Abstract
The identification of mutations and deletions in the SMARCB1 locus in chromosome band 22q11.2 in pediatric rhabdoid tumors provided the first evidence for the involvement of the SWI/SNF chromatin remodeling complex in cancer. Over the last 15 years, alterations in more than 20 members of the complex have been reported in a variety of human tumors. These include germline mutations and copy number alterations in SMARCB1, SMARCA4, SMARCE1, and PBRM1 that predispose carriers to both benign and malignant neoplasms. Somatic mutations, structural abnormalities, or epigenetic modifications that lead to reduced or aberrant expression of complex members have now been reported in more than twenty percent of malignancies, including both solid tumors and hematologic disorders in both children and adults. In this review, we will highlight the role of SMARCB1 in cancer as a paradigm for other tumors with alterations in SWI/SNF complex members and demonstrate the broad spectrum of mutations observed in complex members in different tumor types.
Keywords: SWI/SNF, SMARCB1, SMARCA4, rhabdoid tumor
Recent studies have established that cancer development depends on epigenetic alterations as well as genomic changes [Choi and Lee, 2013; Feinberg, 2014]. Multiple reports have demonstrated roles for altered DNA methylation, histone modifications and microRNA expression in the etiology of a wide variety of human cancers [Sarkar et al., 2013; Waldmann and Schneider, 2013]. The perturbation of SWI/SNF chromatin remodeling complexes is an emerging theme in cancer initiation and progression [Narlikar et al., 2013] and it is now postulated that at least 20% of all human tumors contain mutations in at least one member of the SWI/SNF complex [Kadoch et al., 2013]. In this review, we will specifically highlight those subunits that demonstrate germline alterations and predispose individuals to benign and malignant neoplasms.
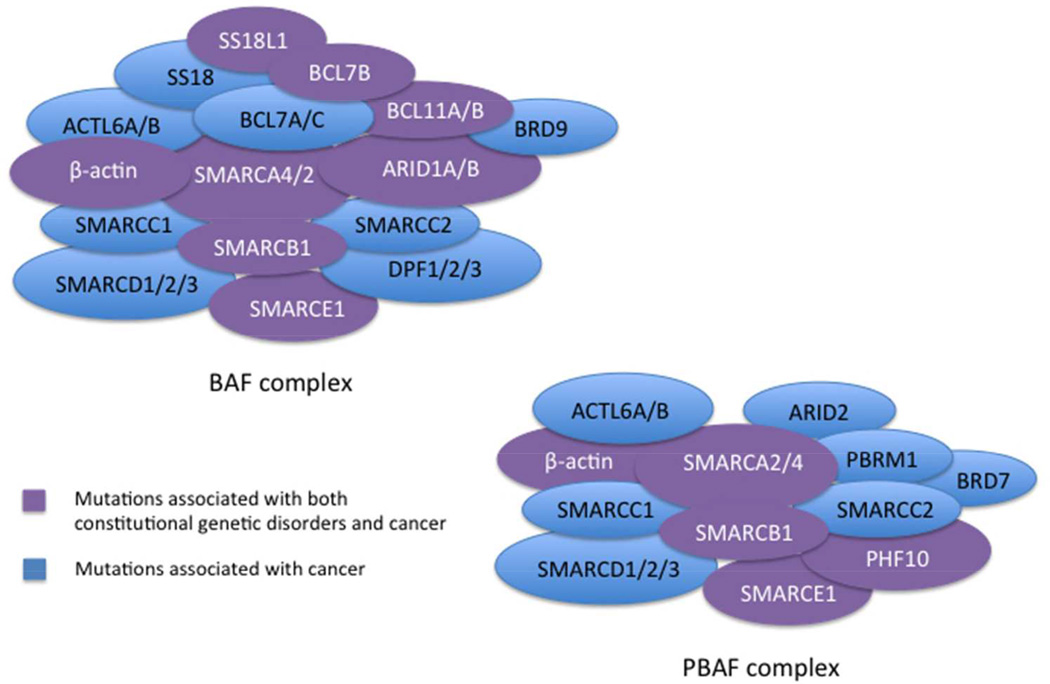
SWI/SNF chromatin remodeling complexes regulate critical cellular processes including cell cycle progression, programmed cell death, differentiation, genomic instability and DNA repair [Narlikar et al., 2013]. The SWI/SNF complex, first discovered in S. cerevisiae, shows strong conservation from yeast to Drosophila to mammals and contains approximately 12–28 components [Kadoch et al., 2013; Yaniv, 2014]. The complex, containing one of 2 mutually exclusive ATPase subunits, BRG1/SMARCA4 or BRM/SMARCA2, physically alters nucleosome positioning using energy generated by ATP hydrolysis. The complexes can be subdivided into two broad categories, BAF or PBAF (Figure 1), based upon the presence of the ARID1A/B subunits, or ARID2 and PBRM1 subunits, respectively [Kadoch et al., 2013; Wei and Weissman, 2014]. Depending upon the configuration of complex components, SWI/SNF complexes can carry out a variety of cellular functions including neural differentiation [Lessard et al., 2007], embryonic stem cell differentiation [Ho et al., 2009], hepatic lipid metabolism [Li et al., 2008], glucose metabolism [Meng et al., 2013] and brain development [Tuoc et al., 2013]. However, the mechanisms that determine the types of cancers associated with inactivation of different complex members and how their losses fuel transformation remain undetermined.
Figure 1.
BAF and PBAF complexes. The subunits specific to the BAF complex including ARID1A and ARID1B and to the PBAF complex including ARID2 and PBRM1 are demonstrated in the cartoon rendition of the complexes. Mutations in subunits colored in blue have been demonstrated in the literature to be associated with cancer, and subunits with mutations that have been associated with either cancer and/or known genetic disorders (i.e. Coffin-Siris syndrome) are colored in purple.
SMARCB1 alterations and rhabdoid tumor
Rhabdoid tumors are rare pediatric malignancies that most often arise in the brain, kidney and soft tissues. The peak incidence is in the first several years of life, although we have studied congenital tumors that arise during fetal development and tumors that have presented in late adulthood. Regardless of the anatomic location, rhabdoid tumors are highly aggressive malignancies, and combined approaches using surgery, chemotherapy and/or radiation are required for treatment [Hilden et al., 2004]. Although the tumors may be rapidly fatal, individual case reports have described patients who have survived for up to 26 years with multiple recurrences [Takahashi-Fujigasaki et al., 2012]. The identification of deletions and mutations in the SMARCB1/INI1/BAF47/hSNF5 locus, hereafter referred to as SMARCB1, in pediatric rhabdoid tumors [Versteege et al., 1998] served as a paradigm for how genetic alterations in chromatin-remodeling complex subunits might play a role in tumor development. The identification of germline mutations in SMARCB1 [Biegel et al., 1999], accompanied by loss of the normal allele, confirmed that rhabdoid tumorigenesis followed the classic two-hit model for a tumor suppressor gene.
Germline and acquired alterations of SMARCB1 and rhabdoid tumor
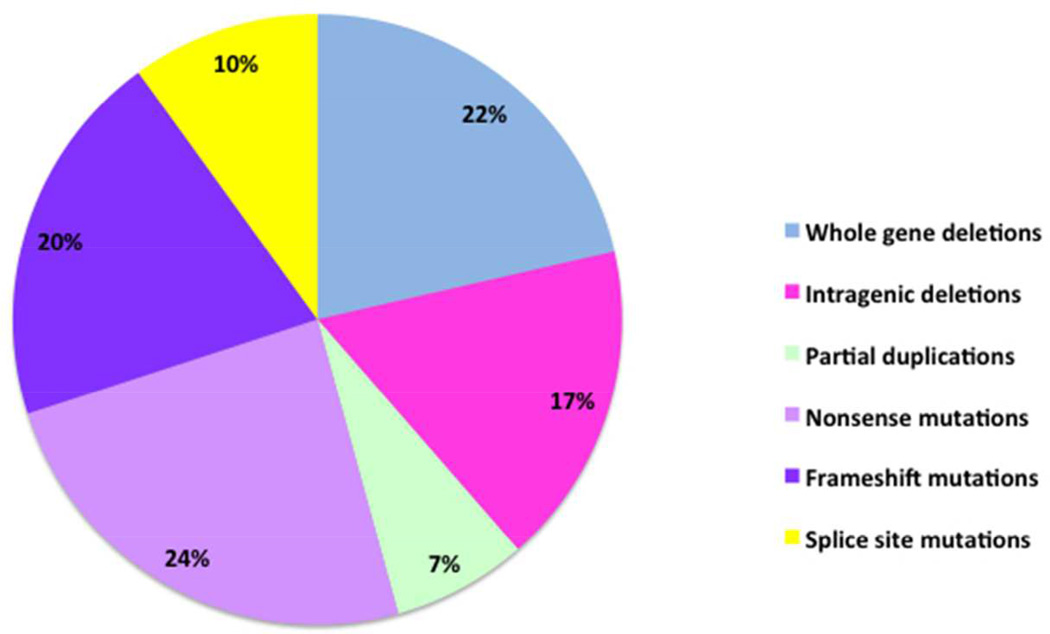
The SMARCB1 locus is located in chromosome band 22q11.2, distal to the region that is typically deleted in DiGeorge and Velo-cardio-facial syndromes. The alterations in SMARCB1 include intragenic and whole gene deletions, intragenic duplications, and mutations. The molecular characterization of two large cohorts of patients with rhabdoid tumors has been reported [Eaton et al., 2011; Bourdeaut et al., 2011] which revealed an overall incidence of germline alterations of SMARCB1 in 35% of patients. Virtually all patients with two primary tumors have a germline mutation or deletion of SMARCB1, however the incidence is also high in children with primary brain tumors (atypical teratoid/rhabdoid tumor; AT/RT) and renal tumors. In contrast, extra-renal rhabdoid tumors are typically associated with bi-allelic, somatic deletions of SMARCB1. A summary of the germline alterations in SMARCB1 in 70 rhabdoid tumor patients studied from one of the author’s laboratory is shown in Figure 2. The percentages of patients with whole gene deletions (22%), intragenic deletions and duplications (24%), nonsense mutations (24%) and frameshift mutations (20%) were similar. However, unlike other tumor suppressor genes, such as TP53 and WT1, there were no missense mutations, and only 10% of the patients had splice-site mutations. The second inactivating event in the tumors from these individuals is typically a deletion of the remaining copy of SMARCB1, or a copy number neutral loss of heterozygosity event that results in duplication of the mutated allele. The types of mutations in sporadic tumors are similar to those observed in the germline; essentially all copy number alterations or truncating mutations. Missense mutations are virtually absent and splice site mutations are uncommon.
Figure 2.
Germline SMARCB1 alterations in patients with rhabdoid tumor. Deletions of SMARCB1, both whole gene (22%) and intragenic (24%), encompass the majority of the germline alterations observed in rhabdoid tumors. A large percentage of the remaining germline alterations observed in SMARCB1 are frameshift mutations (20%) followed by nonsense mutations (17%). A smaller percentage of germline SMARCB1 alterations result from splice site mutations (10%) and partial gene duplications (7%).
The vast majority of germline deletions and mutations appear de novo, due in part to the fatal nature of the disease. Several families have been reported with two or more affected siblings in which there is presumed gonadal mosaicism, and two of our patients demonstrated germline mosaicism for a single exon deletion. While a few multi-generation families have been described with an appearance of some reduced penetrance, no established risk estimates for cancer in SMARCB1 mutation carriers exists. This has become an increasingly difficult issue in providing genetic counseling for families in which an individual is found to have a deletion in SMARCB1 as part of a contiguous 22q11.2 deletion syndrome, but who does not have a tumor. As whole exome and ultimately whole genome sequencing studies become established as clinical diagnostic tests, the identification of SMARCB1 mutations as an incidental finding will increase. Without established risk estimates for cancer in unaffected carriers, it will be challenging to provide guidance for cancer surveillance over an individual’s lifetime. Furthermore, as presented in the accompanying articles in this special issue, and as shown in Figure 1, the number of genes in the SWI/SNF and other chromatin remodeling complexes mutated in genetic disorders such as Coffin-Siris or Nicolaides-Baraitser syndrome, autism spectrum disorders and intellectual disability is also increasing. The current inability to predict the phenotype associated with mutations or deletions in these genes will be particularly challenging in a prenatal or neonatal setting.
SMARCB1 alterations and schwannomatosis
As shown in Table 1 and reviewed by Smith et al. [Smith et al., 2014], germline mutations in SMARCB1 are also seen in association with schwannomatosis, in which affected individuals develop multiple, benign nerve sheath tumors (schwannomas). Approximately 45% of patients with familial schwannomatosis and 9% of patients with apparently sporadic schwannomas have germline mutations in SMARCB1. However, in contrast to the whole gene deletions and truncating mutations observed in in rhabdoid tumor patients, the mutations in schwannomatosis are primarily splice site mutations and missense mutations in exons 1 and the 3’UTR [Hulsebos et al., 2007; Hadfield et al., 2008; Rousseau et al., 2011; Smith et al., 2012; Smith et al., 2014]. To date, one patient with a missense mutation in exon 9 of SMARCB1, who has both schwannomatosis and Coffin-Siris syndrome (Gossai et al, in preparation), has been reported. These studies suggest that loss of function truncating mutations and whole gene deletions are associated with more aggressive tumors, including rhabdoid tumors and malignant peripheral nerve sheath tumors. These tumors most often arise during infancy and early childhood. In contrast, germline missense mutations are more often associated with developmental disorders and late onset, typically benign tumors. We have described several families [Eaton et al., 2011; Carter et al., 2012] in which the identical truncating mutation led to a rhabdoid tumor in a young child, and the development of schwannomas in the adult carrier(s) in the previous generation. This led to the hypothesis that an early developmental window occurs in which the risk for rhabdoid tumors is highest, consistent with the peak incidence at 6 months of age in germline mutation carriers. After three years of age, the incidence dramatically decreases [Eaton et al., 2011]. Therefore, the cells of origin for rhabdoid tumor and schwannoma may differ, with the nature of the SWI/SNF complexes in those cells dictating the morphology and clinical behavior of the resulting neoplasms.
Table 1.
Alterations in SWI/SNF components reported in primary human tumors*
| Subunit | Gene | Cancer Types | Alteration | Germline | Missense | Truncating | Loss | Gene fusion | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|
| Core complex subunits | |||||||||
|
BRM |
SMARCA2 |
Adenoid cystic carcinoma | Mutation, deletion, and amplification | [Ho et al., 2013] | |||||
| Non-melanoma skin cancer | Mutation | [Moloney et al., 2009; Bock et al., 2011] | |||||||
| Hepatocellular carcinoma | Deletion | [Endo et al., 2013] | |||||||
| Head and neck squamous cell carcinoma | Deletion | [Gunduz et al., 2009; Wang et al., 2013] | |||||||
| Gastric cancer | Decreased expression | [Yamamichi et al., 2007] | |||||||
| Clear cell renal cell carcinoma | Decreased expression | [Xia et al., 2014] | |||||||
| Prostate cancer | Decreased expression | [Shen et al., 2008] | |||||||
| Lung cancer (adenocarcinoma and squamous cell carcinoma) | Decreased expression | [Reisman et al., 2003] | |||||||
|
BRG1 |
SMARCA4 |
Small-cell carcinoma of the ovary, hypercalcemic type (malignant rhabdoid tumor of the ovary) | Biallelic inactivation | Yes | [Jelinic et al., 2014; Ramos et al., 2014; Kupryjanczyk et al., 2013; Witkowski et al., 2014] | ||||
| Rhabdoid tumor | Biallelic inactivation | Yes | [Schneppenheim et al., 2010; Hasselblatt et al., 2011; Witkowski et al., 2013] | ||||||
| Medulloblastoma | Mutation | [Parsons et al., 2011; Robinson et al., 2012; Pugh et al., 2012; Jones et al., 2012a] | |||||||
| Lung adenocarcinoma | Mutation and deletion | Rare | [Imielinski et al., 2012; Seo et al., 2012; Medina et al., 2004; Rodriguez-Nieto et al., 2011] | ||||||
| Mantle cell lymphoma | Mutation | [Zhang et al., 2014] | |||||||
| Burkitt lymphoma | Mutation | [Love et al., 2012] | |||||||
| Hepatocellular carcinoma | Mutation and deletion | [Endo et al., 2013] | |||||||
| Esophageal adenocarcinoma | Mutation and translocation | [Dulak et al., 2013] | |||||||
| Melanoma | Mutation | [Hodis et al., 2012] | |||||||
| Non-melanoma skin cancer | Decreased expression | [Bock et al., 2011] | |||||||
| Intraductal papillary mucinous neoplasms of the pancreas | Decreased expression | [Dal Molin et al., 2012] | |||||||
|
BAF47 |
SMARCB1 |
Rhabdoid tumor | Biallelic inactivation | Yes | [Versteege et al., 1998; Biegel et al., 1999] | ||||
| Schwannoma | Biallelic inactivation | Yes | [Hulsebos et al., 2007; Hadfield et al., 2008; Rousseau et al., 2011; Smith et al., 2012] | ||||||
| Meningioma | Biallelic inactivation | Rare | [van den Munckhof et al., 2012; Christiaans et al., 2011; Schmitz et al., 2001] | ||||||
| Epithelioid sarcoma | Biallelic inactivation | Rare | [Le Loarer et al., 2014; Sullivan et al., 2013] | ||||||
| Cribriform neuroepithelial tumor | Biallelic inactivation | [Arnold et al., 2013; Hasselblatt et al., 2009] | |||||||
| Renal medullary carcinoma | Deletion | [Liu et al., 2013; Calderaro et al., 2012] | |||||||
| BAF155 | SMARCC1 | Colorectal carcinoma | Increased expression | [Andersen et al., 2009] | |||||
| Prostate cancer | Increased expression | [Heeboll et al., 2008] | |||||||
| Cervical intraepithelial neoplasia | Increased expression | [Shadeo et al., 2008] | |||||||
| BAF170 | SMARCC2 | Gastric cancer | Mutation | [Kim et al., 2013] | |||||
| Colorectal carcinoma | Mutation | [Kim et al., 2013] | |||||||
| BAF60A | SMARCD1 | Breast cancer | Mutation | [Stephens et al., 2012] | |||||
| Gastric cancer | Mutation | [Wang et al., 2014a] | |||||||
| BAF60B | SMARCD2 | Lung adenocarcinoma | Mutation | [Imielinski et al., 2012] | |||||
| Colon cancer | Mutation | [Seshagiri et al., 2012] | |||||||
| BAF60C | SMARCD3 | Neuroblastoma | Increased expression | [Takita et al., 2004] | |||||
| BAF57 | SMARCE1 | Multiple spinal meningiomas | Mutation and deletion | Yes | [Smith et al., 2013] | ||||
| BAF53A | ACTL6A | Colorectal carcinoma | Mutation | [Cancer Genome Atlas Network, 2012] | |||||
| Lung adenocarcinoma | Mutation | [Imielinski et al., 2012] | |||||||
| BAF53B | ACTL6B | Urothelial cancer | Decreased expression | [Ibragimova et al., 2014] | |||||
| Hepatocellular carcinoma | Decreased expression | [Revill et al., 2013] | |||||||
| Beta-actin | ACTB | Pericytoma with t(7;12) | Translocation | [Bridge et al., 2012; Dahlen et al., 2004b; Dahlen et al., 2004a] | |||||
| Diffuse large B-cell lymphoma | Mutation | [Lohr et al., 2012] | |||||||
| BAF complex subunits | |||||||||
|
BAF45B |
DPF1 |
Esophageal adenocarcinoma | Mutation | [Dulak et al., 2013] | |||||
| Lung adenocarcinoma | Mutation | [Imielinski et al., 2012] | |||||||
| Colon cancer | Mutation | [Seshagiri et al., 2012] | |||||||
|
BAF45C |
DPF3 |
Esophageal adenocarcinoma | Mutation | [Dulak et al., 2013] | |||||
| Lung adenocarcinoma | Mutation | [Imielinski et al., 2012] | |||||||
| Colorectal cancer | Mutation | [Cancer Genome Atlas Network, 2012] | |||||||
|
BAF45D |
DPF2 |
Esophageal adenocarcinoma | Mutation | [Dulak et al., 2013] | |||||
| Lung adenocarcinoma | Mutation | [Imielinski et al., 2012] | |||||||
| Colorectal cancer | Mutation | [Cancer Genome Atlas Network, 2012] | |||||||
|
BAF250A |
ARID1A |
Ovarian clear cell carcinoma | Mutation and deletion | [Wiegand et al., 2010; Jones et al., 2010] | |||||
| Endometrioid ovarian carcinoma | Mutation | [Wiegand et al., 2010] | |||||||
| Endometrial carcinoma | Mutation | [Liang et al., 2012; Guan et al., 2011; Le Gallo et al., 2012] | |||||||
| Cervical carcinoma | Decreased expression | [Katagiri et al., 2012; Cho et al., 2013] | |||||||
| Breast cancer | Mutation and deletion | [Jones et al., 2012b; Stephens et al., 2012; Cornen et al., 2012; Mamo et al., 2012] | |||||||
| Pancreatic ductal adenocarcinoma | Mutation and deletion | [Biankin et al., 2012; Birnbaum et al., 2011] | |||||||
| Pancreatic carcinoma with acinar differentiation | Mutation | [Jiao et al., 2014] | |||||||
| Hepatocellular carcinoma | Mutation and deletion | [Huang et al., 2012; Fujimoto et al., 2012a; Guichard et al., 2012] | |||||||
| Intrahepatic cholangiocarcinomas | Mutation | [Ross et al., 2014; Jiao et al., 2013] | |||||||
| Gastric adenocarcinoma | Mutation and deletion | [Zang et al., 2012; Wang et al., 2011; Wang et al., 2014a] | |||||||
| Esophageal adenocarcinoma | Mutation | [Dulak et al., 2013] | |||||||
| Oesophagogastric junctional adenocarcinoma | Mutation | [Chong et al., 2013] | |||||||
| Colorectal carcinoma | Mutation | [Cancer Genome Atlas Network, 2012; Cajuso et al., 2014] | |||||||
| Renal clear cell carcinoma | Mutation | [Varela et al., 2011] | |||||||
| Transitional cell carcinoma of the bladder | Mutation | [Gui et al., 2011] | |||||||
| Urothelial bladder carcinoma | Mutation | [Balbas-Martinez et al., 2013] | |||||||
| Medulloblastoma | Mutation | [Jones et al., 2012b; Parsons et al., 2011] | |||||||
| Neuroblastoma | Mutation | [Sausen et al., 2013; Pugh et al., 2013] | |||||||
| Lung adenocarcinoma | Mutation and deletion | [Imielinski et al., 2012; Seo et al., 2012] | |||||||
| Pulmonary carcinoids | Mutation | [Fernandez-Cuesta et al., 2014] | |||||||
| Adenoid cystic carcinoma | Mutation and deletion | [Ho et al., 2013] | |||||||
| Prostate cancer | Mutation | [Jones et al., 2012b; Barbieri et al., 2012] | |||||||
| Burkitt lymphoma | Mutation | [Love et al., 2012; Giulino-Roth et al., 2012] | |||||||
| Diffuse large B-cell lymphoma | Mutation | [Zhang et al., 2013] | |||||||
| Follicular lymphoma | Mutation | [Li et al., 2014] | |||||||
| Melanoma | Mutation | [Hodis et al., 2012] | |||||||
|
BAF250B |
ARID1B |
Hepatocellular carcinoma | Mutation and deletion | [Fujimoto et al., 2012a] | |||||
| Colorectal carcinoma | Mutation | [Cajuso et al., 2014] | |||||||
| Breast cancer | Mutation | [Stephens et al., 2012] | |||||||
| Prostate cancer | Mutation | [Barbieri et al., 2012] | |||||||
| Neuroblastoma | Deletion | [Sausen et al., 2013; Pugh et al., 2013] | |||||||
| Melanoma | Mutation | [Hodis et al., 2012] | |||||||
| BCL7A | BCL7A | Pilocytic astrocytoma | Deletion | [Potter et al., 2008] | |||||
| Mycosis fungoides (primary cutaneous T cell lymphoma subtype) | Deletion | [Carbone et al., 2008] | |||||||
| Multiple myeloma | Decreased expression | [Ramos-Medina et al., 2013] | |||||||
| Cutaneous T cell lymphoma | Decreased expression | [van Doorn et al., 2005] | |||||||
| BCL7B | BCL7B | Pilocytic astrocytoma | Deletion | [Potter et al., 2008] | |||||
| Gastric cancer | Mutation | [Wang et al., 2014a] | |||||||
| BCL7C | BCL7C | Gastric cancer | Mutation | [Wang et al., 2014a] | |||||
|
BCL11A |
BCL11A |
Non-small cell lung cancer | Increased expression | [Jiang et al., 2013] | |||||
| Lung squamous cell carcinoma | Copy number amplification | [Boelens et al., 2009; Jiang et al., 2013] | |||||||
| Chronic lymphocytic leukemia | Translocation and copy number gain | [Pfeifer et al., 2007; Ferreira et al., 2008; Yin et al., 2009] | |||||||
| Acute lymphoblastic leukemia | Increased expression | [Agueli et al., 2010] | |||||||
| Acute myeloid leukemia | Translocation | [Trubia et al., 2006] | |||||||
| Low-grade B cell lymphoma | Copy number gain | [Ferreira et al., 2008] | |||||||
| Mediastinal B cell lymphoma | Copy number gain/amplification | [Weniger et al., 2006] | |||||||
| Diffuse large B-cell lymphoma | Copy number gain/amplification | [Fukuhara et al., 2006; Bea et al., 2004] | |||||||
| Marginal zone B cell lymphoma | Copy number gain/amplification | [Flossbach et al., 2013] | |||||||
| Gray zone lymphoma | Copy number gain/amplification | [Eberle et al., 2011] | |||||||
| Classical Hodgkin lymphoma | Copy number gain/amplification | [Martin-Subero et al., 2002] | |||||||
|
BCL11B |
BCL11B |
Acute myeloid leukemia | Focal amplification/ translocation | [Abbas et al., 2014; Bezrookove et al., 2004] | |||||
| T cell acute lymphoblastic leukemia | Deletion, mutation, and gene fusions | [Przybylski et al., 2005; De Keersmaecker et al., 2010; Gutierrez et al., 2011; Kraszewska et al., 2013; Van Vlierberghe et al., 2013] | |||||||
| Mycosis fungoides (primary cutaneous T cell lymphoma subtype) | Increased expression | [Gu et al., 2013] | |||||||
| Adult T cell leukemia/lymphoma | Decreased expression and translocation | [Kurosawa et al., 2013; Fujimoto et al., 2012b] | |||||||
| Head and neck squamous cell carcinoma | Increased expression | [Ganguli-Indra et al., 2009] | |||||||
| BRD9 | BRD9 | Gastric cancer | Mutation | [Wang et al., 2014a] | |||||
| SS18L1 | CREST | Synovial sarcoma | Translocation | [Storlazzi et al., 2003] | |||||
| SS18 | SYT | Synovial sarcoma | Translocation | [Crew et al., 1995; Fligman et al., 1995; Panagopoulos et al., 2001] | |||||
| PBAF complex subunits | |||||||||
| BAF45A | PHF10 | Colon cancer | Mutation | [Seshagiri et al., 2012] | |||||
| Hepatocellular carcinoma | Mutation | [Kan et al., 2013] | |||||||
|
BAF180 |
PBRM1 |
Renal clear cell carcinoma | Biallelic inactivation | Yes | [Varela et al., 2011; Pena-Llopis et al., 2012; Duns et al., 2012; Guo et al., 2011] | ||||
| Intrahepatic cholangiocarcinomas | Mutation | [Jiao et al., 2013] | |||||||
| Gallbladder carcinoma | Mutation | [Jiao et al., 2013] | |||||||
| Breast cancer | Mutation and LOH | [Xia et al., 2008] | |||||||
| Esophageal adenocarcinoma | Mutation | [Dulak et al., 2013] | |||||||
|
BAF200 |
ARID2 |
Hepatocellular carcinoma | Mutation and deletion | [Fujimoto et al., 2012a; Guichard et al., 2012; Li et al., 2011] | |||||
| Pancreatic ductal adenocarcinoma | Mutation and deletion | [Biankin et al., 2012] | |||||||
| Non-small cell lung cancer | Biallelic inactivation | [Manceau et al., 2013] | |||||||
| Colorectal carcinoma | Mutation | [Cajuso et al., 2014] | |||||||
| Esophageal adenocarcinoma | Mutation | [Dulak et al., 2013] | |||||||
| Oral squamous cell carcinoma (gingivo-buccal) | Mutation | [India Project Team of the International Cancer Genome Consortium, 2013] | |||||||
| Breast cancer | Mutation | [Stephens et al., 2012] | |||||||
| Melanoma | Mutation | [Hodis et al., 2012] | |||||||
|
BRD7 |
BRD7 |
Epithelial ovarian carcinoma | Decreased expression | [Park et al., 2014] | |||||
| Colorectal carcinoma | Decreased expression | [Wu et al., 2013] | |||||||
| Nasopharyngeal carcinoma | Decreased expression | [Liu et al., 2008] | |||||||
Data from cell lines and xenografts or single case reports were not included in the table.
Light gray shading; heterozygous alteration, Solid black shading; heterozygous or homozygous alteration
In addition to the germline mutations reported in rhabdoid tumors and schwannomatosis, Van den Munckhof et al. [van den Munckhof et al., 2012] identified SMARCB1 mutations in families with multiple meningiomas of the falx cerebri of the cranium. Loss of function mutations in the SMARCE1 gene have recently been described in several families with multiple meningiomas of the spine [Smith et al., 2013], suggesting that a genotype-phenotype correlation exists between histology, anatomic location and gene mutation. Reports of SMARCE1 mutations in breast cancer cell lines [Kiskinis et al., 2006], as well as in a primary breast tumor [Villaronga et al., 2011], make it likely that germline mutations in SMARCE1 will ultimately arise in other tumor types.
Cancer predisposition associated with SMARCA4 mutations
SMARCB1 is the primary gene associated with rhabdoid tumors of the brain, kidney and extra-renal sites. In fact, homozygous inactivation of this locus appears to be sufficient for tumorigenesis, as whole exome sequencing of primary tumors failed to identify any additional non-random coding sequence mutations [Lee et al., 2012]. A second rhabdoid tumor locus was identified when germline and somatic mutations in SMARCA4 were found in a small number of patients with rhabdoid tumors who did not have SMARCB1 loss [Schneppenheim et al., 2010; Hasselblatt et al., 2011; Witkowski et al., 2013]. As shown in Table 1, a wide variety of solid tumors demonstrate missense and loss of function alterations in SMARCA4. To date, the only other tumor type to demonstrate bi-allelic inactivation of SMARCA4, consistent with a cancer predisposing germline mutation and second somatic alteration, is small cell carcinoma of the ovary, hypercalcemic type (SCCOHT) [Ramos et al., 2014; Witkowski et al., 2014; Jelinic et al., 2014]. Based on the relatively early age at presentation, and the presence of rhabdoid appearing cells by histology, it has been proposed that SCCOHT represents another type of extra-renal rhabdoid tumor [Foulkes et al., 2014]. The germline mutations in SCCOHT included both missense and truncating mutations, typically with loss of the wild-type allele as the second inactivating event in the tumor. In contrast to the typical findings in tumors, and similar to SMARCB1 mutations, the SMARCA4 mutations in developmental disorders are all missense, non-truncating mutations, or in-frame deletions, as described in the accompanying papers in this issue.
Germline mutations in PBRM1 associated with clear cell sarcoma of the kidney
Varela et al. [Varela et al., 2011] were the first to describe PBRM1 mutations in renal cell carcinoma, which were highly associated with the clear cell subtype. Patients with and without germline VHL mutations, which characterize the majority of renal cell carcinomas, demonstrated somatic PBRM1 mutations, initially suggesting that PBRM1 mutations may have been a later genetic event in tumor development. Both genes map to 3p, thus deletions of 3p in tumors would unmask recessive mutations in both VHL and PBRM1. A subsequent report by Pena-Llopis [Pena-Llopis et al., 2012] described germline mutations of PBRM1 in five patients with clear cell renal cell carcinoma, including four frameshift and one missense mutation. Intriguingly, three of these patients also had germline mutations in VHL. The occurrence of germline mutations in two different cancer predisposition genes is highly unusual. The identification of somatic BAP1 mutations in renal cell carcinoma [Pena-Llopis et al., 2012], also in tumors with PBRM1 or VHL mutations, was also interesting given that BAP1 is a cancer predisposition gene. BAP1 and PBRM1 mutations were most often mutually exclusive, however those tumors with PBRM1 and BAP1 mutations had rhabdoid features, and BAP1 loss was associated with a higher grade, compared with those tumors with PBRM1 loss. As shown in Table 1, mutations in PBRM1 have been described in a variety of other carcinomas, raising the possibility that germline alterations could contribute to a broader spectrum of cancers.
Somatic mutations in SWI/SNF complex components
A shown in Table 1 and Figure 1, mutations in multiple members of the SWI/SNF complex have been found in human cancers including non-small cell lung cancer (NSCLC), ovarian carcinomas and renal cell carcinomas [Fukuoka et al., 2004; Medina et al., 2004; Medina et al., 2008; Reisman et al., 2003; Rodriguez-Nieto et al., 2011; Weissman and Knudsen, 2009; Wilson and Roberts, 2011]. Indeed, Kadoch et al. and Wang et al. recently reported that ~20% of all human tumors show mutations in SWI/SNF components [Kadoch et al., 2013; Wang et al., 2014b]. Table 1 provides an overview of the range of mutations found in primary tumors, not including cell lines and xenografts, originating in a broad spectrum of human tissues.
The ubiquitous presence of SWI/SNF complex mutations in human cancer raises several fundamental questions about their roles in human tumor development. For example, in yeast, loss of any complex component yields similar phenotypes [Laurent et al., 1991; Peterson and Herskowitz, 1992; Wang et al., 1996]. However, it seems clear that loss of different SWI/SNF complex members in humans gives rise to different spectra of tumors. Therefore, how does the loss of each individual complex member affect the activity of the remaining SWI/SNF complexes? Potential mechanisms include altered nucleosome positioning induced by absent or aberrant SWI/SNF complex activity that lead to changes in DNA accessibility for RNA polymerase II or transcription factors [Kuwahara et al., 2013; Tolstorukov et al., 2013], secondary effects on histone modifications through altered interactions with histone mark readers, writers and erasers [Hargreaves and Crabtree, 2011], and direct or indirect effects on DNA methylation [Berdasco and Esteller, 2013; Banine et al., 2005]. Once we have a better understanding of these mechanisms, we can address a second important issue- the different effects of complex activities between complete loss of a complex member versus the consequences of missense mutations. Whether the missense mutations in complex members found in developmental disorders represent another mechanism for complete loss of activity or the acquisition of new SWI/SNF functions remains an exciting and open question.
Genetically engineered mouse models of Smarcb1/Snf5 and other complex member loss
Murine models in which Smarcb1/Snf5 was knocked out in various cell types confirmed that homozygous loss of the locus results in embryonic lethality, and that heterozygous carriers developed tumors following somatic loss of the wild-type allele [Guidi et al., 2001; Klochendler-Yeivin et al., 2000; Roberts et al., 2000; Roberts et al., 2002]. Although the tumors formed in the Smarcb1+/− mice resembled the histopathology of human rhabdoid tumors, they occurred after a long latency period (>7 months) with a low penetrance (10–30%) [Guidi et al., 2001; Klochendler-Yeivin et al., 2000; Roberts et al., 2000]. Considering the absence of mutations of well-established oncogenes and tumor suppressor genes in human rhabdoid tumors, additional studies have examined the effects of Smarcb1 inactivation in tandem with other cancer-driving genes [Lee et al., 2012]. While Smarcb1 loss in the absence of the Tp53 tumor suppressor gene resulted in a dramatic acceleration of tumor development, concomitant loss of Smarcb1 and Rb or p16INK4A did not affect MRT development [Isakoff et al., 2005; Klochendler-Yeivin et al., 2006]. However, inactivation of the Rb family of genes gene through the expression of a truncated form of T antigen resulted in an increased tumor penetrance of neural system tumors in spinal cords or brains depending upon the genetic background [Chai et al., 2007; Kuwahara et al., 2012]. In contrast, simultaneous loss of Smarcb1 with Ccnd1, Ezh2 or Smarca4/Brg1 either suppressed or eliminated tumor development in mouse models [Tsikitis et al., 2005; Wang et al., 2009; Wilson et al., 2010]. The virtual loss of tumor development in the absence of EZH2 emphasizes the strong interactions between the Polycomb and SWI/SNF complexes while the effects of SMARCA4 loss suggests that the residual SWI/SNF complexes lacking SMARCB1 may gain oncogenic activity.
Similar to the Smarcb1 genetically engineered mouse model (GEMMs), homozygous knockout of Smarca4, Srg3/Smarcc1/Baf155 and Pbrm1/Baf180 mice show embryonic lethality [Bultman et al., 2000; Han et al., 2008; Wang et al., 1999]. However, rhabdoid tumors have not appeared in knockouts of other family members including Smarca4 [Bultman et al., 2000; Bultman et al., 2007], Brm/Smarca2 [Reyes et al., 1998] or Baf155/Smarcc1/Srg3 [Han et al., 2008]. Instead, loss of Smarca4 in GEMMs contributes to the development of mammary, lung and uterine tumors as well as ovarian cysts [Bultman et al., 2007; Glaros et al., 2008; Serber et al., 2012; von Figura et al., 2014]. Loss of Smarca2 has been associated with lung and prostate tumor development while Smarcc1 inactivation results in mainly sarcomas [Ahn et al., 2010; Glaros et al., 2007; Shen et al., 2008]. A tumor phenotype has not been reported for GEMMs involving Pbrm1/Baf180 inactivation. The reports that heterozygous knockout mice of different SWI/SNF complexes develop divergent tumors appear consistent with the observed differences in tumor specificity found in humans with germline mutations in complex members (see above). The mechanisms that drive these differences remain unknown but may result from the different types of SWI/SNF complexes remaining in the absence of each unique component. The association between complex member loss and the appearance of specific tumors may also reflect their individual roles in the development of specific tissues. This notion would potentially link tumor development with the appearance of developmental disorders found in individuals with germline missense mutations in complex components.
With the development of a GEMM expressing a conditional knockout allele of Smarcb1, attempts were made to define a cell of origin for rhabdoid tumors. However, GEMMs with tissue-restricted inactivation of Smarcb1 either developed aggressive lymphomas [Roberts et al., 2002; Isakoff et al., 2005] or led to developmental blocks [Gresh et al., 2005]. Although these approaches have not yet proved successful, based upon the complex histology and immunophenotypic profiles of the tumors, our current hypothesis proposes that rhabdoid tumors arise from a primitive stem cell.
Future perspectives
The large volume of high impact publications during the past 15 years emphasize that mutations in the SWI/SNF complex play a key role in a broad spectrum of human diseases. Not surprisingly, a complex that regulates central and essential features of cellular functions, including chromatin organization, RNA transcription, DNA damage response and meiosis, may provide a significant challenge for understanding its roles in developmental disorders and cancer. The fact that we have not fully identified the number and composition of SWI/SNF complexes in normal human tissues further exacerbates this problem. However, the studies discussed in this special issue can provide a framework in which to tackle these issues. As more high-throughput gene sequencing and gene expression studies become available along with the identification of protein constituents of individual complexes through proteomic advances, we should gain new insights in the complex’s normal and aberrant activities. Combined with better cell culture models and expansion of GEMMs, we can use these data to develop rational treatments for these important human diseases.
Acknowledgments
The authors would like to thank Dr. Jacquelyn Roth for her help in preparing the manuscript.
The studies described in this review were supported by grants from the National Institutes of Health (CA46274 to J.A.B) and (CA138841 & CA91048 to B.E.W).
The authors are investigators in the Departments of Pathology and Pediatrics, and have investigated the role of several members of the SWI/SNF complex in primary tumors, cell lines, and mouse models with respect to tumor development.
Footnotes
The authors have no conflict of interest to declare.
References
- Abbas S, Sanders MA, Zeilemaker A, Geertsma-Kleinekoort WM, Koenders JE, Kavelaars FG, Abbas ZG, Mahamoud S, Chu IW, Hoogenboezem R, Peeters JK, van Drunen E, van Galen J, Beverloo HB, Lowenberg B, Valk PJ. Integrated genome-wide genotyping and gene expression profiling reveals BCL11B as a putative oncogene in acute myeloid leukemia with 14q32 aberrations. Haematologica. 2014;99:848–857. doi: 10.3324/haematol.2013.095604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agueli C, Cammarata G, Salemi D, Dagnino L, Nicoletti R, La Rosa M, Messana F, Marfia A, Bica MG, Coniglio ML, Pagano M, Fabbiano F, Santoro A. 14q32/miRNA clusters loss of heterozygosity in acute lymphoblastic leukemia is associated with up-regulation of BCL11a. Am J Hematol. 2010;85:575–578. doi: 10.1002/ajh.21758. [DOI] [PubMed] [Google Scholar]
- Ahn J, Ko M, Lee C, Kim J, Yoon H, Seong RH. Srg3, a mouse homolog of BAF155, is a novel p53 target and acts as a tumor suppressor by modulating p21WAF1/CIP1 expression. Oncogene. 2010;30:445–456. doi: 10.1038/onc.2010.424. [DOI] [PubMed] [Google Scholar]
- Andersen CL, Christensen LL, Thorsen K, Schepeler T, Sorensen FB, Verspaget HW, Simon R, Kruhoffer M, Aaltonen LA, Laurberg S, Orntoft TF. Dysregulation of the transcription factors SOX4, CBFB and SMARCC1 correlates with outcome of colorectal cancer. Br J Cancer. 2009;100:511–523. doi: 10.1038/sj.bjc.6604884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold MA, Stallings-Archer K, Marlin E, Grondin R, Olshefski R, Biegel JA, Pierson CR. Cribriform neuroepithelial tumor arising in the lateral ventricle. Pediatr Dev Pathol. 2013;16:301–307. doi: 10.2350/12-12-1287-CR.1. [DOI] [PubMed] [Google Scholar]
- Balbas-Martinez C, Rodriguez-Pinilla M, Casanova A, Dominguez O, Pisano DG, Gomez G, Lloreta J, Lorente JA, Malats N, Real FX. ARID1A alterations are associated with FGFR3-wild type, poor-prognosis, urothelial bladder tumors. PLoS One. 2013;8:e62483. doi: 10.1371/journal.pone.0062483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banine F, Bartlett C, Gunawardena R, Muchardt C, Yaniv M, Knudsen ES, Weissman BE, Sherman LS. SWI/SNF chromatin-remodeling factors induce changes in DNA methylation to promote transcriptional activation. Cancer Res. 2005;65:3542–3547. doi: 10.1158/0008-5472.CAN-04-3554. [DOI] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, Nickerson E, Chae SS, Boysen G, Auclair D, Onofrio RC, Park K, Kitabayashi N, MacDonald TY, Sheikh K, Vuong T, Guiducci C, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Hussain WM, Ramos AH, Winckler W, Redman MC, Ardlie K, Tewari AK, Mosquera JM, Rupp N, Wild PJ, Moch H, Morrissey C, Nelson PS, Kantoff PW, Gabriel SB, Golub TR, Meyerson M, Lander ES, Getz G, Rubin MA, Garraway LA. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bea S, Colomo L, Lopez-Guillermo A, Salaverria I, Puig X, Pinyol M, Rives S, Montserrat E, Campo E. Clinicopathologic significance and prognostic value of chromosomal imbalances in diffuse large B-cell lymphomas. J Clin Oncol. 2004;22:3498–3506. doi: 10.1200/JCO.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Berdasco M, Esteller M. Genetic syndromes caused by mutations in epigenetic genes. Hum Genet. 2013;132:359–383. doi: 10.1007/s00439-013-1271-x. [DOI] [PubMed] [Google Scholar]
- Bezrookove V, van Zelderen-Bhola SL, Brink A, Szuhai K, Raap AK, Barge R, Beverstock GC, Rosenberg C. A novel t(6;14)(q25-q27;q32) in acute myelocytic leukemia involves the BCL11B gene. Cancer Genet Cytogenet. 2004;149:72–76. doi: 10.1016/s0165-4608(03)00302-9. [DOI] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, Chang DK, Cowley MJ, Gardiner BB, Song S, Harliwong I, Idrisoglu S, Nourse C, Nourbakhsh E, Manning S, Wani S, Gongora M, Pajic M, Scarlett CJ, Gill AJ, Pinho AV, Rooman I, Anderson M, Holmes O, Leonard C, Taylor D, Wood S, Xu Q, Nones K, Fink JL, Christ A, Bruxner T, Cloonan N, Kolle G, Newell F, Pinese M, Mead RS, Humphris JL, Kaplan W, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chou A, Chin VT, Chantrill LA, Mawson A, Samra JS, Kench JG, Lovell JA, Daly RJ, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Australian Pancreatic Cancer Genome Initiative. Kakkar N, Zhao F, Wu YQ, Wang M, Muzny DM, Fisher WE, Brunicardi FC, Hodges SE, Reid JG, Drummond J, Chang K, Han Y, Lewis LR, Dinh H, Buhay CJ, Beck T, Timms L, Sam M, Begley K, Brown A, Pai D, Panchal A, Buchner N, De Borja R, Denroche RE, Yung CK, Serra S, Onetto N, Mukhopadhyay D, Tsao MS, Shaw PA, Petersen GM, Gallinger S, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Schulick RD, Wolfgang CL, Morgan RA, Lawlor RT, Capelli P, Corbo V, Scardoni M, Tortora G, Tempero MA, Mann KM, Jenkins NA, Perez-Mancera PA, Adams DJ, Largaespada DA, Wessels LF, Rust AG, Stein LD, Tuveson DA, Copeland NG, Musgrove EA, Scarpa A, Eshleman JR, Hudson TJ, Sutherland RL, Wheeler DA, Pearson JV, McPherson JD, Gibbs RA, Grimmond SM. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
- Birnbaum DJ, Adelaide J, Mamessier E, Finetti P, Lagarde A, Monges G, Viret F, Goncalves A, Turrini O, Delpero JR, Iovanna J, Giovannini M, Birnbaum D, Chaffanet M. Genome profiling of pancreatic adenocarcinoma. Genes Chromosomes Cancer. 2011;50:456–465. doi: 10.1002/gcc.20870. [DOI] [PubMed] [Google Scholar]
- Bock VL, Lyons JG, Huang XX, Jones AM, McDonald LA, Scolyer RA, Moloney FJ, Barnetson RS, Halliday GM. BRM and BRG1 subunits of the SWI/SNF chromatin remodelling complex are downregulated upon progression of benign skin lesions into invasive tumours. Br J Dermatol. 2011;164:1221–1227. doi: 10.1111/j.1365-2133.2011.10267.x. [DOI] [PubMed] [Google Scholar]
- Boelens MC, Kok K, van der Vlies P, van der Vries G, Sietsma H, Timens W, Postma DS, Groen HJ, van den Berg A. Genomic aberrations in squamous cell lung carcinoma related to lymph node or distant metastasis. Lung Cancer. 2009;66:372–378. doi: 10.1016/j.lungcan.2009.02.017. [DOI] [PubMed] [Google Scholar]
- Bourdeaut F, Lequin D, Brugieres L, Reynaud S, Dufour C, Doz F, Andre N, Stephan JL, Perel Y, Oberlin O, Orbach D, Bergeron C, Rialland X, Freneaux P, Ranchere D, Figarella-Branger D, Audry G, Puget S, Evans DG, Pinas JC, Capra V, Mosseri V, Coupier I, Gauthier-Villars M, Pierron G, Delattre O. Frequent hSNF5/INI1 germline mutations in patients with rhabdoid tumor. Clin Cancer Res. 2011;17:31–38. doi: 10.1158/1078-0432.CCR-10-1795. [DOI] [PubMed] [Google Scholar]
- Bridge JA, Sanders K, Huang D, Nelson M, Neff JR, Muirhead D, Walker C, Seemayer TA, Sumegi J. Pericytoma with t(7;12) and ACTB-GLI1 fusion arising in bone. Hum Pathol. 2012;43:1524–1529. doi: 10.1016/j.humpath.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman SJ, Herschkowitz JI, V G, Gebuhr TC, Yaniv M, Perou CM, Magnuson T. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2007;27:460–468. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Cajuso T, Hanninen UA, Kondelin J, Gylfe AE, Tanskanen T, Katainen R, Pitkanen E, Ristolainen H, Kaasinen E, Taipale M, Taipale J, Bohm J, Renkonen-Sinisalo L, Mecklin JP, Jarvinen H, Tuupanen S, Kilpivaara O, Vahteristo P. Exome sequencing reveals frequent inactivating mutations in ARID1A, ARID1B, ARID2 and ARID4A in microsatellite unstable colorectal cancer. Int J Cancer. 2014;135:611–623. doi: 10.1002/ijc.28705. [DOI] [PubMed] [Google Scholar]
- Calderaro J, Moroch J, Pierron G, Pedeutour F, Grison C, Maille P, Soyeux P, de la Taille A, Couturier J, Vieillefond A, Rousselet MC, Delattre O, Allory Y. SMARCB1/INI1 inactivation in renal medullary carcinoma. Histopathology. 2012;61:428–435. doi: 10.1111/j.1365-2559.2012.04228.x. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone A, Bernardini L, Valenzano F, Bottillo I, De Simone C, Capizzi R, Capalbo A, Romano F, Novelli A, Dallapiccola B, Amerio P. Array-based comparative genomic hybridization in early-stage mycosis fungoides: Recurrent deletion of tumor suppressor genes BCL7A, SMAC/DIABLO, and RHOF. Genes Chromosomes Cancer. 2008;47:1067–1075. doi: 10.1002/gcc.20601. [DOI] [PubMed] [Google Scholar]
- Carter JM, O'Hara C, Dundas G, Gilchrist D, Collins MS, Eaton K, Judkins AR, Biegel JA, Folpe AL. Epithelioid malignant peripheral nerve sheath tumor arising in a schwannoma, in a patient with "neuroblastoma-like" schwannomatosis and a novel germline SMARCB1 mutation. Am J Surg Pathol. 2012;36:154–160. doi: 10.1097/PAS.0b013e3182380802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J, Lu X, Godfrey V, Fletcher C, Roberts CW, Van Dyke T, Weissman BE. Tumor-specific cooperation of retinoblastoma protein family and snf5 inactivation. Cancer Res. 2007;67:3002–3009. doi: 10.1158/0008-5472.CAN-06-4207. [DOI] [PubMed] [Google Scholar]
- Cho H, Kim JS, Chung H, Perry C, Lee H, Kim JH. Loss of ARID1A/BAF250a expression is linked to tumor progression and adverse prognosis in cervical cancer. Hum Pathol. 2013;44:1365–1374. doi: 10.1016/j.humpath.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Choi JD, Lee JS. Interplay between epigenetics and genetics in cancer. Genomics Inform. 2013;11:164–173. doi: 10.5808/GI.2013.11.4.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong IY, Cunningham D, Barber LJ, Campbell J, Chen L, Kozarewa I, Fenwick K, Assiotis I, Guettler S, Garcia-Murillas I, Awan S, Lambros M, Starling N, Wotherspoon A, Stamp G, Gonzalez-de-Castro D, Benson M, Chau I, Hulkki S, Nohadani M, Eltahir Z, Lemnrau A, Orr N, Rao S, Lord CJ, Ashworth A. The genomic landscape of oesophagogastric junctional adenocarcinoma. J Pathol. 2013;231:301–310. doi: 10.1002/path.4247. [DOI] [PubMed] [Google Scholar]
- Christiaans I, Kenter SB, Brink HC, van OsTA, Baas F, van den Munckhof P, Kidd AM, Hulsebos TJ. Germline SMARCB1 mutation and somatic NF2 mutations in familial multiple meningiomas. J Med Genet. 2011;48:93–97. doi: 10.1136/jmg.2010.082420. [DOI] [PubMed] [Google Scholar]
- Cornen S, Adelaide J, Bertucci F, Finetti P, Guille A, Birnbaum DJ, Birnbaum D, Chaffanet M. Mutations and deletions of ARID1A in breast tumors. Oncogene. 2012;31:4255–4256. doi: 10.1038/onc.2011.598. [DOI] [PubMed] [Google Scholar]
- Crew AJ, Clark J, Fisher C, Gill S, Grimer R, Chand A, Shipley J, Gusterson BA, Cooper CS. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the kruppel-associated box in human synovial sarcoma. EMBO J. 1995;14:2333–2340. doi: 10.1002/j.1460-2075.1995.tb07228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlen A, Fletcher CD, Mertens F, Fletcher JA, Perez-Atayde AR, Hicks MJ, Debiec-Rychter M, Sciot R, Wejde J, Wedin R, Mandahl N, Panagopoulos I. Activation of the GLI oncogene through fusion with the beta-actin gene (ACTB) in a group of distinctive pericytic neoplasms: Pericytoma with t(7;12) Am J Pathol. 2004a;164:1645–1653. doi: 10.1016/s0002-9440(10)63723-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlen A, Mertens F, Mandahl N, Panagopoulos I. Molecular genetic characterization of the genomic ACTB-GLI fusion in pericytoma with t(7;12) Biochem Biophys Res Commun. 2004b;325:1318–1323. doi: 10.1016/j.bbrc.2004.10.172. [DOI] [PubMed] [Google Scholar]
- Dal Molin M, Hong SM, Hebbar S, Sharma R, Scrimieri F, de Wilde RF, Mayo SC, Goggins M, Wolfgang CL, Schulick RD, Lin MT, Eshleman JR, Hruban RH, Maitra A, Matthaei H. Loss of expression of the SWI/SNF chromatin remodeling subunit BRG1/SMARCA4 is frequently observed in intraductal papillary mucinous neoplasms of the pancreas. Hum Pathol. 2012;43:585–591. doi: 10.1016/j.humpath.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker K, Real PJ, Gatta GD, Palomero T, Sulis ML, Tosello V, Van Vlierberghe P, Barnes K, Castillo M, Sole X, Hadler M, Lenz J, Aplan PD, Kelliher M, Kee BL, Pandolfi PP, Kappes D, Gounari F, Petrie H, Van der Meulen J, Speleman F, Paietta E, Racevskis J, Wiernik PH, Rowe JM, Soulier J, Avran D, Cave H, Dastugue N, Raimondi S, Meijerink JP, Cordon-Cardo C, Califano A, Ferrando AA. The TLX1 oncogene drives aneuploidy in T cell transformation. Nat Med. 2010;16:1321–1327. doi: 10.1038/nm.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, McKenna A, Carter SL, Cibulskis K, Sivachenko A, Saksena G, Voet D, Ramos AH, Auclair D, Thompson K, Sougnez C, Onofrio RC, Guiducci C, Beroukhim R, Zhou Z, Lin L, Lin J, Reddy R, Chang A, Landrenau R, Pennathur A, Ogino S, Luketich JD, Golub TR, Gabriel SB, Lander ES, Beer DG, Godfrey TE, Getz G, Bass AJ. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duns G, Hofstra RM, Sietzema JG, Hollema H, van Duivenbode I, Kuik A, Giezen C, Jan O, Bergsma JJ, Bijnen H, van der Vlies P, van den Berg E, Kok K. Targeted exome sequencing in clear cell renal cell carcinoma tumors suggests aberrant chromatin regulation as a crucial step in ccRCC development. Hum Mutat. 2012;33:1059–1062. doi: 10.1002/humu.22090. [DOI] [PubMed] [Google Scholar]
- Eaton KW, Tooke LS, Wainwright LM, Judkins AR, Biegel JA. Spectrum of SMARCB1/INI1 mutations in familial and sporadic rhabdoid tumors. Pediatr Blood Cancer. 2011;56:7–15. doi: 10.1002/pbc.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle FC, Salaverria I, Steidl C, Summers TA, Jr, Pittaluga S, Neriah SB, Rodriguez-Canales J, Xi L, Ylaya K, Liewehr D, Dunleavy K, Wilson WH, Hewitt SM, Raffeld M, Gascoyne RD, Siebert R, Jaffe ES. Gray zone lymphoma: Chromosomal aberrations with immunophenotypic and clinical correlations. Mod Pathol. 2011;24:1586–1597. doi: 10.1038/modpathol.2011.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Yasui K, Zen Y, Gen Y, Zen K, Tsuji K, Dohi O, Mitsuyoshi H, Tanaka S, Taniwaki M, Nakanuma Y, Arii S, Yoshikawa T. Alterations of the SWI/SNF chromatin remodelling subunit-BRG1 and BRM in hepatocellular carcinoma. Liver Int. 2013;33:105–117. doi: 10.1111/liv.12005. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Epigenetic stochasticity, nuclear structure and cancer: The implications for medicine. J Intern Med. 2014 doi: 10.1111/joim.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Cuesta L, Peifer M, Lu X, Sun R, Ozretic L, Seidel D, Zander T, Leenders F, George J, Muller C, Dahmen I, Pinther B, Bosco G, Konrad K, Altmuller J, Nurnberg P, Achter V, Lang U, Schneider PM, Bogus M, Soltermann A, Brustugun OT, Helland A, Solberg S, Lund-Iversen M, Ansen S, Stoelben E, Wright GM, Russell P, Wainer Z, Solomon B, Field JK, Hyde R, Davies MP, Heukamp LC, Petersen I, Perner S, Lovly CM, Cappuzzo F, Travis WD, Wolf J, Vingron M, Brambilla E, Haas SA, Buettner R, Thomas RK. Frequent mutations in chromatin-remodelling genes in pulmonary carcinoids. Nat Commun. 2014;5:3518. doi: 10.1038/ncomms4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira BI, Garcia JF, Suela J, Mollejo M, Camacho FI, Carro A, Montes S, Piris MA, Cigudosa JC. Comparative genome profiling across subtypes of low-grade B-cell lymphoma identifies type-specific and common aberrations that target genes with a role in B-cell neoplasia. Haematologica. 2008;93:670–679. doi: 10.3324/haematol.12221. [DOI] [PubMed] [Google Scholar]
- Fligman I, Lonardo F, Jhanwar SC, Gerald WL, Woodruff J, Ladanyi M. Molecular diagnosis of synovial sarcoma and characterization of a variant SYT-SSX2 fusion transcript. Am J Pathol. 1995;147:1592–1599. [PMC free article] [PubMed] [Google Scholar]
- Flossbach L, Holzmann K, Mattfeldt T, Buck M, Lanz K, Held M, Moller P, Barth TF. High-resolution genomic profiling reveals clonal evolution and competition in gastrointestinal marginal zone B-cell lymphoma and its large cell variant. Int J Cancer. 2013;132:E116–E127. doi: 10.1002/ijc.27774. [DOI] [PubMed] [Google Scholar]
- Foulkes WD, Clarke BA, Hasselblatt M, Majewski J, Albrecht S, McCluggage WG. No small surprise - small cell carcinoma of the ovary, hypercalcaemic type, is a malignant rhabdoid tumour. J Pathol. 2014 doi: 10.1002/path.4362. [DOI] [PubMed] [Google Scholar]
- Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, Arai Y, Takahashi H, Shirakihara T, Nagasaki M, Shibuya T, Nakano K, Watanabe-Makino K, Tanaka H, Nakamura H, Kusuda J, Ojima H, Shimada K, Okusaka T, Ueno M, Shigekawa Y, Kawakami Y, Arihiro K, Ohdan H, Gotoh K, Ishikawa O, Ariizumi S, Yamamoto M, Yamada T, Chayama K, Kosuge T, Yamaue H, Kamatani N, Miyano S, Nakagama H, Nakamura Y, Tsunoda T, Shibata T, Nakagawa H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012a;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- Fujimoto R, Ozawa T, Itoyama T, Sadamori N, Kurosawa N, Isobe M. HELIOS-BCL11B fusion gene involvement in a t(2;14)(q34;q32) in an adult T-cell leukemia patient. Cancer Genet. 2012b;205:356–364. doi: 10.1016/j.cancergen.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Tagawa H, Kameoka Y, Kasugai Y, Karnan S, Kameoka J, Sasaki T, Morishima Y, Nakamura S, Seto M. Characterization of target genes at the 2p15-16 amplicon in diffuse large B-cell lymphoma. Cancer Sci. 2006;97:499–504. doi: 10.1111/j.1349-7006.2006.00209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka J, Fujii T, Shih JH, Dracheva T, Meerzaman D, Player A, Hong K, Settnek S, Gupta A, Buetow K, Hewitt S, Travis WD, Jen J. Chromatin remodeling factors and BRM/BRG1 expression as prognostic indicators in non-small cell lung cancer. Clin Cancer Res. 2004;10:4314–4324. doi: 10.1158/1078-0432.CCR-03-0489. [DOI] [PubMed] [Google Scholar]
- Ganguli-Indra G, Wasylyk C, Liang X, Millon R, Leid M, Wasylyk B, Abecassis J, Indra AK. CTIP2 expression in human head and neck squamous cell carcinoma is linked to poorly differentiated tumor status. PLoS One. 2009;4:e5367. doi: 10.1371/journal.pone.0005367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulino-Roth L, Wang K, MacDonald TY, Mathew S, Tam Y, Cronin MT, Palmer G, Lucena-Silva N, Pedrosa F, Pedrosa M, Teruya-Feldstein J, Bhagat G, Alobeid B, Leoncini L, Bellan C, Rogena E, Pinkney KA, Rubin MA, Ribeiro RC, Yelensky R, Tam W, Stephens PJ, Cesarman E. Targeted genomic sequencing of pediatric burkitt lymphoma identifies recurrent alterations in antiapoptotic and chromatin-remodeling genes. Blood. 2012;120:5181–5184. doi: 10.1182/blood-2012-06-437624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaros S, Cirrincione GM, Muchardt C, Kleer CG, Michael CW, Reisman D. The reversible epigenetic silencing of BRM: Implications for clinical targeted therapy. Oncogene. 2007;26:7058–7066. doi: 10.1038/sj.onc.1210514. [DOI] [PubMed] [Google Scholar]
- Glaros S, Cirrincione GM, Palanca A, Metzger D, Reisman D. Targeted knockout of BRG1 potentiates lung cancer development. Cancer Res. 2008;68:3689–3696. doi: 10.1158/0008-5472.CAN-07-6652. [DOI] [PubMed] [Google Scholar]
- Gresh L, Bourachot B, Reimann A, Guigas B, Fiette L, Garbay S, Muchardt C, Hue L, Pontoglio M, Yaniv M, Klochendler-Yeivin A. The SWI/SNF chromatin-remodeling complex subunit SNF5 is essential for hepatocyte differentiation. Embo J. 2005;24:3313–3324. doi: 10.1038/sj.emboj.7600802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Wang Y, Zhang G, Li W, Tu P. Aberrant expression of BCL11B in mycosis fungoides and its potential role in interferon-induced apoptosis. J Dermatol. 2013;40:596–605. doi: 10.1111/1346-8138.12160. [DOI] [PubMed] [Google Scholar]
- Guan B, Mao TL, Panuganti PK, Kuhn E, Kurman RJ, Maeda D, Chen E, Jeng YM, Wang TL, Shih I. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol. 2011;35:625–632. doi: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, He M, Li Z, Sun X, Jia W, Chen J, Yang S, Zhou F, Zhao X, Wan S, Ye R, Liang C, Liu Z, Huang P, Liu C, Jiang H, Wang Y, Zheng H, Sun L, Liu X, Jiang Z, Feng D, Chen J, Wu S, Zou J, Zhang Z, Yang R, Zhao J, Xu C, Yin W, Guan Z, Ye J, Zhang H, Li J, Kristiansen K, Nickerson ML, Theodorescu D, Li Y, Zhang X, Li S, Wang J, Yang H, Wang J, Cai Z. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, Clement B, Balabaud C, Chevet E, Laurent A, Couchy G, Letouze E, Calvo F, Zucman-Rossi J. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi CJ, Sands AT, Zambrowicz BP, Turner TK, Demers DA, Webster W, Smith TW, Imbalzano AN, Jones SN. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol Cell Biol. 2001;21:3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunduz E, Gunduz M, Ali MA, Beder L, Tamamura R, Katase N, Tominaga S, Yamanaka N, Shimizu K, Nagatsuka H. Loss of heterozygosity at the 9p21-24 region and identification of BRM as a candidate tumor suppressor gene in head and neck squamous cell carcinoma. Cancer Invest. 2009;27:661–668. doi: 10.1080/07357900802563010. [DOI] [PubMed] [Google Scholar]
- Guo G, Gui Y, Gao S, Tang A, Hu X, Huang Y, Jia W, Li Z, He M, Sun L, Song P, Sun X, Zhao X, Yang S, Liang C, Wan S, Zhou F, Chen C, Zhu J, Li X, Jian M, Zhou L, Ye R, Huang P, Chen J, Jiang T, Liu X, Wang Y, Zou J, Jiang Z, Wu R, Wu S, Fan F, Zhang Z, Liu L, Yang R, Liu X, Wu H, Yin W, Zhao X, Liu Y, Peng H, Jiang B, Feng Q, Li C, Xie J, Lu J, Kristiansen K, Li Y, Zhang X, Li S, Wang J, Yang H, Cai Z, Wang J. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet. 2011;44:17–19. doi: 10.1038/ng.1014. [DOI] [PubMed] [Google Scholar]
- Gutierrez A, Kentsis A, Sanda T, Holmfeldt L, Chen SC, Zhang J, Protopopov A, Chin L, Dahlberg SE, Neuberg DS, Silverman LB, Winter SS, Hunger SP, Sallan SE, Zha S, Alt FW, Downing JR, Mullighan CG, Look AT. The BCL11B tumor suppressor is mutated across the major molecular subtypes of T-cell acute lymphoblastic leukemia. Blood. 2011;118:4169–4173. doi: 10.1182/blood-2010-11-318873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield KD, Newman WG, Bowers NL, Wallace A, Bolger C, Colley A, McCann E, Trump D, Prescott T, Evans DG. Molecular characterisation of SMARCB1 and NF2 in familial and sporadic schwannomatosis. J Med Genet. 2008;45:332–339. doi: 10.1136/jmg.2007.056499. [DOI] [PubMed] [Google Scholar]
- Han D, Jeon S, Sohn DH, Lee C, Ahn S, Kim WK, Chung H, Seong RH. SRG3, a core component of mouse SWI/SNF complex, is essential for extra-embryonic vascular development. Developmental Biology. 2008;315:136–146. doi: 10.1016/j.ydbio.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselblatt M, Gesk S, Oyen F, Rossi S, Viscardi E, Giangaspero F, Giannini C, Judkins AR, Fruhwald MC, Obser T, Schneppenheim R, Siebert R, Paulus W. Nonsense mutation and inactivation of SMARCA4 (BRG1) in an atypical teratoid/rhabdoid tumor showing retained SMARCB1 (INI1) expression. Am J Surg Pathol. 2011;35:933–935. doi: 10.1097/PAS.0b013e3182196a39. [DOI] [PubMed] [Google Scholar]
- Hasselblatt M, Oyen F, Gesk S, Kordes U, Wrede B, Bergmann M, Schmid H, Fruhwald MC, Schneppenheim R, Siebert R, Paulus W. Cribriform neuroepithelial tumor (CRINET): A nonrhabdoid ventricular tumor with INI1 loss and relatively favorable prognosis. J Neuropathol Exp Neurol. 2009;68:1249–1255. doi: 10.1097/NEN.0b013e3181c06a51. [DOI] [PubMed] [Google Scholar]
- Heeboll S, Borre M, Ottosen PD, Andersen CL, Mansilla F, Dyrskjot L, Orntoft TF, Torring N. SMARCC1 expression is upregulated in prostate cancer and positively correlated with tumour recurrence and dedifferentiation. Histol Histopathol. 2008;23:1069–1076. doi: 10.14670/HH-23.1069. [DOI] [PubMed] [Google Scholar]
- Hilden JM, Meerbaum S, Burger P, Finlay J, Janss A, Scheithauer BW, Walter AW, Rorke LB, Biegel JA. Central nervous system atypical teratoid/rhabdoid tumor: Results of therapy in children enrolled in a registry. J Clin Oncol. 2004;22:2877–2884. doi: 10.1200/JCO.2004.07.073. [DOI] [PubMed] [Google Scholar]
- Ho AS, Kannan K, Roy DM, Morris LG, Ganly I, Katabi N, Ramaswami D, Walsh LA, Eng S, Huse JT, Zhang J, Dolgalev I, Huberman K, Heguy A, Viale A, Drobnjak M, Leversha MA, Rice CE, Singh B, Iyer NG, Leemans CR, Bloemena E, Ferris RL, Seethala RR, Gross BE, Liang Y, Sinha R, Peng L, Raphael BJ, Turcan S, Gong Y, Schultz N, Kim S, Chiosea S, Shah JP, Sander C, Lee W, Chan TA. The mutational landscape of adenoid cystic carcinoma. Nat Genet. 2013;45:791–798. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Deng Q, Wang Q, Li KY, Dai JH, Li N, Zhu ZD, Zhou B, Liu XY, Liu RF, Fei QL, Chen H, Cai B, Zhou B, Xiao HS, Qin LX, Han ZG. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44:1117–1121. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- Hulsebos TJ, Plomp AS, Wolterman RA, Robanus-Maandag EC, Baas F, Wesseling P. Germline mutation of INI1/SMARCB1 in familial schwannomatosis. Am J Hum Genet. 2007;80:805–810. doi: 10.1086/513207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibragimova I, Dulaimi E, Slifker MJ, Chen DY, Uzzo RG, Cairns P. A global profile of gene promoter methylation in treatment-naive urothelial cancer. Epigenetics. 2014;9:760–773. doi: 10.4161/epi.28078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H, Banerji S, Zander T, Seidel D, Leenders F, Ansen S, Ludwig C, Engel-Riedel W, Stoelben E, Wolf J, Goparju C, Thompson K, Winckler W, Kwiatkowski D, Johnson BE, Janne PA, Miller VA, Pao W, Travis WD, Pass HI, Gabriel SB, Lander ES, Thomas RK, Garraway LA, Getz G, Meyerson M. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- India Project Team of the International Cancer Genome Consortium. Mutational landscape of gingivo-buccal oral squamous cell carcinoma reveals new recurrently-mutated genes and molecular subgroups. Nat Commun. 2013;4:2873. doi: 10.1038/ncomms3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakoff MS, Sansam CG, Tamayo P, Subramanian A, Evans JA, Fillmore CM, Wang X, Biegel JA, Pomeroy SL, Mesirov JP, Roberts CW. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc Natl Acad Sci U S A. 2005;102:17745–17750. doi: 10.1073/pnas.0509014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinic P, Mueller JJ, Olvera N, Dao F, Scott SN, Shah R, Gao J, Schultz N, Gonen M, Soslow RA, Berger MF, Levine DA. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet. 2014;46:424–426. doi: 10.1038/ng.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang BY, Zhang XC, Su J, Meng W, Yang XN, Yang JJ, Zhou Q, Chen ZY, Chen ZH, Xie Z, Chen SL, Wu YL. BCL11A overexpression predicts survival and relapse in non-small cell lung cancer and is modulated by microRNA-30a and gene amplification. Mol Cancer. 2013;12 doi: 10.1186/1476-4598-12-61. 61-4598-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P, Offerhaus GJ, Roa JC, Roberts LR, Gores GJ, Popescu I, Alexandrescu ST, Dima S, Fassan M, Simbolo M, Mafficini A, Capelli P, Lawlor RT, Ruzzenente A, Guglielmi A, Tortora G, de Braud F, Scarpa A, Jarnagin W, Klimstra D, Karchin R, Velculescu VE, Hruban RH, Vogelstein B, Kinzler KW, Papadopoulos N, Wood LD. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Yonescu R, Offerhaus GJ, Klimstra DS, Maitra A, Eshleman JR, Herman JG, Poh W, Pelosof L, Wolfgang CL, Vogelstein B, Kinzler KW, Hruban RH, Papadopoulos N, Wood LD. Whole-exome sequencing of pancreatic neoplasms with acinar differentiation. J Pathol. 2014;232:428–435. doi: 10.1002/path.4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM, Rausch T, Warnatz HJ, Ryzhova M, Bender S, Sturm D, Pleier S, Cin H, Pfaff E, Sieber L, Wittmann A, Remke M, Witt H, Hutter S, Tzaridis T, Weischenfeldt J, Raeder B, Avci M, Amstislavskiy V, Zapatka M, Weber UD, Wang Q, Lasitschka B, Bartholomae CC, Schmidt M, von Kalle C, Ast V, Lawerenz C, Eils J, Kabbe R, Benes V, van Sluis P, Koster J, Volckmann R, Shih D, Betts MJ, Russell RB, Coco S, Tonini GP, Schuller U, Hans V, Graf N, Kim YJ, Monoranu C, Roggendorf W, Unterberg A, Herold-Mende C, Milde T, Kulozik AE, von Deimling A, Witt O, Maass E, Rossler J, Ebinger M, Schuhmann MU, Fruhwald MC, Hasselblatt M, Jabado N, Rutkowski S, von Bueren AO, Williamson D, Clifford SC, McCabe MG, Collins VP, Wolf S, Wiemann S, Lehrach H, Brors B, Scheurlen W, Felsberg J, Reifenberger G, Northcott PA, Taylor MD, Meyerson M, Pomeroy SL, Yaspo ML, Korbel JO, Korshunov A, Eils R, Pfister SM, Lichter P. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012a;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Li M, Parsons DW, Zhang X, Wesseling J, Kristel P, Schmidt MK, Markowitz S, Yan H, Bigner D, Hruban RH, Eshleman JR, Iacobuzio-Donahue CA, Goggins M, Maitra A, Malek SN, Powell S, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012b;33:100–103. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Wang TL, Shih I, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Jr, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, Gao H, Hao K, Willard MD, Xu J, Hauptschein R, Rejto PA, Fernandez J, Wang G, Zhang Q, Wang B, Chen R, Wang J, Lee NP, Zhou W, Lin Z, Peng Z, Yi K, Chen S, Li L, Fan X, Yang J, Ye R, Ju J, Wang K, Estrella H, Deng S, Wei P, Qiu M, Wulur IH, Liu J, Ehsani ME, Zhang C, Loboda A, Sung WK, Aggarwal A, Poon RT, Fan ST, Wang J, Hardwick J, Reinhard C, Dai H, Li Y, Luk JM, Mao M. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23:1422–1433. doi: 10.1101/gr.154492.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri A, Nakayama K, Rahman MT, Rahman M, Katagiri H, Ishikawa M, Ishibashi T, Iida K, Otsuki Y, Nakayama S, Miyazaki K. Frequent loss of tumor suppressor ARID1A protein expression in adenocarcinomas/adenosquamous carcinomas of the uterine cervix. Int J Gynecol Cancer. 2012;22:208–212. doi: 10.1097/IGC.0b013e3182313d78. [DOI] [PubMed] [Google Scholar]
- Kim SS, Kim MS, Yoo NJ, Lee SH. Frameshift mutations of a chromatin-remodeling gene SMARCC2 in gastric and colorectal cancers with microsatellite instability. APMIS. 2013;121:168–169. doi: 10.1111/j.1600-0463.2012.02953.x. [DOI] [PubMed] [Google Scholar]
- Kiskinis E, Garcia-Pedrero JM, Villaronga MA, Parker MG, Belandia B. Identification of BAF57 mutations in human breast cancer cell lines. Breast Cancer Res Treat. 2006;98:191–198. doi: 10.1007/s10549-005-9149-9. [DOI] [PubMed] [Google Scholar]
- Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klochendler-Yeivin A, Picarsky E, Yaniv M. Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex. Mol Cell Biol. 2006;26:2661–2674. doi: 10.1128/MCB.26.7.2661-2674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraszewska MD, Dawidowska M, Kosmalska M, Sedek L, Grzeszczak W, Kowalczyk JR, Szczepanski T, Witt M Polish Pediatric Leukemia Lymphoma Study Group (PPLLSG) BCL11B, FLT3, NOTCH1 and FBXW7 mutation status in T-cell acute lymphoblastic leukemia patients. Blood Cells Mol Dis. 2013;50:33–38. doi: 10.1016/j.bcmd.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Kupryjanczyk J, Dansonka-Mieszkowska A, Moes-Sosnowska J, Plisiecka-Halasa J, Szafron L, Podgorska A, Rzepecka IK, Konopka B, Budzilowska A, Rembiszewska A, Grajkowska W, Spiewankiewicz B. Ovarian small cell carcinoma of hypercalcemic type - evidence of germline origin and SMARCA4 gene inactivation. a pilot study. Pol J Pathol. 2013;64:238–246. doi: 10.5114/pjp.2013.39331. [DOI] [PubMed] [Google Scholar]
- Kurosawa N, Fujimoto R, Ozawa T, Itoyama T, Sadamori N, Isobe M. Reduced level of the BCL11B protein is associated with adult T-cell leukemia/lymphoma. PLoS One. 2013;8:e55147. doi: 10.1371/journal.pone.0055147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara Y, Mora-Blanco EL, Banine F, Rogers AB, Fletcher C, Sherman LS, Roberts CW, Weissman BE. Establishment and characterization of MRT cell lines from genetically engineered mouse models and the influence of genetic background on their development. Int J Cancer. 2012 doi: 10.1002/ijc.27976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara Y, Wei D, Durand J, Weissman BE. SNF5 reexpression in malignant rhabdoid tumors regulates transcription of target genes by recruitment of SWI/SNF complexes and RNAPII to the transcription start site of their promoters. Mol Cancer Res. 2013;11:251–260. doi: 10.1158/1541-7786.MCR-12-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent BC, Treitel MA, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc. Natl. Acad. Sci. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gallo M, O'Hara AJ, Rudd ML, Urick ME, Hansen NF, O'Neil NJ, Price JC, Zhang S, England BM, Godwin AK, Sgroi DC, NIH Intramural Sequencing Center (NISC) Comparative Sequencing Program. Hieter P, Mullikin JC, Merino MJ, Bell DW. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet. 2012;44:1310–1315. doi: 10.1038/ng.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Loarer F, Zhang L, Fletcher CD, Ribeiro A, Singer S, Italiano A, Neuville A, Houlier A, Chibon F, Coindre JM, Antonescu CR. Consistent SMARCB1 homozygous deletions in epithelioid sarcoma and in a subset of myoepithelial carcinomas can be reliably detected by FISH in archival material. Genes Chromosomes Cancer. 2014;53:475–486. doi: 10.1002/gcc.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, Lawrence MS, Auclair D, Mora J, Golub TR, Biegel JA, Getz G, Roberts CW. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122:2983–2988. doi: 10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Kaminski MS, Li Y, Yildiz M, Ouillette P, Jones S, Fox H, Jacobi K, Saiya-Cork K, Bixby D, Lebovic D, Roulston D, Shedden K, Sabel M, Marentette L, Cimmino V, Chang AE, Malek SN. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123:1487–1498. doi: 10.1182/blood-2013-05-500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhao H, Zhang X, Wood LD, Anders RA, Choti MA, Pawlik TM, Daniel HD, Kannangai R, Offerhaus GJ, Velculescu VE, Wang L, Zhou S, Vogelstein B, Hruban RH, Papadopoulos N, Cai J, Torbenson MS, Kinzler KW. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43:828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu C, Li N, Hao T, Han T, Hill DE, Vidal M, Lin JD. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 2008;8:105–117. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Cheung LW, Li J, Ju Z, Yu S, Stemke-Hale K, Dogruluk T, Lu Y, Liu X, Gu C, Guo W, Scherer SE, Carter H, Westin SN, Dyer MD, Verhaak RG, Zhang F, Karchin R, Liu CG, Lu KH, Broaddus RR, Scott KL, Hennessy BT, Mills GB. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012;22:2120–2129. doi: 10.1101/gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang L, Niu Z, Zhou M, Peng C, Li X, Deng T, Shi L, Tan Y, Li G. Promoter methylation inhibits BRD7 expression in human nasopharyngeal carcinoma cells. BMC Cancer. 2008;8 doi: 10.1186/1471-2407-8-253. 253-2407-8-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Galli S, Srinivasan R, Linehan WM, Tsokos M, Merino MJ. Renal medullary carcinoma: Molecular, immunohistochemistry, and morphologic correlation. Am J Surg Pathol. 2013;37:368–374. doi: 10.1097/PAS.0b013e3182770406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, Novak AJ, Dogan A, Ansell SM, Link BK, Zou L, Gould J, Saksena G, Stransky N, Rangel-Escareno C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Hernandez-Lemus E, Schwarz-Cruz y Celis A, Imaz-Rosshandler I, Ojesina AI, Jung J, Pedamallu CS, Lander ES, Habermann TM, Cerhan JR, Shipp MA, Getz G, Golub TR. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, Lugar PL, Rizzieri DA, Lagoo AS, Bernal-Mizrachi L, Mann KP, Flowers CR, Naresh KN, Evens AM, Chadburn A, Gordon LI, Czader MB, Gill JI, Hsi ED, Greenough A, Moffitt AB, McKinney M, Banerjee A, Grubor V, Levy S, Dunson DB, Dave SS. The genetic landscape of mutations in burkitt lymphoma. Nat Genet. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamo A, Cavallone L, Tuzmen S, Chabot C, Ferrario C, Hassan S, Edgren H, Kallioniemi O, Aleynikova O, Przybytkowski E, Malcolm K, Mousses S, Tonin PN, Basik M. An integrated genomic approach identifies ARID1A as a candidate tumor-suppressor gene in breast cancer. Oncogene. 2012;31:2090–2100. doi: 10.1038/onc.2011.386. [DOI] [PubMed] [Google Scholar]
- Manceau G, Letouze E, Guichard C, Didelot A, Cazes A, Corte H, Fabre E, Pallier K, Imbeaud S, Le Pimpec-Barthes F, Zucman-Rossi J, Laurent-Puig P, Blons H. Recurrent inactivating mutations of ARID2 in non-small cell lung carcinoma. Int J Cancer. 2013;132:2217–2221. doi: 10.1002/ijc.27900. [DOI] [PubMed] [Google Scholar]
- Martin-Subero JI, Gesk S, Harder L, Sonoki T, Tucker PW, Schlegelberger B, Grote W, Novo FJ, Calasanz MJ, Hansmann ML, Dyer MJ, Siebert R. Recurrent involvement of the REL and BCL11A loci in classical hodgkin lymphoma. Blood. 2002;99:1474–1477. doi: 10.1182/blood.v99.4.1474. [DOI] [PubMed] [Google Scholar]
- Medina PP, Carretero J, Fraga MF, Esteller M, Sidransky D, Sanchez-Cespedes M. Genetic and epigenetic screening for gene alterations of the chromatin-remodeling factor, SMARCA4/BRG1, in lung tumors. Genes Chromosomes Cancer. 2004;41:170–177. doi: 10.1002/gcc.20068. [DOI] [PubMed] [Google Scholar]
- Medina PP, Romero OA, Kohno T, Montuenga LM, Pio R, Yokota J, Sanchez-Cespedes M. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat. 2008;29:617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- Meng ZX, Li S, Wang L, Ko HJ, Lee Y, Jung DY, Okutsu M, Yan Z, Kim JK, Lin JD. Baf60c drives glycolytic metabolism in the muscle and improves systemic glucose homeostasis through deptor-mediated akt activation. Nat Med. 2013;19:640–645. doi: 10.1038/nm.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney FJ, Lyons JG, Bock VL, Huang XX, Bugeja MJ, Halliday GM. Hotspot mutation of brahma in non-melanoma skin cancer. J Invest Dermatol. 2009;129:1012–1015. doi: 10.1038/jid.2008.319. [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagopoulos I, Mertens F, Isaksson M, Limon J, Gustafson P, Skytting B, Akerman M, Sciot R, Dal Cin P, Samson I, Iliszko M, Ryoe J, Debiec-Rychter M, Szadowska A, Brosjo O, Larsson O, Rydholm A, Mandahl N. Clinical impact of molecular and cytogenetic findings in synovial sarcoma. Genes Chromosomes Cancer. 2001;31:362–372. doi: 10.1002/gcc.1155. [DOI] [PubMed] [Google Scholar]
- Park YA, Lee JW, Kim HS, Lee YY, Kim TJ, Choi CH, Choi JJ, Jeon HK, Cho YJ, Ryu JY, Kim BG, Bae DS. Tumor suppressive effects of bromodomain-containing protein 7 (BRD7) in epithelial ovarian carcinoma. Clin Cancer Res. 2014;20:565–575. doi: 10.1158/1078-0432.CCR-13-1271. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, Gallia GL, Jallo GI, Binder ZA, Nikolsky Y, Hartigan J, Smith DR, Gerhard DS, Fults DW, VandenBerg S, Berger MS, Marie SK, Shinjo SM, Clara C, Phillips PC, Minturn JE, Biegel JA, Judkins AR, Resnick AC, Storm PB, Curran T, He Y, Rasheed BA, Friedman HS, Keir ST, McLendon R, Northcott PA, Taylor MD, Burger PC, Riggins GJ, Karchin R, Parmigiani G, Bigner DD, Yan H, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, Yamasaki T, Zhrebker L, Sivanand S, Spence P, Kinch L, Hambuch T, Jain S, Lotan Y, Margulis V, Sagalowsky AI, Summerour PB, Kabbani W, Wong SW, Grishin N, Laurent M, Xie XJ, Haudenschild CD, Ross MT, Bentley DR, Kapur P, Brugarolas J. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Pfeifer D, Pantic M, Skatulla I, Rawluk J, Kreutz C, Martens UM, Fisch P, Timmer J, Veelken H. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood. 2007;109:1202–1210. doi: 10.1182/blood-2006-07-034256. [DOI] [PubMed] [Google Scholar]
- Potter N, Karakoula A, Phipps KP, Harkness W, Hayward R, Thompson DN, Jacques TS, Harding B, Thomas DG, Palmer RW, Rees J, Darling J, Warr TJ. Genomic deletions correlate with underexpression of novel candidate genes at six loci in pediatric pilocytic astrocytoma. Neoplasia. 2008;10:757–772. doi: 10.1593/neo.07914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybylski GK, Dik WA, Wanzeck J, Grabarczyk P, Majunke S, Martin-Subero JI, Siebert R, Dolken G, Ludwig WD, Verhaaf B, van Dongen JJ, Schmidt CA, Langerak AW. Disruption of the BCL11B gene through inv(14)(q11.2q32.31) results in the expression of BCL11B-TRDC fusion transcripts and is associated with the absence of wild-type BCL11B transcripts in T-ALL. Leukemia. 2005;19:201–208. doi: 10.1038/sj.leu.2403619. [DOI] [PubMed] [Google Scholar]
- Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, Kim J, Lawrence MS, Lichenstein L, McKenna A, Pedamallu CS, Ramos AH, Shefler E, Sivachenko A, Sougnez C, Stewart C, Ally A, Birol I, Chiu R, Corbett RD, Hirst M, Jackman SD, Kamoh B, Khodabakshi AH, Krzywinski M, Lo A, Moore RA, Mungall KL, Qian J, Tam A, Thiessen N, Zhao Y, Cole KA, Diamond M, Diskin SJ, Mosse YP, Wood AC, Ji L, Sposto R, Badgett T, London WB, Moyer Y, Gastier-Foster JM, Smith MA, Guidry Auvil JM, Gerhard DS, Hogarty MD, Jones SJ, Lander ES, Gabriel SB, Getz G, Seeger RC, Khan J, Marra MA, Meyerson M, Maris JM. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, Greulich H, Lawrence MS, Lennon NJ, McKenna A, Meldrim J, Ramos AH, Ross MG, Russ C, Shefler E, Sivachenko A, Sogoloff B, Stojanov P, Tamayo P, Mesirov JP, Amani V, Teider N, Sengupta S, Francois JP, Northcott PA, Taylor MD, Yu F, Crabtree GR, Kautzman AG, Gabriel SB, Getz G, Jager N, Jones DT, Lichter P, Pfister SM, Roberts TM, Meyerson M, Pomeroy SL, Cho YJ. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos P, Karnezis AN, Craig DW, Sekulic A, Russell ML, Hendricks WP, Corneveaux JJ, Barrett MT, Shumansky K, Yang Y, Shah SP, Prentice LM, Marra MA, Kiefer J, Zismann VL, McEachron TA, Salhia B, Prat J, D'Angelo E, Clarke BA, Pressey JG, Farley JH, Anthony SP, Roden RB, Cunliffe HE, Huntsman DG, Trent JM. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat Genet. 2014;46:427–429. doi: 10.1038/ng.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Medina R, Montes-Moreno S, Maestre L, Canamero M, Rodriguez-Pinilla M, Martinez-Torrecuadrada J, Piris MA, Majid A, Dyer MJ, Pulford K, Roncador G. BCL7A protein expression in normal and malignant lymphoid tissues. Br J Haematol. 2013;160:106–109. doi: 10.1111/bjh.12080. [DOI] [PubMed] [Google Scholar]
- Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: Correlation with poor prognosis. Cancer Res. 2003;63:560–566. [PubMed] [Google Scholar]
- Revill K, Wang T, Lachenmayer A, Kojima K, Harrington A, Li J, Hoshida Y, Llovet JM, Powers S. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology. 2013;145:1424–1435. e1–e25. doi: 10.1053/j.gastro.2013.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) Embo J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci U S A. 2000;97:13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell. 2002;2:415–425. doi: 10.1016/s1535-6108(02)00185-x. [DOI] [PubMed] [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, Chalhoub N, Baker SJ, Huether R, Kriwacki R, Curley N, Thiruvenkatam R, Wang J, Wu G, Rusch M, Hong X, Becksfort J, Gupta P, Ma J, Easton J, Vadodaria B, Onar-Thomas A, Lin T, Li S, Pounds S, Paugh S, Zhao D, Kawauchi D, Roussel MF, Finkelstein D, Ellison DW, Lau CC, Bouffet E, Hassall T, Gururangan S, Cohn R, Fulton RS, Fulton LL, Dooling DJ, Ochoa K, Gajjar A, Mardis ER, Wilson RK, Downing JR, Zhang J, Gilbertson RJ. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Nieto S, Canada A, Pros E, Pinto AI, Torres-Lanzas J, Lopez-Rios F, Sanchez-Verde L, Pisano DG, Sanchez-Cespedes M. Massive parallel DNA pyrosequencing analysis of the tumor suppressor BRG1/SMARCA4 in lung primary tumors. Hum Mutat. 2011;32:E1999–E2017. doi: 10.1002/humu.21415. [DOI] [PubMed] [Google Scholar]
- Ross JS, Wang K, Gay L, Al-Rohil R, Rand JV, Jones DM, Lee HJ, Sheehan CE, Otto GA, Palmer G, Yelensky R, Lipson D, Morosini D, Hawryluk M, Catenacci DV, Miller VA, Churi C, Ali S, Stephens PJ. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. Oncologist. 2014;19:235–242. doi: 10.1634/theoncologist.2013-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau G, Noguchi T, Bourdon V, Sobol H, Olschwang S. SMARCB1/INI1 germline mutations contribute to 10% of sporadic schwannomatosis. BMC Neurol. 2011;11 doi: 10.1186/1471-2377-11-9. 9-2377-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Horn G, Moulton K, Oza A, Byler S, Kokolus S, Longacre M. Cancer development, progression, and therapy: An epigenetic overview. Int J Mol Sci. 2013;14:21087–21113. doi: 10.3390/ijms141021087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, Blackford A, Parmigiani G, Diaz LA, Jr, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE, Hogarty MD. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45:12–17. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz U, Mueller W, Weber M, Sevenet N, Delattre O, von Deimling A. INI1 mutations in meningiomas at a potential hotspot in exon 9. Br J Cancer. 2001;84:199–201. doi: 10.1054/bjoc.2000.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneppenheim R, Fruhwald MC, Gesk S, Hasselblatt M, Jeibmann A, Kordes U, Kreuz M, Leuschner I, Martin Subero JI, Obser T, Oyen F, Vater I, Siebert R. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am J Hum Genet. 2010;86:279–284. doi: 10.1016/j.ajhg.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, Lee J, Jung YJ, Kim JO, Shin JY, Yu SB, Kim J, Lee ER, Kang CH, Park IK, Rhee H, Lee SH, Kim JI, Kang JH, Kim YT. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:2109–2119. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serber DW, Rogala A, Makarem M, Rosson GB, Simin K, Godfrey V, Van Dyke T, Eaves CJ, Bultman SJ. The BRG1 chromatin remodeler protects against ovarian cysts, uterine tumors, and mammary tumors in a lineage-specific manner. PLoS ONE. 2012;7:e31346. doi: 10.1371/journal.pone.0031346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman V, Jaiswal BS, Guillory J, Ha C, Dijkgraaf GJ, Stinson J, Gnad F, Huntley MA, Degenhardt JD, Haverty PM, Bourgon R, Wang W, Koeppen H, Gentleman R, Starr TK, Zhang Z, Largaespada DA, Wu TD, de Sauvage FJ. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadeo A, Chari R, Lonergan KM, Pusic A, Miller D, Ehlen T, Van Niekerk D, Matisic J, Richards-Kortum R, Follen M, Guillaud M, Lam WL, MacAulay C. Up regulation in gene expression of chromatin remodelling factors in cervical intraepithelial neoplasia. BMC Genomics. 2008;9 doi: 10.1186/1471-2164-9-64. 64-2164-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Powers N, Saini N, Comstock CES, Sharma A, Weaver K, Revelo MP, Gerald W, Williams E, Jessen WJ, Aronow BJ, Rosson G, Weissman B, Muchardt C, Yaniv M, Knudsen KE. The SWI/SNF ATPase brm is a gatekeeper of proliferative control in prostate cancer. Cancer Research. 2008;68:10154–10162. doi: 10.1158/0008-5472.CAN-08-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MJ, O'Sullivan J, Bhaskar SS, Hadfield KD, Poke G, Caird J, Sharif S, Eccles D, Fitzpatrick D, Rawluk D, du Plessis D, Newman WG, Evans DG. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet. 2013;45:295–298. doi: 10.1038/ng.2552. [DOI] [PubMed] [Google Scholar]
- Smith MJ, Wallace AJ, Bowers NL, Rustad CF, Woods CG, Leschziner GD, Ferner RE, Evans DG. Frequency of SMARCB1 mutations in familial and sporadic schwannomatosis. Neurogenetics. 2012;13:141–145. doi: 10.1007/s10048-012-0319-8. [DOI] [PubMed] [Google Scholar]
- Smith MJ, Wallace AJ, Bowers NL, Eaton H, Evans DG. SMARCB1 mutations in schwannomatosis and genotype correlations with rhabdoid tumors. Cancer Genetics. 2014 doi: 10.1016/j.cancergen.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J, Easton D, Langerod A, Oslo Breast Cancer Consortium (OSBREAC) Lee MT, Shen CY, Tee BT, Huimin BW, Broeks A, Vargas AC, Turashvili G, Martens J, Fatima A, Miron P, Chin SF, Thomas G, Boyault S, Mariani O, Lakhani SR, van de Vijver M, van 't Veer L, Foekens J, Desmedt C, Sotiriou C, Tutt A, Caldas C, Reis-Filho JS, Aparicio SA, Salomon AV, Borresen-Dale AL, Richardson AL, Campbell PJ, Futreal PA, Stratton MR. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi CT, Mertens F, Mandahl N, Gisselsson D, Isaksson M, Gustafson P, Domanski HA, Panagopoulos I. A novel fusion gene, SS18L1/SSX1, in synovial sarcoma. Genes Chromosomes Cancer. 2003;37:195–200. doi: 10.1002/gcc.10210. [DOI] [PubMed] [Google Scholar]
- Sullivan LM, Folpe AL, Pawel BR, Judkins AR, Biegel JA. Epithelioid sarcoma is associated with a high percentage of SMARCB1 deletions. Mod Pathol. 2013;26:385–392. doi: 10.1038/modpathol.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi-Fujigasaki J, Matumoto M, Kan I, Oka H, Yasue M. Atypical teratoid/rhabdoid tumor with 26-year overall survival: Case report. J Neurosurg Pediatr. 2012;9:400–405. doi: 10.3171/2012.1.PEDS11350. [DOI] [PubMed] [Google Scholar]
- Takita J, Ishii M, Tsutsumi S, Tanaka Y, Kato K, Toyoda Y, Hanada R, Yamamoto K, Hayashi Y, Aburatani H. Gene expression profiling and identification of novel prognostic marker genes in neuroblastoma. Genes Chromosomes Cancer. 2004;40:120–132. doi: 10.1002/gcc.20021. [DOI] [PubMed] [Google Scholar]
- Tolstorukov MY, Sansam CG, Lu P, Koellhoffer EC, Helming KC, Alver BH, Tillman EJ, Evans JA, Wilson BG, Park PJ, Roberts CW. Swi/snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc Natl Acad Sci U S A. 2013;110:10165–10170. doi: 10.1073/pnas.1302209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trubia M, Albano F, Cavazzini F, Cambrin GR, Quarta G, Fabbiano F, Ciambelli F, Magro D, Hernandezo JM, Mancini M, Diverio D, Pelicci PG, Coco FL, Mecucci C, Specchia G, Rocchi M, Liso V, Castoldi G, Cuneo A. Characterization of a recurrent translocation t(2;3)(p15-22;q26) occurring in acute myeloid leukaemia. Leukemia. 2006;20:48–54. doi: 10.1038/sj.leu.2404020. [DOI] [PubMed] [Google Scholar]
- Tsikitis M, Zhang Z, Edelman W, Zagzag D, Kalpana GV. Genetic ablation of cyclin D1 abrogates genesis of rhabdoid tumors resulting from Ini1 loss. Proc Natl Acad Sci U S A. 2005;102:12129–12134. doi: 10.1073/pnas.0505300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuoc TC, Boretius S, Sansom SN, Pitulescu ME, Frahm J, Livesey FJ, Stoykova A. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Developmental Cell. 2013;25:256–269. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- van den Munckhof P, Christiaans I, Kenter SB, Baas F, Hulsebos TJ. Germline SMARCB1 mutation predisposes to multiple meningiomas and schwannomas with preferential location of cranial meningiomas at the falx cerebri. Neurogenetics. 2012;13:1–7. doi: 10.1007/s10048-011-0300-y. [DOI] [PubMed] [Google Scholar]
- van Doorn R, Zoutman WH, Dijkman R, de Menezes RX, Commandeur S, Mulder AA, van der Velden PA, Vermeer MH, Willemze R, Yan PS, Huang TH, Tensen CP. Epigenetic profiling of cutaneous T-cell lymphoma: Promoter hypermethylation of multiple tumor suppressor genes including BCL7a, PTPRG, and p73. J Clin Oncol. 2005;23:3886–3896. doi: 10.1200/JCO.2005.11.353. [DOI] [PubMed] [Google Scholar]
- Van Vlierberghe P, Ambesi-Impiombato A, De Keersmaecker K, Hadler M, Paietta E, Tallman MS, Rowe JM, Forne C, Rue M, Ferrando AA. Prognostic relevance of integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. Blood. 2013;122:74–82. doi: 10.1182/blood-2013-03-491092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, Bignell G, Butler A, Cho J, Dalgliesh GL, Galappaththige D, Greenman C, Hardy C, Jia M, Latimer C, Lau KW, Marshall J, McLaren S, Menzies A, Mudie L, Stebbings L, Largaespada DA, Wessels LF, Richard S, Kahnoski RJ, Anema J, Tuveson DA, Perez-Mancera PA, Mustonen V, Fischer A, Adams DJ, Rust A, Chan-on W, Subimerb C, Dykema K, Furge K, Campbell PJ, Teh BT, Stratton MR, Futreal PA. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- Villaronga MA, Lopez-Mateo I, Markert L, Espinosa E, Fresno Vara JA, Belandia B. Identification and characterization of novel potentially oncogenic mutations in the human BAF57 gene in a breast cancer patient. Breast Cancer Res Treat. 2011;128:891–898. doi: 10.1007/s10549-011-1492-4. [DOI] [PubMed] [Google Scholar]
- von Figura G, Fukuda A, Roy N, Liku ME, Morris Iv JP, Kim GE, Russ HA, Firpo MA, Mulvihill SJ, Dawson DW, Ferrer J, Mueller WF, Busch A, Hertel KJ, Hebrok M. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nature Cell Biology. 2014;16:255–267. doi: 10.1038/ncb2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann T, Schneider R. Targeting histone modifications--epigenetics in cancer. Curr Opin Cell Biol. 2013;25:184–189. doi: 10.1016/j.ceb.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Wang JR, Gramling SJ, Goldstein DP, Cheng D, Chen D, Azad AK, Tse A, Hon H, Chen Z, Mirshams M, Simpson C, Huang SH, Marquez S, O'Sullivan B, Liu FF, Roberts H, Xu W, Brown DH, Gilbert RW, Gullane PJ, Irish JC, Reisman DN, Liu G. Association of two BRM promoter polymorphisms with head and neck squamous cell carcinoma risk. Carcinogenesis. 2013;34:1012–1017. doi: 10.1093/carcin/bgt008. [DOI] [PubMed] [Google Scholar]
- Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, Chan TL, Kan Z, Chan AS, Tsui WY, Lee SP, Ho SL, Chan AK, Cheng GH, Roberts PC, Rejto PA, Gibson NW, Pocalyko DJ, Mao M, Xu J, Leung SY. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S, Chan KH, Chan AS, Tsui WY, Ho SL, Chan AK, Man JL, Foglizzo V, Ng MK, Chan AS, Ching YP, Cheng GH, Xie T, Fernandez J, Li VS, Clevers H, Rejto PA, Mao M, Leung SY. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014a;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, Workman JL, Crabtree GR. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. Embo J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wang X, Haswell JR, Roberts CW. Molecular pathways: SWI/SNF (BAF) complexes are frequently mutated in cancer--mechanisms and potential therapeutic insights. Clin Cancer Res. 2014b;20:21–27. doi: 10.1158/1078-0432.CCR-13-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sansam CG, Thom CS, Metzger D, Evans JA, Nguyen PT, Roberts CW. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res. 2009;69:8094–8101. doi: 10.1158/0008-5472.CAN-09-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZJ, Churchman M, Campbell IG, Xu WH, Yan ZY, McCluggage WG, Foulkes WD, Tomlinson IP. Allele loss and mutation screen at the peutz-jeghers (LKB1) locus (19p13.3) in sporadic ovarian tumours. Br J Cancer. 1999;80:70–72. doi: 10.1038/sj.bjc.6690323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Weissman BE. Genetics and genomics of malignant rhabdoid tumors. Chichester: John Wiley & Sons Ltd; 2014. [Google Scholar]
- Weissman B, Knudsen KE. Hijacking the chromatin remodeling machinery: Impact of SWI/SNF perturbations in cancer. Cancer Res. 2009;69:8223–8230. doi: 10.1158/0008-5472.CAN-09-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weniger MA, Pulford K, Gesk S, Ehrlich S, Banham AH, Lyne L, Martin-Subero JI, Siebert R, Dyer MJ, Moller P, Barth TF. Gains of the proto-oncogene BCL11A and nuclear accumulation of BCL11A(XL) protein are frequent in primary mediastinal B-cell lymphoma. Leukemia. 2006;20:1880–1882. doi: 10.1038/sj.leu.2404324. [DOI] [PubMed] [Google Scholar]
- Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, Heravi-Moussavi A, Giuliany R, Chow C, Fee J, Zayed A, Prentice L, Melnyk N, Turashvili G, Delaney AD, Madore J, Yip S, McPherson AW, Ha G, Bell L, Fereday S, Tam A, Galletta L, Tonin PN, Provencher D, Miller D, Jones SJ, Moore RA, Morin GB, Oloumi A, Boyd N, Aparicio SA, Shih I, Mes-Masson AM, Bowtell DD, Hirst M, Gilks B, Marra MA, Huntsman DG. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, Koellhoffer EC, Pomeroy SL, Orkin SH, Roberts CW. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski L, Carrot-Zhang J, Albrecht S, Fahiminiya S, Hamel N, Tomiak E, Grynspan D, Saloustros E, Nadaf J, Rivera B, Gilpin C, Castellsague E, Silva-Smith R, Plourde F, Wu M, Saskin A, Arseneault M, Karabakhtsian RG, Reilly EA, Ueland FR, Margiolaki A, Pavlakis K, Castellino SM, Lamovec J, Mackay HJ, Roth LM, Ulbright TM, Bender TA, Georgoulias V, Longy M, Berchuck A, Tischkowitz M, Nagel I, Siebert R, Stewart CJ, Arseneau J, McCluggage WG, Clarke BA, Riazalhosseini Y, Hasselblatt M, Majewski J, Foulkes WD. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat Genet. 2014;46:438–443. doi: 10.1038/ng.2931. [DOI] [PubMed] [Google Scholar]
- Witkowski L, Lalonde E, Zhang J, Albrecht S, Hamel N, Cavallone L, May ST, Nicholson JC, Coleman N, Murray MJ, Tauber PF, Huntsman DG, Schonberger S, Yandell D, Hasselblatt M, Tischkowitz MD, Majewski J, Foulkes WD. Familial rhabdoid tumour 'avant la lettre'--from pathology review to exome sequencing and back again. J Pathol. 2013;231:35–43. doi: 10.1002/path.4225. [DOI] [PubMed] [Google Scholar]
- Wu WJ, Hu KS, Chen DL, Zeng ZL, Luo HY, Wang F, Wang DS, Wang ZQ, He F, Xu RH. Prognostic relevance of BRD7 expression in colorectal carcinoma. Eur J Clin Invest. 2013;43:131–140. doi: 10.1111/eci.12024. [DOI] [PubMed] [Google Scholar]
- Xia QY, Rao Q, Cheng L, Shen Q, Shi SS, Li L, Liu B, Zhang J, Wang YF, Shi QL, Wang JD, Ma HH, Lu ZF, Yu B, Zhang RS, Zhou XJ. Loss of BRM expression is a frequently observed event in poorly differentiated clear cell renal cell carcinoma. Histopathology. 2014;64:847–862. doi: 10.1111/his.12334. [DOI] [PubMed] [Google Scholar]
- Xia W, Nagase S, Montia AG, Kalachikov SM, Keniry M, Su T, Memeo L, Hibshoosh H, Parsons R. BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res. 2008;68:1667–1674. doi: 10.1158/0008-5472.CAN-07-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamichi N, Inada K, Ichinose M, Yamamichi-Nishina M, Mizutani T, Watanabe H, Shiogama K, Fujishiro M, Okazaki T, Yahagi N, Haraguchi T, Fujita S, Tsutsumi Y, Omata M, Iba H. Frequent loss of brm expression in gastric cancer correlates with histologic features and differentiation state. Cancer Res. 2007;67:10727–10735. doi: 10.1158/0008-5472.CAN-07-2601. [DOI] [PubMed] [Google Scholar]
- Yaniv M. Chromatin remodeling: From transcription to cancer. Cancer Genet. 2014 doi: 10.1016/j.cancergen.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Yin CC, Lin KI, Ketterling RP, Knudson RA, Medeiros LJ, Barron LL, Huh YO, Luthra R, Keating MJ, Abruzzo LV. Chronic lymphocytic leukemia with t(2;14)(p16;q32) involves the BCL11A and IgH genes and is associated with atypical morphologic features and unmutated IgVH genes. Am J Clin Pathol. 2009;131:663–670. doi: 10.1309/AJCPXLY46UPFLISC. [DOI] [PubMed] [Google Scholar]
- Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, Lim KH, Ong CK, Huang D, Chin SY, Tan IB, Ng CC, Yu W, Wu Y, Lee M, Wu J, Poh D, Wan WK, Rha SY, So J, Salto-Tellez M, Yeoh KG, Wong WK, Zhu YJ, Futreal PA, Pang B, Ruan Y, Hillmer AM, Bertrand D, Nagarajan N, Rozen S, Teh BT, Tan P. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- Zhang J, Grubor V, Love CL, Banerjee A, Richards KL, Mieczkowski PA, Dunphy C, Choi W, Au WY, Srivastava G, Lugar PL, Rizzieri DA, Lagoo AS, Bernal-Mizrachi L, Mann KP, Flowers C, Naresh K, Evens A, Gordon LI, Czader M, Gill JI, Hsi ED, Liu Q, Fan A, Walsh K, Jima D, Smith LL, Johnson AJ, Byrd JC, Luftig MA, Ni T, Zhu J, Chadburn A, Levy S, Dunson D, Dave SS. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jima D, Moffitt AB, Liu Q, Czader M, Hsi ED, Fedoriw Y, Dunphy CH, Richards KL, Gill JI, Sun Z, Love C, Scotland P, Lock E, Levy S, Hsu DS, Dunson D, Dave SS. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood. 2014;123:2988–2996. doi: 10.1182/blood-2013-07-517177. [DOI] [PMC free article] [PubMed] [Google Scholar]