Abstract
Objectives
To investigate how the genetic susceptibility gene DIO2 confers risk to osteoarthritis (OA) onset in humans and to explore whether counteracting the deleterious effect could contribute to novel therapeutic approaches.
Methods
Epigenetically regulated expression of DIO2 was explored by assessing methylation of positional CpG-dinucleotides and the respective DIO2 expression in OA-affected and macroscopically preserved articular cartilage from end-stage OA patients. In a human in vitro chondrogenesis model, we measured the effects when thyroid signalling during culturing was either enhanced (excess T3 or lentiviral induced DIO2 overexpression) or decreased (iopanoic acid).
Results
OA-related changes in methylation at a specific CpG dinucleotide upstream of DIO2 caused significant upregulation of its expression (β=4.96; p=0.0016). This effect was enhanced and appeared driven specifically by DIO2 rs225014 risk allele carriers (β=5.58, p=0.0006). During in vitro chondrogenesis, DIO2 overexpression resulted in a significant reduced capacity of chondrocytes to deposit extracellular matrix (ECM) components, concurrent with significant induction of ECM degrading enzymes (ADAMTS5, MMP13) and markers of mineralisation (ALPL, COL1A1). Given their concurrent and significant upregulation of expression, this process is likely mediated via HIF-2α/RUNX2 signalling. In contrast, we showed that inhibiting deiodinases during in vitro chondrogenesis contributed to prolonged cartilage homeostasis as reflected by significant increased deposition of ECM components and attenuated upregulation of matrix degrading enzymes.
Conclusions
Our findings show how genetic variation at DIO2 could confer risk to OA and raised the possibility that counteracting thyroid signalling may be a novel therapeutic approach.
Introduction
Osteoarthritis (OA) is a prevalent, complex, disabling disease of articular joints, characterised by degradation of articular cartilage and remodelling of the subchondral bone. There is no effective therapy to reverse or slow down the disease except for joint replacement surgery at the end stage. As a result, OA has a large detrimental impact on the quality of life of the elderly and causes a major burden on health and social care.1 To allow development of new therapies, there is an ongoing need for insight into the underlying mechanisms driving OA. Genetic studies provided evidence that genes orchestrating growth plate endochondral ossification play a underlying role in common OA susceptibility,2 hence functional follow-up approaches require focus on both the early developmental and late-acting effects of these OA genes. A notable example is the deiodinase iodothyronine type 2 (D2) gene (DIO2) with the C-allele of the single nucleotide polymorphism (SNP) rs225014 (frequency ∼ 0.35) located in the coding region that conferred consistent risk to OA.3 4 The gene product of DIO2 is responsible for catalysing the conversion of intracellular inactive thyroid hormone (T4) to active thyroid hormone (T3). T3 subsequently signals terminal maturation of growth plate chondrocytes leading to cell hypertrophy, cartilage matrix destruction mediated via upregulation of hypoxia inducible factor-2α (HIF-2α) and runt-related transcription factor-2 (RUNX2),5 6 mineralisation of the cartilage and eventually formation of bone.7 There are striking parallels between the chondrocyte signalling events that take place in the growth plate and those of hypertrophic chondrocytes in OA-affected articular cartilage.5 This has led to the hypothesis that with age and environmental stresses the propensity of the highly specialised, maturational arrested articular chondrocytes is affected by loss of epigenic control. Progression of age and disease could result in reactivation of genes involved in endochondral ossification, leading to loss and mineralisation of articular cartilage, a process known to contribute to OA.3 8–10 Functional genomic studies showed high expression of DIO2 mRNA and D2 protein levels in osteoarthritic as compared with healthy cartilage.5 11 12 Furthermore, DIO2 allelic imbalance was assessed and showed that the OA risk allele ‘C’ was more abundantly present in articular joint tissues than the wildtype allele ‘T’.11 In transgenic rats, DIO2 overexpression conferred risk to articular cartilage destruction.13 The underlying mechanism how the DIO2 SNP confers susceptibility to OA in humans remains, however, to be determined,14 but most likely acts via aberrant upregulation of its expression. The DIO2 locus in humans contains several putative CCCTC-binding factor (CTCF) binding sites, including one that is overlapping with the rs225014 location.15 CTCF is considered to facilitate long-range chromatin interactions in order to insulate gene expression, and distal transcriptional elements on the genome are brought in close proximity to transcriptional start sites (TSSs) of genes to inhibit expression.16
In the current study, we focus on regulatory mechanisms of DIO2 expression in preserved and osteoarthritic human articular cartilage, thereby taking into account the DIO2 risk allele. The direct effect of changes in DIO2 expression on chondrocyte function and human cartilage extracellular matrix (ECM) homeostasis is subsequently studied in human in vitro chondrogenesis models, which should be considered a well-defined system for studying changes in the ECM when chondrocytes differentiate, become hypertrophic and start to exhibit cartilage-debilitating expression patterns.
Materials and methods
The ongoing RAAK study is aimed at the biobanking of joint materials (cartilage, bone and, where available, ligaments) and mesenchymal stem cells and primary chondrocytes of patients in the Leiden University Medical Center. In the current study, we used paired preserved and OA-affected cartilage samples from 52 Caucasian end-stage OA patients undergoing joint replacement surgery for primary OA (23 hips, 29 knees). For additional details on the RAAK study, cell cultures, RNA and DNA extraction, quantitative RT-PCR, ChIP and the data analysis, see online supplemental methods.
Electrophoretic mobility shift assay
For electrophoretic mobility shift assays (EMSAs), synthetic oligonucleotides containing the putative CTCF binding site were 5′-end labelled by γ-32P-ATP and subsequently purified by gel filtration on Sephadex G-25 Medium columns. For additional details, see online supplemental methods.
Quantification of methylation
The methylated fraction of CpG dinucleotides was assessed with MALDI-TOF mass spectrometry (Epityper, Sequenom), a commonly applied method to quantify CpG methylation.17–19 For additional details, see online supplemental methods.
Lentiviral constructs and transduction
C-terminal FLAG-tagged cys-D2 (kindly provided by Prof. Dr. Bianco20) was digested with EcoRI followed by Klenow treatment and digestion with XbaI. Inserts were inserted into the EcoRV-XbaI sites of the pLV-CMV-IRES-eGFP Lentiviral backbone. For additional details, see online supplemental methods.
In vitro chondrogenesis
3D pellets were formed using 2.5×105 human bone marrow-derived mesenchymal stem cells (hBMSCs). Chondrogenesis was initiated in serum-free chondrogenic differentiation medium. From day 14 onwards, cell pellets were maintained either in the standard chondrogenic differentiation medium or in the presence of T3 (10 nM) or IOP (10 μM). For additional details, see online supplemental methods.
Relative pixel intensity
The relative pixel intensity was computed by loading the photos into ImageJ (V.1.47).21–23 For additional details, see online supplemental methods.
Results
Epigenetic regulation of DIO2 expression by CpG methylation in articular cartilage
Online available ChIP-seq data revealed multiple transcription factors to bind the rs225014 locus of which CTCF was predicted to bind with the highest certainty (see online supplementary figure S1). To assess the regulatory properties of the CTCF binding site overlapping rs225014 and to test whether the rs225014 alleles directly affect the binding, we performed an EMSA. The putative CTCF sequence overlapping rs225014 (DIO2-CTCF1) was found not functional nor, for that matter, dependent of the rs225014 alleles (see online supplementary figure S2, lanes 1–6).
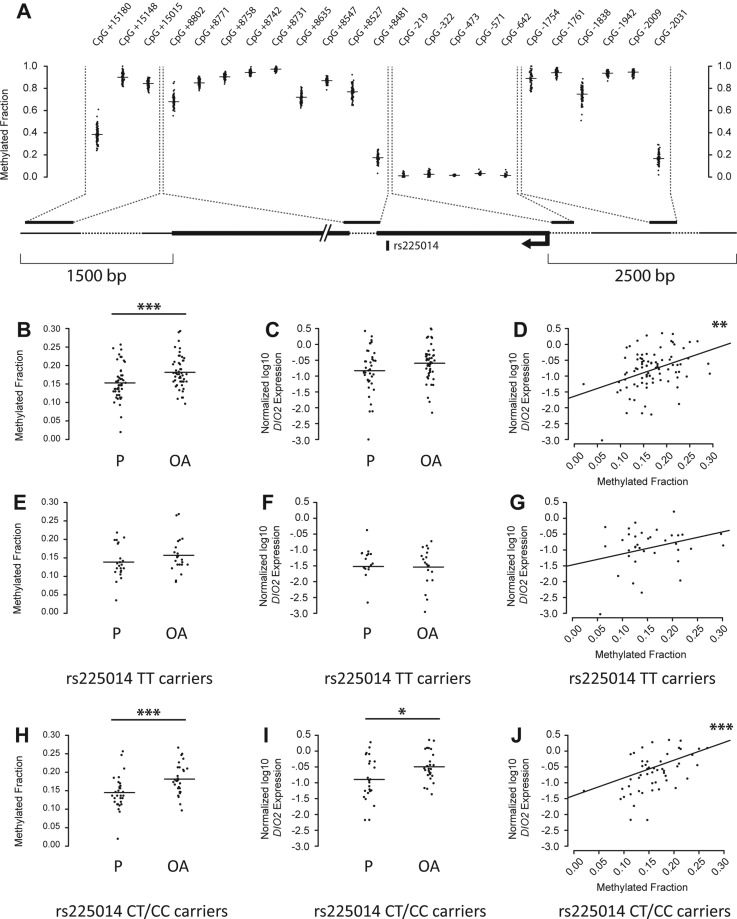
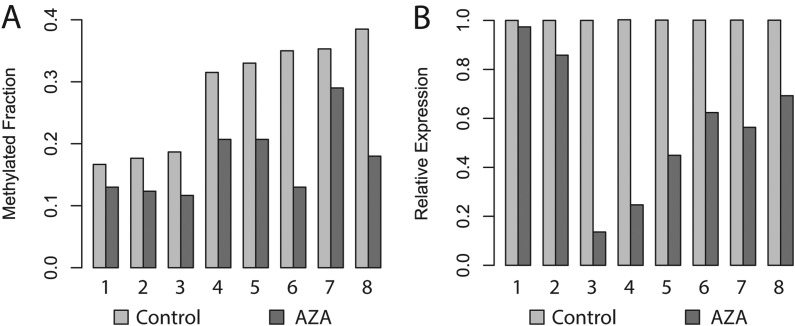
As no further TFs were confidently predicted to bind the rs225014 locus, we set out to elucidate putative regulatory mechanisms of DIO2, independent of the rs225014 base change. We, therefore, quantified expression of DIO2 and methylation of 23 CpG dinucleotides across the DIO2 locus in macroscopically preserved and OA-affected cartilage from joints of patients undergoing total arthroplasty of the knee (N=29) or hip (N=23) (RAAK study; see online supplementary table S1). We found borderline significant upregulation of DIO2 expression (β=0.22, p=0.063, figure 1C) in OA compared with preserved cartilage. Furthermore, we observed several CpG dinucleotides to be differentially methylated between preserved and OA-affected cartilage (see online supplementary table S2). However, only for the CpG site 2031 base pairs upstream of the DIO2 transcription start site (CpG-2031, figure 1A) we observed significant differential methylation between OA and preserved cartilage (β=0.028, p=0.0007, figure 1B) and a significant positive association between methylation and DIO2 expression in all samples (β=4.959, p=0.0016, figure 1D; see online supplementary table S2). To confirm the regulatory properties of CpG-2031 on DIO2 expression, we applied 5-aza-2′-deoxycytidine (AZA), a demethylating agent, to the culture medium in eight primary chondrocyte cultures derived from preserved cartilage of total hip replacement patients. Addition resulted in a decrease in methylation at CpG-2031 (p=0.003, figure 2A) corresponding to downregulation of DIO2 expression (p=0.004, figure 2B). Having observed the association between DIO2 expression and OA-associated methylation at CpG-2031, we applied a multivariate model to assess the individual effects of CpG-2031 methylation, joint site, sex, BMI, age and rs225014 alleles on DIO2 expression in articular cartilage (see online supplementary table S3). We could hereby ratify the association between increased CpG-2031 methylation and DIO2 expression (β=4.008, p=0.019). The most notable observation, however, was the significant independent association of rs225014 genotype on DIO2 expression (β=0.557, p=0.0003), indicating additive effects of both genotype and DNA methylation differences on DIO2 expression. This effect appeared to be mainly driven by the risk allele. Upon stratification by the rs225014 alleles, no significant differences were observed between preserved and OA-affected cartilage among homozygous carriers of the rs225014 wildtype allele T in methylation (β=0.018, p=0.112, figure 1E) or expression (β=−0.017, p=0.929, figure 1F). In carriers of the risk allele, however, an increase was observed in OA cartilage compared with the preserved tissue, both in the difference of CpG-2031 methylation (β=0.034, p=0.00002, figure 1H) and in expression (β=0.35, p=0.012, figure 1I), concomitant with an increased association between methylation and expression (β=6.816, p=0.00001, figure 1J). As expected, this association between expression and methylation was much smaller among homozygous wildtype allele carriers (β=3.863, p=0.050, figure 1G).
Figure 1.
A functional CpG dinucleotide (CpG-2031) significantly modulates DIO2 expression. (A) Schematic overview of the quantified CpG dinucleotides in pooled preserved and paired osteoarthritis (OA)-affected samples across the DIO2 locus. (B) Methylation between preserved and OA-affected samples for CpG dinucleotide located 2031 base pairs upstream (CpG-2031) of the DIO2 transcriptional start site (TSS) (GLMM, N=103, β=0.028, p=0.001, Bonferroni adjusted; see also online supplementary table S2). (C) Real-time qRT-PCR data of DIO2 expression between preserved and OA-affected cartilage (GLMM, N=87, β=0.22, p=0.063). (D) Association between methylation at CpG-2031 and DIO2 expression (GLMM, N=87, β=4.959, p=0.002, Bonferroni adjusted for 23 CpG sites tested). (E) Methylation between preserved and OA-affected cartilage for CpG-2031 among homozygous rs225014 wildtype allele carriers (GLMM, N=44, β=0.018, p=0.112). (F) Real-time qRT-PCR data of DIO2 expression between preserved and OA-affected cartilage among homozygous rs225014 wildtype allele carriers (GLMM, N=36, β=−0.017, p=0.929). (G) Association between methylation at CpG-2031 and DIO2 expression among homozygous rs225014 wildtype allele carriers (GLMM, N=36, β=3.863, p=0.050). (H) Methylation between preserved and OA-affected cartilage for CpG-2031 among the heterozygous and homozygous carriers of the rs225014 risk allele (GLMM, N=59, β=0.034, p=0.00002). (I) Real-time qRT-PCR data of DIO2 expression between preserved and OA-affected cartilage among rs225014 risk allele carriers (GLMM, N=51, β=0.35, p=0.012). (J) Association between methylation at CpG-2031 and DIO2 expression among rs225014 risk allele carriers (GLMM, N=51, β=5.58, p=0.0006). *p<0.05, **p<0.01, ***p< 0.001. OA, osteoarthritic cartilage; P, preserved cartilage.
Figure 2.
Methylation regulates DIO2 expression in articular cartilage. Each pair of bars reflects a unique donor cell culture, derived from total hip replacement patients. (A) Methylation at dinucleotide CpG-2031 without treatment (Control) and after treatment with 1.5 µM of the demethylating agent 5-aza-2-deoxycytidine (AZA). (B) Real-time qRT-PCR data of DIO2 expression without treatment and after AZA treatment.
DIO2 effects on in vitro chondrogenesis; DIO2 overexpression
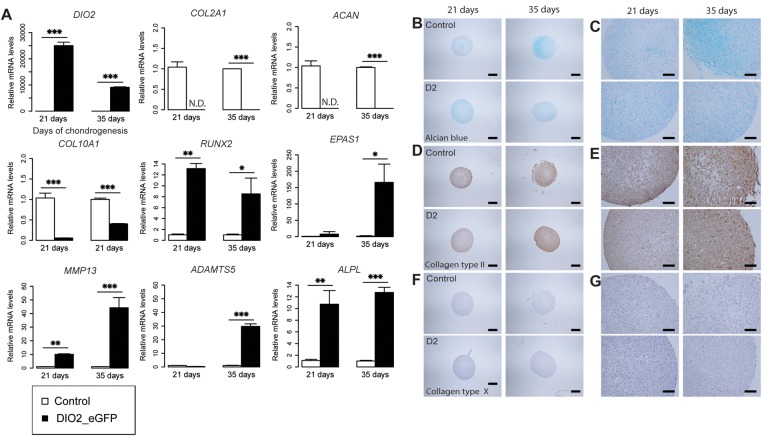
To assess the direct effect of DIO2 upregulation upon cartilage matrix homeostasis, we examined in vitro chondrogenesis of hBMSCs in a pellet culture for five consecutive weeks and generated lentiviral-mediated overexpression of DIO2 (figure 3A and online supplementary figure S3). As a result, we observed a greatly reduced expression of genes encoding the main proteins of articular cartilage ECM, COL2A1, ACAN and COL10A1 by reverse transcriptase qPCR (p<0.0001; figure 3A). In parallel, we observed profound and significant upregulated gene expression of the OA markers of hypertrophy and ECM breakdown ADAMTS5, MMP13, RUNX2 and EPAS1 (encoding HIF-2α) from 3 weeks onwards (p<0.05, figure 3A). Histology confirmed a reduced deposition of glycosaminoglycans (GAGs) (figure 3B,C). Although we did not have data to apply proper statistics to produce a significant number, pixel intensity measurements of the pictures reproduced as in figure 3B,C showed us a 35% difference after 35 days between control and DIO2 overexpressing pellets, concurrent with visual lower collagen type II (COL2) and collagen type X (COL10) protein expression by immunohistochemical staining at consecutive weeks compared with controls (figure 3D–G).
Figure 3.
Overexpressing DIO2 has a detrimental effect on cartilage extracellular matrix homeostasis. (A) qRT-PCR analysis of DIO2, COL2A1, ACAN, COL10A1, RUNX2, EPAS1, MMP13, ADAMTS5 and ALPL at 21 and 35 days of treatment in the chondrogenic human bone marrow-derived stem cells (hBMSC) transduced with respectively control vector (eGFP) and DIO2 (DIO2_eGFP; see online supplementary figure S3). (B–G) Sections comparing control (top) and DIO2 overexpressing (bottom) chondrogenic hBMSC pellets at 21 and 35 days of treatment. (B, C) Alcian blue staining. (D, E) Immunohistochemical staining of collagen type II. (F, G) Immunohistochemical staining of collagen type X. (B, D and F) Scale bar, 400 μm. (C, E and G) Scale bar, 100 μm. The expression levels were arbitrarily defined as ‘1’ in the pellets grown under control conditions (white), and data from the pellets overexpressing DIO2 (black) are given as mean±SEM. *p<0.05, **p<0.01, ***p<0.001. SEM<0.05 are not distinguishable in the figure.
DIO2 effects on in vitro chondrogenesis; thyroid hormone signalling
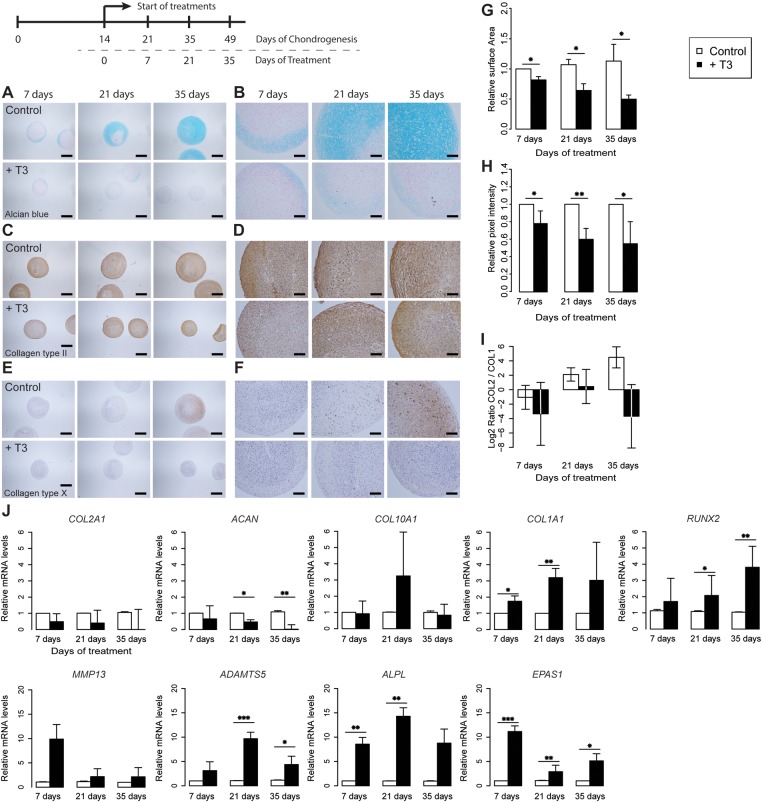
To explore whether the effects of DIO2 overexpression are due to enhanced thyroid signalling, we examined in vitro chondrogenesis of hBMSCs from five different donors (see online supplementary table S4) in pellet cultures for seven consecutive weeks (see online supplementary figure S4) while adding active T3 to the chondrogenic medium from two weeks onwards (figure 4A–F). Excess T3 resulted in a significant, progressive reduction in pellet sizes compared with controls, at consecutive weeks of culturing (p<0.05, figure 4G). Respective expression analyses showed a reduced expression of the ECM genes ACAN and COL2A1 (figure 4J), although the downregulation of COL2A1 appeared not statistically significant. Assessing the chondrogenic potential by measuring the ratio between COL2A1 and COL1A1 expression showed a dramatic effect on the chondrogenic effect of the cell system upon addition of T3 (figure 4I). Moreover, the OA markers of hypertrophy and ECM breakdown genes ADAMTS5, RUNX2 and EPAS1 were consistently upregulated from 3 weeks of treatment onwards across the donor cultures (p<0.05, figure 4J). The differences in mean expression of MMP13 between controls and T3-treated cells across the donors at consecutive time points were not significant. Immunohistochemical studies contributed to the confirmation that excess T3 resulted in a reduced expression of COL2 (figure 4C,D) and COL10 (figure 4E,F) and a pronounced reduction of GAGs (figure 4A,B) as reflected by the reduced Alcian blue staining at consecutive time points with an overall mean decrease of 35% (p=0.0002) as measured by the quantitative pixel intensities (figure 4H).
Figure 4.
Thyroid hormone is believed to be a key regulator in human cartilage development. (A–F) Sections of a representative donor (see online supplementary figure S4) comparing control (top) and T3-treated (bottom) chondrogenic human bone marrow-derived stem cell (hBMSC) pellets at 7, 21 and 35 days of treatment. (A, B) Alcian blue staining. (C, D) Immunohistochemical staining of collagen type II. (E, F) Immunohistochemical staining of collagen type X. (A, C and E) Scale bar, 400 μm. (B, D and F) Scale bar, 100 μm. (G) Mean surface area measurements at 7, 21 and 35 days of treatment with N=3 donors in each group. (H) Mean quantitative pixel-intensity measurements (see online supplementary figure S5) at 7, 21 and 35 days of treatment with N=5 donors in each group. (I) Log2 ratio of COL2A1 and COL1A1 with N=5 donors in each group. (J) Mean real-time qRT-PCR analysis of COL2A1, ACAN, COL10A1, COL1A1, ALPL, ADAMTS5 MMP13, RUNX2 and EPAS1 at 7, 21 and 35 days of treatment in chondrogenic hBMSCs of N=5 donors in each group. The surface area measurement, pixel intensities and level of expression were arbitrarily defined as ‘1’ in the pellets grown under control conditions (white) and the data from the pellets grown in the addition of excess T3 (black) are given as mean±SEM. *p<0.05, **p<0.01, ***p<0.001. SEM<0.05 are not distinguishable in the figure.
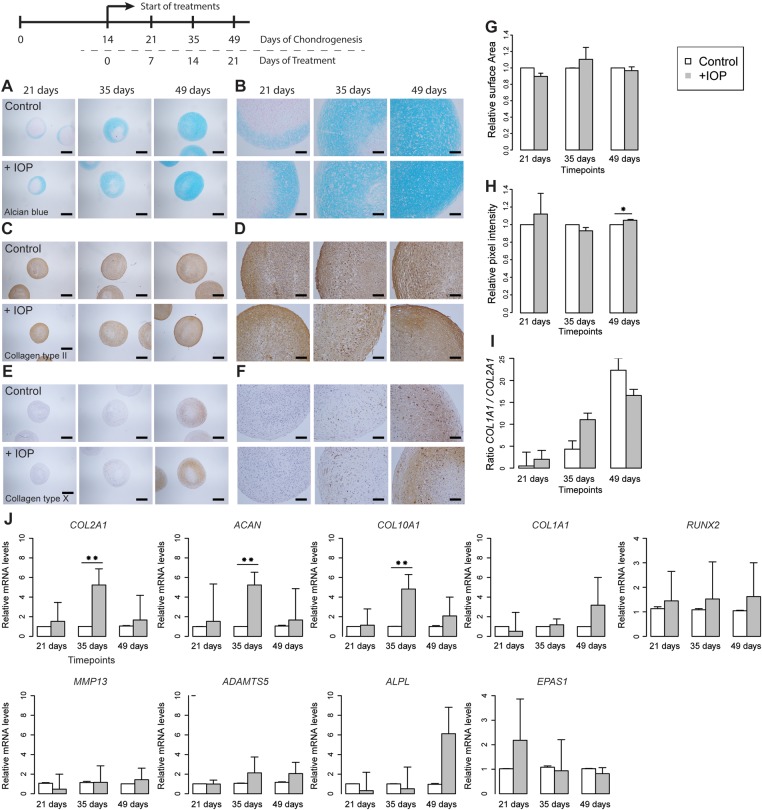
Pharmacological inhibition of D2
To investigate whether thyroid signalling blockade attenuates the detrimental effect on cartilage matrix homeostasis, pellet cultures were treated with the pharmacological deiodinase inhibitor iopanoic acid (IOP) during in vitro chondrogenesis of hBMSCs of the five donors. Quantification of surface areas indicated increasing sizes at consecutive time points in the IOP-treated pellets comparable to non-treated, control pellets (figure 5G). Respective expression analyses showed a significantly increased expression of the ECM genes ACAN, COL2A1 and COL10A1 after 3 weeks of treatment (p<0.01, figure 5J), whereas the OA markers of hypertrophy and ECM breakdown genes ADAMTS5, RUNX2 and EPAS1 were similar to control cultures (figure 5J). Furthermore, we found no significant difference in chondrogenic potential between control and IOP-treated pellets when assessing the ratio between COL2A1 and COL1A1 expression (figure 5I). Following these expression patterns, histological analysis of matrix components showed slightly higher levels of COL2 and COL10 staining (figure 5C–F) and higher levels of GAGs as reflected by a significant 4% increase in Alcian blue staining at week 5 of treatment (p=0.018; figure 5A,B and H). Of note is the denser cartilage matrix structure with less cellular lacunae at week 5 of treatment in cells treated with IOP compared with controls (figure 5A). By discriminating ‘cartilage’ from the ‘lacunae’ using the ImageJ data, we could show that administration of IOP resulted in a 10.92% decrease of lacunae (p=4.27×10−5) on day 35 of treatment.
Figure 5.
Counteracting thyroid signalling by inhibiting deiodinases in human cartilage development. (A–F) Sections of a representative donor (see online supplementary figure S4) comparing control (top) and IOP-treated (bottom) chondrogenic human bone marrow-derived stem cell (hBMSC) pellets at 7, 21 and 35 days of treatment. (A, B) Alcian blue staining. (C, D) Immunohistochemical staining of collagen type II. (E, F) Immunohistochemical staining of collagen type X. (A, C and E) Scale bar, 400 μm. (B, D and F) Scale bar, 100 μm. (G) Mean surface area measurements at 7, 21 and 35 days of treatment with N=3 donors in each group. (H) Mean quantitative pixel-intensity measurements (see online supplementary figure S5) at 7, 21 and 35 days of treatment with N=5 donors in each group. (I) Log2 ratio of COL2A1 and COL1A1 with N=5 donors in each group. (J) Mean real-time qRT-PCR analysis of COL2A1, ACAN, COL10A1, COL1A1, ALPL, ADAMTS5 MMP13, RUNX2 and EPAS1 at 7, 21 and 35 days of treatment in chondrogenic hBMSCs of N=5 donors in each group. The surface area measurement, pixel intensities and level of expression were arbitrarily defined as ‘1’ in the pellets grown under control conditions (white) and the data from the pellets grown in the addition of IOP (grey) are given as mean±SEM. *p<0.05, **p<0.01, ***p<0.001. SEM<0.05 are not distinguishable in the figure.
Discussion
In the current study, we provided insights into how genetic variation at the DIO2 locus confers risk to OA. As a result of OA-related changes in articular cartilage, loss of epigenetic silencing results in upregulation of DIO2 expression among DIO2 rs225014 risk allele carriers (figures 1 and 2). By applying an in vitro chondrogenesis model with genetically modified hBMSC, it was subsequently shown that genetic upregulation of DIO2 expression resulted in a marked reduction of the capacity of chondrocytes to deposit ECM components, concurrent with induction of OA-specific markers of cartilage matrix degeneration (ADAMTS5 and MMP13) and mineralisation (ALPL) (figure 3). Given their concurrent upregulation, this process is likely mediated via HIF-2α/RUNX2 signalling, a hallmark of the OA disease process.6 24–26 Moreover, we show that the detrimental effects of DIO2 upregulation are a result of increased T3 synthesis as reflected by the identical results when adding T3 to the culture medium (figure 4). Given that the effects observed in both treatments are very similar and the fact that the specific downstream effect of D2 action is the conversion of inactive T4 to active T3, we are confident that the reported effects in both experiments are reflecting the same mechanism, although we did not directly assess the levels of the trace element T3 in our in vitro chondrogenesis model. Together our data are in line with our previous observations that carriers of the DIO2 risk allele are prone to improper endochondral ossification and respective skeletal morphogenesis that could result in subtle malformations of joints or articular cartilage ECM composition.27
In contrast, we showed that inhibiting deiodinases by addition of IOP contributed to prolonged ‘healthy’ cartilage homeostasis by virtue of attenuated upregulation of matrix degrading enzymes, a constant COL2A1/COL1A1 ratio, denser cartilage matrix structure with significant less cellular lacunae, which indicates a reduced propensity of chondrocytes to enter the terminal maturational process (figure 5). In view of these findings, we advocate that attenuation of thyroid signalling by, for example, inhibiting deiodinases could contribute to novel therapeutic options of OA or could improve outcomes of cartilage tissue-engineering approaches. Nevertheless, given that T3 has many and various biological functions, both in the circulation and in a tissue-specific manner, local administration of a thyroid-blocking agent is likely necessary and a challenging aspect. Furthermore, the effect of, for example, IOP on other joint tissues (eg, ligament and synovium) requires investigation. It should be noted that IOP is a general inhibitor of deiodinases and as such could have also inhibited D1 and D3 action. In this respect, expression of DIO1, being important mainly in the circulation, is likely absent in cartilage tissues. Furthermore, by inhibiting D3, we prevented the conversion of active T3 to inactive T4, thereby ruling out the effect of T3 depletion due to inactive D2. Despite the beneficial effects of IOP, we observed upregulation of COL10 similar to control cultures reflecting the normal initiation of chondrocyte hypertrophy (figure 5J). Vice versa, we showed that upon addition of T3 chondrocytes directly enter the terminal maturational process towards bone, as reflected by the upregulation of enzymatic breakdown (ADAMTS5) and mineralisation (ALPL, COL1A1), this without significant induction of COL10 deposition (figure 4E,F). Together, these data indicate that chondrocyte hypertrophy in our model was not necessarily detrimental to cartilage homeostasis in contrast to DIO2 upregulation. Our data, therefore, indicate that upregulation of DIO2 does not affect the ‘early’ hypertrophic expression of COL10A1 but induces the later ‘progression stage’ marker MMP13, and the ‘late stage’ markers ALPL and COL1A1.28 In our model, we used the expression of EPAS1 and RUNX2 to show downstream effects of upregulation of DIO2 and observed a significant positive association between T3 and EPAS1. Although such an observation based on association does not imply a direct causal relationship, recent data of Chatonnet et al,29 showed, in a ChIP-seq analysis in mouse C17.2 neural progenitor cells, that EPAS1 harbours specific thyroid hormone nuclear receptor (THR) binding sites and is directly reactive to thyroid hormone. In view of these data, we advocate that active thyroid hormone, likely by local DIO2 action, could have an important impact on EPAS1 upregulation during the pathophysiology of OA. Additional studies are, however, necessary to elucidate whether T3 is directly affecting expression of EPAS1, or RUNX2 for that matter, by binding to a positional thyroid receptor in humans.
Despite the fact that the direction of gene expression changes of COL2A1 and MMP13 upon addition of T3 to the culture media appeared consistent with our overall results (figure 4J), the mean differences in expression were not statistically significant across the donors. Most likely this was the result of the considerable heterogeneity in the differential gene expression patterns of these specific genes across donors, especially with respect to timing the respective downregulation and upregulation at consecutive time points (see online supplementary figure S4), a phenomenon generally recognised in the in vitro chondrogenesis models of primary hBMSCs.30
We detected several CpG dinucleotides across the DIO2 locus that were differentially methylated between preserved and OA-affected cartilage. Although we have only verified their functionality with respect to DIO2 expression, we cannot exclude that methylation at these sites regulates expression of more distal genes. We found a consistent positive correlation between methylation at CpG-2031 and DIO2 expression in articular cartilage among carriers of the rs225014 risk allele, which may not comply with the conventional inverse relation between CpG methylation and gene expression. However, in recent genome-wide approaches, it has been recognised that this conventional relation primarily holds among CpG dinucleotides residing in CpG islands and proximal promoters, whereas gene body and distal enhancer methylation, as is the case for CpG-2031, has been shown to correlate in either direction with gene expression.31–33
With respect to our observations of DIO2 expression in preserved and osteoarthritic cartilage, it should be noted that, in contrast to previously reported high upregulation of DIO2 expression in osteoarthritic compared with healthy cartilage,5 11 we showed a moderate upregulation in osteoarthritic compared with preserved cartilage of the same joint only among carriers of the rs225014 risk allele (figure 1I). This difference suggests that upregulation of DIO2 expression may be an early event in OA pathophysiology and might be set to continue progressively among rs225014 risk allele carriers.
We were unable to validate the functionality of a putative CTCF binding site directly at the rs225014 locus nor for that matter the causality of the previously assessed consistent allelic imbalance.11 Possibly, the rs225014 SNP affects three-dimensional chromatin conformations underlying the relation between the rs225014 tagged allelic imbalance and methylation-dependent upregulation of DIO2 among rs225014 riskallele carriers.
In conclusion, our data provide evidence in humans that genetic predisposition combined with early OA-related changes results in loss of epigenetic silencing of DIO2, which likely induces EPAS1 and RUNX2 mediated upregulation of cartilage matrix degrading enzymes (ADAMTS5 and MMP13) and mineralisation of matrix (ALPL and COL1A1), thereby driving the OA process most distinctly among DIO2 risk allele carriers. Furthermore, our data show that counteracting the thyroid signalling by inhibiting deiodinases could contribute to needed novel therapeutic approaches of OA.
Supplementary Material
Acknowledgments
We thank all participants of the PAPRIKA and RAAK study. The PAPRIKA and RAAK studies were supported by the Leiden University Medical Centre, the Dutch Arthritis Association (DAA 101-402 and Reumafonds LRR) and the Centre of Medical System Biology and Netherlands Consortium for Healthy Aging both in the framework of the Netherlands Genomics Initiative (NGI). Furthermore, we acknowledge support by TreatOA and IDEAL, which are funded by the European Union’s Seventh Framework Program (FP7/2007-2011) under respective grant agreement nos. 200800 and 259679. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Contributors: NB, WdH, YFMR, FJV, PES and IM conceived and designed the experiments. NB, WdH, YFMR, RvdB, NL, AEvE and AD performed the experiments. NB, WdH, YFMR, AEvE and AD analysed the data. SDB, BAP, BJD and RGHHN contributed reagents/materials/analysis tools. NB, WdH, YFMR and IM wrote the manuscript. All authors critically reviewed the manuscript.
Competing interests: None.
Patient consent: Obtained.
Ethics approval: LUMC.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Woolf A, Pfleger B. Burden of major musculoskeletal conditions. Bull World Health Organ 2003;81:646–56. [PMC free article] [PubMed] [Google Scholar]
- 2.Reynard LN, Loughlin J. The genetics and functional analysis of primary osteoarthritis susceptibility. Expert Rev Mol Med 2013;15:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meulenbelt I, Min JL, Bos Set al. Identification of DIO2 as a new susceptibility locus for symptomatic osteoarthritis. Hum Mol Genet 2008;17:1867–75. [DOI] [PubMed] [Google Scholar]
- 4.Meulenbelt I, Bos SD, Chapman Ket al. Meta-analyses of genes modulating intracellular T3 bio-availability reveal a possible role for the DIO3 gene in osteoarthritis susceptibility. Ann Rheum Dis 2011;70:164–7. [DOI] [PubMed] [Google Scholar]
- 5.Ijiri K, Zerbini LF, Peng Het al. Differential expression of GADD45beta in normal and osteoarthritic cartilage: potential role in homeostasis of articular chondrocytes. Arthritis Rheum 2008;58:2075–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Husa M, Liu-Bryan R, Terkeltaub R. Shifting HIFs in osteoarthritis. Nat Med 2010;16:641–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldring MB, Goldring SR. Articular cartilage and subchondral bone in the pathogenesis of osteoarthritis. Ann N Y Acad Sci 2010;1192:230–7. [DOI] [PubMed] [Google Scholar]
- 8.Reynard LN, Bui C, Canty-Laird EG, et al. Expression of the osteoarthritis-associated gene GDF5 is modulated epigenetically by DNA methylation. Hum Mol Genet 2011;20:3450–60. [DOI] [PubMed] [Google Scholar]
- 9.Young DA, Bui C, Barter MJ. Understanding CpG methylation in the context of osteoarthritis. Epigenomics 2012;4:593–5. [DOI] [PubMed] [Google Scholar]
- 10.Young DA. More evidence for a role of CpG methylation in the pathogenesis of osteoarthritis. Arthritis Rheum 2012. [DOI] [PubMed] [Google Scholar]
- 11.Bos SD, Bovee JV, Duijnisveld BJet al. Increased type II deiodinase protein in OA-affected cartilage and allelic imbalance of OA risk polymorphism rs225014 at DIO2 in human OA joint tissues. Ann Rheum Dis 2012;71:1254–8. [DOI] [PubMed] [Google Scholar]
- 12.Karlsson C, Dehne T, Lindahl Aet al. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthritis Cartilage 2010;18:581–92. [DOI] [PubMed] [Google Scholar]
- 13.Nagase H, Nagasawa Y, Tachida Yet al. Deiodinase 2 upregulation demonstrated in osteoarthritis patients cartilage causes cartilage destruction in tissue-specific transgenic rats. Osteoarthritis Cartilage 2013;21:514–23. [DOI] [PubMed] [Google Scholar]
- 14.Valdes AM, Spector TD. The clinical relevance of genetic susceptibility to osteoarthritis. Best Pract Res Clin Rheumatol 2010;24:3–14. [DOI] [PubMed] [Google Scholar]
- 15.Rosenbloom KR, Dreszer TR, Long JCet al. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res 2012;40:D912–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holwerda S, de LW. Chromatin loops, gene positioning, and gene expression. Front Genet 2012;3:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrich M, Nelson MR, Stanssens Pet al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci USA 2005;102:15785–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talens RP, Boomsma DI, Tobi EWet al. Variation, patterns, and temporal stability of DNA methylation: considerations for epigenetic epidemiology. FASEB J 2010;24:3135–44. [DOI] [PubMed] [Google Scholar]
- 19.Tobi EW, Lumey LH, Talens RPet al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet 2009;18:4046–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gereben B, Goncalves C, Harney JW, et al. Selective proteolysis of human type 2 deiodinase: a novel ubiquitin-proteasomal mediated mechanism for regulation of hormone activation. Mol Endocrinol 2000;14:1697–708. [DOI] [PubMed] [Google Scholar]
- 21.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012;9:671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwabe T, Neuert H, Clandinin T. A network of cadherin-mediated interactions polarizes growth cones to determine targeting specificity. Cell 2013;154:351–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Meth 2012;9:671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bos SD, Slagboom PE, Meulenbelt I. New insights into osteoarthritis: early developmental features of an ageing-related disease. Curr Opin Rheumatol 2008;20:553–9. [DOI] [PubMed] [Google Scholar]
- 25.Saito T, Fukai A, Mabuchi Aet al. Transcriptional regulation of endochondral ossification by HIF-2[alpha] during skeletal growth and osteoarthritis development. Nat Med 2010;16:678–86. [DOI] [PubMed] [Google Scholar]
- 26.Yang S, Kim J, Ryu JHet al. Hypoxia-inducible factor-2[alpha] is a catabolic regulator of osteoarthritic cartilage destruction. Nat Med 2010;16:687–93. [DOI] [PubMed] [Google Scholar]
- 27.Waarsing JH, Kloppenburg M, Slagboom PE, et al. Osteoarthritis susceptibility genes influence the association between hip morphology and osteoarthritis. Arthritis Rheum 2011;63:1349–54. [DOI] [PubMed] [Google Scholar]
- 28.van der Kraan PM, van den Berg WB. Chondrocyte hypertrophy and osteoarthritis: role in initiation and progression of cartilage degeneration? Osteoarthritis Cartilage 2012;20:223–32. [DOI] [PubMed] [Google Scholar]
- 29.Chatonnet F, Guyot R, Benoît G, et al. Genome-wide analysis of thyroid hormone receptors shared and specific functions in neural cells. Proc Natl Acad Sci 2013;110:E766–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hellingman CA, Davidson EN, Koevoet Wet al. Smad signaling determines chondrogenic differentiation of bone-marrow-derived mesenchymal stem cells: inhibition of Smad1/5/8P prevents terminal differentiation and calcification. Tissue Eng Part A 2011;17:1157–67. [DOI] [PubMed] [Google Scholar]
- 31.Bell J, Pai A, Pickrell Jet al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol 2011;12:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang D, Cheng L, Badner JAet al. Genetic Control of Individual Differences in Gene-Specific Methylation in Human Brain. Am J Hum Genet 2010;86:411–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibbs JR, van der Brug MP, Hernandez DGet al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet 2010;6:e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.