followed by association of CD3γε and ζ–ζ.
The TM lysine residue of the TCRβ chain was identified as a key contact site for the CD3γε dimer and mutation of this basic residue did not affect assembly of TCR with CD3δε (Call et al., 2002). The four-chain complex of TCRαβand CD3δε assembled efficiently in the absence of CD3γ and ζ chains, indicating that it represents a relatively early assembly intermediate. Mutation of the lysine in the TM of the TCRα chain abrogated formation of this assembly intermediate while mutation of the other two basic TM residues had no effect. These experiments thus demonstrated that each basic residue is essential for assembly of one of the three signaling dimers with TCR: the two TCR lysine residues that are positioned within the center of the TCRα and β TM domains are required for assembly with the CD3δε and CD3γε dimers, respectively, while the TCRα TM arginine that is located in the upper third of the TCRα TM segment represents an interaction point for the ζ–ζ dimer (Fig. 3). These data are in agreement with prior work in which Brenner and colleagues had demonstrated an interaction between TCRβ and CD3γ using crosslinking techniques (Brenner et al., 1985), as well as studies by Geisler and colleagues who had demonstrated that the TCRαβ-CD3δε intermediate was formed in a Jurkat cell line deficient in expression of the CD3γ chain (Geisler, 1992).
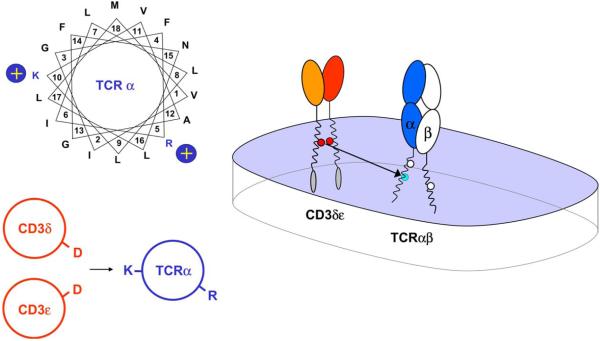
We analyzed the interaction between the TCR and CD3δε in detail since it represents a relevant early step in the assembly process (Fig. 4). Assembly was observed both in the presence and absence of TCRβ, indicating that CD3δε assembles with TCR solely through interaction with the a chain (Call et al., 2002). Both the four-chain TCRαβ-CD3δε and the three-chain TCRα-CD3δε intermediates have been identified in cellular systems (Geisler, 1992; Kearse et al., 1995). The interaction was also observed with a truncated TCRα chain in which only the TCRα TM domain and several flanking residues were present (Call et al., 2002), indicating that assembly is driven by an interaction between the TCRα TM domain and CD3δε. The TCRα TM lysine located in the center of the TM domain serves a critical function in this assembly step and its interaction with CD3δε is highly specific since it could not be substituted by arginine, histidine, glutamine or asparagine. A helical wheel representation of the TCRα TM domain (Fig. 4) indicates that it is located on the opposite face of the TM helix relative to the arginine, explaining how the two basic TM residues can serve as interaction sites for two signaling dimers.
Fig. 4.
Assembly of TCRα with CD3δε through interactions between three TM helices. A helical wheel projection illustrates that the two basic TCRα TM residues are located on approximately opposite faces of the TM helix, demonstrating how they can mediate distinct assembly events. Systematic mutagenesis demonstrated that only the lysine residue (K) in the TM domain of TCRα is required for assembly with CD3δε and that the other two basic residues do not participate in this assembly step. The interaction between the TM lysine and CD3δε is highly specific since the lysine could not be substituted even by arginine. Both acidic TM residues of the CD3δε dimer are critical since mutation of either aspartic acid residue (D) greatly reduced complex formation. Assembly occurred with equal efficiency in the presence and absence of TCRβ, indicating that this assembly step results in the formation of a three-helix TM interface between TCRα and CD3δε. Such a three-helix interface can effectively shield the three ionizable residues from the lipid. This arrangement does not necessarily lead to a charge imbalance since partial or complete protonation of the acidic residues could reduce the average net charge for the aspartic acid pair.
How does CD3δε interact with the TM domain of TCRα? Mutagenesis of the two aspartic acid TM residues of the CD3δε dimer demonstrated that an alanine substitution at either site greatly reduced assembly with TCRα (Call et al., 2002), indicating that both acidic residues play a critical role. The conservative substitutions from aspartic acid to asparagine were particularly informative since the side chains have the same size and both aspartic acid and asparagine can serve as hydrogen bond donors and acceptors. Substitution of either aspartic acid to asparagine reduced assembly to a similar intermediate level (20–30% compared to wild type) while simultaneous substitution at both sites abrogated assembly. These data demonstrate that both aspartic acid residues are involved in this assembly step and that they play a similar role. This assembly step thus results in the formation of a three-helix transmembrane bundle, with each helix contributing one ionizable residue to the interface between the three helices (Fig. 4).
Interestingly, the same mechanism was found to be responsible for the assembly of CD3γε and ζ–ζ with TCR. Mutagenesis experiments targeting the lysine in the TM domain of TCRβ and the two aspartic acid TM residues of CD3γε indicated that this step is also dependent on the three ionizable TM residues located on the interacting TM helices. The interaction sites between TCRα-CD3δε and TCRβ-CD3γε are very similar in the membrane since both assembly steps involve a lysine located in the center of the respective TCR chain and the pair of acidic TM residues on the interacting CD3 dimer. The similarity between these two assembly events in the membrane was further illustrated by an experiment in which the TM domain of CD3γ was exchanged with that of CD3δ. This chimeric CD3γ chain was functional, yielding fully assembled complexes in which the TM domains of the interacting CD3 dimers were identical at the TCRα and TCRβ interaction sites (unpublished data). The extracellular domain of CD3γ contributes specificity since a chimeric protein with the extracellular domain of CD3δ and the TM and cytoplasmic domains of CD3γ failed to assemble with TCRβ.
Formation of a three-helix interface in the membrane is also critical for the interaction of the ζ–ζ dimer with TCR. Since ζ–ζ forms a disulfide-linked homodimer, we added different C-terminal epitope tags to wild-type and mutant chains and selected dimers by sequential non-denaturing IP in which only one aspartic acid residue was mutated (Call et al., 2002). Again, the conservative asparagine mutation of one of the two aspartic acid TM residues reduced the yield of assembled complex to ~20%, while a non-conservative substitution abrogated assembly.
6. Organization of the assembly process
Formation of the TCR–CD3 complex thus depends on the proper placement of three basic and six acidic transmembrane residues and the complex is not formed even when only one of these nine ionizable residues is mutated. The same principal mechanism serves to organize the three major assembly events in which the CD3δε, CD3γε and ζ–ζ signaling dimers interact with the TCR through one of the three basic TCR TM residues. This arrangement is surprising since basic and acidic residues typically form pairwise interactions in an aqueous environment. However, the environment in the membrane is very distinct and can result in protein–protein interactions that are not or only rarely observed in an aqueous environment. Elegant work on model transmembrane helices has demonstrated that dimers and trimers can form when a single aspartic acid or glutamic acid residue is placed within a hydrophobic polyleucine helix (Gratkowski et al., 2001; Zhou et al., 2001). Such structures are also formed by TM helices with a single asparagine or glutamine residue (Choma et al., 2000; Zhou et al., 2000, 2001; Gratkowski et al., 2001; Zhou et al., 2001), suggesting that hydrogen bonds between these polar residues mediate assembly. It is thus possible that the two acidic residues located in the TM domains of each signaling dimer interact, and that the basic TCR TM residue contacts this pair of acidic TM residues. The protonation state of the aspartic acid/glutamic acid TM residues is not known, but a charge imbalance may not necessarily be present (Engelman, 2003). For example, the aspartic acid pair could be partially protonated so that the average charge is −1 rather than −2.
The high-resolution crystal structure of bacteriorhodopsin (Luecke et al., 1999b), an integral membrane protein, has demonstrated an unusual interaction between acidic and basic groups that may be relevant for the TCR–CD3 complex. Bacteriorhodopsin is a light driven proton pump and the proton is generated deep within the TM domain by photoisomerization of the retinal. A critical water molecule is coordinated between one basic group (the Schiff base of the retinal) and two aspartic acid residues (D85 and D212). The interaction between this water molecule and the Schiff base is lost following the light-induced conformational change of the retinal, which destabilizes the water molecule (Luecke et al., 1999a). The resulting proton is then transported through a complex set of interactions from the center of the membrane to the cytoplasmic surface. This example demonstrates an unusual interaction between one basic and two acidic groups in an integral membrane protein involving a water molecule, and it is thus possible that the observed interactions between basic and acidic TM residues in the TCR involve structural water molecule(s) located at protein–protein interfaces.
Assembly of signaling dimers with the TCR occurs in a preferential sequence (CD3δε, CD3γε, ζζ), and these higher-order assembly steps are primarily dependent on protein interactions in the membrane (Fig. 5). Interactions among the extracellular domains are important in the formation of the individual TCR, CD3γε and CD3δε dimers, and both TCR and CD3γε have been expressed as soluble dimers without their TM domains (Garboczi et al., 1996; Garcia et al., 1996; Sun et al., 2001). In the case of TCRα-CD3δε assembly, we have shown directly that interactions among extracellular immunoglobulin domains are not required for formation of this trimeric complex (Call et al., 2002). Given the similarities among the TCRα-CD3δε and TCRβ-CD3γε interaction sites in the membrane, the ectodomains may contribute to complex formation by preventing association of two CD3δε or CD3γε heterodimers with TCR (Fig. 6).
Fig. 5.
Organization of TCR–CD3 assembly. Each of the three major assembly steps involves a basic TCR TM residue and a pair of acidic TM residues on the interacting signaling dimer. Formation of the correct receptor structure thus depends on the proper placement of a total of nine ionizable TM residues. Higher-order assembly is thus organized in three discrete steps based on the same principle mechanism. Each assembly event leads to the creation of a three-helix interface at which a basic and two acidic residues are shielded from the surrounding lipid.
Fig. 6.
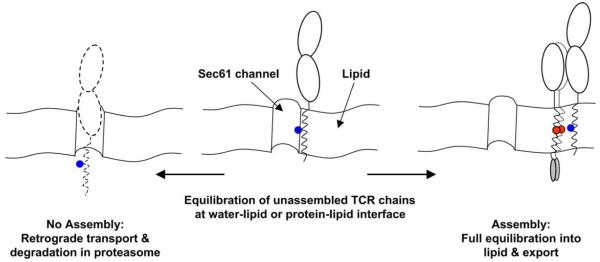
Model for coordination of assembly, export and degradation by ionizable TM residues. Work by Rapoport and colleagues (Heinrich et al., 2000) has demonstrated that TM helices with exposed basic residues are not stably integrated into the lipid bilayer. Such chains have a higher propensity to maintain an interaction with the Sec61p channel through which TM domains gain access to the hydrophobic interior of the lipid bilayer. A dynamic equilibration model can thus explain integration of TM domains into the lipid bilayer: highly hydrophobic TM domains fully equilibrate into the lipid phase, while TM domains with exposed hydrophilic residues have a greater propensity to equilibrate at the water-lipid or protein–lipid interfaces created in the vicinity of the channel. The Sec61p channel is also used for retrograde transport of proteins out of the ER, leading to their destruction in the proteasome. The dynamic equilibration model can thus explain the retention of unassembled TCR–CD3 components in the ER as well as the short half-life of individual chains in which ionizable TM residues are exposed. Assembly effectively shields these ionizable TM residues from the lipid and thus permits full equilibration of the complex into the lipid phase and export from the ER. Assembly also shields other ER retention signals, such as the retention motifs located in the C-terminal segment of the cytoplasmic domains of CD3γ and δ (Engel et al., 1992).
7. Coordination of TCR assembly, export and degradation by ionizable TM residues
Unassembled TCRα and β chains are retained in the ER and degraded within a relatively short time following synthesis (Chen et al., 1988; Bonifacino et al., 1989). ER retention and rapid degradation are due to the presence of basic TM residues, as shown elegantly by Klausner and colleagues (Bonifacino et al., 1990, 1991). Transfer of the transmembrane domain of TCRα to another type 1 membrane protein that is efficiently transported to the cell surface in its wild-type form (Tac, CD25) resulted in ER retention and rapid degradation (t1/2 of 10 min for the chimeric protein). The two basic TM residues of TCRα were responsible for this phenotype since substitution of both basic residues by leucine yielded a protein that was transported out of the ER with the efficiency and kinetics of wild-type Tac. Rapid degradation with a t1/2 of 10–15 min was also observed for a chimeric protein that carried the TM domain of TCRβ, indicating that the same mechanism operates for both TCRα and β chains. Disulfide-linked TCR heterodimer and unassembled TCRα chain were found to be degraded at the same rate (t1/2 of 35–45 min, somewhat longer than for the chimeric Tac proteins), indicating that interaction of the two TCR chains is not sufficient to prevent rapid degradation. However, assembly of TCRα and β chains with the CD3γε dimer inhibited degradation (Wileman et al., 1990), indicating that shielding of basic TM residues by the creation of protein–protein interfaces prevents degradation. Assembly of the TCR with signaling dimers thus masks the signals that promote degradation of unassembled or partially assembled receptor components.
Interestingly, ER retention and degradation result from introduction of either basic or acidic residues into the TM segment. Placement of either an arginine or an aspartic acid residue into the TM domain of Tac created a protein with a short half-life (t1/2 of 10–20 min) (Bonifacino et al., 1991). Importantly, Tac mutants carrying single arginine or aspartic acid residues in the center of the TM domain were glycosylated, indicating proper targeting to ER membranes. However, treatment of membranes containing normal or mutant forms of Tac at an alkaline pH extracted a significant fraction of mutant proteins, but not wild-type Tac, indicating that the proteins had not fully equilibrated into the lipid bilayer. The half-life of the Tac mutants was dependent on the position of the introduced basic or acidic residues, and rapid degradation resulted when the basic/acidic residues were placed in the central segment of the TM domain (positions 5, 8 or 10 of the TM domain for arginine and 8, 10, 13, and 15 for aspartic acid). Interestingly, these correspond to the positions of basic residues in the TM domains of the TCR: both lysine residues are located in the center of the TM domains of TCRα and β, and the arginine is located at position 5 of the predicted TCRα TM domain. The acidic residues of CD3γ, δ and ε are located in the center of the TM domain, while the aspartic acid in the TM domain of ζ is positioned in the upper third of the TM segment (see Fig. 1).
These experiments could explain why unassembled TCRα, TCRβ, CD3γ and CD3δ chains have a short half-life when expressed alone in transfectants. When the murine CD3γ chain is expressed alone, it is retained in the ER and degraded, but when it is coexpressed with CD3ε it forms a complex with a long half-life that resides in the ER (Bonifacino et al., 1989). The model in which the two acidic TM residues of a signaling dimer interact may help to explain these results. Formation of the signaling dimer may partially shield the two acidic residues from the lipid by creation of a protein–protein interface in the membrane. Only assembly with the TCR buries the acidic residues deeply in the interior of a three-helix protein interface and creates a structure that is energetically more favorable and thus more stable. This may also explain why the ζ chain has a longer half-life than CD3γ or CD3δ when expressed alone (Chen et al., 1988; Bonifacino et al., 1989): ζ forms a homodimer and thus does not require expression of a second protein to partially shield the TM aspartic acid residue from the lipid.
Integration of membrane proteins into the lipid bilayer is dependent on the Sec61p channel in the ER membrane which allows TM domains to come into contact with the hydrophobic interior of the membrane (Gorlich and Rapoport, 1993; Matlack et al., 1998). This channel provides a site in the membrane through which a TM domain can dynamically equilibrate between the lipid and aqueous phases (Borel and Simon, 1996; Heinrich et al., 2000). The regulation of TCR–CD3 assembly as well as retention and degradation of free chains and partial complexes can be explained based on a lipid partioning model in which the Sec61p channel represents the site where unassembled chains dynamically equilibrate between the lipid and aqueous phases based on exposed basic and acidic TM residues. Assembly with other components of the complex shields these polar groups from the lipid at protein–protein interfaces and thus permits full equilibration into the lipid bilayer and export from the ER, while a failure of assembly leads to retrograde transport through the channel and degradation in the proteasome. Retrograde transport through the Sec61p channel represents the initial step in the destruction of ER proteins and requires movement of TM domains from the hydrophobic interior of the membrane into the channel (Plemper and Wolf, 1999).
Experimental support for the lipid equilibration model comes from studies with a model type 1 membrane protein whose interaction with the Sec61p channel was studied by introduction of a photoreactive amino acid in the TM domain (Heinrich et al., 2000). ER targeting was shown to result in a transient interaction of the TM domain with Sec61a and Sec61)' chains. In addition, a direct interaction of the TM domain with lipids could be visualized based on lipid crosslinks. Introduction of one or two arginine residues in the center of the TM domain yielded proteins that were properly targeted into ER vesicles and glycosylated. They were, however, not efficiently integrated into the lipid phase since a large percentage of the chains could be extracted by treatment with high pH, with alkali extractability being more pronounced for the protein with the two basic TM residues. The mutant proteins showed stronger crosslinks to the Sec61a and Sec61)' chains and additional crosslinks to a channel associated protein, TRAM. Lipid crosslinks were identified, indicating that these TM domains also interacted with surrounding lipids. These experiments thus demonstrated that the wild-type TM domain moved away from the channel, while TM domains with basic amino acid sidechain(s) had a greater propensity to remain at the interface of channel and lipid. These TM domains also contacted the TRAM protein which may thus be involved in retention at the translocation site of proteins whose TM domain is not sufficiently hydrophobic to fully equilibrate into the lipid phase. The amphipathic environment in the vicinity of the Sec61p channel thus creates an environment for dynamic equilibration of membrane proteins with exposed polar TM residues and may allow TCR–CD3 components to assemble into a structure in which basic and acidic TM residues are shielded away from the lipid.
The basic and acidic TM residues of the TCR–CD3 complex are thus not only critical for the assembly of the correct receptor structure, but also provide the molecular signals for export of intact receptors and degradation of receptor components that have failed to assemble into the correct structure. Formation of this intricate receptor structure thus represents a highly organized set of events that can now be described in biophysical and structural terms.
Acknowledgements
We thank Jason Pyrdol for outstanding technical contributions to the experimental work described in this review. This work was supported by a grant from the NIH to K.W.W. (RO1 AI 054520).
References
- Alcover A, Mariuzza RA, Ermonval M, Acuto O. Lysine 271 in the transmembrane domain of the T-cell antigen receptor beta chain is necessary for its assembly with the CD3 complex but not for alpha/beta dimerization. J. Biol. Chem. 1990;265:4131–4135. [PubMed] [Google Scholar]
- Berkhout B, Alarcon B, Terhorst C. Transfection of genes encoding the T cell receptor-associated CD3 complex into COS cells results in assembly of the macromolecular structure. J. Biol. Chem. 1988;263:8528–8536. [PubMed] [Google Scholar]
- Bijlmakers MJ, Neefjes JJ, Wojcik-Jacobs EH, Ploegh HL. The assembly of H2-Kb class I molecules translated in vitro requires oxidized glutathione and peptide. Eur. J. Immunol. 1993;23:1305–1313. doi: 10.1002/eji.1830230618. [DOI] [PubMed] [Google Scholar]
- Bijlmakers MJ, Benaroch P, Ploegh HL. Assembly of HLA DR1 molecules translated in vitro: binding of peptide in the endoplasmic reticulum precludes association with invariant chain. EMBO J. 1994;13:2699–2707. doi: 10.1002/j.1460-2075.1994.tb06560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg RS, Alarcon B, Sancho J, McDermott FV, Lopez P, Breitmeyer J, Terhorst C. Assembly and function of the T cell antigen receptor. Requirement of either the lysine or arginine residues in the transmembrane region of the alpha chain. J. Biol. Chem. 1990;265:14036–14043. [PubMed] [Google Scholar]
- Bonifacino JS, Chen C, Lippincott-Schwartz J, Ashwell JD, Klausner RD. Subunit interactions within the T-cell antigen receptor: clues from the study of partial complexes. Proc. Natl. Acad. Sci. U.S.A. 1988;85:6929–6933. doi: 10.1073/pnas.85.18.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Suzuki CK, Lippincott-Schwartz J, Weissman AM, Klausner RD. Pre-Golgi degradation of newly synthesized T-cell antigen receptor chains: intrinsic sensitivity and the role of subunit assembly. J. Cell Biol. 1989;109:73–83. doi: 10.1083/jcb.109.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Cosson P, Klausner RD. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell. 1990;63:503–513. doi: 10.1016/0092-8674(90)90447-m. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Cosson P, Shah N, Klausner RD. Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J. 1991;10:2783–2793. doi: 10.1002/j.1460-2075.1991.tb07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel AC, Simon SM. Biogenesis of polytopic membrane proteins: membrane segments assemble within translocation channels prior to membrane integration. Cell. 1996;85:379–389. doi: 10.1016/s0092-8674(00)81116-2. [DOI] [PubMed] [Google Scholar]
- Brenner MB, Trowbridge IS, Strominger JL. Cross-linking of human T cell receptor proteins: association between the T cell idiotype beta subunit and the T3 glycoprotein heavy subunit. Cell. 1985;40:183–190. doi: 10.1016/0092-8674(85)90321-6. [DOI] [PubMed] [Google Scholar]
- Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW. The organizing principle in the formation of the T cell receptor–CD3 complex. Cell. 2002;111:967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KS, Backstrom BT, Tiefenthaler G, Palmer E. CART: a conserved antigen receptor transmembrane motif. Semin. Immunol. 1994;6:393–410. doi: 10.1006/smim.1994.1049. [DOI] [PubMed] [Google Scholar]
- Carr PD, Gustin SE, Church AP, Murphy JM, Ford SC, Mann DA, Woltring DM, Walker I, Ollis DL, Young IG. Structure of the complete extracellular domain of the common beta subunit of the human GM-CSF, IL-3, and IL-5 receptors reveals a novel dimer configuration. Cell. 2001;104:291–300. doi: 10.1016/s0092-8674(01)00213-6. [DOI] [PubMed] [Google Scholar]
- Chen C, Bonifacino JS, Yuan LC, Klausner RD. Selective degradation of T cell antigen receptor chains retained in a pre-Golgi compartment. J. Cell Biol. 1988;107:2149–2161. doi: 10.1083/jcb.107.6.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choma C, Gratkowski H, Lear JD, DeGrado WF. Asparagine-mediated self-association of a model transmembrane helix. Nat. Struct. Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- Cosson P, Lankford SP, Bonifacino JS, Klausner RD. Membrane protein association by potential intramembrane charge pairs. Nature. 1991;351:414–416. doi: 10.1038/351414a0. [DOI] [PubMed] [Google Scholar]
- Dave VP, Cao Z, Browne C, Alarcon B, Fernandez-Miguel G, Lafaille J, de la Hera A, Tonegawa S, Kappes DJ. CD3 delta deficiency arrests development of the alpha beta but not the gamma delta T cell lineage. EMBO J. 1997;16:1360–1370. doi: 10.1093/emboj/16.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I, Letourneur F, Houston JT, Ottenhoff TH, Klausner RD. T cell receptor structure and function: analysis by expression of portions of isolated subunits. Adv. Exp. Med. Biol. 1992;323:1–7. doi: 10.1007/978-1-4615-3396-2_1. [DOI] [PubMed] [Google Scholar]
- Engelman DM. Electrostatic fasteners hold the T cell receptor–CD3 complex together. Mol. Cell. 2003;11:5–6. doi: 10.1016/s1097-2765(03)00016-9. [DOI] [PubMed] [Google Scholar]
- Exley M, Terhorst C, Wileman T. Structure, assembly and intracellular transport of the T cell receptor for antigen. Semin. Immunol. 1991;3:283–297. [PubMed] [Google Scholar]
- Exley M, Wileman T, Mueller B, Terhorst C. Evidence for multivalent structure of T-cell antigen receptor complex. Mol. Immunol. 1995;32:829–839. doi: 10.1016/0161-5890(95)00046-h. [DOI] [PubMed] [Google Scholar]
- Fernandez-Miguel G, Alarcon B, Iglesias A, Bluethmann H, Alvarez-Mon M, Sanz E, de la Hera A. Multivalent structure of an alphabeta T cell receptor. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1547–1552. doi: 10.1073/pnas.96.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- Geisler C. Failure to synthesize the CD3-gamma chain. Consequences for T cell antigen receptor assembly, processing, and expression. J. Immunol. 1992;148:2437–2445. [PubMed] [Google Scholar]
- Geisler C, Kuhlmann J, Rubin B. Assembly, intracellular processing, and expression at the cell surface of the human alpha beta T cell receptor/CD3 complex, function of the CD3-zeta chain. J. Immunol. 1989;143:4069–4077. [PubMed] [Google Scholar]
- Gorlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- Gratkowski H, Lear JD, DeGrado WF. Polar side chains drive the association of model transmembrane peptides. Proc. Natl. Acad. Sci. U.S.A. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C, Berkhout B, Alarcon B, Sancho J, Wileman T, Terhorst C. Requirements for cell surface expression of the human TCR/CD3 complex in non-T cells. Int. Immunol. 1991;3:359–368. doi: 10.1093/intimm/3.4.359. [DOI] [PubMed] [Google Scholar]
- He X, Chow D, Martick MM, Garcia KC. Allosteric activation of a spring-loaded natriuretic peptide receptor dimer by hormone. Science. 2001;293:1657–1662. doi: 10.1126/science.1062246. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Zhang JX, Helenius A. Protein folding and maturation in a cell-free system. Biochem. Cell Biol. 1998;76:867–873. doi: 10.1139/bcb-76-5-867. [DOI] [PubMed] [Google Scholar]
- Heinrich SU, Mothes W, Brunner J, Rapoport TA. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- Huppa JB, Ploegh HL. In vitro translation and assembly of a complete T cell receptor–CD3 complex. J. Exp. Med. 1997;186:393–403. doi: 10.1084/jem.186.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H. Pre-TCR/CD3 and TCR/CD3 complexes: decamers with differential signalling properties. Immunol. Today. 1997;18:565–569. [PubMed] [Google Scholar]
- John S, Banting GS, Goodfellow PN, Owen MJ. Surface expression of the T cell receptor complex requires charged residues within the alpha chain transmembrane region. Eur. J. Immunol. 1989;19:335–339. doi: 10.1002/eji.1830190218. [DOI] [PubMed] [Google Scholar]
- Kane LP, Lin J, Weiss A. Signal transduction by the TCR for antigen. Curr. Opin. Immunol. 2000;12:242–249. doi: 10.1016/s0952-7915(00)00083-2. [DOI] [PubMed] [Google Scholar]
- Kearse KP, Roberts JL, Singer A. TCR alpha-CD3 delta epsilon association is the initial step in alpha beta dimer formation in murine T cells and is limiting in immature CD4+ CD8+ thymocytes. Immunity. 1995;2:391–399. doi: 10.1016/1074-7613(95)90147-7. [DOI] [PubMed] [Google Scholar]
- Klausner RD, Lippincott-Schwartz J, Bonifacino JS. The T cell antigen receptor: insights into organelle biology. Annu. Rev. Cell Biol. 1990;6:403–431. doi: 10.1146/annurev.cb.06.110190.002155. [DOI] [PubMed] [Google Scholar]
- Koning F, Maloy WL, Coligan JE. The implications of subunit interactions for the structure of the T cell receptor–CD3 complex. Eur. J. Immunol. 1990;20:299–305. doi: 10.1002/eji.1830200211. [DOI] [PubMed] [Google Scholar]
- Livnah O, Stura EA, Middleton SA, Johnson DL, Jolliffe LK, Wilson IA. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science. 1999;283:987–990. doi: 10.1126/science.283.5404.987. [DOI] [PubMed] [Google Scholar]
- Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution. Science. 1999a;286:255–261. doi: 10.1126/science.286.5438.255. [DOI] [PubMed] [Google Scholar]
- Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol. 1999b;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- Manolios N, Letourneur F, Bonifacino JS, Klausner RD. Pairwise, cooperative and inhibitory interactions describe the assembly and probable structure of the T-cell antigen receptor. EMBO J. 1991;10:1643–1651. doi: 10.1002/j.1460-2075.1991.tb07687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie-Cardine A, Schraven B. Coupling the TCR to downstream signalling pathways: the role of cytoplasmic and transmembrane adaptor proteins. Cell Signal. 1999;11:705–712. doi: 10.1016/s0898-6568(99)00047-9. [DOI] [PubMed] [Google Scholar]
- Matlack KE, Mothes W, Rapoport TA. Protein translocation: tunnel vision. Cell. 1998;92:381–390. doi: 10.1016/s0092-8674(00)80930-7. [DOI] [PubMed] [Google Scholar]
- Plemper RK, Wolf DH. Retrograde protein translocation: eradication of secretory proteins in health and disease. Trends Biochem. Sci. 1999;24:266–270. doi: 10.1016/s0968-0004(99)01420-6. [DOI] [PubMed] [Google Scholar]
- Punt JA, Roberts JL, Kearse KP, Singer A. Stoichiometry of the T cell antigen receptor (TCR) complex: each TCR/CD3 complex contains one TCR alpha, one TCR beta, and two CD3 epsilon chains. J. Exp. Med. 1994;180:587–593. doi: 10.1084/jem.180.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy I, Wilson IA, Michnick SW. Erythropoietin receptor activation by a ligand-induced conformation change. Science. 1999;283:990–993. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- Ribaudo RK, Margulies DH. Independent and synergistic effects of disulfide bond formation, beta 2-microglobulin, and peptides on class I MHC folding and assembly in an in vitro translation system. J. Immunol. 1992;149:2935–2944. [PubMed] [Google Scholar]
- Ribaudo RK, Margulies DH. Polymorphism at position nine of the MHC class I heavy chain affects the stability of association with beta 2-microglobulin and presentation of a viral peptide. J. Immunol. 1995;155:3481–3493. [PubMed] [Google Scholar]
- Rutledge T, Cosson P, Manolios N, Bonifacino JS, Klausner RD. Transmembrane helical interactions: zeta chain dimerization and functional association with the T cell antigen receptor. EMBO J. 1992;11:3245–3254. doi: 10.1002/j.1460-2075.1992.tb05402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Jose E, Sahuquillo AG, Bragado R, Alarcon B. Assembly of the TCR/CD3 complex: CD3 epsilon/delta and CD3 epsilon/gamma dimers associate indistinctly with both TCR alpha and TCR beta chains. Evidence for a double TCR heterodimer model. Eur. J. Immunol. 1998;28:12–21. doi: 10.1002/(SICI)1521-4141(199801)28:01<12::AID-IMMU12>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Sun ZJ, Kim KS, Wagner G, Reinherz EL. Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer. Cell. 2001;105:913–923. doi: 10.1016/s0092-8674(01)00395-6. [DOI] [PubMed] [Google Scholar]
- Sussman JJ, Bonifacino JS, Lippincott-Schwartz J, Weissman AM, Saito T, Klausner RD, Ashwell JD. Failure to synthesize the T cell CD3-zeta chain: structure and function of a partial T cell receptor complex. Cell. 1988;52:85–95. doi: 10.1016/0092-8674(88)90533-8. [DOI] [PubMed] [Google Scholar]
- Walter P, Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- Weissman AM, Frank SJ, Orloff DG, Mercep M, Ashwell JD, Klausner RD. Role of the zeta chain in the expression of the T cell antigen receptor: genetic reconstitution studies. EMBO J. 1989;8:3651–3656. doi: 10.1002/j.1460-2075.1989.tb08539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wileman T, Carson GR, Concino M, Ahmed A, Terhorst C. The gamma and epsilon subunits of the CD3 complex inhibit pre-Golgi degradation of newly synthesized T cell antigen receptors. J. Cell Biol. 1990;110:973–986. doi: 10.1083/jcb.110.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DS, Keefe AD, Szostak JW. The use of mRNA display to select high-affinity protein-binding peptides. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3750–3755. doi: 10.1073/pnas.061028198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FX, Cocco MJ, Russ WP, Brunger AT, Engelman DM. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat. Struct. Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- Zhou FX, Merianos HJ, Brunger AT, Engelman DM. Polar residues drive association of polyleucine transmembrane helices. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2250–2255. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]