Abstract
Bacterial vaginosis is a common reproductive infection in which commensal vaginal lactobacilli are displaced by a mixed population of pathogenic bacteria. Bacterial vaginosis increases susceptibility to HIV, and it has been suggested that host innate immune responses to pathogenic bacteria contribute to enhanced infection, yet the cellular mechanisms mediating the increased HIV susceptibility remain uncharacterized.
We evaluated the HIV-enhancing effects of bacterial vaginosis by inoculating endocervical epithelia with Atopobium vaginae, a bacterial vaginosis-associated bacteria, and assaying secreted factors for HIV-enhancing activity. When epithelia and A. vaginae were cocultured, we observed increased HIV-enhancing activity mediated by secreted low molecular weight factors. From this complex mixture we identified several upregulated host proteins, which functioned in combination to enhance HIV infection.
These studies suggest that the host immune response to bacterial vaginosis-associated bacteria results in the release of HIV-enhancing factors. The combined activity of bacterial vaginosis-induced proteins likely mediates HIV enhancement.
Keywords: Atopobium vaginae, bacterial vaginosis, female reproductive tract, heterosexual HIV transmission, innate immunity, mucosal immunology
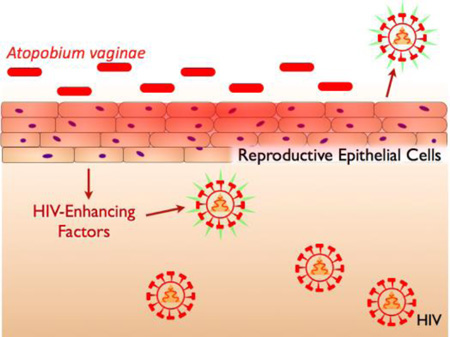
Graphical Abstract
Bacterial vaginosis-associated bacteria Atopobium vaginae stimulates endocervical epithelia to secrete factors that enhance downstream HIV infection.
1. Introduction
Bacterial vaginosis (BV) is a microbial shift condition in which commensal vaginal lactobacilli are displaced by an overgrowth of mixed pathogenic bacterial populations [1]. BV is the most common infection of the female reproductive tract (FRT) for which clinical help is sought, affecting 20–60% of women [2, 3, 4]. Importantly, BV increases a woman’s susceptibility to HIV by 60% [4]. As the mechanism by which BV increases HIV acquisition remains enigmatic, it is critical to characterize the interactions between the FRT and BV-associated bacteria (BVAB).
The FRT possesses multiple mechanisms of innate defense, the foremost of which is the protective epithelial cell layer that lines the FRT, and represents the first point of contact for invading pathogens [5]. In addition to acting as a physical barrier, FRT epithelia also secrete antimicrobial proteins, including host defense peptides, into the cervicovaginal fluid [6, 7, 8]. Among these host defense peptides are the alpha- and beta-defensins (e.g. HNP5, hBD2) [9, 10], and peptides belonging to the whey acid protein family (e.g. SLPI, trappin- 2/elafin) [11, 12]. As a result, cervicovaginal fluid exhibits antibacterial and antiviral activity, including potent anti-HIV activity, which is accomplished by the synergistic contributions of individual peptides [13].
FRT epithelia also perform a critical role in immune surveillance. Pathogens introduced to the FRT will first encounter the epithelia, and these cells are equipped with immune sensory mechanisms that initiate a response characterized by the release of soluble cytokines and antimicrobial proteins [14, 15]. This response serves to combat pathogens while simultaneously initiating additional host immune responses. In the case of BV, the immune response initiated in FRT epithelia upon stimulation with BVAB is implicated in increasing HIV infection by multiple mechanisms. First, hBD2 upregulation in FRT epithelium likely mediates the recruitment of lymphocytes, target cells for HIV infection [16]. Second, intruding BVAB induce an inflammatory response that could increase HIV susceptibility by activating NF-κB, a major transcription factor driving HIV genomic replication [15, 17]. Each of these host immune responses represents a possible mechanism by which BV enhances HIV susceptibility, making it a high priority to characterize the interactions between BVAB and host immunity.
We previously evaluated the immune interactions between FRT epithelia and vaginal bacteria [16]. We observed that endocervical epithelia are highly responsive to BVAB, and can be used as a sensitive indicator of pathogenic interactions. At the same time, we demonstrated that of 10 tested bacterial species, one in particular was a strong stimulator of host response: Atopobium vaginae. This species is a highly specific marker of BV, and additionally has been demonstrated to elicit a potent immune response in host cells and tissues [16, 18–20]. In this report, we describe how such host-pathogen immune interactions affect downstream HIV infection. We demonstrate that soluble factors secreted upon inoculation of endocervical epithelia with A. vaginae can increase HIV infection. Further characterization suggested that these HIV-enhancing proteins likely work in concert to increase HIV susceptibility as a consequence of the immune interaction between reproductive epithelia and BVAB.
2. Materials and methods
2.1. Epithelial cultures
The human endocervical epithelial line End1 (CRL-2615) was purchased from American Type Culture Collection (ATCC).This culture was maintained according to ATCC instructions. TZM-bl cells (Dr. John C. Kappes, Dr. Xiaoyun Wu, Tranzyme Inc.) were acquired from the National Institutes of Health AIDS Research and Reference Reagent Program, and were maintained in DMEM (Mediatech Inc.) 10% Fetal Bovine Serum (FBS, Gemini Bio-Products).
2.2. Bacterial cultures
The bacterial culture Atopobium vaginae (BAA-55) was purchased from ATCC. A. vaginae was grown in tryptic soy broth with 5% defibrinated rabbit blood or on equivalent agar plates in anaerobic GasPaks (Becton, Dickinson and Company) at 37 °C. To achieve consistency in bacterial preparations, maintenance cultures were aliquoted and snap frozen by submerging in liquid nitrogen for 2 hr, then transferred to −80 °C until use.
2.3. Inoculation of epithelia with A. vaginae
Epithelia were grown to confluency on 100 mm tissue culture-treated plates (Techno Plastic Products) as instructed by ATCC. To prepare inocula for experiments, snap-frozen aliquots of A. vaginae were thawed, and desired volumes were centrifuged. Supernatants were aspirated, and bacteria were resuspended in Ham’s F12 media (Mediatech Inc.). The maintenance keratinocyte serum free media (KSFM, Invitrogen Life Technologies) was aspirated from each plate of epithelia, and 8 ml of the inoculum was applied to each plate. Plates were returned to 37°C/5% CO2 for the duration of the experiment. In all experiments, epithelia were inoculated at an average multiplicity of infection of 3.3. The number of bacterial CFUs was determined by serially diluting the inocula and plating on appropriate media for back-calculation of density.
2.4. Conditioned media preparation
At the experimental endpoint, 24 ml conditioned media (CM) from three 100 mm dishes were pooled and filtered through a 30 Kda molecular weight cut off (MWCO) filter (Millipore) at 4 °C to remove cell debris. Clarified CM were stored at −20 °C until desalting. To desalt, pooled CM was subjected to filtration through an Amicon 3 KDa MWCO. The sample was centrifuged on the filter to concentrate, then diluted with water to reduce salt concentration in accordance manufacturer’s instructions. This procedure was repeated until the final salt concentration was negligible (0.01% of the original composition, allowing it to be analyzed in cell culture and by electrophoresis). The desalted CM was equilibrated to 1 ml with water and stored at −20 °C for subsequent experiments. For downstream cell culture experiments, equal volumes of 24X desalted CM were combined with 2X DMEM to achieve 1X DMEM equivalent in cell culture. For Bio-plex analysis, 24X CM samples were diluted 1:10 in DPBS (Mediatech Inc.) and assayed according to manufacturer’s instructions.
2.5. HIV enhancement assay
TZM-bl cells were seeded at 5,000 cells per well in a 96-well black-wall, clear-bottom plate. Cells were grown for 48 hr in DMEM 10% FBS prior to experiment. For treatment, media were aspirated from each well, and replaced with 1% FBS, CM or recombinant protein, and HIV-1 BaL inoculum at 4 ng/ml p24 (MOI ~0.02), all prepared in DMEM. Treatments were incubated at 37 °C/5% CO2 for 4 hr, after which wells were aspirated, and treatment media were replaced with identical FBS and CM/recombinant protein conditions, but without virus. At 24 hr post-infection, media were aspirated from all wells, and 100 µl Glo Lysis Buffer (Promega) was applied to each well. Plates were frozen at −80°C, and later thawed at room temperature. For the assay, 100 µl BrightGlo Reagent (Promega) was added to each well and luciferase activity was quantified. An uninfected control well was also measured, and the background relative light units (RLUs) subtracted from all experimental wells.
2.6. Trypan blue viability assay
TZM-bl cells were seeded and treated as in the HIV Enhancement Assay, except that 1X DMEM was added in place of viral inoculum. At 24 hr post-treatment, cells were lifted with warm 0.05% trypsin/EDTA, and trypan blue was added to each sample. This resuspension was loaded onto a hemocytometer, and live cells were identified by their exclusion of trypan blue.
2.7. Tricine-SDS PAGE for whole protein visualization and immunoblotting
For protein visualization, 24X desalted CM was further concentrated and resolved by Tricine-SDS PAGE, and proteins were visualized by silver nitrate staining. For immunoblotting, the following recombinant proteins were used as standards: Lipocalin-2 (Abcam ab95007); Cyclophilin-A (Abcam ab86219); Trappin-2 (Novoprotein CA39); HE4 (Novoprotein C550). Gels were transferred to PVDF membranes and immunoblotted with the following rabbit polyclonal antibodies: anti-Lipocalin-2 (Abcam ab41105); anti-Cyclophilin-A (Millipore 07–313); anti-Trappin-2 (Santa Cruz Biotehcnology sc-20637); anti-HE4 (Abcam ab85179).
2.8. Mass spectrometry
For MS analysis, 1 ml of 24X CM was concentrated and resuspended in 50 mM NH4CO3. Subsequent reduction, alkylation, and trypsin digestion were also performed in 50 mM NH4CO3. Sample clean-up was performed on MacroSpin Silica C18 Columns (The Nest Group) according to manufacturer’s instructions. Eluted peptides were analyzed on a nanoflow HPLC QSTAR-ELITE quadrupole TOF system. The resulting MS/MS spectra were searched against an IPI_human database assuming the digestion enzyme trypsin.
2.9. Statistical analyses
For viral enhancement assays of CM at different concentrations, two-way ANOVA with Bonferroni posttests were used to compare fold infection in each epithelial condition to the no epithelial condition, or to compare RLUs in the mock condition to the Atopobium condition. For cytokine analysis, paired t-tests were used to directly compare mock and A. vaginae-inoculated conditions for each cytokine. For TZM-bl trypan assays, unpaired t-tests were used to compare viability in treated conditions to media controls. For recombinant protein viral enhancement assays, one-tailed unpaired t-tests were used to compare treated conditions to untreated controls.
3. Results
3.1. A. vaginae stimulation increases HIV-enhancing activity of FRT epithelial conditioned media
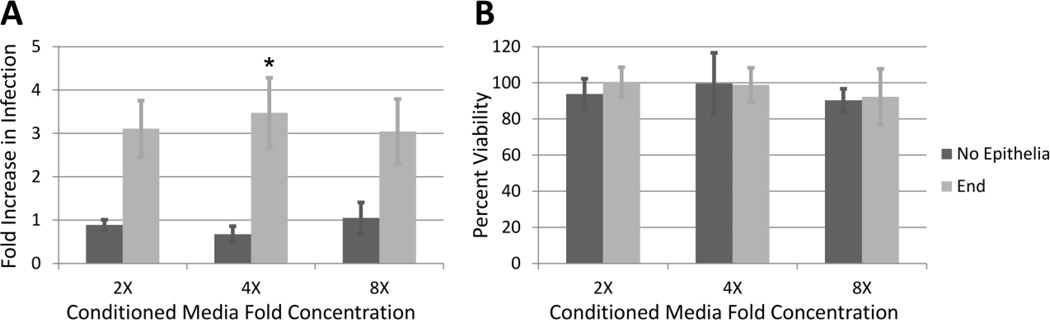
As many low molecular weight cytokines and antimicrobial peptides regulated by the epithelial innate immune response also alter HIV susceptibility [16, 17, 21], we sought to characterize the effects of these factors on HIV infection. To isolate the secreted, low molecular weight factors produced by reproductive epithelial cells in response to the BVAB, we inoculated End1 cells with the BVAB Atopobium vaginae. Matched mock-inoculated conditions received a media change, and bacteria-only conditions included inoculum but no epithelia. After a 24 hr co-incubation, the conditioned media (CM) was collected, desalted, and assayed for HIV-enhancing activity using well-characterized HIV reporter TZM-bl cells. Fig. 1 panel A shows the fold-enhancement of HIV-infection induced by CM collected from A. vaginae-inoculated End1 cells relative to mock-inoculated End1 cells. The CM generated from End1 cells inoculated with A. vaginae exhibited significant HIV-enhancing effects compared to the mock CM condition, inducing a 3–4-fold increase in infection over the mock-inoculated condition at all concentrations tested. The fold-difference in activity was greatest when treatments were added at 4X concentration, and this effect decreased at higher concentrations as the mock-inoculated CM began to exhibit slight viral enhancement, thereby decreasing the fold-difference. Importantly, we did not observe increased infection for the A. vaginae-inoculated/no epithelia condition over the mock-inoculated/no epithelia condition, indicating that CM obtained from A. vaginae bacteria alone did not increase HIV infection.
Figure 1. A. vaginae Stimulation Significantly Increases the HIV-Enhancing Activity of Endocervical Epithelial CM.
A) TZM-bl cells were infected with HIV-1 in the presence of CM applied at final concentrations of 2–8X. Graphed values are the fold increase in infection of the Atopobium-treated condition divided by the mock-treated condition, where a value of 1 represents no increase in infection of the Atopobium condition compared to mock. Shown are data for CM obtained from inoculated End1 cells, or from bacteria-alone conditions as a control (No Epithelia). n = 3–8. Asterisk (*=p<0.05) indicates an increase in infection of the End1-A. vaginae condition compared to No Epithelia-A. vaginae condition. B) In parallel, TZM-bl cells were treated with CM and endpoint viability was quantified by trypan blue and normalized to a media-only control condition. n = 3–4 for each condition.
To ensure that the observed activity was a specific HIV-enhancing activity and not due to changes in cell viability, we measured cell viability in uninfected TZM-bl cells treated with CM. Fig. 1 panel B demonstrates that the CM treatment did not induce changes in TZM-bl viability, suggesting that the observed HIV-enhancing activity was likely on account of differences in infection, rather than differences in the number of cells producing luciferase. Therefore, we sought to identify the HIV-enhancing factors in this sample.
3.3. Mass spectrometry reveals potential enhancers of HIV infection
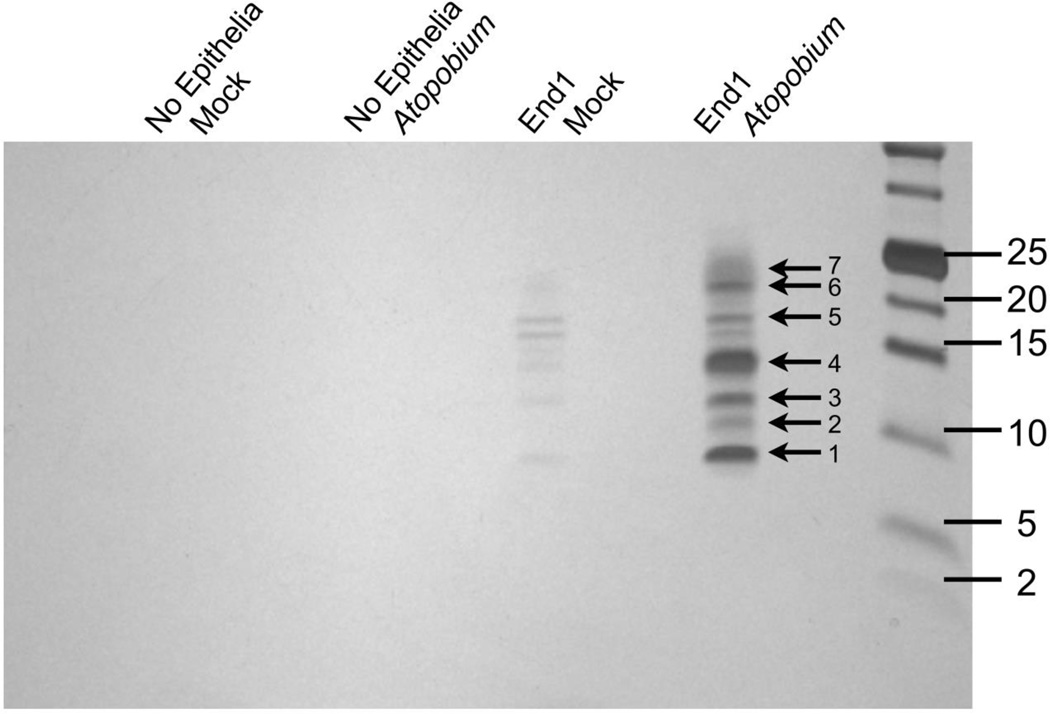
To characterize potential HIV-enhancing factors, CM samples generated from media control alone, A. vaginae alone, End1 cells alone, or End1 cells inoculated with A. vaginae were resolved by PAGE and visualized by silver stain. Fig. 2 demonstrates that no proteins could be visualized in the media control, or A. vaginae alone conditions. The End1 condition did contain low molecular weight proteins that were just discernible by silver stain. In contrast, the End1 cells inoculated with A. vaginae produced pronounced bands. The pattern observed in this sample mirrored that of the End1 alone condition, but staining intensity suggested upregulation of these proteins in response to bacterial inoculation.
Figure 2. Several Proteins are Enriched in HIV-Enhancing CM Fraction.
CM samples were concentrated and resolved by Tricine SDS-PAGE, then visualized by silver stain. Indicated bands were excised for MS analysis. Gel image is one representative of three independent experiments.
To identify these factors, the CM fraction from End1 cells inoculated with A. vaginae was concentrated and subjected to ESI-QUAD-TOF analysis. Sixty-six protein identities were returned (Table 1). To narrow these results, we selected proteins of interest based on their previously detection in the female reproductive tract, indicating physiological relevance, and their previous implication in innate immunity or HIV infection. These requirements allowed us to select four proteins to investigate further: lipocalin 2 (LCN2 gene), cyclophilin A (PPIA gene), trappin-2 (PI3 gene), and HE4 (WFDC2 gene) [12, 13, 22, 23].
Table 1. Proteins identified by MS analysis of soluble 3–30 KDa CM fraction.
Shown are protein identities obtained by analyzing the 3–30 KDa CM fraction by mass spectrometry, and referencing against a human database. For each entry, the gene symbol(s), description, and molecular weight are given, followed by Mascot score, which was used to order the protein identities. Proteins chosen for subsequent studies are shown in bold and highlighted.
| Gene Symbol | Description | MW [Da] | Score |
|---|---|---|---|
| BASP1 | Isoform 1 of Brain acid soluble protein 1 | 22680 | 225 |
| TPI1; TPI1P1 | Triosephosphate isomerase isoform 2 | 31057 | 204 |
| WFDC2 | Isoform 1 of WAP four-disulfide core domain protein 2 | 13953 | 186 |
| ACTB | Actin, cytoplasmic 1 | 42052 | 126 |
| IGF2; INS; INS-IGF2 | Insulin | 12315 | 122 |
| SOD1 | Superoxide dismutase [Cu-Zn] | 16154 | 111 |
| TPM2 | Isoform 2 of Tropomyosin beta chain | 33027 | 102 |
| SH3BGRL3 | Putative uncharacterized protein | 24108 | 100 |
| KRT17 | Keratin, type I cytoskeletal 17 | 48361 | 100 |
| MT1G | Isoform 1 of Metallothionein-1G | 7277 | 89 |
| VGF | Neurosecretory protein VGF | 67275 | 86 |
| YBX1 | Nuclease-sensitive element-binding protein 1 | 35903 | 84 |
| LCN2 | Isoform 1 of Neutrophil gelatinase-associated lipocalin | 22745 | 84 |
| RPS27A | Ubiquitin-40S ribosomal protein S27a | 18296 | 83 |
| PPIA | Peptidyl-prolyl cis-trans isomerase A | 18229 | 82 |
| PFN1 | Profilin-1 | 15216 | 78 |
| TXN | Thioredoxin | 12015 | 77 |
| PKM2 | Isoform M1 of Pyruvate kinase isozymes M1/M2 | 58538 | 77 |
| RPS28 | 40S ribosomal protein S28 | 7893 | 74 |
| (TPM1) | cDNA FLJ16459 fis, clone BRCAN2002473, moderately similar to Tropomyosin, fibroblast isoform 2 | 37487 | 73 |
| TPM2 | Isoform 3 of Tropomyosin beta chain | 28666 | 71 |
| SDC4 | Syndecan-4 | 21628 | 68 |
| KRT5 | Protein | 23267 | 68 |
| (KRT5) | cDNA FLJ54081, highly similar to Keratin, type II cytoskeletal 5 | 59042 | 68 |
| ENO2 | Gamma-enolase | 47581 | 68 |
| SCG5 | Uncharacterized protein | 21486 | 67 |
| UCHL1 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | 25151 | 65 |
| MARCKS | Myristoylated alanine-rich C-kinase substrate | 31707 | 65 |
| SUMO4 | Small ubiquitin-related modifier 4 | 10735 | 64 |
| LOC728825; SUMO2 | Small ubiquitin-related modifier 2 | 10889 | 64 |
| CFL1 | Cofilin-1 | 18719 | 64 |
| CANX | Calnexin | 67982 | 63 |
| SFN | Isoform 1 of 14-3-3 protein sigma | 27871 | 63 |
| VCP | Transitional endoplasmic reticulum ATPase | 89950 | 61 |
| ISG15 | Ubiquitin-like protein ISG15 | 17933 | 60 |
| HMGA1 | Isoform HMG-I of High mobility group protein HMG-I/HMG-Y | 51230 | 58 |
| PEBP1 | Phosphatidylethanolamine-binding protein 1 | 21158 | 56 |
| HSPA5 | 78 kDa glucose-regulated protein | 72402 | 56 |
| GAL | Galanin peptides | 13294 | 54 |
| (PPIA) | cDNA FLJ75025, highly similar to Homo sapiens peptidylprolyl isomerase A (cyclophilin A) | 11582 | 53 |
| LRRFIP1 | Isoform 2 of Leucine-rich repeat flightless-interacting protein 1 | 86979 | 51 |
| PARK7 | Protein DJ-1 | 20050 | 49 |
| TPM3 | 33 kDa protein | 33430 | 48 |
| CALM3; CALM2; CALM1 | Calmodulin | 16827 | 47 |
| ACTBL2 | Beta-actin-like protein 2 | 42318 | 47 |
| KRT79 | Keratin, type II cytoskeletal 79 | 58085 | 46 |
| KRT2 | Keratin, type II cytoskeletal 2 epidermal | 66110 | 46 |
| PGK1 | Phosphoglycerate kinase 1 | 44985 | 45 |
| PTMS | Parathymosin | 11523 | 45 |
| HNRNPK | Isoform 1 of Heterogeneous nuclear ribonucleoprotein K | 51230 | 45 |
| SCG2 | Secretogranin-2 | 70897 | 44 |
| AVP | Vasopressin-neurophysin 2-copeptin | 17325 | 43 |
| OXT | Oxytocin-neurophysin 1 | 13739 | 43 |
| DSC3 | Isoform 3A of Desmocollin-3 | 101218 | 43 |
| EIF4B | Eukaryotic translation initiation factor 4B | 69167 | 42 |
| ZNF510 | Zinc finger protein 510 | 73710 | 41 |
| PI3 | Elafin | 12718 | 41 |
| EIF3J | Eukaryotic translation initiation factor 3 subunit J | 29159 | 41 |
| TUBA4A | Tubulin alpha-4A chain | 50634 | 39 |
| PTMA | Protein | 15849 | 39 |
| MIR1244-3; MIR1244-2; MIR1244-1 | Prothymosin alpha | 11804 | 39 |
| TRIM29 | Isoform Alpha of Tripartite motif-containing protein 29 | 66478 | 38 |
| TMSB10 | Thymosin beta-10 | 5023 | 38 |
| KCNG2 | Potassium voltage-gated channel subfamily G member 2 | 52176 | 38 |
| SNORA6; RPSAP15; SNORA62; RPSA | Laminin receptor-like protein LAMRL5 | 33089 | 38 |
| ACO2 | Aconitase (Fragment) | 65934 | 38 |
3.4. Immunoblot confirms upregulation of putative HIV-enhancing proteins in the 3–30 KDa CM fraction
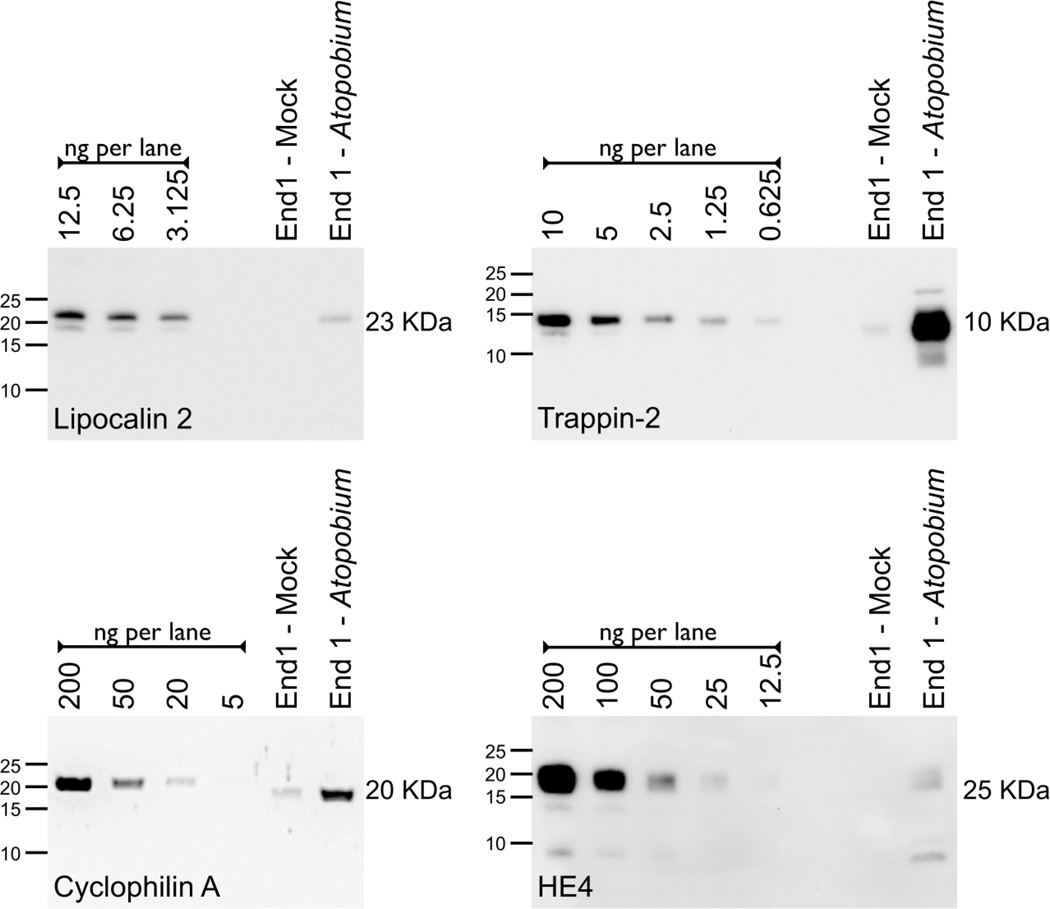
To confirm the upregulation of lipocalin 2, cyclophilin A, trappin-2 and HE4, we performed immunoblots on the CM sample from either mock-inoculated or A. vaginae-inoculated End1 cells. Recombinant protein standards were analyzed alongside the CM in order to validate commercial antibodies and to estimate the protein content. Fig 3 demonstrates the upregulation of each of these four proteins in the A. vaginae-inoculated condition compared to a matched mock-inoculated fraction.
Figure 3. Several Proteins are Upregulated by End1 Epithelia in Response to A. vaginae stimulation.
The following volumes of CM were concentrated, resolved, and immunoblotted for protein detection: 300 µl for Lipocalin 2; 400 µl for Cyclophilin A; 450 µl for Trappin-2 and 2000 µl for HE4. Recombinant standards (labeled as ng/lane) were resolved alongside CM to estimate protein content. One representative of three independent experiments for each analyte is shown.
3.5. HIV enhancement is mediated by the combined activity of upregulated proteins
Having observed that each of the selected proteins was upregulated by A. vaginae-inoculation of End1 cells, we sought to determine whether any one of these proteins might be responsible for the corresponding HIV-enhancing activity. Semi-quantitative immunblotting revealed that each of the selected proteins in the A. vaginae-inoculated End1 sample had been present at concentrations of 2–40 ng/ml in prior TZM-bl assays. Therefore, recombinant preparations of each protein were added to a TZM-bl infection at concentrations as high as 100 ng/ml to gauge viral enhancement activity. Surprisingly, none of the four proteins altered HIV infection (data not shown).
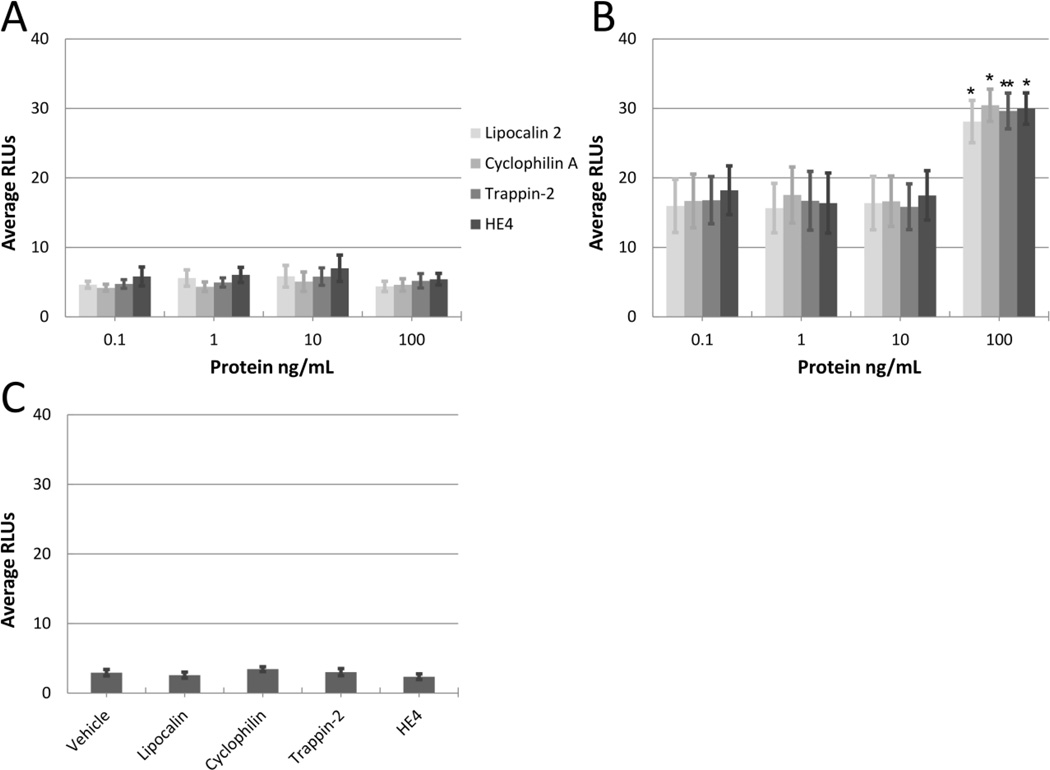
We next considered that the HIV-enhancing activity is likely a combined effect of host proteins, and that the addition of any single element might not recapitulate the viral enhancement observed in the complex CM sample. To evaluate this possibility, we again performed the TZM-bl assay, this time enriching each recombinant protein in the mock-inoculated or A. vaginae-inoculated End1 CM. Fig. 4 panel A demonstrates a similar lack of HIV-enhancing activity observed for any of the four recombinant proteins in the mock-inoculated End1 condition. Fig. 4 panel B demonstrates a significant increase in HIV-enhancing activity exerted by each of the four proteins of interest when applied at 100 ng/ml in the A. vaginae-inoculated CM composition.
Figure 4. Recombinant Proteins Enhance HIV Infection in the Presence of A. vaginae-Inoculated End1 CM.
Recombinant proteins lipocalin 2, cyclophilin A, trappin-2 or HE4 were added to the TZM-bl reporter assay at 0.1–100 ng/ml, in the presence of A) mock-inoculated End1 CM, or B) A. vaginae-inoculated End1 CM. CM was added at 4X concentration in all conditions. Asterisks (*=p<0.05, **=p<0.01) indicate an increase in RLUs compared to control condition containing CM without recombinant protein. n = 3–5. C) Individual proteins were assayed at 100 ng/ml in the presence of 10 µg/ml BSA. n = 3.
As we had previously observed an increased total protein content in the A. vaginae-inoculated End1 CM fraction, we considered that the HIV-enhancing activity of this fraction, with or without added recombinant proteins, could be a nonspecific artifact of increased protein concentration in the TZM-bl reporter assay. To confirm that the viral enhancement was specific, we also assayed each recombinant protein in the presence of a protein not known to enhance viral infection. Bicinchoninic acid protein quantification revealed that when applied at 4X final concentration, the A. vaginae-inoculated End1 sample contributed 3 µg/ml total protein to the TZM-bl assay. Therefore, we added bovine serum albumin (BSA) at an excess concentration of 10 µg/ml to examine potential HIV-enhancing activity by nonspecific effects of the increased protein content. Fig. 4 panel C shows that BSA failed to exhibit HIV-enhancing activity on its own, and also failed to facilitate HIV-enhancing activity of any recombinant protein, confirming that the observed HIV enhancing activity was specific to the A. vaginae-inoculated End1 CM composition. These data suggested that the HIV-enhancing activity of the A. vaginae-inoculated End1 sample is specific, and that one or more of the upregulated proteins likely function in combination to enhance HIV infection.
4. Discussion
In this study, we evaluated the immune response initiated by FRT epithelia upon stimulation with BVAB, and showed that the interaction between host and bacterial cells resulted in the secretion of HIV-enhancing molecules that increased downstream HIV infection. We found that End1 cells produced HIV-enhancing CM after inoculation with A. vaginae. This observation is in line with our previous characterization, which revealed End1 cells as a highly responsive cell type in response to inoculation with BVAB, secreting increased concentrations of cytokines and host defense peptides [16].
Our proteomic analysis of the stimulatory CM revealed a complex mixture. In contrast to our prior studies, we did not detect inflammatory cytokines in our conditioned media preparations. We determined that this was not due to the failure of epithelium to secrete cytokines, but rather due to the retention of cytokines on the filters that were used to process our CM samples. This led us to pursue other secreted factors that stimulate viral infection in downstream target cells.
The incubation duration utilized in the TZM-bl assay is intended to evaluate factors influencing a single round of viral infection. Thus we expect that the increased infection induced by CM is due to events occurring at early infectious stages, from viral entry to proviral transcription. It is highly likely that our observations are due to effects of CM on viral entry, as this is a common means by which host factors may influence infection in the TZM-bl assay. In support of this, other groups have identified host-derived factors that influence viral attachment, retrograde transport of viral-containing vesicles, and viral fusion [24]. It is also possible that proviral transcription is the mechanism by which our treatments enhanced infection. This is because activation of intracellular inflammatory cascades initiated in response to paracrine signaling often activates NF-κB, a strong transcriptional activator of the HIV proviral genome [17]. In support of this model, other groups have identified human serum components that enhance HIV transcription [25]. Despite the absence of cytokines from our analyzed active sample, it is possible that other secreted epithelial factors initiate signaling pathways by uncharacterized mechanisms, and so we cannot discount this potential mechanism.
From the factors that were identified, we chose four potential HIV-enhancing proteins to research further. Interestingly, while the secreted epithelial molecules studied here increased viral infection, other groups have previously demonstrated anti-HIV activity of one of the four peptides evaluated: trappin-2/elafin [27]. Trappin-2 and elafin are post-translational processing variants of the PI3 gene product, with elafin (5.9 KDa) representing the cleavage product of its precursor, trappin-2 (9.9KDa) [28]. The anti-HIV activity of the PI3 gene product was demonstrated in a similar TZM-bl system using similar peptide concentrations (0.01 – 10 ng/ml). However, the assay conditions were considerably different, including a preincubation of virus with peptide prior to addition to TZM-bl cells. This preincubation was subsequently shown to be essential for observing anti-HIV activity [28]. Further, it was demonstrated that the anti-HIV activity of elafin is more potent than that of its precursor, trappin-2, which we utilized here [28]. Our utilization of trappin-2 coincided with the ion sequences detected by mass spectrometry, and with the apparent size of the major immunoreactive band detected in our 3–30 KDa CM fraction by immunoblot. These treatment and cleavage distinctions could account for the observed differences in activity.
In addition to trappin-2, the other three proteins were confirmed by immunoblot to be upregulated in A. vaginae-inoculated End1 CM in comparison to the mock-inoculated condition. Interestingly, each of the four proteins enhanced infection only in the presence of the A. vaginae-inoculated End1 CM. These results suggest that another component in the active fraction functions as a major HIV-enhancing factor in a protein concentration-dependent manner, and that just as antiviral proteins work in concert to inhibit HIV infection [13], so might HIV-enhancing factors function in combination to enhance viral infection. This possible scenario supports further evaluation of additional proteins identified in the CM from stimulated FRT epithelia.
Conclusion
The mechanism by which BV predisposes women to HIV acquisition is not well understood. Here we demonstrate that the epithelial immune response to BVAB, which is characterized by an upreglation of secreted proteins, increases downstream HIV infection. Our investigation did not identify a single factor that exhibited HIV-enhancing activity, but instead we present a collection of proteins that may act together to increase viral infection. These proteins are likely mediators of increased viral susceptibility in vivo, and could represent molecular targets for combating heterosexual HIV acquisition in women.
Acknowledgements
Conflict of interest. This work was supported by the National Institutes of Health [grant numbers AI052017, AI082623, AI082693 to AMC].
List of abbreviations
- BV
Bacterial vaginosis
- BVAB
Bacterial vaginosis associated bacteria
- CM
Conditioned media
- FRT
Female reproductive tract
- BSA
Bovine serum albumin
Footnotes
Author contributions were as follows-designed research: CRE, ALC and AMC; performed research: CRE, CD and SC; analyzed data and wrote manuscript: CRE and AMC.
References
- 1.Sobel JD. Bacterial Vaginosis. Annu. Rev. Med. 2000;51:349–356. doi: 10.1146/annurev.med.51.1.349. [DOI] [PubMed] [Google Scholar]
- 2.Koumans EH, Sternberg M, Bruce C, McQuillan G, Kendrick J, Sutton M, Markowitz LE. The prevalence of bacterial vaginosis in the United States, 2001–2004; associations with symptoms, sexual behaviors, and reproductive health. Sex. Transm. Dis. 2007;34:864–869. doi: 10.1097/OLQ.0b013e318074e565. [DOI] [PubMed] [Google Scholar]
- 3.Martin HL, Richardson BA, Nyange PM, Lavreys L, Hillier SL, Chohan B, Mandaliya K, Ndinya-Achola JO, Bwayo J, Kreiss J. Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J. Infect. Dis. 1999;180:1863–1868. doi: 10.1086/315127. [DOI] [PubMed] [Google Scholar]
- 4.Atashili J, Poole C, Ndumbe PM, Adimora AA, Smith JS. Bacterial vaginosis and HIV acquisition: a meta-analysis of published studies. AIDS. 2008;22:1493–1501. doi: 10.1097/QAD.0b013e3283021a37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wira CR, Grant-Tschudy KS, Crane-Godreau MA. Epithelial cells in the female reproductive tract: a central role as sentinels of immune protection. Am. J. Reprod. Immunol. 2005;53:65–76. doi: 10.1111/j.1600-0897.2004.00248.x. [DOI] [PubMed] [Google Scholar]
- 6.Wira CR, Patel MV, Ghosh M, Mukura L, Fahey JV. Innate Immunity in the Human Female Reproductive Tract: Endocrine Regulation of Endogenous Antimicrobial Protection Against HIV and Other Sexually Transmitted Infections. Am. J. Reprod. Immunol. 2011;65:196–211. doi: 10.1111/j.1600-0897.2011.00970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole AM, Cole AL. Antimicrobial polypeptides are key anti-HIV-1 effector molecules of cervicovaginal host defense. Am. J. Reprod. Immunol. 2008;59:27–34. doi: 10.1111/j.1600-0897.2007.00561.x. [DOI] [PubMed] [Google Scholar]
- 8.Shukair SA, Allen SA, Cianci GC, Stieh DJ, Anderson MR, Baig SM, Gioia CJ, Spongberg EJ, Kauffman SM, McRaven MD, Lakougna HY, Hammond C, Kiser PF, Hope TJ. Human cervicovaginal mucus contains an activity that hinders HIV-1 movement. Mucosal Immunology. 2013;6:427–434. doi: 10.1038/mi.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quayle AJ, Porter EM, Nussbaum AA, Wang YM, Brabec C, Yip KP, Mok SC. Gene expression, immunolocalization, and secretion of human defensin-5 in human female reproductive tract. Am. J. Pathol. 1998;152:1247–1258. [PMC free article] [PubMed] [Google Scholar]
- 10.Fan SR, Liu XP, Liao QP. Human defensins and cytokines in vaginal lavage fluid of women with bacterial vaginosis. Int. J. Gynaecol. Obstet. 2008;103:50–54. doi: 10.1016/j.ijgo.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 11.King AE, Critchley HO, Kelly RW. Presence of secretory leukocyte protease inhibitor in human endometrium and first trimester decidua suggests an antibacterial protective role. Mol. Hum. Reprod. 2000;6:191–196. doi: 10.1093/molehr/6.2.191. [DOI] [PubMed] [Google Scholar]
- 12.King AE, Critchley HO, Sallenave JM, Kelly RW. Elafin in human endometrium: an anti- protease and anti-microbial molecule expressed during menstruation. J. Clin. Endocrinol. Metab. 2003;88:4426–4431. doi: 10.1210/jc.2003-030239. [DOI] [PubMed] [Google Scholar]
- 13.Venkataraman N, Cole AL, Svoboda P, Pohl J, Cole AM. Cationic polypeptides are required for anti-HIV-1 activity of human vaginal fluid. J. Immunol. 2005;175:7560–7567. doi: 10.4049/jimmunol.175.11.7560. [DOI] [PubMed] [Google Scholar]
- 14.Fazeli A, Bruce C, Anumba DO. Characterization of Toll-like receptors in the female reproductive tract in humans. Hum. Reprod. 2005;20:1372–1378. doi: 10.1093/humrep/deh775. [DOI] [PubMed] [Google Scholar]
- 15.Akira S, Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 16.Eade CR, Diaz C, Wood MP, Anastos K, Patterson BK, Gupta P, Cole AL, Cole AM. Identification and characterization of bacterial vaginosis-associated pathogens using a comprehensive cervical-vaginal epithelial coculture assay. PLoS ONE. 2012;7:e50106. doi: 10.1371/journal.pone.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osborn L, Kunkel S, Nabel GJ. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc. Natl. Acad. Sci. USA. 1989;86:2336–2340. doi: 10.1073/pnas.86.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradshaw CS, Tabrizi SN, Fairley CK, Morton AN, Rudland E, Garland SM. The association of Atopobium vaginae and Gardnerella vaginalis with bacterial vaginosis and recurrence after oral metronidazole therapy. J. Infect. Dis. 2006;194:828–836. doi: 10.1086/506621. [DOI] [PubMed] [Google Scholar]
- 19.Ferris MJ, Masztal A, Aldridge KE, Fortenberry JD, Fidel PL, Jr, Martin DH. Association of Atopobium vaginae, a recently described metronidazole resistant anaerobe, with bacterial vaginosis. BMC Infect. Dis. 2004;4:5. doi: 10.1186/1471-2334-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Libby EK, Pascal KE, Mordechai E, Adelson ME, Trama JP. Atopobium vaginae triggers an innate immune response in an in vitro model of bacterial vaginosis. Microbes Infect. 2008;10:439–446. doi: 10.1016/j.micinf.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Klotman ME, Rapista A, Teleshova N, Micsenyi A, Jarvis GA, Lu W, Porter E, Chang TL. Neisseria gonorrhoeae-induced human defensins 5 and 6 increase HIV infectivity: role in enhanced transmission. J. Immunol. 2008;180:6176–6185. doi: 10.4049/jimmunol.180.9.6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zegels G, Van Raemdonck GA, Coen EP, Tjalma WA, Van Ostade XW. Comprehensive proteomic analysis of human cervical-vaginal fluid using colposcopy samples. Proteome Sci. 2009;7:17. doi: 10.1186/1477-5956-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pushkarsky T, Zybarth G, Dubrovsky L, Yurchenko V, Tang H, Guo H, Toole B, Sherry B, Bukrinsky M. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl. Acad. Sci. USA. 2001;98:6360–6365. doi: 10.1073/pnas.111583198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durum SK, Schmidt JA, Oppenheim JJ. Interleukin 1: an immunological perspective. Annu. Rev. Immunol. 1985;3:263–287. doi: 10.1146/annurev.iy.03.040185.001403. [DOI] [PubMed] [Google Scholar]
- 25.Harada A, Sekido N, Akahoshi T, Wada T, Mukaida N, Matsushima K. Essential involvement of interleukin-8 IL-8 in acute inflammation. J. Leukocyte Biol. 1994;56:559–564. [PubMed] [Google Scholar]
- 26.Krueger J, Ray A, Tamm I, Sehgal PB. Expression and function of interleukin-6 in epithelial cells. J. Cell Biochem. 1991;45:327–334. doi: 10.1002/jcb.240450404. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh M, Shen Z, Fahey JV, Cu-Uvin S, Mayer K, Wira CR. Trappin-2/Elafin: a novel innate anti-human immunodeficiency virus-1 molecule of the human female reproductive tract. Immunology. 2010;129:207–219. doi: 10.1111/j.1365-2567.2009.03165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drannik AG, Nag K, Yao XD, Henrick BM, Jain S, Ball TB, Plummer FA, Wachihi C, Kimani J, Rosenthal KL. Anti-HIV-1 activity of elafin is more potent than its precursor's, trappin-2, in genital epithelial cells. J. Virol. 2012;86:4599–4610. doi: 10.1128/JVI.06561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]