Abstract
Plasmids are transmissible, extrachromosomal genetic elements that are often responsible for environmental or host-specific adaptations. In order to identify the forces driving the evolution of these important molecules, we determined the complete nucleotide sequence of the five-plasmid complement of the radish and Arabidopsis pathogen Pseudomonas syringae pv. maculicola ES4326 and conducted an intraspecific comparative genomic analysis. To date, this is the most complex fully sequenced plasmid complement of any gram-negative bacterium. The plasmid complement comprises two pPT23A-like replicons, pPMA4326A (46,697 bp) and pPMA4326B (40,110 bp); a pPS10-like replicon, pPMA4326C (8,244 bp); and two atypical, replicase-deficient replicons, pPMA4326D (4,833 bp) and pPMA4326E (4,217 bp). A complete type IV secretion system is found on pPMA4326A, while the type III secreted effector hopPmaA is present on pPMA4326B. The region around hopPmaA includes a shorter hopPmaA homolog, insertion sequence (IS) elements, and a three-element cassette composed of a resolvase, an integrase, and an exeA gene that is also present in several human pathogens. We have also identified a novel genetic element (E622) that is present on all but the smallest plasmid (pPMA4326E) that has features of an IS element but lacks an identifiable transposase. This element is associated with virulence-related genes found in a wide range of P. syringae strains. Comparative genomic analyses of these and other P. syringae plasmids suggest a role for recombination and integrative elements in driving plasmid evolution.
Plasmids are portable genetic arsenals that facilitate the adaptation of their bacterial hosts to new environmental pressures. These independently evolving accessory molecules house a wealth of genes that function in resistance, metabolism, symbiosis, and plasmid maintenance (43). Plasmid lability, autonomy, and interspecific transmissibility contribute substantially to the dissemination of virulence and resistance genes and to the emergence and reemergence of infectious bacterial diseases.
Considerable advances have been made in understanding the mechanisms of plasmid maintenance and transmission and their roles in the dissemination of important genes. Surprisingly, though, we know almost nothing about the processes and pressures that drive the evolution of individual plasmids or the whole plasmid complement. An understanding of evolutionary dynamics and processes that drive plasmid genesis, accretion, degeneration, and transmission requires whole-genome, intraspecific, comparative sequence data.
One of the most promising systems for the study of plasmid evolution is the plant pathogen Pseudomonas syringae. P. syringae is a widespread plant-associated γ-proteobacterium that is perhaps the most intensively studied bacterial model system in plant pathology (33). P. syringae is a highly tractable model organism that is amenable to genetic manipulation and both qualitative and quantitative multihost assays. Additionally, valuable genomic resources that include one completed and two partially completed whole-genome sequences are available for this species. The recently completed genome of P. syringae pv. tomato DC3000 contributed two plasmids, of 67.5 and 73.7 kb (11), bringing the total number of fully sequenced P. syringae plasmids to four (11, 64, 77). All are members of the pPT23A-like replicon family (71).
P. syringae plasmids appear capable of providing considerable advantages to their phytopathogenic hosts. For example, plasmids commonly encode resistance to copper and streptomycin, two bactericidal agents that are used extensively for disease control (74, 75). UV light resistance is also frequently plasmid encoded and may facilitate epiphytic bacterial growth (76). More importantly, many P. syringae plasmids have been directly implicated in disease. Numerous plasmid-encoded virulence genes have been identified, including the gene cluster that directs the biosynthesis of the phytotoxin coronatine. Coronatine enables P. syringae to grow to higher densities in planta and move systemically (7, 8, 14, 50, 52, 81). P. syringae plasmids can also encode secreted virulence proteins referred to as type III effectors (2, 11, 13, 30, 39, 45, 64). These proteins are channeled directly into eukaryotic hosts via a specialized type III protein secretion system encoded by a chromosomally localized hrp/hrc gene cluster (37, 41, 84). Frequently, these host-determining type III effectors are found in clusters on plasmids along with insertion sequence (IS) elements and transposon remnants, giving plasmids the capacity to define bacterial host specificity and range (3, 39).
We have sequenced the complete five-plasmid complement from the highly virulent radish and Arabidopsis thaliana pathogen P. syringae pv. maculicola ES4326. This is the largest and most complex fully sequenced plasmid complement of any gram-negative bacterium thus far. This study provides a rare glimpse into the forces that drive plasmid evolution and provides a means of identifying the specific determinants that may promote plasmid compatibility.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains were grown overnight with continuous shaking in 5 ml of King's medium B at 30°C for Pseudomonas or in 5 ml of Luria-Bertani medium at 37°C for Escherichia coli. When necessary, antibiotics were added to the culture medium at the following concentrations: streptomycin, 100 μg/ml; kanamycin, 50 μg/ml; and ampicillin, 100 μg/ml.
Plasmid DNA extraction and isolation.
Plasmid DNAs were extracted from 1.5-ml samples of overnight cultures by either a boiling lysis (65) or a modified TENS (85) protocol. Large-scale plasmid extractions were performed on 500-ml cultures in King's medium B by use of a modified alkaline lysis Midi-prep kit (Qiagen, Valencia, Calif.). Whole plasmids of <10 kb were isolated with a QIAquick PCR purification kit (Qiagen) from 1× Tris-borate-EDTA (TBE)-0.7% agarose gels run at 6 V/cm for 2 h. Plasmids of >10 kb were separated by pulsed-field gel electrophoresis in a 0.5× TBE-1% agarose gel run at 200 V for 15 h with a 70-s pulse time, followed by 11 h with a 120-s pulse time. Plasmids were then isolated from the agarose gel by the use of Nanosep MF and Nanosep 30K columns (Pall Corp., East Hills, N.Y.) according to the manufacturer's instructions.
Shotgun cloning, DNA sequencing, and assembly.
For plasmids up to 10 kb, partial digestion was achieved with a DNase Shotgun cleavage kit (Novagen, Madison, Wis.). Up to 2 μg of DNA was partially digested with a 1/350 dilution of DNase I for 1 min. The ends were blunted by the use of T4 DNA polymerase and were dephosphorylated by the use of calf intestinal phosphatase (New England Biolabs, Beverly, Mass.). For plasmids larger than 10 kb, the DNA was partially digested with Bsh1236I and the fragment ends were dephosphorylated by the use of shrimp alkaline phosphatase (MBI Fermentas, Burlington, Ontario, Canada). Fragments were purified by the use of a QIAQuick PCR purification kit and were shotgun cloned into the pCR 4Blunt-TOPO vector (Invitrogen Corp., Carlsbad, Calif.) according to the manufacturer's instructions. The resulting ligation mixtures were transformed into chemically competent E. coli DH5α cells.
Individual colonies were picked and dipped into a standard 25-μl PCR mixture containing 0.1 μM M13F (5′-GTAAAACGACGGCCAGT-3′) and 0.1 μM M13R (5′-CAGGAAACAGCTATGACCATG-3′). The cycling conditions were 1 cycle of 94°C for 5 min; 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2.5 min; and 1 cycle of 72°C for 5 min. Fragments larger than 600 bp were cleaned enzymatically with 0.2 μl of calf intestinal phosphatase and 0.2 μl of exonuclease (New England Biolabs) and then incubated at 37°C for 30 min followed by 85°C for 15 min.
Sequencing was performed with a CEQ dye terminator cycle sequencing quick start kit (Beckman Coulter Canada, Inc., Mississauga, Ontario, Canada) on a Beckman CEQ2000 XL sequencer. A single 10-μl cycle sequencing reaction contained 2.5 μl of PCR product, 2.5 μl of Beckman dye terminator cycle sequencing mix, 1.5 μl of 1× PCR buffer with MgCl2, 3 μl of water, and either 0.5 μl (0.5 μM) of modified T3 primer (5′-GCCAAGCTCAGAATTAACCCTCACTAAAGG-3′) or 0.5 μl (0.5 μM) of modified T7 primer (5′-CGACGGCCAGTGAATTGTAATACGACTC-3′). The cycling conditions were 96°C for 20 s, 55°C for 20 s, and 60°C for 4 min, repeated 50 times. After ethanol precipitation, the products were resuspended in Beckman sample loading solution. Chromatogram sequence data were viewed and edited with BioEdit (v. 5.0.9) and were subsequently imported into the contig assembly program Sequencher (Gene Codes Corp., Ann Arbor, Mich.). Contigs were connected by primer amplification from contig ends. A minimum of threefold coverage was obtained for all plasmids, with an average coverage of sixfold.
Bioinformatic and phylogenetic analyses.
Annotation was achieved with open reading frame (ORF) prediction algorithms Glimmer2 (15), Genemark (9), and FrameD (67) and was cross-referenced with BLAST outputs (http://www.ncbi.nlm.nih.gov/BLAST/). Conserved domains and motifs were identified with the protein analysis tools of Pfam, Interpro, and Swissprot, available through the Expasy web site (http://ca.expasy.org/), and of the Conserved Domain Database at the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov/). IS elements and their borders were identified with the IS Finder database (http://www-is.biotoul.fr/is.html).
Amino acid alignments used for phylogenetic analyses were made with ClustalX, v. 1.83 (80), using default parameters. Neighbor-joining trees were made with PHYLIP, v. 3.6a (18), using the SEQBOOT, PROTDIST, NEIGHBOR, and CONSENSE algorithms. Protein distances were calculated based on the Jones-Taylor-Thornton protein weight matrix with 1,000 bootstrap replicates. All other variables were set to their defaults. Trees were visualized with Treeview, v. 1.6.6 (56).
Nucleotide sequence accession numbers.
Plasmid sequences were deposited under GenBank accession numbers AY603979 to AY603983.
RESULTS AND DISCUSSION
We present here the complete nucleotide sequence of the five-plasmid complement of the highly virulent radish and Arabidopsis pathogen P. syringae pv. maculicola ES4326. P. syringae pv. maculicola ES4326 carries five plasmids, named sequentially pPMA4326A to pPMA4326E, with sizes of 46,697, 40,110, 8,244, 4,833, and 4,217 bp, respectively (Fig. 1). There are three replicon types belonging to three distinct homology groups. pPMA4326A and pPMA4326B belong to the established pPT23A-like family of replicons (71), pPMA4326C belongs to the pPS10-like replicons (54), and pPMA4326D and pPMA4326E are both representatives of an atypical, replicase-deficient replicon. While the selective advantage conferred by pPMA4326C, pPMA4326D, and pPMA4326E is unknown, pPMA4326A and pPMA4326B possess loci with known functions and may provide a selective advantage to their bacterial host under certain environmental conditions.
FIG. 1.
(A to E) Genetic maps of pPMA4326A, pPMA4326B, pPMA4326C, pPMA4326D, and pPMA4326E. ORFs and features are color coded according to their putative functions. IS element borders are shown as black vertical lines and remnant IS element borders are indicated with gray vertical lines. Hrp boxes are shown as open squares, and inverted repeats with a stem of ≥8 bp are shown as open circles. (F) Genetic redundancy within the plasmid complement. Each color indicates homologous genes or regions among the five plasmids. These colors do not correspond to the functional categories used for panels A to E. pPMA4326A, pPMA4326B, pPMA4326C, pPMA4326D, and pPMA4326E are shown as the outermost to the innermost circles, respectively. Plasmids are not drawn to scale.
pPMA4326A and pPMA4326B. (i) Maintenance, stability, and transmission.
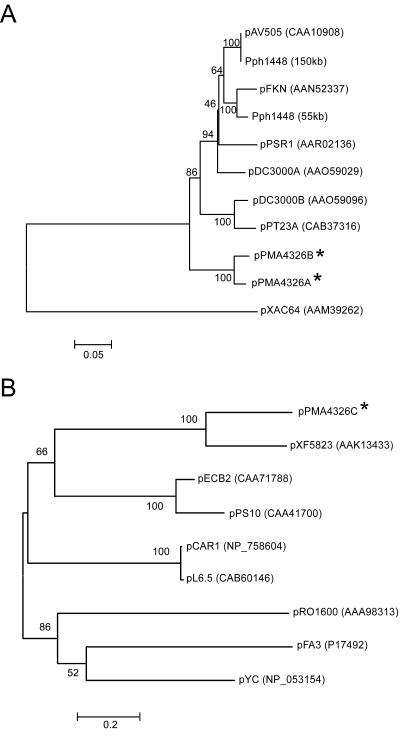
The predicted ORFs and features of pPMA4326A and pPMA4326B are shown in Tables 1 and 2, respectively. ORFs A01 and B01 of pPMA4326A and pPMA4326B, respectively, encode putative replication proteins of the pPT23A-like family (repA). This family includes the replication proteins of P. syringae plasmids pDC3000A and pDC3000B, pFKN, pAV505, and pPSR1, which are carried by P. syringae pv. tomato DC3000, P. syringae pv. maculicola M6, P. syringae pv. phaseolicola HRI1302A, and P. syringae pv. syringae A2, respectively (11, 25, 64, 77). This replicon type is broadly distributed within the P. syringae complex and has been shown to be closely related to the ColE2 replicons of E. coli (25). The repA proteins of pPMA4326A and pPMA4326B share 96% amino acid (aa) identity, with the majority of variation being present at the C terminus. A phylogenetic analysis of the protein sequences places the pPMA4326A and pPMA4326B replication proteins in their own clade (Fig. 2A).
TABLE 1.
Predicted ORFs and features of pPMA4326A
| ORF or featurea | Position | Size (aa)b | %GC | Homology (BLAST)c | Accession no. | Score/E value; aa identity (no. with identity/total [%]) |
|---|---|---|---|---|---|---|
| A01 | 1-1311 | 437 | 60 | Replication protein (repA); PtoDC3000 (pDC3000A) | AAO59029 | 775/0.0; 388/437 (88) |
| A02 | 1514-1801 | 96 | 53 | ssDNA-binding protein (ssb); PtoDC3000 | AAO54791 | 165/5e−41; 86/93 (92) |
| A03 | 1946-2251 | 102 | 49 | Acetyltransferase; P. fluorescens | ZP_00086296 | 88.6/8e−18; 46/98 (46) |
| MFA01 | 2630-3091 | 462 bp | 56 | rulB; PtoDC3000 (pDC3000A) | AE016855 | 288/7e−78; 145/152 (95) |
| A04 | 3897-3427 | 157 | 50 | Hypothetical | ||
| MFA02 | 4209-4827 | 619 bp | 49 | Element 622 (IS-like element, no transposase) | ||
| A05 | 4895-5314 | 140 | 60 | Hypothetical | ||
| A06 | 5804-6127 | 108 | 50 | Conserved hypothetical; Photorhabdus luminescens | NP_931526 | 72/8e−13; 38/61 (62) |
| A07 | 6160-6585 | 142 | 60 | Conserved hypothetical; E. coli O157:H7 | BAB35465 | 77/2e−14; 42/134 (31) |
| A08 | 7102-7464 | 121 | 47 | Hypothetical | ||
| A09 | 7637-7945 | 103 | 55 | IS222 transposase orfA; Pseudomonas sp. strain ND6 | AAP44234 | 178/5e−45; 94/110 (85) |
| A10 | 8128-8799 | 224 | 55 | IS222 transposase orfB; PtoDC3000 | AAO56533 | 420/1e−116; 198/223 (88) |
| A11 | 9541-9338 | 68 | 53 | Hypothetical | ||
| A12 | 10649-9618 | 344 | 51 | Conserved hypothetical; PtoDC3000 (intergenic space) | AE016856 | 707/0.0; 47/197 (23) |
| A13 | 11647-10727 | 307 | 50 | Conserved hypothetical; PtoDC3000 | AAO53752 | 512/1e−162; 256/292 (87) |
| A14 | 12436-11795 | 214 | 58 | Partitioning/stability (stbA); PtoPT23 (pPT23B) | AAB81646 | 362/2e−99; 188/205 (91) |
| A15 | 12794-13447 | 218 | 49 | Partitioning/stability (yafB/parA); E. coli (R721) | BAB12593 | 239/8c−63; 129/217 (59) |
| A16 | 13469-13807 | 113 | 47 | Partitioning/stability (yafA); E. coli (R721) | BAB12592 | 65/9e−11; 38/87 (43) |
| A17 | 14012-14989 | 326 | 58 | IS1406 transposase; PtoDC3000 | AAO58162 | 639/0.0; 320/325 (98) |
| A18 | 15724-15338 | 129 | 46 | Hypothetical | ||
| A19 | 16436-15966 | 157 | 38 | Pyocin S3 immunity (bip); P. aeruginosa B56394 | AAR02169 | 304/1c−82; 151/156 (96) |
| A20 | 16874-16524 | 117 | 51 | Transcriptional regulator (pbsX); PtoDC3000 (pDC3000B) | AAO59134 | 78.2/1e−14; 40/109 (36) |
| A21 | 17139-17666 | 176 | 42 | Transcriptional regulator; PsyA2 (pPSR1) | AAR02171 | 326/4e−89; 170/175 (97) |
| A22 | 17732-17947 | 72 | 42 | Conserved hypothetical; PtoDC3000 (pDC3000B) | AAO59136 | 44.7/1e−04; 21/52 (40) |
| A23 | 17976-18674 | 233 | 60 | Conjugal transfer (virB1); PsyA2 (pPSR1) | AAR02172 | 421/1e−117; 212/235 (90) |
| A24 | 18671-19009 | 113 | 62 | Conjugal transfer (virB2); PsyA2 (pPSR1) | AAR02173 | 211/8e−55; 102/112 (91) |
| A25 | 19016-19498 | 161 | 56 | Conjugal transfer (virB3); PsyA2 (pPSR1) | AAR02174 | 313/3e−85; 154/158 (97) |
| A26 | 19359-21911 | 851 | 58 | Conjugal transfer (virB4); PsyA2 (pPSR1) | AAR02175 | 1617/0.0; 795/848 (93) |
| A27 | 21908-22594 | 229 | 58 | Conjugal transfer (virB5); PsyA2 (pPSR1) | AAR02176 | 407/1e−113; 204/228 (89) |
| A28 | 22897-23130 | 78 | 46 | Hypothetical | ||
| A29 | 23141-24073 | 311 | 54 | Conjugal transfer (virB6); PsyA2 (pPSR1) | AAR02177 | 531/1e−150; 268/310 (86) |
| A30 | 24141-24455 | 104 | 66 | Conjugal transfer (virB7); PsyA2 (pPSR1) | AAR02178 | 159/9e−39; 85/105 (80) |
| A31 | 24492-25280 | 263 | 58 | Conjugal transfer (virB8); PsyA2 (pPSR1) | AAR02179 | 489/1e−138; 242/262 (92) |
| A32 | 25270-26079 | 270 | 62 | Conjugal transfer (virB9); PsyA2 (pPSR1) | AAR02180 | 500/1e−141; 245/269 (91) |
| A33 | 26204-27415 | 404 | 61 | Conjugal transfer (virB10); PsyA2 (pPSR1) | AAR02181 | 705/0.0; 355/406 (87) |
| A34 | 27425-28483 | 353 | 62 | Conjugal transfer (virB11); PsyA2 (pPSR1) | AAR02182 | 620/1e−177; 294/341 (86) |
| A35 | 28493-28858 | 122 | 60 | Conserved hypothetical; PgyPG4180 | AF346402 | 287/6e−77; 111/118 (94) |
| A36 | 29219-29485 | 89 | 54 | Hypothetical | ||
| MFA03 | 29919-30008 | 90 bp | 59 | virD4; PsyA2 (pPSR1) | AAR02183 | 50.4/3e−06; 27/30 (90) |
| A37 | 30074-31090 | 339 | 51 | Hypothetical | ||
| A38 | 31626-33299 | 558 | 59 | Conjugal transfer (virD4); PsyA2 (pPSR1) | AAR02183 | 848/0.0; 418/555 (75) |
| A39 | 33340-33702 | 121 | 62 | Killer protein (kikA); E. coli (R721) | BAB12662 | 100/1e−21; 48/97 (49) |
| A40 | 33730-35895 | 722 | 61 | Conjugal transfer (traE); PsyA2 (pPSR1) | AAR02184 | 1355/0.0; 684/721 (94) |
| A41 | 35892-36446 | 185 | 58 | Hypothetical | ||
| A42 | 36474-37034 | 187 | 61 | ssDNA-binding protein (ssb); PtoDC3000 | AAO54198 | 360/3e−99; 175/189 (92) |
| MFA04 | 37088-37285 | 198 bp | 67 | Conserved hypothetical; P. aeruginosa PA01 (phage Pf1 PA0719) | AAG04108 | 60/2e−09; 39/66 (59) |
| A43 | 37376-38095 | 240 | 47 | Partitioning/stability (stbB); PsyA2 (pPSR1) | AAR02186 | 385/1e−106; 189/238 (79) |
| A44 | 38092-38664 | 191 | 48 | Conserved hypothetical; E. amylovora (pEU30) | AAQ97965 | 82/2e−15; 52/180 (28) |
| A45 | 39102-38683 | 140 | 51 | Conserved hypothetical (yciA); E. coli (R721) | BAB12622 | 43:1/3e−04; 24/67 (35) |
| A46 | 39444-40169 | 242 | 49 | Conjugal transfer (relaxosome component); PsyA2 (pPSR1) | AAR02187 | 445/1e−124; 222/241 (92) |
| A47 | 40159-43893 | 1245 | 58 | Conjugal transfer (relaxase); PsyA2 (pPSR1) | AAR02188 | 2224/0.0; 1114/1250 (89) |
| A48 | 44073-44228 | 52 | 55 | Conserved hypothetical; PtoDC3000 | AAO58724 | 79.3/5e−15; 37/53 (69) |
| A49 | 44373-44801 | 143 | 62 | Transcriptional regulator (gntR); PtoDC3000 (pDC3000A) | AAO59095 | 261/2e−69; 129/142 (90) |
| A50 | 45137-46057 | 307 | 57 | Conserved hypothetical; P. aeruginosa PA14 | AAP84187 | 195/4e−49; 104/189 (55) |
MF, miscellaneous feature.
Unless otherwise indicated.
Abbreviations: Psy, P. syringae pv. syringae; Pto, P. syringae pv. tomato; Pma, P. syringae pv. maculicola.
TABLE 2.
Predicted ORFs and features of pPMA4326B
| ORF or featurea | Position | Size (aa)b | %GC | Homology (BLAST)c | Accession no. | Score/E value; aa identity (no. with identity/total [%]) |
|---|---|---|---|---|---|---|
| B01 | 1-1302 | 434 | 61 | Replication protein (repA); PtoDC3000 (pDC3000A) | AAO59029 | 771/0.0; 386/437 (88) |
| B02 | 1451-1744 | 98 | 54 | ssDNA-binding protein (ssb); PtoDC3000 | AAO54791 | 162/4e−40; 83/93 (89) |
| B03 | 1816-2016 | 67 | 57 | Conserved hypothetical; E. coli O157:H7 CP-933P | AAK16987 | 82.4/6e−16; 40/56 (71) |
| B04 | 2016-2309 | 98 | 51 | parE; E. coli O157:H7 | BAB35704 | 143/3e−34; 66/92 (71) |
| B05 | 2562-2975 | 138 | 51 | Hypothetical | ||
| MFB01 | 3181-4741 | 1,561 bp | 59 | IS801 transposase; PmaM6 (pFKN) | AAK49542 | 541/1e−153; 249/284 (87), 141/145 (97) |
| MFB02 | 5256-5094 | 163 bp | 52 | ISPsy transposase; PtoDC3000 | AAO56690 | 64.3/2e−16; 31/33 (93), 17/21 (80) |
| B06 | 5391-6809 | 473 | 59 | MFS sugar transporter; PtoDC3000 | AAO56003 | 881/0.0; 433/472 (91) |
| B07 | 8291-7062 | 410 | 61 | IS801 transposase; PmaM6 (pFKN) | AAK49542 | 828/0.0; 390/407 (95) |
| B08 | 9426-8608 | 273 | 53 | hopPmaA-like; PmaES4326 (candidate type III effector) | AAL84239 | 412/1e−114; 210/229 (91) |
| MFB03 | 9898-10196 | 299 bp | 48 | E622 (IS-like element, no transposase); PmaES4326 | ||
| B09 | 10161-10766 | 202 | 51 | Resolvase; S. enterica serovar Typhimurium LT2 (pSLT) | AAL23535 | 277/3e−74; 140/179 (78) |
| B10 | 10750-12408 | 553 | 58 | Integrase; S. enterica serovar Typhimurium LT2 (pSLT) | AAL23534 | 832/0.0; 393/555 (70) |
| B11 | 12398-13369 | 324 | 58 | exeA; Pseudomonas sp. strain TW3 | AAF23986 | 613/1e−175; 311/323 (96) |
| B12 | 15934-13610 | 775 | 55 | hopPmaA; PmaES4326 | AAL84239 | 1532/0.00; 774/774 (100) |
| MFB04 | 16247-16449 | 203 bp | 52 | E622 (IS-like element, no transposase); PmaES4326 | ||
| MFB05 | 17872-17513 | 360 bp | 56 | ISPsy12 transposase orfB; PtoDC3000 | AAO56533 | 224/3e−57; 104/120 (86) |
| B13 | 18973-18293 | 227 | 52 | Conserved hypothetical; Streptomyces violaceoruber (pSV2) | AAO50163 | 97.4/7e−20; 65/198 (32) |
| B14 | 19433-19161 | 91 | 47 | Conserved hypothetical; Listonella anguillarum (pEIB1, intergenic) | AY255699 | 159/2e−38; 64/91 (70) |
| B15 | 20162-19527 | 212 | 58 | Partitioning/stability (stbA); PtoPT23 (pPT23B) | AAB81646 | 359/9e−99; 187/204 (91) |
| B16 | 20570-21268 | 233 | 52 | Partitioning/stability (parA); Shewanella oneidensis MR-1 (megaplasmid) | AAN52997 | 157/7e−38; 89/223 (39) |
| B17 | 21261-21584 | 108 | 49 | Hypothetical | ||
| MFB06 | 22492-21711 | 782 bp | 59 | IS801 transposase; PmaM6 (pFKN) | AAK49542 | 204/5e−60; 88/97 (90), 97/104 (93) |
| MFB07 | 23698-22809 | 890 bp | 59 | Membrane protein; PtoDC3000 (pDC3000B) | AAO59103 | 338/1e−144; 181/187 (96), 95/98 (96) |
| B18 | 23898-25313 | 472 | 60 | MFS sugar transporter; PtoDC3000 (pDC3000B) | AAO59104 | 930/0.0; 470/471 (99) |
| MFB08 | 25401-25895 | 495 bp | 57 | rulB; PsyA2 (pPSR1) | AAC44639 | 295/6e−80; 145/165 (87) |
| B19 | 25979-26119 | 47 | 50 | Hypothetical | ||
| B20 | 27284-26316 | 323 | 55 | ISPssy transposase; PtoDC3000 | AAO58997 | 600/1e−171; 298/325 (91) |
| MFB09 | 27442-27634 | 193 bp | 61 | Transcriptional regulator (lysR); PsyB728a | ZP_00126456 | 70/1e−17; 32/43 (74%), 17/22 (77) |
| B21 | 28765-27641 | 375 | 56 | Conserved hypothetical; PtoDC3000 (pDC3000B) | AAO59105 | 699/0.0; 352/352 (100) |
| B22 | 29483-29010 | 158 | 47 | Conserved hypothetical; PtoDC3000 (pDC3000B) | AAO59098 | 301/8e−82; 141/157 (89) |
| B23 | 30490-29708 | 261 | 61 | Conserved hypothetical; PtoDC3000 (pDC3000B) | AAO59106 | 541/1e−153; 257/260 (98) |
| MFB10 | 30585-30673 | 89 bp | 57 | ISXc5 transposase; Xanthomonas campestris TAP44-3 (pKLH443) | CAB65711 | 53.9/3e−07; 24/29 (82) |
| B24 | 30768-31841 | 358 | 54 | ISPsy1 transposase; Pgy PG4180 (p4180A) | AAD50908 | 663/0.0; 328/357 (91) |
| MFB11 | 32002-32280 | 279 bp | 55 | ISPs1 transposase; PtoDC3000 | AAO57780 | 176/2e−44; 84/93 (90) |
| B25 | 32371-33348 | 326 | 55 | ISPssy transposase; PtoDC3000 | AAO58997 | 611/1e−174; 301/325 (92) |
| B26 | 33565-37038 | 1,157 | 59 | Relaxase/mobilization protein; PsyA2 (pPSR1) | AAR02188 | 1946/0.0; 973/1163 (83) |
| B27 | 37215-37415 | 67 | 52 | Conserved hypothetical; PtoDC3000 | AAO58724 | 68.6/1e−11; 31/41 (75) |
| MFB12 | 37480-37617 | 138 bp | 38 | Relaxase/mobilization protein; PsyA2 (pPSR1) | AAM64140 | 72.4/7e−13; 30/46 (65) |
| B28 | 37785-38213 | 143 | 62 | Transcriptional regulator (gntR); PtoDC3000 (pDC3000A) | AAO59095 | 261/5e−70; 129/142 (90) |
| B29 | 38549-39469 | 307 | 57 | Conserved hypothetical; P. aeruginosa PA14 | AAP84187 | 195/4e−49; 104/189 (55), 58/150 (38) |
MF, miscellaneous feature.
Unless otherwise stated.
Abbreviations: Psy, P. syringae pv. syringae; Pto, P. syringae pv. tomato; Pma, P. syringae pv. maculicola.
FIG. 2.
(A) Gene genealogy of P. syringae pPT23A-like replication proteins, rooted with RepA of pXAC64 (Xanthomonas axonopodis pv. citri 306). Replication proteins from the 150- and 55-kb plasmids of P. syringae pv. phaseolicola 1448 were obtained from the partially completed genome sequence available at The Institute for Genomic Research web site (http://www.tigr.org). (B) Midpoint-rooted gene genealogy of pPS10-like replication proteins. Protein-based genealogies were generated with PHYLIP (18) by using the neighbor-joining method with the Jones-Thornton-Taylor protein matrix and 1,000 bootstrap replicates. Replication proteins from P. syringae pv. maculicola ES4326 are indicated with an asterisk. The plasmids shown are from the following organisms: pRO1600, P. aeruginosa; pYC, Yersinia pestis; pFA3, Neisseria gonorrhoeae; pCAR1, Pseudomonas resinovorans, pL6.5, Pseudomonas fluorescens; pXF5823, X. fastidiosa; pPS10, P. syringae pv. savastanoi; and pECB2, Pseudomonas alcaligenes.
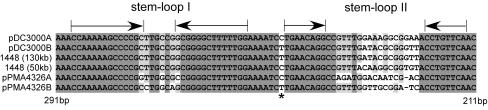
The putative replication origin, which has been shown to lie upstream of repA, is characterized by the presence of a highly conserved stem-loop structure (stem-loop I) (25, 77) (Fig. 3). Adjacent to stem-loop I is a second putative stem-loop structure (stem-loop II) that was identified through a comparative analysis of coresident plasmids pPMA4326A and pPMA4326B, pDC3000A and pDC3000B, and the 130- and 55-kb plasmids of P. syringae pv. phaseolicola 1448A. Despite an overall high nucleotide conservation of the origin among pPT23A-like plasmids, the region comprising the loop portion of stem-loop II is hypervariable and different for coresident plasmids. In addition to this loop sequence, a single nucleotide position flanking stem-loop II is also different in coresident plasmids. These two features may function as a plasmid-specific recognition site for the replication protein (discussed below).
FIG. 3.
Alignment of a possible incompatibility region within the putative replication origin of coresident pPT23A-like plasmids between bases 291 and 211 upstream of the replicase gene. Arrows indicate inverted repeats that may form stem-loop structures. The putative loop sequence of stem-loop II is variable between coresident (compatible) plasmids. A single nucleotide position, indicated by an asterisk, varies between coresident plasmids.
Immediately downstream of repA on both plasmids is a single-stranded DNA-binding protein (encoded by ssb) which is absent from other fully sequenced P. syringae plasmids (11, 64, 77). Some DNA-binding proteins have been implicated in the initiation of plasmid replication, and in some cases, in plasmid copy control (34, 58).
Active plasmid partitioning commonly requires the activity of at least two proteins, typically ParA and ParB, which are responsible for the equal allocation of plasmids during cell division (10). Multimer resolution systems, also encoded by some plasmids, facilitate stability by increasing the number of independently segregating plasmids for partitioning (87). pPMA4326A and pPMA4326B each carry three putative partitioning determinants, including a putative parA gene (ORFs A15 and B16, respectively) and a resolvase gene (ORFs A14 and B15, respectively). The latter has homology to stability determinant stbA from plasmid pPT23A of P. syringae pv. tomato PT23. A third ORF (ORFs A16 and B17) flanks the putative parA genes on both plasmids. On pPMA4326B, this ORF overlaps parA, suggesting possible involvement in the partitioning process. Of the three putative partitioning genes, only the resolvase genes are homologous, perhaps implying that pPMA4326A and pPMA4326B did not originate via duplication (discussed below).
Plasmid stability is frequently promoted by postsegregational killing systems. Plasmid pPMA4326B appears to possess a parDE-like operon, which contributes to plasmid maintenance and stability in the broad-host-range plasmid RK2 (42). B04 encodes a member of the ParE family of plasmid stabilization proteins, identified by a match to PFAM entry pfam05016 (E = 6 × 10−12). The reading frame of parE overlaps that of B03 by a single nucleotide; however, B03 shows no homology to parD but is similar to genes for several conserved hypothetical proteins found in E. coli. pPMA4326A does not appear to have a stability system, although ORF A39 is similar to kikA, a gene whose expression is lethal in Klebsiella species (31, 35). There is no obvious plasmid-encoded antidote protein, suggesting that either the antidote is chromosomally encoded, the kikA homolog is not functional, or it has another role on this plasmid.
Both pPMA4326A and pPMA4326B appear to be transmissible. pPMA4326B possesses a mobilization-like protein (Mob) that shares 92% aa identity with the putative relaxase of pPMA4326A (A47). pPMA4326A, on the other hand, may be self-transmissible, as it possesses a complete suite of genes that are necessary for the synthesis of a conjugative pilus and the subsequent transfer of plasmid DNA. ORFs A23 to A48 form the transfer (tra) region, which resembles most closely the tra gene clusters of pPSR1 (P. syringae pv. syringae A2) and pF3028 (Haemophilus influenzae). Due to nonuniformity and inconsistencies in tra gene nomenclature, putative tra genes will be referred to by virB homolog designations. ORFs A23 to A34 are virB1 to virB11 homologs which are involved in structural formation of the conjugative apparatus. The virB1-to-virB11 cluster is adjacent to a second cluster of genes that are involved in DNA processing and transfer (ORFs A38 to A48). These two clusters are typically continuous; however, on pPMA4326A, they are separated by ORFs A35, A36, and A37, encoding three hypothetical proteins which have no known function. ORF A38 is homologous to the gene for VirD4, a protein involved in coupling and transfer of the nucleoprotein complex through the conjugative pilus (51). ORFs A40, A42, and A43 are homologous to genes previously identified in the tra gene clusters of pPSR1 and pF3028 and may play a role in the DNA transfer process. A46 and A47 are predicted to be involved in the formation of the relaxosome structure, a multiprotein complex that performs a site-specific nick at the transfer origin (oriT), replicates the plasmid, and aids in the transfer of the DNA strand to the recipient bacterium (28). To identify candidate oriT sequences, we aligned the regions upstream of the putative relaxosome components from pPMA4326A, pPSR1, and the E. coli plasmid R721. This highlighted several conserved regions between these plasmids, including the sequence 5′-ACAGGCACG-3′, whose two guanine residues serve as the nick site on plasmid R721 (48). This sequence, which may form part of the oriT, is located 159 bp upstream of the putative relaxosome component (A46), near an inverted repeat.
The pPMA4326A tra cluster, in particular virB1, virB4, and the relaxase, provides an unusual perspective on the evolution of plasmids and new genes. pFKN from P. syringae pv. maculicola M6 was purported to lack a transfer or mobilization mechanism; however, ORF 28 of this plasmid (AAK49560) has a domain with a similarity to mobilization/nuclease proteins (64). The gene appears to be composed of partial virB1 and virB4 genes that have been fused to a mobilization/nuclease domain of the relaxase gene. Rohmer et al. (64) suggested that ORF 28 is a remnant of a tra gene cluster. In fact, of the entire 2,707 bp of ORF 28, the first 560 bp share 91% nucleotide identity with a portion of the pPMA4326A virB1 gene, the central 1,013-bp portion shares 88% nucleotide identity with a portion of virB4, and the last 1,037 bp share 84% nucleotide identity with a portion of the putative relaxase gene (Fig. 4). This strongly suggests that pFKN may have once possessed a tra gene cluster homologous to that of pPMA4326A and that two deletion events may have created this fused gene. Given that pFKN is capable of integrase-mediated integration and excision from the P. syringae pv. maculicola M6 genome, these deletions may have occurred during one of these events. ORF 28 is expressed under standard culture conditions (64), and as the nuclease/mobilization domain is still conserved, the protein may still be capable of providing mobilization functions.
FIG. 4.
Possible origin of the mobilization-like gene ORF 28 (pFKN, from P. syringae pv. maculicola M6 [accession no. AAK49560]). Nucleotide identities between different portions of ORF 28 and the pPMA4326A tra cluster are shown beneath each segment.
(ii) Virulence-associated genes and mobile elements.
pPT23A-like plasmids are of particular agricultural importance, as they commonly encode virulence and virulence-associated genes such as type III effectors (2, 13, 39, 45, 70, 82). Plasmid pPMA4326A does not appear to encode any identifiable virulence genes, but it does possess several putative transposases. ORFs A09 and A10 appear to constitute an IS3-like element that is most closely related to IS222 from the D3 bacteriophage of Pseudomonas aeruginosa (49), and ORF A17 encodes an IS1406 transposase of the IS5 family. The putative borders of both IS elements encompass only their respective transposases.
A notable feature on pPMA4326A is a 622-bp putatively noncoding region (MFA02) that has features of an IS element and is found on all P. syringae pv. maculicola ES4326 plasmids except for pPMA4326E. We have designated this element E622. E622 has a GC content of 49% and possesses 60-bp inverted repeats on each end that are 97% identical. Within each of these inverted repeats are two perfect tandem repeats of 12 bp (Fig. 5A). The element lacks an identifiable transposase, but the region between nucleotides 148 and 318 has a weak similarity (BLAST expect value [E] > 1) to Tn3-like transposases. E622 is often found near putative virulence or virulence-associated genes. For example, the 622-bp sequence, with large central portions deleted, is situated adjacent to the type III effector hopPsyH on pPSR1 of P. syringae pv. syringae A2 (77). Small fragments of E622 are present in two regions near virulence-associated genes on pPMA4326B and in a noncoding region flanking the pathogenicity island of plasmid pAV511 of P. syringae pv. phaseolicola 1449B (39). E622 is also found on the P. syringae pv. tomato DC3000 plasmid pDC3000B. On the genome of P. syringae pv. tomato DC3000, this element flanks the virulence gene holPtoQ as well as virulence-associated resolvase, transposase, and coronafacic acid synthetase genes (11). In several instances, E622 constitutes part of the ORFs of nearby genes. It makes up the last 60 bp of a gene for a conserved domain protein of P. syringae pv. tomato DC3000 (accession no. AAO59108) and was predicted by gene-finding algorithms to make up the first 48 bp of the predicted mob gene (D03) of pPMA4326D. E622 constitutes the last 60 bp of candidate type III effector holPtoAA of P. syringae pv. tomato DC3000 (AF458394) and the first 36 bp of the virulence-associated resolvase gene (B09) on pPMA4326B. The presence of E622 within and near unrelated sequences suggests movement via illegitimate recombination; however, it does not appear to code for its own transposase. This suggests that E622 is either transactivated or is merely a remnant of an as yet unidentified mobile element.
FIG. 5.
(A) Alignment of closely related E622 homologs illustrating the level of sequence conservation. (B) Alignment of conserved terminal ends of E622 (pPMA4326B) and more divergent homologs from Pantoea stewartii subsp. stewartii (AF282857) and plasmid pCC7120Δ of Nostocales sp. (AP003604). Solid boxes, direct repeats; arrows, inverted repeats; dashed box, Hrp box motif.
A candidate Hrp box, a regulatory sequence associated with type III effectors, is found within E622. The candidate Hrp box sequence, 5′-GGAATTGACTGCGAGTGAGCGCACGCC-3′, contains several of the conserved residues (underlined) that characterize the Hrp box. The presence of this putative regulatory sequence within the widely distributed E622 element suggests a mechanism for the independent movement of type III regulatory elements that may ultimately contribute to the evolution of new type III effectors.
Divergent homologs of E622 have also been identified, including a 522-bp E622 homolog on pCC7120Δ of the cyanobacterium Anabaena sp. (46). This homolog has a GC content of 37% and has perfect inverted terminal repeats that show 80% nucleotide identity to E622 (Fig. 5B). An E622 homolog is also present in the noncoding region of the Pantoea stewartii subsp. stewartii hrp/hrc pathogenicity cluster (AF282857) (20). This 830-bp homolog has a GC content of 41% and possesses the characteristic terminal inverted and direct repeats (Fig. 5B). The conservation between distant homologs is particularly noticeable, suggesting the persistence of E622 through evolutionary time.
Plasmid pPMA4326B can be designated a virulence plasmid because it possesses multiple virulence-associated elements. ORFs B07 to B12, which include the type III effector hopPmaA, an exeA-like gene, an integrase gene, a resolvase gene, a shorter hopPmaA homolog, and an IS801 element, constitute a “fitness island” (61). ORF B12 is hopPmaA (AAL84239), a gene that was previously isolated and identified as a plasmid-borne virulence gene by Guttman et al. (30) in a genetic screen designed to identify type III secreted effectors in P. syringae pv. maculicola ES4326. The N-terminal region shares 60% aa identity with the putative type III effector HolPsyAE from P. syringae pv. syringae B728a (26), while the C-terminal region of HopPmaA shares 30% aa identity with hypothetical proteins from E. coli O157:H7 EDL933 (59). A Hrp box (5′-GGAACCCACGGGCCCTTGTGGTCACACATA-3′) and a ribosome binding site (RBS) with the sequence 5′-TAAGGAA-3′ have already been identified (30). HopPmaA, the only plasmid-encoded type III effector in P. syringae pv. maculicola ES4326, was recovered seven times in this screen, whereas most other effectors were only recovered once or twice. This high rate of recovery may be due to pPMA4326B being a multicopy plasmid or to the nearly optimal RBS of hopPmaA, which could contribute to a high translational efficiency. A second Hrp box, 5′-GGAACCGATCCCGATGAGTAGCCACACA-3′, is situated 922 bp upstream of and on the same strand as the hopPmaA Hrp box. This second Hrp box may be orphaned, as suggested by its proximity to a truncated IS element.
hopPmaA is adjacent to a three-gene cassette composed of overlapping resolvase (B09), integrase (B10), and exeA-like (B11) genes. The cassette can also be found on the plasmids and chromosomes of various bacteria, including the human pathogens Salmonella enterica and P. aeruginosa (Fig. 6). While there are remnants of the integrase and exeA-like genes flanking virPphA on pAV511 and on the chromosomes of P. syringae pv. tomato DC3000 and P. syringae pv. syringae B728A, this is the first instance of this full cassette in a plant pathogen. The organization of this cassette is consistent with that of the class II Tn3-like transposons, which, in addition to other accessory ORFs, encode a resolvase and a site-specific recombinase (27, 78). The integrase and resolvase genes of this cassette may be integral components for mobility, while the function of the exeA gene is unclear. ExeA proteins have been implicated in type II secretion, and more specifically, in the secretion of the virulence-related aerolysin toxin by Aeromonas hydrophila; however, this secretion is dependent on a second factor, encoded by exeB (4, 36, 40, 68), which is not present within the P. syringae pv. maculicola ES4326 cassette.
FIG. 6.
Interspecific genomic comparison of the organization of the cassette comprising overlapping resolvase, integrase, and exeA genes. Closely related cassette homologs, both intact and degenerate, are distributed predominantly in Pseudomonas and Salmonella strains. Distant homologs of the integrase-exeA gene are found linked (but not overlapping) in a broad range of bacteria. Frame shifts are shown as diagonal lines, gene fragments are shown as rectangles, and stop codons are indicated with solid black circles. GenBank accession numbers are indicated in brackets.
The flanking regions of the element were scanned for inverted repeats to define possible boundaries for the cassette. The hopPmaA, exeA, integrase, and resolvase genes are bounded by 201-bp inverted repeats (MFB03 and MFB04), which are much longer than the characteristic 50-bp repeats described for Tn3 (27, 78). These repeats share high nucleotide identities (90 and 92%, respectively) with E622. If these inverted repeats do define the borders, then this result suggests that this potentially mobile cassette introduced hopPmaA into pPMA4326B. The homologs of the resolvase-integrase-exeA cassette in other species, however, do not appear to have these inverted repeats, implying that the presence of E622 around this three-gene element in P. syringae pv. maculicola ES4326 is most likely the result of cassette integration into E622.
Flanking this cassette is an ORF for a smaller 273-aa hopPmaA homolog (B08) which shares 91% aa identity with the first 229 aa of the 775-aa HopPmaA protein. As noted for the protein encoded by full-length hopPmaA, this candidate effector shares 47% identity with the first 230 aa of the putative type III effector encoded by holPsyAE (26). Immediately upstream of the putative start codon is an RBS that is identical to that found near hopPmaA, along with a putative Hrp box with the sequence 5′-TGAACTTTTCTGGTTGGAGCACCACTCA-3′. It is unknown whether this candidate effector is functional, but its presence on this fitness island may provide insight into effector origins and their evolution.
Type III effectors have been described as being modular, and they possess an N-terminal secretion and translocation signal along with a C-terminal recognition domain (29). New effectors may evolve through the fusion of proteins to an appropriate N-terminal secretion and translocation sequence. Such an evolutionary mechanism is supported by the proteins encoded by hopPmaA and holPsyAE, which have N termini that are similar to that of the hopPmaA-like candidate effector, but whose C termini are variable. Thus, this effector candidate may eventually serve as a precursor to a new effector that is constructed during random genetic rearrangement leading to its fusion with another gene.
Adjacent to this candidate effector is a putative IS801 transposase gene (B07) that shares 95% aa identity with IS801 on the pathogenicity island of plasmid pFKN from P. syringae pv. maculicola M6 (64). The borders of this element are clearly identifiable and do not appear to encompass any additional ORFs. There are two additional IS801 transposase genes, which appear to be degenerate (MFB01 and MFB06), and although they may not be active, their border sequences are still present, yielding a total of two IS801 right border and three IS801 left border sequences on pPMA4326B. The two right border sequences differ by only two nucleotides, while the three left border sequences differ by three nucleotides. The high conservation of these borders has broad evolutionary implications, as IS801 is capable of recognizing transposon borders and initiating transposition even when the transposase is expressed in trans (63). The recognition of these borders in different combinations could result in the transposition of larger plasmid segments; however, such events would be dependent on the specificity of the IS801 transposase, which is currently unknown. Some transposases, such as that of Tn5, have been shown to have low specificities (53, 86), while others, such as that of transposon Tn10, are capable of excision irrespective of the position or orientation of their terminal repeats (12). Thus, transposase transactivation and border specificity may play an important role in plasmid evolution.
Two distinct regions on plasmid pPMA4326B point to an IS-element-mediated exchange of gene cassettes among P. syringae plasmids. A 3.3-kb region encompassing MFB09, B21, B22, B23, and MFB10 exhibits 99% nucleotide identity to a region on plasmid pDC3000B of P. syringae pv. tomato DC3000, while a 2.5-kb region that includes MFB07 and B18 exhibits 98% nucleotide identity to pDC3000B. Both segments are bounded by IS elements, suggesting either illegitimate recombination through transposition or homologous recombination facilitated by common IS element sequences. This stepwise mechanism of plasmid accretion through the acquisition of cassettes is supported by previous work (1, 32, 79).
(iii) Other genes of interest.
pPMA4326A and pPMA4326B both encode gntR-like regulators (ORFs B28 and A49, respectively) that share 100% nucleotide identity. On all fully sequenced P. syringae plasmids, a gntR-like regulator is present proximal to both the replication and putative relaxase/mobilization genes (11, 64, 77), suggesting its involvement in plasmid replication or transmission. pPMA4326A also possesses two other regulators of different families, encoded by ORFs A20 and A21.
Several genes have predicted functions that may provide a selective advantage under specific environmental conditions. ORF A03 is predicted to be a member of the gcn5-related acetyltransferase (GNAT) protein family, identified by its similarity to COG3153 (E = 9 × 10−12). The specific acetylation target of A03 is not known, although some acetyltransferases have been shown to be involved in the detoxification of aminoglycoside antibiotics (5). ORF A17 is similar to a bacteriocin immunity protein (bip) from pPSR1 and P. aeruginosa that confers pyocin S3 immunity; however, the pyocin biosynthetic gene that is normally linked to the immunity gene is absent (17). This plasmid may therefore provide a selective advantage in cases where pyocin S3 would otherwise compromise the bacterial host.
Plasmid pPMA4326B encodes two transporters (B06 and B18), both of which are putative sugar transporters of the major facilitator superfamily (MFS). MFS transporters are responsible for the transport and uptake of various compounds, including simple sugars, antibiotics, amino acids, and metabolites (57). B06 shares 91% aa identity with a chromosomally encoded MFS transporter of P. syringae pv. tomato DC3000, whereas B18 shares 100% aa identity with the MFS transporter of pDC3000B.
ORF B22 has a chromosomally carried homolog in P. syringae pv. tomato DC3000 which has been suggested to be virulence associated since it possesses a potential Hrp box (60). B22, however, has no identifiable Hrp box within 500 bp of its initiation codon. ORF B14 is homologous to a nonannotated region on pEIB1 from Listonella anguillarum (83) and possesses a candidate Hrp box just upstream of the ORF (5′-GGAATTTTCGGCATTTTAGGCGTACAACCA-3′).
(iv) Miscellaneous features (pseudogenes/gene remnants).
MFA01 of plasmid pPMA4326A appears to be a remnant of the UV light resistance protein gene rulB (76). This truncated homolog exhibits 90% nucleotide identity to the 3′ end of the 1,299-bp full-length rulB gene, from nucleotides 829 to 1294. MFB08 of pPMA4326B is also a truncated rulB gene that shows, in its entirety, 93% nucleotide identity to the 5′ end of the full 1,299-bp rulB gene, from nucleotides 331 to 826. Effectively, a single rulB gene appears to have been divided into two segments, with pPMA4326A and pPMA4326B each receiving one portion. This may have occurred during a past recombination event between the two plasmids.
pPMA4326C.
pPMA4326C is 8,244 bp, has a GC content of 52.9%, and carries 10 predicted ORFs (C01 to C10) that account for 74% of the plasmid (Table 3). Four of the ORFs appear to encode common plasmid maintenance determinants, including a replication protein, a mobilization protein, and a parDE-like operon. The replication protein encoded by ORF C01 is most closely related to the replication proteins of pXF5823 from Xylella fastidiosa LAR20 (62) and pPS10 from P. syringae pv. savastanoi PS93 (54) (Fig. 2B). A phylogenetic analysis indicates that the pPMA4326C replicase mostly clusters with Pseudomonas plasmids, although it is most closely related to pXF5823. The presence of the pXF5823 plasmid repB gene within the predominantly Pseudomonas clade points to horizontal transfer of this plasmid from the Pseudomonas group into X. fastidiosa. The replication promoter, the regulatory sequences, and their organization resemble those that were previously described for plasmid pPS10 and include a dnaA box (5′-CTATCCACA-3′), iterons made up of three direct repeats of 5′-TAACTGTGCCTTTTTTCCGGT-3′, and an AT-rich region with two direct repeats of 5′-TTTCTTGTTT-3′ (23, 24, 54). The iterons show 76% nucleotide identity to those of pXF5823.
TABLE 3.
Predicted ORFs and features of pPMA4326C
| ORF or featurea | Position | Size (aa)b | %GC | Homology (BLAST)c | Accession no. | Score/E value; aa identity (no. with identity/total [%]) |
|---|---|---|---|---|---|---|
| C01 | 174-1028 | 285 | 49 | Replication protein (repA); X. fastidiosa LAR20 (pXF5823) | AAK13433 | 340/9e−93; 166/268 (61) |
| C02 | 1098-1328 | 77 | 51 | Hypothetical | ||
| C03 | 1411-1686 | 92 | 46 | Hypothetical | ||
| C04 | 1754-2410 | 219 | 53 | Conjugal transfer (virB5); PsyA2 (pPSR1) | AAR02176 | 340/1e−91; 173/219 (78) |
| C05 | 2431-2736 | 102 | 40 | Hypothetical | ||
| C06 | 2737-3662 | 309 | 47 | Conjugal transfer (virB6); PsyA2 (pPSR1) | AAR02177 | 316/1e−84; 153/302 (50) |
| MFC01 | 3763-4384 | 622 bp | 49 | E622 (IS-like element, no transposase); PmaES4326 | ||
| C07 | 4400-6505 | 702 | 60 | Mobilization protein; Rhodobacter blasticum (pMG160) | BAC54124 | 150/3e−35; 88/202 (43) |
| C08 | 6737-6982 | 82 | 59 | Hypothetical | ||
| C09 | 7339-7538 | 67 | 56 | Conserved hypothetical; P. fluorescens | ZP_00084371 | 73.6/3e−13; 30/59 (50) |
| C10 | 7539-7817 | 93 | 59 | parE; E. coli O157:H7 | A90914 | 97.8/1e−20; 44/91 (48) |
MF, miscellaneous feature.
Unless otherwise stated.
Abbreviations: Psy, P. syringae pv. syringae; Pto, P. syringae pv. tomato; Pma, P. syringae pv. maculicola.
The stability of pPMA4326C may be mediated through a protein-based postsegregational killing system homologous to that of pPMA4326B. ORF C10, which overlaps with C09, belongs to the toxin-encoding ParE family of plasmid stabilization proteins. The transmission of this plasmid is likely facilitated by ORF C07, which is homologous to mob genes from pMG160 and pTF1 from Rhodobacter blasticus and Acidithiobacillus ferrooxidans, respectively (16, 38). Immediately downstream of mob is E622 (MFC01), which in turn is flanked by 44- and 23-bp stem-loop structures that may form an oriT.
ORFs C04 and C06 are homologous to conjugation genes virB5 and virB6 of pPSR1 and pPMA4326A. Since virB5 and virB6 alone are insufficient for conjugal transfer, they may serve an alternate function. Assuming that these genes were acquired as a single unit, this cassette is unlikely to have been acquired from either pPMA4326A or pPSR1, as the predicted hypothetical protein whose gene is between the pPMA4326C virB5 and virB6 homologs is not related to the hypothetical protein whose gene is between the virB5 and virB6 homologs of either pPMA4326A or pPSR1.
pPMA4326D and pPMA4326E.
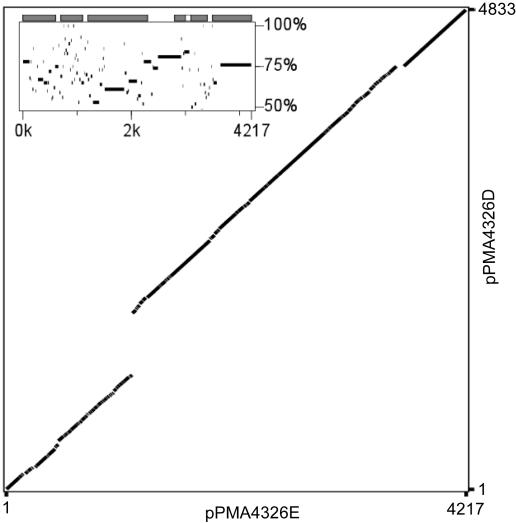
pPMA4326D and pPMA4326Eshare a high nucleotide identity and a similar genetic organization (Fig. 7). Each plasmid contains six putative ORFs (Tables 4 and 5), none of which appears to code for a replication protein. A TBLASTX search of the plasmids revealed that the same region on both plasmids shares homology with the experimentally determined minimal replicon of plasmid pH1 from Acidithiobacillus ferrooxidans TFN (44). The putative minimal replicons of pPMA4326D and pPMA4326E are denoted MFD02 and MFE01, respectively. Within the minimal replicons of pPMA4326D, pPMA4326E, and pH1 are three inverted repeats that are highly conserved across all three plasmids and may therefore have a direct role in replication. Nonetheless, putative dnaA boxes identified within the minimal replicon of pH1 by Kalyaeva et al. (44) are not conserved in either pPMA4326D or pPMA4326E, suggesting that these may not be true dnaA binding sites. Nonetheless, these plasmids are likely theta-replicons that are unrelated to replication protein-deficient ColE1 (44).
FIG. 7.
Dot plot of pPMA4326D and pPMA4326E created with Pipmaker (69). The large gap in the plot corresponds to the novel element E622 that is present on pPMA4326D, but not pPMA4326E. (Inset) Percent identity plot indicating nucleotide identity between plasmids.
TABLE 4.
Predicted ORFs and features of pPMA4326D
| ORF or featurea | Position | Size (aa)b | %GC | Homology (BLAST)c | Accession no. | Score/E value; aa identity (no. with identity/total [%]) |
|---|---|---|---|---|---|---|
| D01 | 1-183 | 61 | 61 | Hypothetical | ||
| D02 | 659-237 | 141 | 62 | Hypothetical | ||
| MFD01 | 1184-1805 | 622 bp | 49 | E622 (IS-like element, no transposase); PmaES4326 | ||
| D03 | 1812-3215 | 467 | 59 | Mobilization protein; Sinorhizobium meliloti (pRm1132f) | AAG59888 | 133/3e−30; 109/389 (38) |
| D04 | 3297-3521 | 75 | 36 | Hypothetical | ||
| MFD02 | 3543-4171 | 629 bp | 58 | Replication domain; Acidithiobacillus ferrooxidans TFN (pH1) | AJ251742 | 47/2e−11; 27/56 (48); 21/28 (75) |
| D05 | 4458-4279 | 60 | 66 | Hypothetical | ||
| D06 | 4685-4455 | 77 | 65 | Conserved hypothetical; PtoDC3000 | AAO53570 | 70.5/2e−12; 34/75 (45) |
MF, miscellaneous feature.
Unless otherwise stated.
Abbreviations: Pma, P. syringae pv. maculicola; Pto, P. syringae pv. tomato.
TABLE 5.
Predicted ORFs and features of pPMA4326E
| ORF or featurea | Position | Size (aa)b | %GC | Homology (BLAST)c | Accession no. | Score/E value; a identity (no. with identity/total [%]) |
|---|---|---|---|---|---|---|
| E01 | 1-135 | 45 | 60 | Hypothetical | ||
| E02 | 693-259 | 145 | 55 | Hypothetical | ||
| E03 | 1167-2402 | 412 | 59 | Mobilization protein; Sinorhizobium meliloti (pRm1132f) | AAG59888 | 113/2e−24; 117/405 (28%) |
| E04 | 2599-2823 | 75 | 36 | Hypothetical | ||
| MFE01 | 2846-3473 | 628 bp | 55 | Replication domain; Acidithiobacillus ferrooxidans TFN (pH1) | AJ251742 | 91.8/5e−26; 50/90 (55%) |
| E05 | 3815-3612 | 68 | 65 | Hypothetical | ||
| E06 | 4042-3812 | 77 | 62 | Conserved hypothetical; PtoDC3000 | AAO53570 | 66.6/3e−11; 34/75 (45%) |
MF, miscellaneous feature.
Unless otherwise stated.
Pto, P. syringae pv. tomato.
The mobilization of these plasmids is probably effected by ORFs D03 and E03, which show homology to a mob gene of the Sinorhizobium meliloti cryptic plasmid pRm1132f (6). Just upstream of the putative mob gene of pPMA4326E is a putative oriT, characterized by numerous inverted and direct repeats that are commonly involved in relaxase recognition and termination of plasmid DNA transfer (21, 22). E622 lies between the putative mob and oriT genes on pPMA4326D. ORF prediction algorithms identified part of E622 as making up the first 48 bp of the pPMA4326D mob gene; however, because no RBS could be identified for this ORF, a nested methionine with an appropriate RBS (AGAGG) was used as a putative start codon.
Plasmid origins and incompatibility.
Plasmid incompatibility describes the inability of two plasmids employing similar maintenance or transmission mechanisms to coexist in the absence of strong selection pressure (55, 71). Plasmids pPMA4326A and pPMA4326B and plasmids pPMA4326D and pPMA4326E represent two pairs of plasmids that clearly challenge the laws of plasmid incompatibility.
The replication gene and origin are often strong incompatibility determinants (10, 25, 55, 71). The repA genes of pPMA4326A and pPMA4326B share 96% aa identity, while the replication origins of the two plasmids, believed to lie upstream of repA, share 94% nucleotide identity over a 638-bp stretch. While this suggests that pPMA4326A and pPMA4326B may have originated from a common ancestral replicon via plasmid duplication, the overall similarity of their genetic composition is quite low. They share only seven homologous genes, six of which are within an 8-kb segment bounded by the relaxase/mob and ssb genes (flanking repA). The nucleotide identity between the plasmids in this region is 90%. While plasmid duplication can explain this apparent similarity, it fails to explain the overall poor similarity between the two plasmids and the nonhomologous parA genes. The localization of the homologous genes in a single 8-kb unit could imply that recombination between the plasmids led to the exchange of this region. The exchange of repA genes among plasmids has significant repercussions for the inference of plasmid evolutionary relationships, as replication gene phylogenies are currently the basis for many of these analyses (25, 72, 73). Extensive exchange among replication genes would invalidate many of these phylogenies.
Homologous exchange of the replication protein would impose strong incompatibility on pPMA4326A and pPMA4326B unless selective pressure for the maintenance of both plasmids was present. This selective pressure would necessitate changes in both the replication protein and the origin to create sufficient variation to permit the coexistence of these two plasmids within P. syringae pv. maculicola ES4326. We identified a putative stem-loop structure (stem-loop II) through a comparison of coresident pPT23A-like plasmid replication origins that may be an important incompatibility determinant. The 19-bp loop sequence, situated 221 bp upstream of the replicase gene, differs substantially for compatible plasmids and may be complementary to the replication protein C terminus, which exhibits more sequence variation than the N terminus.
The high degree of similarity in the replication region of these coresident plasmids could be the result of plasmid duplication or recombination. We believe that recombination is more likely than duplication because two of the three putative partitioning genes are not homologous between the two plasmids.
Plasmids pPMA4326D and pPMA4326E are strikingly similar replicons, which suggests that they originated from a recent common ancestor. The presence of these two related plasmids within P. syringae pv. maculicola ES4326 may have occurred via several evolutionary paths. After the acquisition of one plasmid by P. syringae pv. maculicola ES4326, changes in plasmid incompatibility determinants may have resulted in plasmid duplication. The primary incompatibility determinant for these plasmids is most likely the putative replication region. In fact, pPMA4326E and pH1 share more nucleotide sequence identity in this region than do pPMA4326E and pPMA4326D. The relatively low sequence identity between pPMA4326D and pPMA4326E in this putative replication region could explain their apparent compatibility. An alternative possibility to plasmid duplication within P. syringae pv. maculicola ES4326 is the independent acquisition of pPMA4326D and pPMA4326E from two separate sources. Horizontal acquisition of this plasmid type is highly probable, given that it possesses a mobA-like gene. The %GC contents of these plasmids are 56 and 57%, respectively, which correlates fairly well with the 58 to 59% GC content of P. syringae (19), suggesting either that these have been resident within the strain long enough to have acclimated to the host %GC content or that they were acquired from a bacterium with a similar %GC content. Interestingly, Acidithiobacillus ferrooxidans, from which the closely related pH1 replicon was isolated, has a GC content of 59% (47). The maintenance of both pPMA4326D and pPMA4326E suggests either a strong selective advantage of redundancy of this particular plasmid type or that these plasmids employ a postsegregational killing system that promotes their stability.
Conclusion.
The complexity and diversity of the P. syringae pv. maculicola ES4326 plasmids have provided a glimpse into both the evolution of single plasmids and the phenomenon of plasmid incompatibility. The plasmid complement of this strain appears to have been strongly influenced by horizontal plasmid transfer, as suggested by the presence of atypical, replicase-deficient replicons, but there is insufficient information as to whether these represent interspecific or intraspecific acquisitions. The plasmid complement of the very closely related strain P. syringae pv. maculicola YM7930 (66) further supports horizontal acquisition by P. syringae pv. maculicola ES4326. This strain clearly lacks pPMA4326A, pPMA4326C, pPMA4326D, and pPMA4326E, but it has a plasmid that is very similar to pPMA4326B (J. Stavrinides and D. S. Guttman, unpublished data). The presence of redundant IS elements and their derivatives suggests an important role for mobile elements in creating alternate genetic combinations and in driving plasmid rearrangement. Additional plasmid genomic data will help us to uncover the precise role of these IS elements and to determine whether their influence is largely through legitimate or illegitimate recombination. Knowledge of the underlying mechanisms driving the evolution of plasmids will provide vital information about the genetic potential of these minigenomes and, ultimately, how they impact the evolution of their bacterial hosts.
Acknowledgments
We thank Sara Sarkar and Pauline Wang for insightful discussions, advice, suggestions, and comments. We thank Susan Gropp and Wenbo Ma for reviewing the manuscript, Andrew Szto and Chris Grouios for their assistance, and Jack Hsu for his exceptional programming skills. We are extremely grateful to Carol Bender and Lori Burrows for providing strains, George Sundin for providing his pPSR1 manuscript prior to its publication, and TIGR (http://www.tigr.org) for providing genome sequence data prior to its official release.
D.S.G. was supported by grants from the Natural Sciences and Engineering Research Council of Canada and the Canadian Foundation for Innovation. J.S. is supported by a Canada Graduate Scholarship.
REFERENCES
- 1.Alfano, J. R., A. O. Charkowski, W. L. Deng, J. L. Badel, T. Petnicki-Ocwieja, K. van Dijk, and A. Collmer. 2000. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. USA 97:4856-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, D. L., R. W. Jackson, A. J. Fillingham, S. C. Goss, J. D. Taylor, J. W. Mansfield, and A. Vivian. 2001. Highly conserved sequences flank avirulence genes: isolation of novel avirulence genes from Pseudomonas syringae pv. pisi. Microbiology 147:1171-1182. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, D. L., A. Pitman, and R. W. Jackson. 2003. Pathogenicity and other genomic islands in plant pathogenic bacteria. Mol. Plant Pathol. 4:407-420. [DOI] [PubMed] [Google Scholar]
- 4.Ast, V. M., I. C. Schoenhofen, G. R. Langen, C. W. Stratilo, M. D. Chamberlain, and S. P. Howard. 2002. Expression of the ExeAB complex of Aeromonas hydrophila is required for the localization and assembly of the ExeD secretion port multimer. Mol. Microbiol. 44:217-231. [DOI] [PubMed] [Google Scholar]
- 5.Azucena, E., and S. Mobashery. 2001. Aminoglycoside-modifying enzymes: mechanisms of catalytic processes and inhibition. Drug Resist. Updat. 4:106-117. [DOI] [PubMed] [Google Scholar]
- 6.Barran, L. R., N. Ritchot, and E. S. P. Bromfield. 2001. Sinorhizobium meliloti plasmid pRm1132f replicates by a rolling-circle mechanism. J. Bacteriol. 183:2704-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender, C. L., H. E. Stone, J. J. Sims, and D. A. Cooksey. 1987. Reduced pathogen fitness of Pseudomonas syringae pathovar tomato Tn5 mutants defective in coronatine production. Physiol. Mol. Plant Pathol. 30:273-284. [Google Scholar]
- 8.Bender, C. L., S. A. Young, and R. E. Mitchell. 1991. Conservation of plasmid DNA sequences in coronatine-producing pathovars of Pseudomonas syringae. Appl. Environ. Microbiol. 57:993-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bignell, C., and C. M. Thomas. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91:1-34. [DOI] [PubMed] [Google Scholar]
- 11.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. W. Zhou, J. Liu, Q. P. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Y. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalmers, R. M., and N. Kleckner. 1996. IS10 Tn10 transposition efficiently accommodates diverse transposon end configurations. EMBO J. 15:5112-5122. [PMC free article] [PubMed] [Google Scholar]
- 13.Cournoyer, B., J. D. Sharp, A. Astuto, M. J. Gibbon, J. D. Taylor, and A. Vivian. 1995. Molecular characterization of the Pseudomonas syringae pv. pisi plasmid-borne avirulence gene avrPpiB which matches the R3 resistance locus in pea. Mol. Plant-Microbe Interact. 8:700-708. [DOI] [PubMed] [Google Scholar]
- 14.Cuppels, D. A., and T. Ainsworth. 1995. Molecular and physiological characterization of Pseudomonas syringae pv. tomato and Pseudomonas syringae pv. maculicola strains that produce the phytotoxin coronatine. Appl. Environ. Microbiol. 61:3530-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drolet, M., P. Zanga, and P. C. K. Lau. 1990. The mobilization and origin of transfer regions of a thiobacillus-ferrooxidans plasmid-relatedness to plasmid-Rsf1010 and plasmid-Psc101. Mol. Microbiol. 4:1381-1391. [DOI] [PubMed] [Google Scholar]
- 17.Duport, C., C. Baysse, and Y. Michelbriand. 1995. Molecular characterization of pyocin S3, a novel S-type pyocin from Pseudomonas aeruginosa. J. Biol. Chem. 270:8920-8927. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package), version 3.5c. Department of Genetics, University of Washington, Seattle.
- 19.Fouts, D. E., R. B. Abramovitch, J. R. Alfano, A. M. Baldo, C. R. Buell, S. Cartinhour, A. K. Chatterjee, M. D'Ascenzo, M. L. Gwinn, S. G. Lazarowitz, N. C. Lin, G. B. Martin, A. H. Rehm, D. J. Schneider, K. van Dijk, X. Y. Tang, and A. Collmer. 2002. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. USA 99:2275-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frederick, R. D., M. Ahmad, D. R. Majerczak, A. S. Arroyo-Rodriguez, S. Manulis, and D. L. Coplin. 2001. Genetic organization of the Pantoea stewartii subsp. stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN, and wtsE operons. Mol. Plant-Microbe Interact. 14:1213-1222. [DOI] [PubMed] [Google Scholar]
- 21.Furuya, N., and T. Komano. 1997. Mutational analysis of the R64 oriT region: requirement for precise location of the NikA binding sequence. J. Bacteriol. 179:7291-7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuya, N., and T. Komano. 1995. Specific binding of the NikA protein to one arm of 17-base-pair inverted repeat sequences within the OriT region of plasmid R64. J. Bacteriol. 177:46-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia de Viedma, D., R. Giraldo, M. J. Ruizechevarria, R. Lurz, and R. Diazorejas. 1995. Transcription of repA, the gene of the initiation protein of the Pseudomonas plasmid pps10, is autoregulated by interactions of the RepA protein at a symmetrical operator. J. Mol. Biol. 247:211-223. [DOI] [PubMed] [Google Scholar]
- 24.Garcia de Viedma, D., A. Serrano-Lopez, and R. Diaz-Orejas. 1995. Specific binding of the replication protein of plasmid pPS10 to direct and inverted repeats is mediated by an HTH motif. Nucleic Acids Res. 23:5048-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibbon, M. J., A. Sesma, A. Canal, J. R. Wood, E. Hidalgo, J. Brown, A. Vivian, and J. Murillo. 1999. Replication regions from plant-pathogenic Pseudomonas syringae plasmids are similar to ColE2-related replicons. Microbiology 145:325-334. [DOI] [PubMed] [Google Scholar]
- 26.Greenberg, J. T., and B. A. Vinatzer. 2003. Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Curr. Opin. Microbiol. 6:20-28. [DOI] [PubMed] [Google Scholar]
- 27.Grinsted, J., F. Delacruz, and R. Schmitt. 1990. The Tn21 subgroup of bacterial transposable elements. Plasmid 24:163-189. [DOI] [PubMed] [Google Scholar]
- 28.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guttman, D. S., and J. T. Greenberg. 2001. Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Mol. Plant-Microbe Interact. 14:145-155. [DOI] [PubMed] [Google Scholar]
- 30.Guttman, D. S., B. A. Vinatzer, S. F. Sarkar, M. V. Ranall, G. Kettler, and J. T. Greenberg. 2002. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295:1722-1726. [DOI] [PubMed] [Google Scholar]
- 31.Hengen, P. N., D. Denicourt, and V. N. Iyer. 1992. Isolation and characterization of KikA, a region on IncN group plasmids that determines killing of Klebsiella oxytoca. J. Bacteriol. 174:3070-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hensel, M., T. Nikolaus, and C. Egelseer. 1999. Molecular and functional analysis indicates a mosaic structure of Salmonella pathogenicity island 2. Mol. Microbiol. 31:489-498. [DOI] [PubMed] [Google Scholar]
- 33.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiszczynska-Sawicka, E., and J. Kur. 1997. Effect of Escherichia coli IHF mutations on plasmid p15A copy number. Plasmid 38:174-179. [DOI] [PubMed] [Google Scholar]
- 35.Holcik, M., and V. N. Iyer. 1996. Structure and mode of action of KikA, a genetic region lethal to Klebsiella oxytoca and associated with conjugative antibiotic-resistance plasmids of the IncN group. Plasmid 35:189-203. [DOI] [PubMed] [Google Scholar]
- 36.Howard, S. P., H. G. Meiklejohn, D. Shivak, and R. Jahagirdar. 1996. A TonB-like protein and a novel membrane protein containing an ATP-binding cassette function together in exotoxin secretion. Mol. Microbiol. 22:595-604. [DOI] [PubMed] [Google Scholar]
- 37.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inui, M., K. Nakata, J. H. Roh, A. A. Vertes, and H. Yukawa. 2003. Isolation and molecular characterization of pMG160, a mobilizable cryptic plasmid from Rhodobacter blasticus. Appl. Environ. Microbiol. 69:725-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson, R. W., E. Athanassopoulos, G. Tsiamis, J. W. Mansfield, A. Sesma, D. L. Arnold, M. J. Gibbon, J. Murillo, J. D. Taylor, and A. Vivian. 1999. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc. Natl. Acad. Sci. USA 96:10875-10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jahagirdar, R., and S. P. Howard. 1994. Isolation and characterization of a second exe operon required for extracellular protein secretion in Aeromonas hydrophila. J. Bacteriol. 176:6819-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin, Q. L., R. Thilmony, J. Zwiesler-Vollick, and S. Y. He. 2003. Type III protein secretion in Pseudomonas syringae. Microb. Infect. 5:301-310. [DOI] [PubMed] [Google Scholar]
- 42.Johnson, E. P., A. R. Strom, and D. R. Helinski. 1996. Plasmid RK2 toxin protein ParE: purification and interaction with the ParD antitoxin protein. J. Bacteriol. 178:1420-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kado, C. I. 1998. Origin and evolution of plasmids. Antonie Leeuwenhoek 73:117-126. [DOI] [PubMed] [Google Scholar]
- 44.Kalyaeva, E., I. Bass, G. Kholodii, and V. Nikiforov. 2002. A broad host range plasmid vector that does not encode replication proteins. FEMS Microbiol. Lett. 211:91-95. [DOI] [PubMed] [Google Scholar]
- 45.Kamiunten, H. 1999. Isolation and characterization of virulence gene psvA on a plasmid of Pseudomonas syringae pv. eriobotryae. Ann. Phytopathol. Soc. Jpn. 65:501-509. [Google Scholar]
- 46.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing Cyanobacterium anabaena sp. strain PCC 7120. DNA Res. 8:205-213. [DOI] [PubMed] [Google Scholar]
- 47.Kelly, D. P., and A. P. Wood. 2000. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 50:511-516. [DOI] [PubMed] [Google Scholar]
- 48.Komano, T., S. Fujitani, N. Funayama, A. Kanno, and T. Nisioka. 1987. Genetic and physical characterization of the plasmid R721. Jpn. J. Genet. 62:528-535.
- 49.Kropinski, A. M. 2000. Sequence of the genome of the temperate, serotype-converting, Pseudomonas aeruginosa bacteriophage D3. J. Bacteriol. 182:6066-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang, L. Z., P. Sobiczewski, J. M. Paterson, and A. L. Jones. 1994. Variation in virulence, plasmid content, and genes for coronatine synthesis between Pseudomonas syringae pv. morsprunorum and P.s. syringae from Prunus. Plant Dis. 78:389-392. [Google Scholar]
- 51.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 52.Mittal, S., and K. R. Davis. 1995. Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol. Plant-Microbe Interact. 8:165-171. [DOI] [PubMed] [Google Scholar]
- 53.Naumann, T. A., and W. S. Reznikoff. 2002. Tn5 transposase with an altered specificity for transposon ends. J. Bacteriol. 184:233-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nieto, C., R. Giraldo, E. Fernandeztresguerres, and R. Diaz. 1992. Genetic and functional analysis of the basic replicon of pps10, a plasmid specific for Pseudomonas isolated from Pseudomonas syringae pathovar savastanoi. J. Mol. Biol. 223:415-426. [DOI] [PubMed] [Google Scholar]
- 55.Novick, R. P. 1987. Plasmid incompatibility. Microbiol. Rev. 51:381-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 57.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park, K., S. Mukhopadhyay, and D. K. Chattoraj. 1998. Requirements for and regulation of origin opening of plasmid P1. J. Biol. Chem. 273:24906-24911. [DOI] [PubMed] [Google Scholar]
- 59.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Limk, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Y. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 60.Petnicki-Ocwieja, T., D. J. Schneider, V. C. Tam, S. T. Chancey, L. Shan, Y. Jamir, L. M. Schechter, M. D. Janes, C. R. Buell, X. Y. Tang, A. Collmer, and J. R. Alfano. 2002. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 99:7652-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Preston, G. M., B. Haubold, and P. B. Rainey. 1998. Bacterial genomics and adaptation to life on plants: implications for the evolution of pathogenicity and symbiosis. Curr. Opin. Microbiol. 1:589-597. [DOI] [PubMed] [Google Scholar]
- 62.Qin, X. T., and J. S. Hartung. 2001. Construction of a shuttle vector and transformation of Xylella fastidiosa with plasmid DNA. Curr. Microbiol. 43:158-162. [DOI] [PubMed] [Google Scholar]
- 63.Richter, G. Y., K. Bjorklof, M. Romantschuk, and D. Mills. 1998. Insertion specificity and trans-activation of IS801. Mol. Gen. Genet. 260:381-387. [DOI] [PubMed] [Google Scholar]
- 64.Rohmer, L., S. Kjemtrup, P. Marchesini, and J. L. Dangl. 2003. Nucleotide sequence, functional characterization and evolution of pFKN, a virulence plasmid in Pseudomonas syringae pathovar maculicola. Mol. Microbiol. 47:1545-1562. [DOI] [PubMed] [Google Scholar]
- 65.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 66.Sarkar, S. F., and D. S. Guttman. 2004. The evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70:1999-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schiex, T., J. Gouzy, A. Moisan, and Y. de Oliveira. 2003. FrameD: a flexible program for quality check and gene prediction in prokaryotic genomes and noisy matured eukaryotic sequences. Nucleic Acids Res. 31:3738-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schoenhofen, I. C., C. Stratilo, and S. P. Howard. 1998. An ExeAB complex in the type II secretion pathway of Aeromonas hydrophila: effect of ATP-binding cassette mutations on complex formation and function. Mol. Microbiol. 29:1237-1247. [DOI] [PubMed] [Google Scholar]
- 69.Schwartz, S., Z. Zhang, K. A. Frazer, A. Smit, C. Riemer, J. Bouck, R. Gibbs, R. Hardison, and W. Miller. 2000. PipMaker-A Web server for aligning two genomic DNA sequences. Genome Res. 10:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sesma, A., M. T. Aizpun, A. Ortiz-Barredo, D. Arnold, A. Vivian, and J. Murillo. 2001. Virulence determinants other than coronatine in Pseudomonas syringae pv. tomato PT23 are plasmid-encoded. Physiol. Mol. Plant Pathol. 58:83-93. [Google Scholar]
- 71.Sesma, A., G. W. Sundin, and J. Murillo. 1998. Closely related plasmid replicons coexisting in the phytopathogen Pseudomonas syringae show a mosaic organization of the replication region and altered incompatibility behavior. Appl. Environ. Microbiol. 64:3948-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sesma, A., G. W. Sundin, and J. Murillo. 2000. Phylogeny of the replication regions of pPT23A-like plasmids from Pseudomonas syringae. Microbiology 146:2375-2384. [DOI] [PubMed] [Google Scholar]
- 73.Silva, F. J., R. van Ham, B. Sabater, and A. Latorre. 1998. Structure and evolution of the leucine plasmids carried by the endosymbiont (Buchnera aphidicola) from aphids of the family Aphididae. FEMS Microbiol. Lett. 168:43-49. [DOI] [PubMed] [Google Scholar]
- 74.Sundin, G. W., and C. L. Bender. 1993. Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 59:1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sundin, G. W., and C. L. Bender. 1994. Relative fitness in-vitro and in planta of Pseudomonas syringae strains containing copper and streptomycin resistance plasmids. Can. J. Microbiol. 40:279-285. [Google Scholar]
- 76.Sundin, G. W., S. P. Kidambi, M. Ullrich, and C. L. Bender. 1996. Resistance to ultraviolet light in Pseudomonas syringae: sequence and functional analysis of the plasmid-encoded rulAB genes. Gene 177:77-81. [DOI] [PubMed] [Google Scholar]
- 77.Sundin, G. W., C. T. Mayfield, Y. Zhao, T. S. Gunasekera, G. L. Foster, and M. S. Ullrich. 2004. Complete nucleotide sequence and analysis of pPSR1 (72,601 bp), a pPT23A-family plasmid from Pseudomonas syringae pv. syringae A2. Mol. Gen. Genomics 270:462-475. [DOI] [PubMed] [Google Scholar]
- 78.Tan, H. M. 1999. Bacterial catabolic transposons. Appl. Microbiol. Biotechnol. 51:1-12. [DOI] [PubMed] [Google Scholar]
- 79.Tauschek, M., R. A. Strugnell, and R. M. Robins-Browne. 2002. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 44:1533-1550. [DOI] [PubMed] [Google Scholar]
- 80.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ullrich, M., S. Bereswill, B. Volksch, W. Fritsche, and K. Geider. 1993. Molecular characterization of field isolates of Pseudomonas syringae pv. glycinea differing in coronatine production. J. Gen. Microbiol. 139:1927-1937. [DOI] [PubMed] [Google Scholar]
- 82.Vivian, A., J. Murillo, and R. W. Jackson. 2001. The roles of plasmids in phytopathogenic bacteria: mobile arsenals? Microbiology 147:763-780. [DOI] [PubMed] [Google Scholar]
- 83.Wu, H. Z., H. Z. Zhang, C. X. Lu, N. Liang, H. Y. Jin, Y. Ma, and Y. X. Zhang. 2003. DNA sequencing of a plasmid with virulence from marine fish pathogen Vibrio anguillarum. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 35:956-959. [PubMed] [Google Scholar]
- 84.Zaharik, M. L., S. Gruenheid, A. J. Perrin, and B. B. Finlay. 2002. Delivery of dangerous goods: type III secretion in enteric pathogens. Int. J. Med. Microbiol. 291:593-603. [DOI] [PubMed] [Google Scholar]
- 85.Zhou, C., Y. J. Yang, and A. Y. Jong. 1990. Mini-prep in 10 minutes. BioTechniques 8:172-173. [PubMed] [Google Scholar]
- 86.Zhou, M., A. Bhasin, and W. S. Reznikoff. 1998. Molecular genetic analysis of transposase-end DNA sequence recognition: cooperativity of three adjacent base-pairs in specific interaction with a mutant Tn5 transposase. J. Mol. Biol. 276:913-925. [DOI] [PubMed] [Google Scholar]
- 87.Zielenkiewicz, U., and P. Ceglowski. 2001. Mechanisms of plasmid stable maintenance with special focus on plasmid addiction systems. Acta Biochim. Pol. 48:1003-1023. [PubMed] [Google Scholar]