Abstract
Background
Heart development is tightly regulated by signaling events acting upon a defined number of progenitor and differentiated cardiac cells. While loss-of-function of these signaling pathways leads to congenital malformation, the consequences of cardiac progenitor cell (CPC) or embryonic cardiomyocyte loss are less clear. In this study, we tested the hypothesis that embryonic mouse hearts exhibit a robust mechanism for regeneration following extensive cell loss.
Methods and Results
By combining a conditional cell ablation approach with a novel blastocyst complementation strategy, we generated murine embryos that exhibit a full spectrum of CPC or cardiomyocyte ablation. Remarkably, ablation of up to 60% of CPCs at embryonic day 7.5 was well-tolerated and permitted embryo survival. Ablation of embryonic cardiomyocytes to a similar degree (50-60%) at embryonic day 9.0 could be fully rescued by residual myocytes with no obvious adult cardiac functional deficit. In both ablation models, an increase in cardiomyocyte proliferation rate was detected and accounted for at least some of the rapid recovery of myocardial cellularity and heart size.
Conclusions
Our study defines the threshold for cell loss in the embryonic mammalian heart and reveals a robust cardiomyocyte compensatory response that sustains normal fetal development.
Keywords: cardiac development, cardiac progenitor cells, cardiomyocyte, regeneration, stem cell plasticity
INTRODUCTION
The development of the mammalian four-chambered heart requires a close interplay between extracellular morphogens and signals (e.g. Wnt, TGF-β, FGF, Notch, and Hedgehog) and cell-intrinsic effectors such as transcription factors and epigenetic modifiers1, 2. Some of these factors regulate early lineage specification, while others mediate cellular expansion and maturation. Pioneering work on model organisms such as Drosophila, Xenopus, chick, zebrafish, and mouse has revealed the exquisite sensitivity of the early embryo to perturbations in these pathways that result in morphological defects and subsequent early embryonic lethality3. Disruption in these formative events during embryonic development also has a profound impact on human life, as congenital heart disease remains the most common cause of birth defects world-wide4.
While cardiogenesis is in many ways a delicate process, the heart is nonetheless able to withstand injury and regenerate throughout the lifespan of some animal species as demonstrated by studies in amphibians and lower vertebrates such as the newt and zebrafish5, 6. In these organisms, it has been proposed that an increased rate of proliferation by preexisting cardiomyocytes is the predominant endogenous mechanism responsible for cardiac injury repair7-9. Collectively, these non-mammalian models have motivated efforts to define the extent of heart regeneration present in mice and humans and to identify underlying repair mechanisms that could represent potential targets for therapeutic intervention. Indeed, recent reports suggest that the neonatal murine heart can regenerate after experimental injury in a process that depends on Meis1-regulated cardiomyocyte proliferation10, 11. Furthermore, a prior study from Drenckhahn and colleagues found that healthy embryonic cardiomyocytes can augment their rate of proliferation to compensate for the reduced proliferation rate of diseased cardiomyocytes that are deficient in holocytochrome-c synthase, an enzyme that is essential for mitochondrial respiration12. While this approach and others examined the role of cell competition during cardiac development13, they did not directly address the consequences of cardiomyocyte loss. Accordingly, the ability of an embryonic mammalian heart to regenerate following rapid and extensive cell loss has yet to be addressed.
In this study, we used a novel blastocyst complementation strategy to test the ability of a mammalian embryonic heart to compensate for rapid cardiac cell loss and to quantitatively define the extent of cardiac lineage cell ablation that is compatible with normal development. We hypothesized that genetically-mediated cell loss in the mouse heart can be fully compensated by residual non-ablated cells if a robust regenerative mechanism exists at the cardiac progenitor cell (CPC) or immature cardiomyocyte (CM) stage of development and found that ablation of up to 50-60% of CPCs or immature CMs is well-tolerated and supports live birth to adulthood. Furthermore, we found that proliferation of residual cardiomyocytes is involved in the cell replacement process. Our findings encourage further investigation of the endogenous mechanisms responsible for replacing lost CPCs and CMs in the embryonic heart. In the context of adult mammalian cardiac injury, advances in this area may lead to therapies that direct the post-injury response towards cellular regeneration rather than fibrosis.
METHODS
An expanded Methods section is available in the Supplemental Materials.
Experimental Animals
An Nkx2.5Cre/+ knock-in mouse line was kindly provided by Dr. Robert Schwartz14. An αMHCCre/+ transgenic mouse line was kindly provided by Dr. E. Dale Abel15. ROSA26eGFP-DTA and ROSA26LacZ mouse lines were purchased from the Jackson Laboratory16, 17. Experimental animal protocols were approved by the Institutional Animal Care and Use Committees of Massachusetts General Hospital and Stanford University. All experiments were performed on somite-matched embryos or sex-matched adult mice.
Establishment of ESC Lines
Derivation of the V6.518 and R119 ESC lines has been described previously. For generation of Nkx2.5Cre/+;ROSA26eGFP-DTA and αMHCCre/+;ROSA26eGFP-DTA compound transgenic ESC lines, timed matings were performed between male Nkx2.5Cre/+ mice or αMHCCre/+ mice with female ROSA26eGFP-DTA mice. At 3.5 days post-coitum (dpc), females were sacrificed and blastocysts were flushed from the uterine horns with M2 medium (Sigma-Aldrich, M7167) and washed several times. Using a mouth pipette with a pulled glass capillary, blastocysts were plated individually onto 24-well gelatin-coated plates containing mitomycin-C (Sigma-Aldrich, M4287) inactivated mouse embryonic fibroblast (MEF) feeder layers in ESC Derivation Media and cultured, undisturbed, at 37°C in 5% CO2 in humidified air for 5–7 days without media changes. As blastocysts hatched from their zona pellucidae, the inner cell mass (ICM) outgrowth was identified and transferred into 200 μL of 0.25% trypsin-EDTA solution (Life Technologies, 25200) for 5 min at 37°C and gently dissociated by pipetting. Trypsin was inactivated with fetal bovine serum (FBS, Atlanta Biologicals, S11550), and the ICM cells were centrifuged and reseeded onto fresh MEFs in ESC Maintenance Media supplemented with 2i20, 21. Undifferentiated ES colonies were then gradually expanded to establish ESC lines. Lines were selected for further use based on undifferentiated morphology, the presence of the Cre transgene and Y chromosome by PCR, and expression of eGFP. Primer sequences used for genotyping are listed in Supplementary Table 1. ESC Derivation and Maintenance Media compositions are reported in Supplementary Methods.
Chimera Production
Embryos were staged by vaginal plugging of the mother, with noon on the day of appearance of the plug designated as embryonic day (E) 0.5. For the initial studies, approximately 10-20 low passage (P5-P10) Nkx2.5Cre/+;ROSA26eGFP-DTA or αMHCCre/+;ROSA26eGFP-DTA ESCs were microinjected into E3.5 blastocysts from superovulated CD-1 females (Charles River Laboratories). For the reverse complementation studies, P15-P25 V6.5 or R1 ESCs were microinjected into E3.5 blastocysts from superovulated ROSA26eGFP-DTA females which had been mated to Nkx2.5Cre/+ males. For both approaches, the injected blastocysts were subsequently transferred into the uterus of 2.5 dpc pseudopregnant 6-8-week-old CD-1 foster mothers previously mated with vasectomized males22. Genotype was identified based upon expression of eGFP and the presence of the Cre transgene by PCR. Chimeric contribution was determined by flow-cytometric analysis as described in Supplementary Methods.
Ex vivo single-cell clonal analysis
Details regarding single-cell culture studies can be found in the Supplementary Methods.
Immunohistochemistry
For cell culture experiments in 96-well plates, cells were fixed for 5 minutes in 4% paraformaldehyde and washed with PBS. E8.5 embryos were fixed for 10 minutes in 4% paraformaldehyde and embedded in OCT compound on dry ice. E9.5-E10.5 embryos and live-born mouse tissues were fixed overnight in 4% paraformaldehyde and embedded in paraffin. Tissue blocks were cut at 5 μm thickness. Paraffin-embedded sections underwent antigen retrieval in a pressure cooker for 3 minutes in citrate-based unmasking solution, pH 6.0 (Vector Labs, H-3300). Plated cells and tissue sections were blocked and permeabilized with 10% goat serum, 1% BSA, and 0.1% saponin in PBS for 1 hour and then incubated overnight at 4° C with the primary antibody. Primary antibodies utilized are listed in Supplementary Table 2. Samples were then washed with PBS and incubated at room temperature for 1 hour with corresponding secondary antibodies conjugated to Alexa Fluor 488, Alexa Fluor 546, or Alexa Fluor 647 (Invitrogen, 1:400 dilution). Samples were subsequently rinsed in PBS and stained with NucBlue Fixed Cell ReadyProbes DAPI Reagent (Life Technologies, R37606). Slides were mounted in ProLong Gold Antifade Reagent (Life Technologies, P-36930). For tissue sections, epifluorescence images were acquired on a Zeiss AxioImager M1 upright widefield fluorescence/DIC microscope with dual CCD cameras. Immunostaining was performed on 3-5 embryos/group. For cell culture experiments, each well of a 96-well plate was individually imaged using a BioTek Cytation 3 Cell Imaging Multi-Mode Reader.
Quantification of Cardiomyocyte Proliferation
For assessment of cardiomyocyte proliferation, embryo tissue sections were prepared as described in Immunohistochemistry using antibodies to cTnT, CD31, and pH3 or Ki-67. 1 mm z-stack immunofluorescence images of each section were acquired on a Zeiss LSM710 confocal microscope at 20× magnification to confirm that each DAPI+ cell nucleus counted was associated with a cardiomyocyte (if surrounded in three dimensions by cTnT stain) or an endocardial cell (if surrounded in three dimensions by a CD31 stain).
Echocardiography
Echocardiography was performed on 4-5 week-old mice using a 13-MHz linear-array transducer with a digital ultrasound system (Vivid 7; GE Medical Systems, Milwaukee, WI). The mice were lightly sedated using an intraperitoneal injection of ketamine (50 mg/kg). Heart rate, left ventricular internal diameter at end-diastole (LVIDED), and left ventricular internal diameter at end-systole (LVIDES) were measured, and the LV fractional shortening was calculated by using an M-Mode echocardiogram obtained at the midpapillary level. For each mouse, all values were averaged from measurements obtained from 9 cardiac cycles. All measurements and calculations were performed by an operator blinded to the mouse genotype.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism version 6.02 for Windows (GraphPad Software, San Diego, California, USA) and RStudio version 0.98.1102 for Windows (Boston, Massachusetts, USA). For comparisons between groups where multiple measurements per mouse were recorded (embryonic cell proliferation, embryonic CM size, and adult CM size), a likelihood ratio test via ANOVA was computed on a linear mixed-effects model using RStudio. For comparisons between three groups (cardiomyocyte binucleation, adult heart weight ratios, left ventricular dimensions, and left ventricular fractional shortening), a one-way ANOVA was used with adjustment for multiplicity using Dunnett's multiple comparison test, or, when data did not conform to a normal distribution (ex vivo cardiomyocyte colony sizes), the Kruskal-Wallis test was used with Dunn's correction for multiple comparisons. A p-value of <0.05 was considered significant.
RESULTS
Fractional ablation of embryonic CPCs by chimeric complementation
The myocardial lineage of the heart arises from first and second heart field cells that express Nkx2.5, a key transcription factor in early heart formation (Figure S1)23. We performed targeted genetic ablation of Nkx2.5+ cardiac progenitor cells during embryonic development in order to examine the innate recovery response by the remaining non-ablated cells. By crossing a previously described Nkx2.5-Cre knock-in mouse line (Nkx2.5Cre/+)14 with a mouse line that expressed diphtheria toxin A (DTA) following Cre-dependent excision of a constitutively active eGFP transgene (ROSA26eGFP-DTA)16, we show that cardiac cells were abolished entirely in double transgenic embryos (Figure 1A).
Figure 1.
Fractional ablation of embryonic CPCs by chimeric complementation. (A) Schematic diagram of the eGFP/DTA construct before and after Nkx2.5-Cre-mediated excision of the floxed eGFP transgene (upper panel). Brightfield and GFP fluorescence images of E9.5 Nkx2.5Cre/+, ROSA26eGFP-DTA, and Nkx2.5Cre/+;ROSA26eGFP-DTA transgenic embryos (lower panel). Note the absence of a heart in the compound transgenic embryo (arrow). (B) Schematic diagram of fractional ablation of CPCs by injection of Nkx2.5Cre/+;ROSA26eGFP-DTA ESCs into wild-type CD-1 blastocysts. (C) Generation of Nkx2.5Cre/+;ROSA26eGFP-DTA ESC lines. Brightfield and GFP fluorescence microscopy images are shown for the three ESC lines used. (D) Summary table demonstrating the complementation efficiency for each ESC line based upon the total number of animals collected. (E) Anticipated progression of CPC loss and regeneration in rescued embryos at approximately 50% ESC chimerism. (F) Brightfield and GFP images of E10.5 chimeric Nkx2.5Cre/+;ROSA26eGFP-DTA embryos with varying degrees of CPC ablation. The embryo with ~68% CPC ablation shows a pericardial effusion and developmental arrest. Arrow: heart. Scale bars: 500 μm for (A); 200 μm for (C); 1 mm for (F).
While the complete ablation of CPCs demonstrates the essential requirement of these cells in normal embryogenesis, it does not lend itself to a strategy for examining the regenerative potential of the fetal heart. One solution may be to use a drug-inducible cardiac-specific Cre model; however, our prior experience with either doxycycline or tamoxifen-inducible mice suggests too much inter-embryo variability in drug metabolism in utero for that strategy to be reliable for our purposes24. Hence, we employed a novel strategy that involves injection of double transgenic Nkx2.5Cre/+;ROSA26eGFP-DTA embryonic stem cells (ESCs) into wild-type eGFP− blastocysts to ablate CPCs in quantifiable fractions (Figure 1B). Since ESC injection into mouse blastocysts is well-known to generate chimeric embryos and mice with a wide spectrum of ESC contribution25, this assay enabled us to determine the expected percentage of CPC ablation by assessing the overall contribution of the eGFP+ Nkx2.5Cre/+;ROSA26eGFP-DTA ESCs to the embryo.
We first generated several independent ESC lines carrying both the Nkx2.5Cre/+ and ROSA26eGFP-DTA alleles (Figure 1C). These ES clones were eGFP+ under fluorescence microscopy, indicating the presence and expression of the floxed eGFP transgene. Three such clones (Nkx-DTA1, Nkx-DTA2, and Nkx-DTA3) that exhibited normal proliferation rates and an otherwise healthy in vitro phenotype were subsequently injected into wild-type eGFP− blastocysts and fostered in pseudopregnant female mice. Following injection of 602 wild-type blastocysts, we obtained 414 embryos and mice with a broad range of eGFP+ ESC contribution (Figure 1D). Within each chimeric embryo, CPCs derived from injected eGFP+ ESCs were expected to die due to Cre-mediated expression of DTA so that directly assessing the percentage of cardiac contribution by eGFP+ ESCs would not have been possible (Figure 1E). To overcome this limitation, we examined the expected variance in ESC contribution to the heart relative to other portions of the embryo body (e.g. head, limbs, tail, etc.) by injecting wild-type eGFP− ESCs into eGFP+ mouse blastocysts or red fluorescent tdTomato+ ESCs into wild-type mouse blastocysts. By flow cytometric (FACS) analysis, we found that 97% (29/30) of embryos showed a difference of ≤12% in ESC contribution to the heart compared to their mean contribution to the rest of the embryo (Figure S2A-D). Remarkably, our analysis of the embryonic day 10.5 (E10.5) chimeric embryos from Nkx2.5Cre/+;ROSA26eGFP-DTA ESC injections showed that the ablation of up to ~60% of CPCs, as determined by FACS, appears compatible with normal development, while degrees of ablation >60% lead to overall developmental arrest (Figure 1F).
Immunohistological analysis of chimeric embryos showed that eGFP+ cell contribution was homogenously distributed throughout the embryo, except for the heart region where eGFP+ cells were completely absent within the cardiac troponin T+ (cTnT) cell population (Figure 2A). Consistent with this, the hearts of chimeric embryos showed an absence of eGFP fluorescence (arrows), supporting the high efficiency of CPC ablation and compensation from adjacent non-ablated eGFP− cardiac cells (Figure 1F). Of note, sporadic eGFP+ cells could be found in the endocardium, consistent with the previously described incomplete expression of Nkx2.5 in these cells26 (Figure 2A). Interestingly, the gross morphology and myocardial cellularity were similar between surviving embryos with up to 60% ablation and their stage-matched littermate controls without ESC chimerism, supporting the presence of a regenerative process to compensate for lost CPCs and their myocardial descendants (Figure 2A).
Figure 2.
Histological analysis of CPC-ablated embryos shows a rapid recovery in the heart. (A) eGFP, troponin T (cTnT), and DAPI staining of E8.5 and E10.5 chimeric embryos with up to ~50% CPC ablation. By E8.5, all eGFP+ myocardial cells have been ablated in the heart. Rare, scattered eGFP+ endocardial cells can be identified (arrow). Cardiac chamber morphology and cellularity in the compact and trabecular compartments are similar between control and ablated hearts suggesting a rapid restoration of myocardial cell number after ablation. Note: eGFP+ staining in the atria of the E10.5 control embryo represents auto-fluorescent blood cells. (B-J) Immunostaining of control and ablated hearts. Cardiac transcription factor expression is shown at E8.5 for Gata4 (B) and Mef2c (C), and E9.5 for Islet-1 (D). Expression of genes regulating cardiomyocyte proliferation is shown at E8.5 for Yap (E), nuclear β-catenin (F), and total β-catenin (G). Sarcomeric protein expression is shown at E8.5 for α-actinin (H) and cTnT (I), and at E9.5 for Mlc2v (J). Nuclei are stained with DAPI and cardiomyocytes are identified by cTnT (A, B, C, E, G) or Mlc2v (D and F). For (B-J): degree of ablation in CPC-ablated sections = 41 ± 9% (mean ± SD). ca, common atrium; cv, common ventricle; oft, outflow tract. Scale bars: 100 μm for (A-G); 20 μm for (H-J).
To elucidate potential compensatory processes that might account for the recovery of lost CPCs, we performed immunostaining for transcription factors (i.e. Gata4, Mef2c, Islet-1) and signaling mediators (i.e. β-catenin and Yap) that have been reported to regulate CPC number or myocardial growth27-29 (Figure 2B-2G). However, these proteins exhibited high baseline expression during this early stage of development in control embryos; thus, no qualitative difference in expression could be observed in the ablated embryo heart by immunofluorescence staining. We also examined whether the expression of sarcomeric proteins (i.e. cTnT, α-actinin, myosin light chain 2v) varied between ablated and control hearts and found no difference in these parameters (Figure 2H-2J). Together, these data support an efficient compensatory response by residual un-ablated CPCs and/or their myocardial descendants to rapidly recover full myocardial cellularity in the ablated heart.
Since the robustness of our experimental conclusions depends heavily on having an accurate assessment of the degree of ESC contribution to the hearts of chimeric embryos, we sought to exclude the possibility that Nkx2.5Cre/+;ROSA26eGFP-DTA ESCs harbor an unexpected defect in their ability to contribute to heart-forming cells. To address this, we developed a complementary strategy involving the injection of wild-type eGFP− ESCs into blastocysts derived from the mating of Nkx2.5Cre/+ and ROSA26eGFP-DTA mice (Figure S3A). In particular, we assessed chimeric embryos or mice that are eGFP+ [i.e. derived from Nkx2.5Cre/+;ROSA26eGFP-DTA (ablated) or Nkx2.5+/+;ROSA26eGFP-DTA (control) blastocysts]. Following injection of 639 blastocysts using two different wild-type ESC lines (V6.5 and R1), we obtained 394 embryos or mice, with roughly half (226) being eGFP+. Similar to our Nkx2.5Cre/+;ROSA26eGFP-DTA ESC injection results above, we found that wild-type V6.5 and R1 ESCs exhibit a broad range of contribution (Figure S3B). The total number of recovered control ROSA26eGFP-DTA embryos and mice (144) was greater than the number of ablated Nkx2.5Cre/+;ROSA26eGFP-DTA animals (82), presumably due to lethality in ablated embryos with insufficient wild-type ESC contribution. Consistent with our earlier findings, embryos with up to 60% CPC loss were able to maintain normal embryonic development (Figure S3C). When all surviving embryos and mice were stratified by their degree of wild-type ESC chimerism, it was apparent that up to 60% ablation of CPCs could be tolerated (Figure S3D-S3E, Figure S4A-S4C). As we expected, no eGFP+ cells were present in the cTnT+ myocardium in the ablated mice (Figure S3F). Taken together, these findings support a robust compensatory mechanism in embryonic mouse hearts following CPC ablation.
Fractional ablation of embryonic CMs by chimeric complementation
While our CPC ablation model demonstrates loss of CPCs with exquisite temporal resolution, it does not reveal whether the regenerative response to ablation requires CPCs or whether developing cardiomyocytes are capable of responding to the loss of cardiac cells during development. To address this, we performed fractional ablation of developing immature cardiomyocytes using a previously published α-myosin heavy chain-Cre (αMHCCre/+) transgenic mouse line (Figure S5)15. Embryos with compound αMHCCre/+ and ROSA26eGFP-DTA transgenes have a well-formed heart tube at E8.0, but exhibit a hypocellular looped heart at E9.5 and the absence of a heart with pericardial edema and developmental arrest at E10.5 (Figure 3A). We generated several αMHCCre/+;ROSA26eGFP-DTA ESC lines and selected three (MHC-DTA1, MHC-DTA2, and MHC-DTA3) for further study based upon their capacity to contribute to chimeric embryos and mice with a broad range of efficiency (Figure 3B-3C). Interestingly, we found that these chimeric embryos were also able to tolerate up to 50-60% ablation of their immature myocytes (Figure 3D and Figure S4D-S4E). Immunohistological staining for eGFP and cTnT expression showed a progressive decrease in myocardial cellularity in E9.5 chimeric embryos with higher than 50% contribution by αMHCCre/+;ROSA26eGFP-DTA ESCs (Figure 3E). A rare number of eGFP+ CMs could be observed which may represent residual non-ablated CMs or CMs that were dying but had not been completely eliminated. Additional immunostaining for apoptotic cells confirmed that at least a fraction of these eGFP+ CMs underwent cleavage of caspase-3 (cCsp3) or were positive for TdT-mediated dUTP nick end labeling (TUNEL) (Figure S6A-S6B). Similar to embryos that had undergone CPC ablation, there was no apparent phenotypic difference after E10.5 between surviving embryos with a high degree of ablation (i.e. 50-60%) compared to those with less fractional ablation. Taken together, these results demonstrate a robust myocardial compensatory response when cell ablation occurs during the immature cardiomyocyte stage of development.
Figure 3.
Fractional ablation of embryonic CMs by chimeric complementation. (A) Schematic diagram of the eGFP/DTA construct before and after αMHC-Cre-mediated recombination (left panel). Brightfield and GFP fluorescence images of ROSA26eGFP-DTA control and αMHCCre/+;ROSA26eGFP-DTA transgenic embryos (right panel) at E8.5, E9.5, and E10.5 of development. Note the compound transgenic embryo shows a progressive defect in cardiac morphogenesis by E9.5 and is completely missing a heart structure at E10.5. (B) Generation of αMHCCre/+;ROSA26eGFP-DTA ESC lines. Brightfield and GFP fluorescence microscopy images are shown for the three ESC lines used. (C) Fractional CM ablation by injection of αMHCCre/+;ROSA26eGFP-DTA ESCs into wild-type CD-1 blastocysts (upper panel). The summary table demonstrates the complementation efficiency for each ESC line tested based upon the total number of animals collected (lower panel). (D) Brightfield and GFP fluorescence images of E10.5 chimeric αMHCCre/+;ROSA26eGFP-DTA embryos. Note that embryos with >50% ablation are developmentally arrested and exhibit impaired cardiac morphogenesis. A pericardial effusion is occasionally observed. (E) eGFP, troponin T (cTnT), and DAPI staining of E9.5 chimeric embryos with progressively increasing amounts of cardiomyocyte ablation. Some residual unablated eGFP+ cardiomyocytes remain in the hearts at this stage. Scale bars: 500 μm for (A); 200 μm for (B and E); 1 mm for (D).
Cell ablation triggers cardiomyocyte expansion ex vivo
Since the overall cardiac morphology in both ablation models was largely indistinguishable between surviving <50-60% ablated and control embryos, we examined whether there is a proliferative response to cell ablation by surveying the behavior of residual un-ablated cardiac cells ex vivo. We first dissected the heart tubes from E9.5 un-ablated control embryos, CPC-ablated embryos, and CM-ablated embryos. Next, we FACS-purified residual eGFP− cardiac cells, which are presumably responsible for recovering myocardial cellularity after ablation (Figure S7A), deposited one cell per well into a 96-well plate, and cultured the cells ex vivo for 7 or 14 days (Figure 4A). Approximately 9.5% of 5605 plated cells survived single-cell cloning. Examination of cells within each well revealed several patterns of specification. 36% of single-cell clones from control hearts and 20% and 44% of CPC or CM-ablated clones, respectively, were classified as cardiomyocytes (Figure 4B), exhibiting spontaneous beating and well-organized Z-lines marked by sarcomeric α-actinin expression (Figure 4C). Cardiomyocyte binucleation was also observed, but did not significantly differ among groups (Figure S7B). A small minority of clones (2% of control, 1% of CPC-ablated, and 1% CM-ablated clones) expressed CD31 and were highly proliferative, possessing a polygonal morphology, consistent with an endocardial/endothelial cell phenotype (Figure 4D). The remaining majority of clones (62% of control, 79% of CPC-ablated, and 54% CM-ablated clones) lacked significant sarcomeric α-actinin or CD31 expression (Figure 4E). These might have represented hematopoietic, epicardial, endocardial cushion mesenchyme, or neural crest-derived cells. No clones expressed smooth muscle myosin heavy chain, the definitive marker for the smooth muscle lineage.
Figure 4.
Embryonic CM proliferation is involved in myocardial recovery. (A) Schematic diagram of single-cell isolation and culture of E9.5 cardiac cells. For (B-J): experimental variables are represented as follows: [group: # of mice examined, mean percent ablation ± SD]. For (B-F): [control: n=9; CPC ablation: n=6, 45 ± 11%; CM ablation: n=6, 50 ± 12%]. (B) Percentage of single-cell derived colonies classified as cardiomyocytes, endocardial/endothelial cells, or other cells from dissociated E9.5 embryo hearts. (C-E) Histological analysis of single-cell derived colonies from E9.5 hearts. (C) Sarcomeric α-actinin+ cardiomyocytes showing well-organized Z-lines and sarcomeric structure. (D) CD31+ endocardial/endothelial cells. (E) Non-cardiomyocyte/non-endothelial cell lacking significant sarcomeric α-actinin or CD31 expression. (F) Box-and-whisker plot demonstrating median cell number and interquartile range among wells classified as containing cardiomyocyte colonies. Whiskers span the 10-90% range of values. Statistical comparisons were performed using the Kruskal-Wallis test with Dunn's correction for multiple comparisons. For (G-J): [control: n=4; CPC ablation: n=4, 41 ± 9%; CM ablation: n=4, 53 ± 12%]. Immunofluorescence confocal microscopy with 1 mm z-stack imaging for proliferative cardiomyocytes and endocardial cells in CPC-ablated (G and H) or CM-ablated (I and J) hearts at E8.5 or E9.5, respectively. The percentage of phospho-histone H3+ (pH3) (G and I) or Ki-67+ (H and J) cardiomyocytes in control and ablated hearts (filled arrows) was defined by co-localization with a DAPI+ nucleus that is circumscribed in all dimensions by a troponin T+ (cTnT) cell. Similarly, the percentage of phospho-histone H3+ or Ki-67+ endocardial cells (thin arrows) was defined by co-localization with a DAPI+ nucleus that is circumscribed in all dimensions with a CD31+ cell. Each symbol in the dot plot represents the fraction of cTnT+ or CD31+ cells staining positive for pH3 or Ki-67 in a single cardiac histology section; each symbol shape represents an independent mouse; a horizontal bar indicates the mean value for each group. 100-600 cTnT+ cells and 50-300 CD31+ cells were counted per section. Statistical comparisons were performed using a likelihood ratio test on a linear mixed-effects model. NS, non-significant. Scale bars: 100 μm for (C-E); 50 μm for (G-J).
To examine the effect of CPC or CM ablation on clonal cell growth potential, we counted the cardiomyocyte colony sizes resulting from single-cell plating on day 7 or 14. A normality plot indicated the colony size distribution deviated significantly from a Gaussian distribution, necessitating a nonparametric analysis of the data (Figure S7C). The presence of rare colonies with many cardiomyocytes suggests a hierarchical contribution to normal myocardial development and to the recovery of cardiomyocytes after ablation. On day 7, the median cardiomyocyte number per colony was significantly greater in colonies derived from CM-ablated embryos compared to un-ablated controls (5 versus 3) (Figure 4F). In contrast, no statistically significant difference was observed in cardiomyocyte number per colony between control and CPC-ablated embryos, although there was a trend toward greater colony size in CPC-ablated embryos at day 14 (Figure 4F). The lack of a significant difference in cardiomyocyte colony size between CPC-ablated and control embryos was most likely because CPC-ablated embryos had largely recovered their myocardial cellularity by E9.5, while the CM-ablated embryos were still actively recovering. These results indicate that CM ablation triggers a modest increase in cellular proliferation at E9.5 to increase cardiomyocyte number ex vivo.
Embryonic CM proliferation is involved in myocardial regeneration after ablation in vivo
We next examined whether there is a proliferative response in vivo to compensate for the loss of cardiomyocytes, as suggested by our ex vivo experiments, or whether the recovery following cell ablation is due merely to a hypertrophic response in residual un-ablated CMs30. In the case of hypertrophy, the individual cardiomyocyte size should increase without a corresponding change in myocardial cell proliferation rate, whereas in the case of proliferation, cardiomyocyte size should remain constant or decrease accompanied by an increase in the rate of proliferation. We first performed immunostaining for proliferation markers in embryo sections from both CPC or CM ablation models using the mitosis marker phosphorylated histone H3 (pH3), and the cell proliferation marker Ki-67 (Figure 4G-4J). We found that embryos from both CPC and CM ablation models exhibited increases in expression of both proliferation markers within their cTnT+ CMs. To see whether this correlated with changes in cardiomyocyte size, we quantified the ratio of myocardial area to the number of nuclei within cTnT+ CMs. We found, however, the CM area per nucleus was not statistically different between ablated embryos and controls despite a trend towards smaller cell size (Figure S8A-S8B). This suggests that an increase in proliferation is the prevailing mechanism involved in the morphological recovery of the ablated heart.
In light of the interdependence between the myocardium and neighboring endocardium to maintain normal cardiac development31, we also examined whether the development of the endocardium is affected by ablation of CPCs or CMs. At E8.5 in the CPC ablation model and at E9.5 in the CM ablation model, the endocardium in ablated embryos exhibited similar CD31 expression relative to controls consistent with normal endocardial differentiation (Figure 4G-4J). In addition, we quantified the degree of endocardial cell proliferation in CPC or CM-ablated embryos by assessing the percentage of Ki-67+ or phospho-histone H3+ endocardial cell nuclei. However, we did not observe a consistent difference in endocardial cell proliferation between ablated embryos and controls (Figure 4G-4J).
Since cardiomyocyte maturation is accompanied by changes in structure of the contractile unit including greater myofibril density, more organized alignment, and an increase in sarcomere length32, we examined whether CM maturation was delayed in ablated embryos at the ultrastructural level by transmission electron microscopy (Figure S8C-S8E). We found that at E10.5, hearts from CPC-ablated embryos exhibited a modest decrease in sarcomere thickness despite a lack of change in sarcomere length. This suggests that compensating cardiomyocytes may be phenotypically less mature due to their need for an increased rate of cell proliferation.
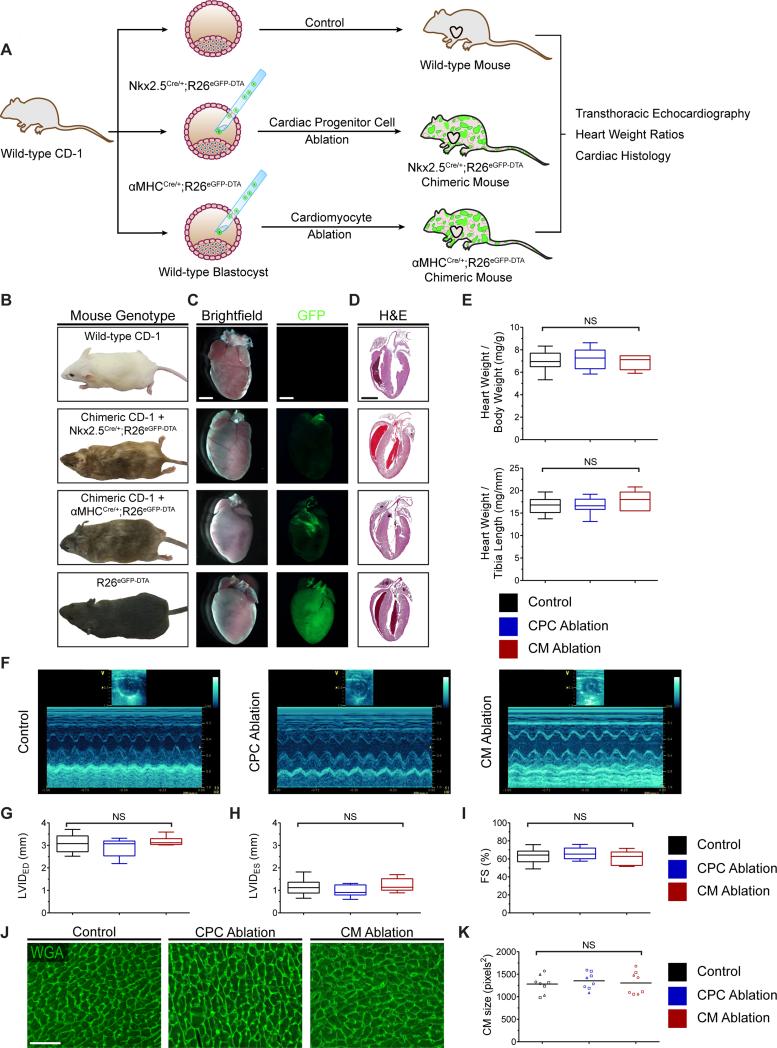
CPC and CM-ablated embryos can survive into adulthood with no overt cardiac defect
The above results demonstrate that a majority of embryonic CPCs or CMs can be ablated without triggering gross embryonic defects. To rule out more subtle congenital phenotypes in CPC- or CM-ablated embryos that would not manifest until late gestation or after birth, we collected chimeric live-born mice that had undergone an average of approximately 40% CPC or CM ablation and assessed their cardiac structure by histology and their cardiac function by echocardiography at 4-5 weeks of age (Figure 5A). We found that these chimeric mice displayed the expected coat color chimerism (Figure 5B) with few to no residual eGFP+ cells in the heart (Figure 5C). Compared to control un-ablated mice, their hearts showed similar gross overall size at the whole organ level (Figure 5C-5D) and had similar heart weights (Figure 5E), LV chamber dimensions (Figure 5F-5H), and contractile function (Figure 5I) as measured by echocardiography. We also assessed whether a hypertrophic response occurred in ablated hearts, which could indicate whether there had been a transient history of heart failure with remodeling changes. We found, instead, no evidence of hypertrophic changes as measured by mean cardiomyocyte size in ablated versus control hearts (Figure 5J-5K). These findings further support our conclusion that acute CPC or CM loss during early embryogenesis is well-tolerated in mice due to a rapid and efficient compensatory proliferative response that supports the development of normal organ size, CM number, and contractile function into early adulthood.
Figure 5.
CPC and CM-ablated embryos can survive into adulthood with no overt cardiac defect. (A) Strategy used for generation and analysis of 4-5 week-old mice undergoing CPC or CM ablation. (B) Representative images of control and ablated chimeric adult mice. (C) Brightfield and GFP fluorescence images of the dissected whole hearts. GFP signal is nearly absent in the hearts that have undergone ablation. (D) H&E staining of representative heart sections from control and ablated hearts. For (E-K): experimental variables are represented as follows: [group: # of mice examined, mean percent ablation ± SD]. For (E-I): results are displayed using box-and-whisker plots demonstrating median value and interquartile range. Whiskers span the minimum and maximum range of values. (E) Heart weight to body weight ratio and heart weight to tibia length ratio in wild-type and ablated hearts [control: n=28; CPC ablation: n=11, 39 ± 9%; CM ablation: n=6, 42 ± 11%] (F-I) Representative M-mode echocardiography images (F) and left ventricular dimensions (G and H) and function (I) quantified by fractional shortening [control: n=16; CPC ablation: n=9, 40 ± 10%; CM ablation: n=7, 41 ± 10%]. (J and K) Wheat germ agglutinin (WGA) staining of control and ablated hearts (J) and dot plot (K) displaying mean CM size. Each symbol in the dot plot represents the mean CM size in an individual heart section; a minimum of three 32× fields per section were counted; each symbol shape represents an independent mouse; a horizontal bar indicates the mean value for each group. [control: n=3; CPC ablation: n=3, 51 ± 6%; CM ablation: n=3, 51 ± 7%]. FS, fractional shortening; LVIDED, left ventricular internal diameter at end-diastole; LVIDES, left ventricular internal diameter at endsystole; NS, non-significant. Scale bars: 2 mm for (C and D); 50 μm for (J).
DISCUSSION
In this study, we developed a novel strategy to quantitatively examine the ability of a mammalian embryonic heart to compensate for rapid cell loss. Specifically, we made use of a blastocyst complementation assay that gives rise to chimeric embryos with fractional CPC or immature CM ablation during embryonic development. We found that embryos with up to 60% CPC loss were fully viable and could survive to adulthood. The compensatory process following CPC ablation at E7.5 was rapid and complete with little to no evidence of molecular alteration or cardiac cellular deficiency by E10.5. To examine the capacity of immature cardiomyocytes to respond to cell loss, we induced CM ablation at E9.0 and found, with remarkable consistency, that the loss of 50-60% of immature CMs was well-tolerated during embryonic development and that adult heart size and function in these animals were completely normal. Consistent with findings from newt and zebrafish cardiac regeneration, increased CM proliferation was evident in both CPC and CM ablation models. These data illustrate the significant capacity for myocardial injury repair in the embryonic mouse heart and raise the prospect that postnatal cardiac regeneration may be achievable in the mammalian heart if key mechanisms for triggering the expansion of proliferation-competent cardiomyocytes can be identified for therapeutic gain.
Our study employed a Cre-mediated conditional cell ablation strategy in mice to examine the response of an embryonic heart to fractional cell loss. Earlier studies have shown the remarkable ability of an adult zebrafish heart to replenish as much as a 70-80% loss of cardiomyocytes33, 34. While the study by Drenckhahn and colleagues illustrated the ability of an embryonic mammalian heart to increase cardiomyocyte proliferation in healthy cells to compensate for impaired proliferation in cardiomyocytes with metabolic derangement, the approach used was associated with an absence of apoptosis in the diseased cells and their subsequent retention.12 To our knowledge, no study has demonstrated complete myocardial regeneration in the embryonic mouse heart following extensive and temporally-controlled cell ablation. However, it should be recognized that fetal cardiomyocytes are proliferative at baseline and the ablation studies described here are likely an assessment of both the normal generative potential of fetal cardiomyocytes during cardiac development, as well as their regenerative potential given the increase in cardiomyocyte and/or cardiomyocyte precursor proliferation in the setting of cardiac cell ablation. Regardless, our data supports the remarkable capacity of the fetal heart to recover from experimental cell ablation during embryonic development.
We found that the embryonic mouse heart can replace as much as 50-60% of developing CPCs or immature CMs after Cre-mediated ablation and that proliferation of immature cardiomyocytes is involved in the compensatory response. In the absence of cell ablation, we observed expression by the majority of embryonic day 8.5-9.5 cardiomyocytes of the proliferation marker Ki-67, which is detectable during the active stages of the cell-cycle, including late G1, S, G2 and M phases, but which is absent in resting cells (G0)35. While this supports that cardiomyocytes are highly proliferative during this stage of development, we found that both CPC and CM ablation triggered a further increase in myocyte proliferative capacity. Likewise, histone H3 phosphorylation was also found to be increased upon CPC or CM ablation. However, its expression was only observed in a small fraction of developing myocytes, consistent with the fact that its phosphorylation is limited primarily to mitosis (M phase)36 (Figure 4G-4J). Due to the rapid differentiation of CPCs into immature cardiomyocytes at embryonic day 7.5 to 8.5 of development, we were unable to document an increase in the number of Isl-1+ CPCs in the residual heart after cell ablation (Figure 2D). Further studies using a combination of inducible cell ablation and blastocyst complementation assays will be necessary to specifically address the role of first or second heart field CPCs in the regenerative process.
Our findings also raise questions regarding the potential mechanisms involved in replacing the number of CPCs and CMs following ablation. While we believe that CPC and CM ablation by intracellular DTA expression should be a relatively cell-autonomous process without an exogenous inflammatory response from circulating leukocytes, differences in the amount of locally secreted signals from non-ablated cardiac cells may trigger the observed proliferative response. In addition, cell-cell contact has been well-described to regulate the rate of cell division37. The acute loss of CPCs or CMs may result in de-repression of this intrinsic negative regulator of cell proliferation. Furthermore, since the developing heart at this stage is continually receiving a new influx of cells from the second heart field (SHF), it is possible that an increase in migration and proliferation of SHF-derived cells may be involved in myocardial regeneration in the right ventricle and outflow tract. Further work will be necessary to clarify the precise mechanisms responsible for embryonic myocardial regeneration at distinct stages of development and within each heart field.
One important caveat of our findings that should be considered carefully is the potential role of other non-myocardial cells (e.g. endocardial and/or epicardial cells) in the compensatory process. It is well-known that signaling factors released from endocardial and epicardial cells play important roles in myocardial development38, 39. Our CPC ablation model depends on the expression of Cre from the endogenous Nkx2.5 promoter, which is known to express in pharyngeal endoderm at later stages of development, as well as endocardial and epicardial cells26. Consequently, this model may underestimate the degree of regeneration possible since it may ablate both the CPCs involved in myocardial formation as well as the endocardial/epicardial cells that regulate the compensatory process.
In summary, we found that quantitative and fractional ablation of developing mouse CPCs and immature CMs leads to cardiac cell loss that can be compensated for by residual unablated cells. This process is efficient and can rescue normal fetal development in hearts that have undergone up to 50-60% CPC or CM ablation. The mechanism for this compensatory response involves, in part, increased proliferation from residual immature CMs. Identification of the mechanisms involved in responding to the loss of CPC and CM number by triggering their increase in proliferation may enable us to devise improved therapies for heart failure due to myocardial cell loss.
Supplementary Material
CLINICAL PERSPECTIVE.
A wealth of studies support the importance of developmental signaling molecules to promote proper heart formation from lineage commitment to cellular expansion and maturation. Disruption in these formative events during embryonic development has a profound impact on human life, as congenital heart disease remains the most common cause of birth defects worldwide. However, whether there is a requirement for a specific number of progenitor cells to fashion a normal mammalian heart and the potential mechanisms that exist to compensate for unexpected cell loss during embryonic development have not been explored. To investigate whether a mechanism is present in the developing heart to compensate for the loss of cardiac cells during the earliest stages of heart formation, we employed a novel cell ablation strategy that could temporally ablate cardiac progenitor cells or immature cardiomyocytes in quantifiable fractions. By doing so, we found that, remarkably, mouse embryos could compensate for up to a 60% loss of cardiac cells without obvious morphological or functional defect. Furthermore, we show that increased proliferation of residual immature cardiomyocytes is at least partly responsible for this regenerative activity. These data provide direct evidence for the existence of a robust regenerative mechanism in the embryonic heart to compensate for cardiac progenitor cell and immature cardiomyocyte loss during development. In addition, the strategy we developed for fractional lineage-specific cell ablation should also be applicable for investigators in other organ systems to address the role of progenitor cell number in development and disease and their mechanism of regeneration.
Acknowledgments
We thank Dr. Robert J. Schwartz at the University of Houston for providing us with the Nkx2.5Cre/+ knock-in mice, Dr. E. Dale Abel at the University of Iowa for providing us with the αMHCCre/+ transgenic mice, Dorothy Hu and Kimberly Atkin McCurdy at the MGH Endocrine Histology Core for assistance with tissue sectioning, Andrew Olson at the Stanford Neuroscience Microscopy Service for assistance with microscopy, Laura Prickett-Rice at the Harvard Stem Cell Institute Flow Cytometry Core Facility and Patty Lovelace and Jennifer Ho at the Stanford Stem Cell FACS Core for assistance with FACS analysis, Anusha Kumar for assistance with cell culture and manuscript critique, R. Sharon Chinthrajah and Peter Sturzu for assistance with blinded cell counting, and David Knowles and Joseph Garner at Stanford University for assistance with statistical analysis. Author contributions: A.C.S. and S.M.W. conceived the design of the project. A.C.S performed the experiments, analyzed the data, and prepared the figures. K.R. provided assistance with ESC derivation and ESC culture. D.P. assisted with tissue sectioning. K.P. and A.F.X. assisted with immunohistochemistry, blinded measurements, mouse colony management, and figure preparation. A.R. performed blastocyst microinjection and transfer surgery. A.C.S. and T.C.T. acquired echocardiographic images and T.C.T. performed related measurements. A.S. assisted with figure preparation and plate reader assays. M.C.E. assisted with ESC culture. R.F. assisted with mouse colony management and ESC culture. G.L. assisted with FACS analysis. A.C.S., M.K.S., and R.G. prepared sections for electron microscopy and acquired images. K.D.R. assisted with immunohistochemistry. A.C.S. and S.M.W. wrote the manuscript, and it was edited by M.S.C. and I.J.D.
Funding Sources: This work was supported in part by a Harvard Stem Cell Institute Seed Grant, an NIH Director's New Innovator Award (DP2OD000041), the NIH/NHLBI Progenitor Cell Biology Consortium (U01HL009976), the California Institute for Regenerative Medicine (RB3-05129), an Endowed Facular Scholar Award from the Lucile Packard Foundation for Children/Child Health Research Institute at Stanford (to S.M.W.), the NIH T32 (HL007208 and HL94274), and a Postdoctoral Fellowship Award from the American Heart Association Founders Affiliate and the Lawrence J. & Florence A. DeGeorge Charitable Trust (to A.C.S.).
Footnotes
Disclosures: None.
References
- 1.Srivastava D. Making or breaking the heart: From lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Vincent SD, Buckingham ME. How to make a heart: The origin and regulation of cardiac progenitor cells. Curr Top Dev Biol. 2010;90:1–41. doi: 10.1016/S0070-2153(10)90001-X. [DOI] [PubMed] [Google Scholar]
- 3.Evans SM, Yelon D, Conlon FL, Kirby ML. Myocardial lineage development. Circ Res. 2010;107:1428–1444. doi: 10.1161/CIRCRESAHA.110.227405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- 5.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 6.Oberpriller JO, Oberpriller JC. Response of the adult newt ventricle to injury. J Exp Zool. 1974;187:249–259. doi: 10.1002/jez.1401870208. [DOI] [PubMed] [Google Scholar]
- 7.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, MacRae CA, Stainier DYR, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jopling C, Sleep E, Raya M, Marti M, Raya A, Belmonte JCI. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bettencourt-Dias M, Mittnacht S, Brockes JP. Heterogeneous proliferative potential in regenerative adult newt cardiomyocytes. J Cell Sci. 2003;116:4001–4009. doi: 10.1242/jcs.00698. [DOI] [PubMed] [Google Scholar]
- 10.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahmoud AI, Kocabas F, Muralidhar SA, Kimura W, Koura AS, Thet S, Porrello ER, Sadek HA. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drenckhahn J-D, Schwarz QP, Gray S, Laskowski A, Kiriazis H, Ming Z, Harvey RP, Du X J, Thorburn DR, Cox TC. Compensatory growth of healthy cardiac cells in the presence of diseased cells restores tissue homeostasis during heart development. Dev Cell. 2008;15:521–533. doi: 10.1016/j.devcel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Villa del Campo C, Claveria C, Sierra R, Torres M. Cell competition promotes phenotypically silent cardiomyocyte replacement in the mammalian heart. Cell Rep. 2014;8:1741–1751. doi: 10.1016/j.celrep.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an nkx2-5/cre gene using rosa26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 15.Abel ED, Kaulbach HC, Tian R, Hopkins JC, Duffy J, Doetschman T, Minnemann T, Boers ME, Hadro E, Oberste-Berghaus C, Quist W, Lowell BB, Ingwall JS, Kahn BB. Cardiac hypertrophy with preserved contractile function after selective deletion of glut4 from the heart. J Clin Invest. 1999;104:1703–1714. doi: 10.1172/JCI7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanova A, Signore M, Caro N, Greene ND, Copp AJ, Martinez-Barbera JP. In vivo genetic ablation by cre-mediated expression of diphtheria toxin fragment a. Genesis. 2005;43:129–135. doi: 10.1002/gene.20162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soriano P. Generalized lacz expression with the rosa26 cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 18.Eggan K, Akutsu H, Loring J, Jackson-Grusby L, Klemm M, Rideout WM, 3rd, Yanagimachi R, Jaenisch R. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci U S A. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gertsenstein M, Nutter LM, Reid T, Pereira M, Stanford WL, Rossant J, Nagy A. Efficient generation of germ line transmitting chimeras from c57bl/6n es cells by aggregation with outbred host embryos. PLoS One. 2010;5:e11260. doi: 10.1371/journal.pone.0011260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogan B. Manipulating the mouse embryo : A laboratory manual. Cold Spring Harbor Laboratory Press; Plainview, N.Y.: 1994. [Google Scholar]
- 23.Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. Nkx-2.5: A novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:969. doi: 10.1242/dev.119.3.969. [DOI] [PubMed] [Google Scholar]
- 24.Chen WP, Wu SM. Small molecule regulators of postnatal nkx2.5 cardiomyoblast proliferation and differentiation. J Cell Mol Med. 2012;16:961–965. doi: 10.1111/j.1582-4934.2011.01513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saburi S, Azuma S, Sato E, Toyoda Y, Tachi C. Developmental fate of single embryonic stem cells microinjected into 8-cell-stage mouse embryos. Differentiation. 1997;62:1–11. doi: 10.1046/j.1432-0436.1997.6210001.x. [DOI] [PubMed] [Google Scholar]
- 26.Stanley EG, Biben C, Elefanty A, Barnett L, Koentgen F, Robb L, Harvey RP. Efficient cre-mediated deletion in cardiac progenitor cells conferred by a 3′utr-ires-cre allele of the homeobox gene nkx2-5. Int J Dev Biol. 2002;46:431–439. [PubMed] [Google Scholar]
- 27.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. Hippo pathway inhibits wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qyang Y, Martin-Puig S, Chiravuri M, Chen S, Xu H, Bu L, Jiang X, Lin L, Granger A, Moretti A, Caron L, Wu X, Clarke J, Taketo MM, Laugwitz KL, Moon RT, Gruber P, Evans SM, Ding S, Chien Kenneth R. The renewal and differentiation of isl1+ cardiovascular progenitors are controlled by a wnt/beta-catenin pathway. Cell Stem Cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 29.Zeisberg EM, Ma Q, Juraszek AL, Moses K, Schwartz RJ, Izumo S, Pu WT. Morphogenesis of the right ventricle requires myocardial expression of gata4. J Clin Invest. 2005;115:1522–1531. doi: 10.1172/JCI23769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28:1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 31.Tian Y, Morrisey EE. Importance of myocyte-nonmyocyte interactions in cardiac development and disease. Circ Res. 2012;110:1023–1034. doi: 10.1161/CIRCRESAHA.111.243899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregorio CC, Antin PB. To the heart of myofibril assembly. Trends Cell Biol. 2000;10:355–362. doi: 10.1016/s0962-8924(00)01793-1. [DOI] [PubMed] [Google Scholar]
- 33.Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DY. Conditional targeted cell ablation in zebrafish: A new tool for regeneration studies. Dev Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Panakova D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, Werdich AA, Yelon D, Macrae CA, Poss KD. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scholzen T, Gerdes J. The ki-67 protein: From the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 36.Hans F, Dimitrov S. Histone h3 phosphorylation and cell division. Oncogene. 2001;20:3021–3027. doi: 10.1038/sj.onc.1204326. [DOI] [PubMed] [Google Scholar]
- 37.Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 38.Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived fgf signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama YS, Chen H, Shou W, Ballestar E, Esteller M, Rojas A, Perez-Pomares JM, de la Pompa JL. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12:415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.