Abstract
Genomics research provides an unprecedented opportunity for us to probe into the pathogenicity and evolution of the world's most deadly pathogenic bacterium, Yersinia pestis, in minute detail. In our present work, extensive microarray analysis in conjunction with PCR validation revealed that there are considerable genome dynamics, due to gene acquisition and loss, in natural populations of Y. pestis. We established a genomotyping system to group homologous isolates of Y. pestis, based on profiling or gene acquisition and loss in their genomes, and then drew an outline of parallel microevolution of the Y. pestis genome. The acquisition of a number of genomic islands and plasmids most likely induced Y. pestis to evolve rapidly from Yersinia pseudotuberculosis to a new, deadly pathogen. Horizontal gene acquisition also plays a key role in the dramatic evolutionary segregation of Y. pestis lineages (biovars and genomovars). In contrast to selective genome expansion by gene acquisition, genome reduction occurs in Y. pestis through the loss of DNA regions. We also theorized about the links between niche adaptation and genome microevolution. The transmission, colonization, and expansion of Y. pestis in the natural foci of endemic plague are parallel and directional and involve gradual adaptation to the complex of interactions between the environment, the hosts, and the pathogen itself. These adaptations are based on the natural selections against the accumulation of genetic changes within genome. Our data strongly support that the modern plague originated from Yunnan Province in China, due to the arising of biovar orientalis from biovar antiqua rather than mediaevalis.
Yersinia pestis, the causative agent of bubonic and pneumonic plagues, is thought to be one of the most dangerous pathogens in the world. There have been three recorded human plague pandemics, which have claimed hundreds of thousands of lives. Areas where this disease is endemic exist widely in Asia, Africa, and the Americas, where occasional epizootics of animal plague pose great threats to public health (18). Plague has been classified as a reemerging disease by the World Health Organization due to the worldwide increasing incidence of human plague.
Y. pestis can be divided into three biovars, i.e., antiqua, mediaevalis, and orientalis, according to their ability to reduce nitrate and utilize glycerol (1). These three biovars are thought to be responsible for the three major plague pandemics: the Justinian plague, the Black Death, and the modern plague, respectively (1). The third plague pandemic was believed to have originated from Yunnan Province, China, in 1855. It then spread around the world with the aid of modern transportation (25). Human plague has been successfully controlled since the 1950s in China. However, 11 natural plague foci still remain in China, covering more than 277 counties in 19 provinces with an area of more than 1 million km2 (10, 13).
We assumed that the host niche, compound interactions between the environment, the reservoirs, the vectors, and the pathogen, would determine the traits (host range, virulence, biochemical features, genetics contents, etc.) of Y. pestis in a specific geographic region and that strains of Y. pestis from different origins should slightly differ in genome content. However, the challenge was how to track down the genetic differences, how to use these differences as markers for genomotyping, and how to illustrate genomotypic and phenotypic microevolution of Y. pestis. The recently decoded whole-genome sequences of Y. pestis CO92 (17), KIM (5), and 91001 (Y. Song and R. Yang, unpublished data) (accession numbers AE017042, AE017043, AE017044, AE017045, and AE017046) provide the unprecedented opportunity to overcome this challenge. Here, we report our results on the considerable genome dynamics in natural populations of Y. pestis due to gene acquisition and loss, as determined by using DNA microarray-based comparative genomic analysis in conjunction with PCR-based screening. Further, we outlined the parallel microevolution of the Y. pestis genome and propose its intimate link with niche adaptation of Y. pestis in natural foci.
MATERIALS AND METHODS
Bacterial strains.
Forty-three strains were used in microarray hybridization (Tables 1 and 2). Thirty-six of them are Y. pestis strains that were isolated from 10 plague foci in China and were selected to represent the most abundant Y. pestis diversity associated with adaptive evolution in plague foci. In addition, seven Yersinia pseudotuberculosis strains were included as controls. Two natural isolates of Y. pestis, 91001 and 82009, were used as reference strains in microarray analysis. Y. pestis 91001, a human avirulent strain of biovar mediaevalis, was isolated from a Microtus-related plague focus (focus L [see below]) in China. Y. pestis 82009, a fully virulent strain of biovar orientalis, was isolated from a house mouse-related plague focus (focus F) and was used as an alternative to strain CO92, which is also an orientalis strain. In addition, a total of 260 isolates of Y. pestis (including the previous 36 isolates) from the 10 plague foci were used in PCR analysis.
TABLE 1.
Natural isolates of Y. pestis used in microarray analysis
| Designation in this study | Original strain no. | Natural focusa | Biovar | Reservoir of isolation |
|---|---|---|---|---|
| YPe01 | 41001 | A | antiqua | Marmota caudate |
| YPe02 | 41007 | |||
| YPe03 | 49002 | B | antiqua | Spermophilus undulates |
| YPe04 | 49006 | Marmota baibacina | ||
| YPe05 | 45062 | Spermophilus undulates | ||
| YPe06 | 40019 | Marmota baibacina | ||
| YPe07 | 42044 | Spermophilus undulates | ||
| YPe08 | 45099 | Marmota baibacina | ||
| YPe09 | 347001 | G | antiqua | Marmota himalayana |
| YPe10 | 348002 | |||
| YPe11 | 27002 | D | antiqua | Marmota himalayana |
| YPe12 | 02052 | |||
| YPe13 | 25009 | |||
| YPe14 | 77022 | |||
| YPe15 | 11001 | C | antiqua | Marmota himalayana |
| YPe16 | 30014 | |||
| YPe17 | 31004 | |||
| YPe18 | 71001 | |||
| YPe19 | 09001 | |||
| YPe20 | 70006 | |||
| YPe21 | 84004 | E | antiqua | Eothenomys miletus |
| YPe22 | 84038 | Apodemus chevrieri | ||
| YPe23 | 114001 | F | orientalis | Rattus flavipectus |
| YPe24 | 82009 | |||
| YPe25 | 96006 | H | antiqua | Spermophilus dauricus |
| YPe26 | 54006 | |||
| YPe27 | 147001 | I | mediaevalis | Meriones unguiculatus |
| YPe28 | 140007 | |||
| YPe29 | 132002 | J | mediaevalis | Spermophilus dauricus alaschanicus |
| YPe30 | 126002 | |||
| YPe31 | 47001 | K | mediaevalis | Marmota himalayana |
| YPe32 | 47004 | |||
| YPe33 | 90001 | L | microtus | Microtus brandti |
| YPe34 | 91001 | |||
| YPe35 | N010001 | M | microtus | Microtus fuscus |
| YPe36 | 18014 |
Focus A, Marmota caudate plague focus of the Pamirs Plateau; focus B, Marmota baibacina-S. undulates plague focus of the Tianshan Mountains; focus C, Marmota himalayana plague focus of the Qinghai-Gansu-Tibet Grassland; focus D, Marmota himalayana plague focus of the Oilian Mountain; focus E, A. chevrieri-E. miletus plague focus of the highland of northwestern Yunnan Province; focus G, Marmota himalayana plague focus of the Gangdisi Mountains, focus F, R. flavipectus plague focus of Yunnan, Guangdong, and Fujian Provinces; focus H, S. dauricus plague focus of the Song-Liao Plain; focus I, Meriones unguiculatus plague focus of the Inner Mongolian Plateau; focus J, S. dauricus alaschanicus plague focus of the Loess Plateau in Gansu and Ningxia Provinces; focus K, Marmota himalayana plague focus of the Kunlun Mountains; focus L, Microtus brandti plague focus of the Xilin Gol Grassland; focus M, Microtus fuscus plague focus of the Qinghai-Tibet Plateau; focus N, Marmota sibirica plague focus of the Hulun Buir Plateau of Inner Mongolia. There are 11 natural plague foci in China (foci C, D, G, and K are the subfoci of the Marmota himalayana plague focus of the Qinghai-Tibet Plateau). Focus N was discovered as early as 1911, but no strain of Y. pestis could be isolated from this focus since the 1950s, and there is no collection of bacterial strains from this focus in China at present. The Y. pestis strains used in the microarray analysis and the PCR analysis are from all 10 foci, A to M.
TABLE 2.
Y. pseudotuberculosis strains used as controls in this study
| Designation | Strain | Biogroup | Source |
|---|---|---|---|
| YPsI | CMCC53518 | I | China Medical Culture Collection |
| YPsII | CMCC53519 | II | China Medical Culture Collection |
| YPsIII | CMCC53520 | III | China Medical Culture Collection |
| YPsIV | CMCC53521 | IV | China Medical Culture Collection |
| YPsV | CMCC53522 | V | China Medical Culture Collection |
| YPs01 | ATCC 29833 | American Type Culture Collection | |
| YPs02 | CMCC53502 | China Medical Culture Collection |
Microarray analysis.
In the present work, 4,005 annotated open reading frames (genes) were amplified successfully from Y. pestis 82009 or 91001 by using gene-specific primer pairs. These 4,005 genes included nearly all of the CO92 genes and the genes unique to 91001 (Table 3) after the exclusion of genes encoding insertion sequence protein, integrase, and transposase. The purified PCR products were spotted in duplicate on CSS-1000 silylated glass slides (CEL) by using a SpotArray72 microarray printing system (Perkin-Elmer Life Sciences) to construct the DNA microarrays. A mixture of equal quantities of 91001 and 82009 genomic DNAs was used as reference DNA. Genomic DNA from each of the natural isolates studied was referred to as test DNA. Cy3- or Cy5-labeled probes were generated by priming of the reference or test DNA with random hexamers and extension with Klenow polymerase (2). The labeled reference and test DNAs were combined to hybridize with the microarrays by dual-fluorescence hybridization (2). All hybridizations were performed in triplicate. The hybridized slides were scanned by using a GenePix 4100A personal microarray scanner (Axon Instruments). The scanning images were processed and the data were further analyzed by using GenePix Pro 4.1 software (Axon Instruments) combined with Microsoft Excel software.
TABLE 3.
Genes unique to Y. pestis strain 91001 represented on the microarray
| Gene identification | Gene name | Gene length (bp) | Annotated function |
|---|---|---|---|
| pCRY01 | repA | 714 | Putative RepA protein |
| pCRY02 | 252 | Hypothetical protein | |
| pCRY03 | hipB1 | 357 | Putative transcriptional regulators |
| pCRY04 | nusG | 456 | Transcription antiterminator |
| pCRY05 | 270 | Hypothetical protein | |
| pCRY06 | 219 | Putative ATP/GTP-binding protein remnant | |
| pCRY07 | virB1 | 711 | Type IV secretory pathway, VirB1 components |
| pCRY08 | virB2 | 306 | Type IV secretory pathway, VirB2 component, putative mating pair formation protein TraC |
| pCRY09 | virB4 | 2,685 | Type IV secretory pathway, VirB4 components |
| pCRY10 | virB5 | 705 | Type IV secretion system, VirB5 component |
| pCRY11 | 228 | Hypothetical protein | |
| pCRY12 | virB6 | 1,074 | Type IV secretory pathway, VirB6 components |
| pCRY13 | virB8 | 684 | Type IV secretion system, VirB8 component |
| pCRY14 | virB9 | 909 | Type IV secretory pathway, VirB9 components |
| pCRY15 | virB10 | 1,251 | Type IV secretory pathway, VirB10 components |
| pCRY16 | virB11 | 1,026 | Type IV secretory pathway, VirB11 components, and related ATPases involved in archaeal flagellar biosynthesis |
| pCRY17 | 399 | Hypothetical protein | |
| pCRY18 | 306 | Hypothetical protein | |
| pCRY19 | 306 | Putative dopa decarboxylase protein remnant | |
| pCRY20 | 294 | Hypothetical protein | |
| pCRY21 | 345 | Hypothetical protein | |
| pCRY22 | 1,752 | Putative mobilization protein MobC | |
| pCRY23 | 768 | Putative mobilization protein MobC | |
| pCRY24 | 474 | Micrococcal nuclease (thermonuclease) homologs | |
| pCRY25 | 342 | Putative membrane protein | |
| pCRY26 | 234 | Hypothetical protein | |
| pCRY27 | parA | 648 | ATPase involved in chromosome partitioning |
| pCRY28 | mpr | 861 | Zinc metalloproteinase Mpr protein |
| pCRY29 | hipB2 | 282 | Predicted transcriptional regulators |
| pCRY30 | 360 | Putative membrane protein | |
| pMT044 | 312 | Hypothetical protein | |
| pMT045 | 489 | Hypothetical protein | |
| pMT046 | 1,143 | Putative ribonucleoside-diphosphate reductase beta subunit | |
| pMT047 | 2,316 | Putative ribonucleoside-diphosphate reductase alpha subunit | |
| pMT086 | 306 | Putative C-type natriuretic protein | |
| pMT087 | 153 | Hypothetical protein | |
| pMT088 | 216 | Hypothetical protein | |
| pMT089 | 177 | Hypothetical protein | |
| pMT090 | 1,323 | Putative DNA ligase | |
| pMT091 | 255 | Hypothetical protein | |
| pMT092 | 477 | Hypothetical protein | |
| pMT093 | 342 | Hypothetical protein | |
| pMT094 | 192 | Hypothetical protein (pseudogene) | |
| pMT127 | 723 | Hypothetical protein (pseudogene) | |
| pMT128 | 606 | Hypothetical protein (pseudogene) | |
| YP0966 | rhlE | 918 | Probable ATP-dependent RNA helicase (pseudogene) |
| YP0969 | 375 | Hypothetical protein | |
| YP0970 | dnaJ1 | 555 | Molecular chaperones (contain C-terminal Zn finger domain) |
| YP0971 | 201 | Hypothetical protein | |
| YP0973 | srmB1 | 1,413 | Superfamily II DNA and RNA helicases |
| YP0974 | acrR3 | 711 | a1 regulator |
| YP0975 | acrA4 | 987 | Membrane fusion protein |
| YP0976 | ccmA2 | 1,743 | ABC-type multidrug transport system, ATPase component |
| YP0977 | 1,269 | ABC-type multidrug transport system, permease component | |
| YP0978 | 1,107 | ABC-type multidrug transport system, permease component | |
| YP0979 | 330 | Hypothetical protein | |
| YP0980 | 228 | Coenzyme F420-dependent N5,N10-methylene tetrahydromethanopterin reductase and related flavin-dependent oxidoreductases | |
| YP0981 | lysR4 | 912 | Putative transcriptional regulator |
| YP0982 | nemA1 | 1,110 | NADH:flavin oxidoreductases, old yellow enzyme family |
| YP0983 | atoC1 | 1,293 | Response regulator containing CheY-like receiver, AAA-type ATPase, and DNA-binding domains |
| YP0984 | baeS2 | 1,824 | Signal transduction histidine kinase |
| YP0985 | 474 | Hypothetical protein | |
| YP0986 | xapB | 609 | Xanthosine permease (pseudogene) |
An intensity ratio (test DNA normalized intensity/reference DNA normalized intensity) was recorded for each spot and then was converted to log2.5. The hexa-ratios of each gene were averaged. Spots displaying low hybridization signals (the lowest 10% based on Cy3-normalized medians) were filtered out; spots with bad data because of slide abnormalities were discarded as well. The efficacy of the DNA microarrays was further assessed by the control hybridizations of 82009 DNA versus reference DNA, 91001 DNA versus reference DNA, and reference DNA versus reference DNA. A log value of lower than −1 was taken as defining the absence of a gene in the relevant strains. Ninety-nine percent of the spots gave correct predictions of the presence or absence of the corresponding genes. The remaining 1% of the spots gave false predictions and were rejected from the analysis. In the end, 3,661 genes were included in the data sets, and a log ratio of −1 was taken as the cutoff value throughout the experiments.
PCR analysis.
All of the difference region (DFR) genes (see below) were used in PCR amplification to validate the deletion events, identified by the microarray methods, in the 36 Y. pestis isolates listed in Table 1. Then, one or more genes were chosen from each DFR to stand for the corresponding DFR, and PCR amplification of the selected genes was performed on the 260 isolates of Y. pestis to screen the distribution of DFRs in these strains. All of the DNAs to be tested were arrayed in 96-well PCR plates. Each primer pair was pretested, with the genomic DNA of strain 91001 or CO92 as a template, to ensure successful amplification. PCR products were analyzed by 1.2% agarose gel electrophoresis with ethidium bromide staining.
RESULTS AND DISCUSSION
DFRs in natural populations of Y. pestis.
Twenty-two genomic regions that were absent in at least one of strains studied were identified by extensive microarray analysis (Table 4). Each of these regions is referred to as a DFR, a term used in a previous study of genome plasticity in Y. pestis (20). The DFR profiles of the 260 isolates of Y. pestis tested can be assigned to 14 groups; each group was termed a genomovar (Table 5). In this way, the genomic variability in natural populations of Y. pestis was tracked down successfully and was further used as markers to rationally group homologous isolates of Y. pestis.
TABLE 4.
DFRs in the genome of Y. pestis
| DFR | Gene region | Annotated function | Gene(s) selected for PCR-based screening |
|---|---|---|---|
| 01 | 91001-pMT044-047 | Ribonucleoside-diphosphate reductase and hypothetical proteins | 91001-pMT045 |
| 02 | 91001-pMT086-094 | C-type natriuretic protein and hypothetical proteins | 91001-pMT092 |
| 03 | CO92-YPMT1.03c-1.12c | Prophage | CO92-YPMT1.07c |
| 04 | 91001-YP0966-YP0986 | RNA helicase, multidrug transport system, two-component regulatory system, and transcriptional regulators | 91001-YP0975 |
| 05 | CO92-YPO0621-0636 | Regulatory proteins, hypothetical proteins, and aminotransferase | CO92-YPO0626 |
| 06 | CO92-YPO0738-0739 | Flagellins | CO92-YPO0739 |
| 07 | CO92-YPO0740-0754 | Flagellins and membrane proteins | CO92-YPO0744 |
| 08 | CO92-YPO0988-0989 | Membrane protein | CO92-YPO0988 |
| 09 | CO92-YPO0998-1007 | Membrane proteins, autotransporter, antigenic leucine-rich repeat proteins, and drug efflux proteins | CO92-YPO1003 |
| 10 | CO92-YPO1165-1172 | Dehydrogenase, regulatory proteins, xanthosine utilization | CO92-YPO1167 |
| 11 | CO92-YPO1986-1987 | Exported protein | CO92-YPO1987 |
| 12 | CO92-YPO2096-2135 | Prophage | CO92-YPO2098, YPO2115 |
| 13 | CO92-YPO2271-2281 | Prophage | CO92-YPO2273 |
| 14 | CO92-YPO2286-2287 | Transport system permease | CO92-YPO2287 |
| 15 | CO92-YPO2315 | Exported protein | CO92-YPO2315 |
| 16 | CO92-YPO2375-2376 | Aldo/keto reductase | CO92-YPO2375 |
| 17 | CO92-YPO2380 | Insecticial toxin | CO92-YPO2380 |
| 18 | CO92-YPO2469 | Conserved hypothetical protein | CO92-YPO2469 |
| 19 | CO92-YPO2487-2489 | Membrane protein and hypothetical proteins | CO92-YPO2489 |
| 20 | CO92-YPO3046-3047 | Sulfatase | CO92-YPO3047 |
| 21 | CO92-YPO3674 | Insecticidal toxin | CO92-YPO3674 |
| 22 | CO92-YPO4012-4045 | Two-component regulatory system, membrane proteins, amino acid utilization, iron transport system, regulatory proteins, sugar transport system, and fimbriae | CO92-YPO4019, -YPO4040 |
TABLE 5.
Genomovars of Yersinia pestis based on DFR profiling
| Genomovara | DFR profileb
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 02 | 03 | 04 | 05 | 06 | 07 | 08 | 09 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
| Yps ancestor | − | − | − | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + |
| Ype ancestor | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + |
| 01 | − | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + |
| 02 | − | + | + | + | + | + | + | + | + | − | + | + | − | + | + | + | + | + | + | + | + | + |
| 03 | − | + | + | − | + | + | + | + | + | − | + | + | − | + | + | + | + | + | + | + | − | + |
| 04 | − | + | + | + | + | + | + | + | − | + | + | + | − | + | + | + | + | + | + | + | + | + |
| 05 | − | − | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + |
| 06 | − | − | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + | + | + | + | − |
| 07 | − | − | + | − | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + |
| 08 | − | − | − | − | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + |
| 09 | − | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 10 | − | + | + | + | + | − | − | + | + | + | + | + | − | + | − | − | + | + | + | + | + | + |
| 11 | − | + | + | + | + | − | − | + | + | + | + | + | − | + | − | − | − | + | + | + | + | + |
| 12 | − | + | + | + | − | − | − | + | + | + | + | + | − | + | − | − | − | + | + | + | + | + |
| 13 | − | + | + | + | + | − | − | − | + | + | + | + | − | + | − | − | − | + | + | + | + | + |
| 14 | + | + | + | + | + | − | + | + | + | + | − | − | − | + | + | + | + | − | − | − | + | + |
Yps ancestor is a presumed genomovar of the ancestral strains of Y. pseudotuberculosis. Ype ancestor is a presumed genomovar of the ancestral strains of Y. pestis.
+, present in the genome; −, absent from the genome.
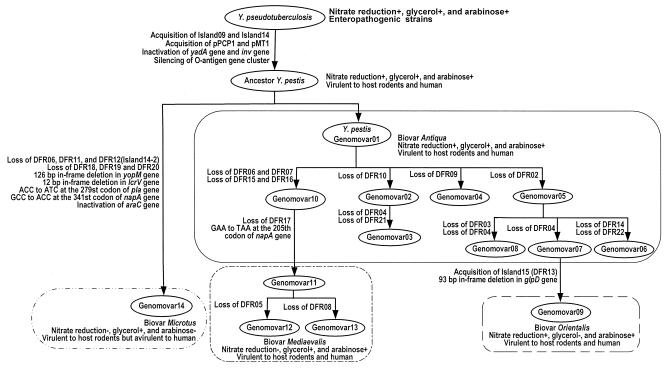
The DFRs represent the dynamic regions of the Y. pestis genome in natural populations, demonstrating its characteristics of acquisition or deletion in the adaptive evolution of Y. pestis in natural plague foci. Here we present an overview of the parallel microevolution of the Y. pestis genome in natural populations, with evidence of genome content flux through the acquisition or loss of plasmids and chromosomal segments (Fig. 1).
FIG. 1.
Deduced pattern of microevolution of Y. pestis based on DFR profiling. A phylogenetic tree of the 14 genomovars of Y. pestis (Table 5) was constructed by using the PHYLIP Mix algorithm with the genomovar of the Ype ancestor (Table 5) as an outgroup (data not shown). The tree was then used as a backbone to develop the evolutionary relationships among the genomovars under the assumption that it is most likely that as few DFR alleles as possible are changed at a time. The figure shows the loss or acquisition of DFRs and islands and the conversion of genomovars and biovars. In addition, strains from Microtus foci, belonging to biovar mediaevalis according to the traditional biovar assignment, were proposed as a new biovar, microtus, because of their unique pathogenic, biochemical, and molecular features. The figure also shows some point mutations leading to the inactivation of certain genes, which most likely accounts for the metabolic variations between the four Y. pestis biovars; these results will be interpreted in detail in another study (Zhou and Yang, unpublished data).
Speciation of Y. pestis from Y. pseudotuberculosis.
Y. pestis is a clone that evolved from Y. pseudotuberculosis 1,500 to 20,000 years ago, shortly before the first known pandemics of human plague (1). Y. pestis has acquired two unique virulence plasmids (pPCP1 and pMT1) in the process of speciation (25). Plasmid pPCP1 encodes the plasminogen activator (Pla), a putative invasin/adhesin that is essential for virulence by the subcutaneous route (4, 12). Plasmid pMT1 encodes murine toxin (Ymt), which has a role in the transmission of plague (9, 22), and the F1 capsular protein provides an additional mechanism to block phagocytosis; this mechanism works differently from that of the type III secretion system encoded by the pCD1 plasmid (6).
There are 21 genomic islands (including the virulence-related pgm locus), probably acquired from other organisms through horizontal gene transfer, on the chromosome of strain CO92 (17). Our experimental data indicate that 18 of them are harbored in both Y. pseudotuberculosis and Y. pestis, while the other 3 (island 09, island 14, and island 15) are present only in Y. pestis (Table 6). Apparently, these three islands were acquired by Y. pestis during the course of speciation (Fig. 1). All three of these islands encode prophages. Some bacteriophages encode bacterial proteins that enable the bacteria to invade host tissues, avoid the host immune defense, and damage host cells (3). With the integration of the bacteriophage genome into the bacterial chromosome, the virulence factors encoded by the prophage can convert their bacterial host from a nonpathogenic strain to a virulent one or to a strain with increased virulence (3).
TABLE 6.
Distribution of genomic islands of Y. pestis in Y. pseudotuberculosis
| Island designation in this studya | Range in CO92 chromosome | Annotated functions | Presence in Y. pseudotuberculosisb | Gene(s) selected for PCR-based screeningc |
|---|---|---|---|---|
| 01 | YPO0255-0273 | Type III secretion system | Yes | |
| 02 | YPO0335-0340 | Insect viral enhancin homologue | Yes | |
| 03 | YPO0590-0642 | Adhesin, autotransporter, secreted proteins, and protein kinase | Yes | |
| 04 | YPO0684-0697 | Adherence proteins | Yes | |
| 05 | YPO0770-0778 | Siderophore biosynthesis | Yes | |
| 06 | YPO0803-0818 | Type II secretion system | Yes | |
| 07 | YPO0871-0887 | Bacteriocin and prophage | Yes | |
| 08 | YPO0961-0995 | Quorum sensing system and siderophore biosynthesis | Yes | |
| 09 | YPO1083-1098 | Prophage | No | YPO1091 |
| 10 | YPO1224-1259 | Outer membrane protease and prophage | Yes | |
| 11 | YPO1448-1480 | Cytotoxic nectotizing factor homologue | Yes | |
| 12 | YPO1900-1917 | Yersiniabactin biosynthesis (HPI) | Yes | |
| 13 | YPO1951-1954 | Hemin storage system (Hms) | Yes | |
| 14-1 | YPO2087-2095 | Prophage | No | YPO2089 |
| 14-2 (DFR 12) | YPO2096-2135 | Prophage | No | YPO2098 and YPO2115 |
| 15 (DFR 13) | YPO2271-2280 | Prophage | No | YPO2273 |
| 16 | YPO2311-2321 | Insecticidal toxin | Yes | |
| 17 | YPO2434-2443 | Iron transport and antibiotic resistance | Yes | |
| 18 | YPO2863-2868 | Membrane proteins | Yes | |
| 19 | YPO2934-2948 | Chaperone/usher fimbrial system | Yes | |
| 20 | YPO3673-3682 | Insecticidal toxin | Yes | |
| 21 | YPO4014-4033 | Iron transport | Yes |
Island 14 was divided into two subislands (island 14-1 and island 14-2) because island 14-1 is present in all of the Y. pestis strains tested but absent from Y. pseudotuberculosis strains, whereas island 14-2 (DFR 12) was found to be absent from both Microtus strains and Y. pseudotuberculosis strains.
Based on microarray analysis of 36 isolates of Y. pestis and 7 strains of Y. pseudotuberculosis.
These genes were selected as target genes in PCR analysis to represent the relevant islands; PCR screening of 260 isolates of Y. pestis and 7 strains of Y. pseudotuberculosis confirmed the microarray results.
The acquisition of a number of plasmids and islands may have induced Y. pestis to evolve rapidly from Y. pseudotuberculosis to a newly emerged pathogen that not only is able to parasitize insects in part of its life cycle but also is highly virulent to rodents and humans, causing pandemics of a systemic and often fatal disease. It differs dramatically from its ancestor Y. pseudotuberculosis, which causes only nonfatal gastrointestinal disease in similar hosts. Y. pseudotuberculosis, harboring the hemin storage (hms) locus, insecticidal toxins, iron uptake systems, and secretion systems, has the potential to attack mammals, causing systemic infection, and to be transmitted by fleas. At a certain stage of human history, a change of the natural, social, or economic environment, probably caused by a change in human population or behaviors, might have led to a dramatic increase of the population size of a certain rodent (1). This boom in the rodent population might have triggered the speciation of Y. pestis from Y. pseudotuberculosis as a directional natural selection. Y. pseudotuberculosis is found widely in the environment and is a common cause of animal infections. The bacteria can invade rodents suffering from cold, famine, or illness due to drastic in-species competition or unfavorable environment, and then it enters into the bodies of fleas through flea biting (1). Y. pseudotuberculosis shares a niche with other microorganisms in rodents and fleas, and thus horizontal gene transfer may occur randomly and the beneficial events of gene transfer would be stabilized by vertical inheritance under natural selection. The stepwise acquisition of several genomic islands and plasmids ignited the emergence of Y. pestis in the end.
There was an accumulation of pseudogenes in Y. pestis during the course of speciation (Fig. 1), which is the outcome of the switch of Y. pestis from an enteric lifestyle to a mammalian blood-borne lifestyle (17). For example, yadA and inv are both inactive in Y. pestis but encode functional adhesin and invasion in Y. pseudotuberculosis, enabling the enteropathogen to specifically adhere to surfaces of the host intestines and invade the lining epithelial cells (21-24). The lipopolysaccharide (LPS) of Y. pseudotuberculosis has been shown to possess an O antigen, which is an essential virulence determinant (19), while Y. pestis expresses rough LPS lacking the O antigen, due to the inactivation of several genes in the O-antigen gene cluster (11). Smooth LPS production may be unnecessary for Y. pestis virulence, and the metabolic burden has been alleviated by the inactivation of the O-antigen biosynthetic operon (16).
Parallel microevolution of the Y. pestis genome.
The third human plague pandemic, beginning in the mid-19th century in China's Yunnan Province and then spreading globally, eventually affected more than 60 countries and regions in Asia, Europe, the Americas, and Africa (1). It is believed that the third pandemic was caused by the emergence of the orientalis strains (25). Indeed, all of the strains isolated from plague focus F in Yunnan Province and its neighboring regions are orientalis strains. All of the orientalis strains tested in this study fell into genomovar 09, which has evolved from genomovar 07 (biovar antiqua) by acquiring DFR 13, which is specific for the orientalis strains (Fig. 1). This strongly supports the notion that biovar orientalis directly evolved from antiqua rather than arising from mediaevalis (11). This fact also suggests that horizontal gene acquisition may play a key role in dramatic evolutionary segregation within species.
For reasons that are probably due to human population density and animal husbandry practices, Y. pestis strains causing the modern plague have been considered to have emerged from China. The data presented here confirm that the modern plague originated from Yunnan Province in China, due to the arising of strains of biovar orientalis from biovar antiqua rather than mediaevalis. Genomovar 09 of Y. pestis seems to be the oldest ancestor of biovar orientalis strains.
DFR 13 (island 15) encodes a filamentous prophage that is integrated into the chromosomal dif locus (7). This conserved prophage was also found in the high-virulence clone Escherichia coli O18:K1:H7, with high homology at nucleotide level (7). E. coli O18:K1:H7, the most virulent among all K1 strains in animal models of sepsis and meningitis, is responsible for almost all cases of neonatal meningitis in the United States and for the preponderance of uncomplicated cystitis cases in North American women. The acquisition of this prophage seems to be linked to the differential disease potential of E. coli O18:K1:H7 (7). Likewise, Y. pestis biovar orientalis, believed to be newly emerged and to be responsible for the third human plague pandemic, possesses major genomic differences compared with its antiqua progenitor.
In contrast to selective genome expansion by horizontal gene acquisition, genome reduction occurs through the loss of DFRs (Fig. 1). The lost genes are always nonessential to bacterial survival (unable to provide a selective benefit of bacterial growth efficiency or fitness in the host) in a specific host niche; that is, if genes are rendered useless due to redundancy within the host niche, then loss bias occurs (14). The parallel loss of DFRs in Y. pestis genomes leads to the discrete segregation between the progenitor and offspring strains, and this genome reduction gradually causes the offspring strains to inhabit a more specific host niche, not overlapping with its progenitor.
Transmission and expansion of Y. pestis in natural foci.
In our study, each genomovar is confined in a specific geographic region, commonly a plague focus or a part of a focus with a unique set of natural environment, reservoirs, and vectors (Table 7). Most of the geographic regions with different primary reservoirs have unique genomovars. Sometimes there is more than one genomovar in a single focus within a single primary reservoir, but each of the genomovars corresponds to a unique set of natural environment and primary vector(s). The microevolution of the genomovars is consistent with the expansion of plague foci. Hence, we present a paradigm of the transmission, colonization, and expansion of Y. pestis in China (Fig. 2). Y. pestis strains from Central Asia and the Far East of Russia may have migrated into the Xinjiang-Tibet region and northeast China, respectively, and then separated into three branches with the expansion of plague foci.
TABLE 7.
Distribution of genomovars in natural plague foci
| Focus | Biovar | Genomovar | No. of isolates tested |
|---|---|---|---|
| A | antiqua | 04 | 11 |
| B | antiqua | 01 | 7 |
| 02 | 13 | ||
| 03 | 10 | ||
| C | antiqua | 05 | 21 |
| 06 | 2 | ||
| 08 | 14 | ||
| D | antiqua | 08 | 16 |
| E | antiqua | 07 | 10 |
| G | antiqua | 10 | 11 |
| F | orientalis | 09 | 22 |
| K | antiqua | 05 | 4 |
| mediaevalis | 11 | 9 | |
| H | antiqua | 10 | 30 |
| I | mediaevalis | 11 | 19 |
| 12 | 2 | ||
| J | mediaevalis | 11 | 9 |
| 13 | 11 | ||
| L | microtus | 14 | 19 |
| M | 20 |
FIG. 2.
Deduced transmission and expansion of Y. pestis in China. Biovar antiqua strains of Y. pestis from Central Asia and the Far East of Russia might have migrated into the Xinjiang-Tibet region and northeast China, respectively, and then separated into three branches with the expansion of plague foci. Branch 1 contains genomovars 01 to 10, including the expansion of foci A to G and focus K. Biovar antiqua strains evolved into orientalis strains in focus F. Branch 2 contains genomovars 10 to 13, including the expansion of foci H to K. Biovar antiqua strains evolved into mediaevalis strains in focus I. Branch 3, the unique one, contains only genomovar 14 (biovar microtus), including two distantly separated plague foci (foci L and M). Yunnan Province is the birthplace of biovar orientalis strains causing the modern plague, and Hong Kong is where the modern plague spread globally via marine shipping during the 1890s.
Links between bacterial genome microevolution and niche adaptation.
Plague is a typical natural focus-based disease. The long-term existence of Y. pestis in a natural focus is accompanied by its interactions with the animal reservoirs and flea vectors. There is a specific natural environment in a defined geographic region (a plague focus or a part of the focus), which ultimately determines the food chain-based relationship of Y. pestis, reservoirs, and vectors. The unique set of hosts (reservoirs and vectors) in the specific natural environment and their interactions with Y. pestis, commonly termed the host niche, determine the existence and also the type of Y. pestis.
It makes sense to say that the origin of ancestral Y. pestis is associated with only one kind of rodent, but Y. pestis has the potential to be transmitted to other species of animals. Once the bacteria are exposed to new animals in new geographic regions by animal contact or vector-borne routes, new host niches come gradually into being. The genetic variations, including gene acquisition, gene loss, point mutation, and genome rearrangement, occur randomly in the genome of Y. pestis. When Y. pestis temporarily colonizes a new host niche, the specific host niche acts as a constant and directional natural selection, leading to the stabilization and vertical inheritance of the beneficial genetic variations in the genome of the Y. pestis colonized in it, which we may define as directional microevolution of the genome. A specific host niche determines not only the long-term existence of Y. pestis but also the genomovar of Y. pestis itself; that is, the expansion of plague foci is a course of the stepwise adaptation of Y. pestis to the new host niches.
The host niches in different natural plague foci, each as a unique natural selection, direct the parallel adaptation of Y. pestis to the corresponding hosts and environment, which is a course of pathogen generalization from a newly emerged single-host species to a multihost pathogen. We can also say that the parallel adaptation to various niches drives Y. pestis strains to diversify into different biovars or genomovars. Certain genomovars are limited to certain geographic regions (or host niches) through a course of within-species segregation, or so-called pathogen specialization, with the advantage of avoiding niche overlapping.
Concluding remarks.
Our study revealed that the genome of Y. pestis, a newly emerged pathogen, is at an intermediate stage of genetic flux, with evidence of selective genome expansion by horizontal acquisition of plasmids or chromosomal islands and genome reduction by loss of DNA regions. The revealed genome dynamics in natural populations of Y. pestis offer an unprecedented opportunity to establish the link between bacterial genome microevolution and niche adaptation under a Darwinian framework. The transmission, colonization, and expansion of Y. pestis in natural foci constitute a parallel, directional, and gradual process of adaptation to the complex of interactions between the environment, the hosts, and the pathogen, which is based on natural selection against the accumulation of small changes within genome.
While this study was in preparation, Hinchliffe et al. described genomic comparisons of Y. pestis and Y. pseudotuberculosis strains performed by using a CO92 gene-specific microarray (8). They identified dozens of DNA loci that were absent or divergent in more than one of the tested strains of Y. pestis and Y. pseudotuberculosis. Quite a number of these results are shared by our study; e.g., DFR 06, DFR 07, DFR 09, DFR 10, DFR 13, DFR 16, and DFR 17 were also found here to be absent from the relevant Y. pestis strains tested by Hinchliffe et al. One major difference of our study is that we selected isolates representing the natural populations of Y. pestis in confined areas that cover a number of typical plague foci. The extensive distribution of typical natural foci, the long-term collection of Y. pestis isolates, and the existing work on the ecology and epidemiology of plague in China enable us to theorize on the bacterial genome microevolution and its links to niche adaptation, taking advantage of the descriptive interpretation of the genomic differences in strains of Y. pestis and Y. pseudotuberculosis from diverse origins. Another difference is that the whole-genome sequencing data for Y. pestis 91001 allowed us to construct a mixed DNA microarray containing a number of genes that are absent in strain CO92, enabling us to capture many more details in the process of genome microevolution.
Acknowledgments
We are grateful to Qi Guo for very fruitful discussions and suggestions. We express our respect to Chinese researchers for their excellent work on the ecology and epidemiology of the plague in China.
Financial support for this work came from the National High Technology Research and Development Program of China (Program 863, no. 2001-AA223061) and the National Natural Science Foundation of China (no. 30371284).
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, E. F., and H. Brussow. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521-529. [DOI] [PubMed] [Google Scholar]
- 4.Cowan, C., H. A. Jones, Y. H. Kaya, R. D. Perry, and S. C. Straley. 2000. Invasion of epithelial cells by Yersinia pestis: evidence for a Y. pestis-specific invasin. Infect. Immun. 68:4523-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng, W., V. Burland, G. Plunkett, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du, Y., R. Rosqvist, and A. Forsberg. 2002. Role of fraction 1 antigen of Yersinia pestis in inhibition of phagocytosis. Infect. Immun. 70:1453-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez, M. D., C. A. Lichtensteiger, R. Caughlan, and E. R. Vimr. 2002. Conserved filamentous prophage in Escherichia coli O18:K1:H7 and Yersinia pestis biovar orientalis. J. Bacteriol. 184:6050-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinchliffe, S. J., K. E. Isherwood, R. A. Stabler, R. A. Stabler, M. B. Prentice, A. Rakin, R. A. Nichols, P. C. F. Oyston, J. Hinds, R. W. Titball, and B. W. Wren. 2003. Application of DNA microarray to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 13:2018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinnebusch, B. J., A. E. Rudolph, P. Cherepanov, J. E. Dixon, T. G. Schwan, and A. Forsberg. 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296:733-735. [DOI] [PubMed] [Google Scholar]
- 10.Ji, S., J. He, W. Bai, Y. Teng, X. Zhan, and C. Lei. 1990. The discovery and research of plague natural foci. Chin. J. Epidemiol. 11(Suppl.):1-42. (In Chinese.) [Google Scholar]
- 11.Kurnik, M., A. Peippo, and E. Ervela. 2000. Characterization of the O-antigen gene clusters of Yersinia pseudotuberculosis and the cryptic O-antigen gene cluster of Yersinia pestis shows that the plague bacillus is most closely related to and has evolved from Y. pseudotuberculosis serotype O:1b. Mol. Microbiol. 37:316-330. [DOI] [PubMed] [Google Scholar]
- 12.Lahteenmaki, K., M. Kukkonen, and T. K. Korhonen. 2001. The Pla surface protease/adhesin of Yersinia pestis mediates bacterial invasion into human endothelial cells. FEBS Lett. 504:69-72. [DOI] [PubMed] [Google Scholar]
- 13.Liu, Z., R. Hai, and R. Li. 2001. The discovery and study of Microtus fuscus natural plague foci in Qinghai-Tibet plateau. Chin. J. Control Endem. Dis. 16:321-327. (In Chinese.) [Google Scholar]
- 14.Moran, N. A. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583-586. [DOI] [PubMed] [Google Scholar]
- 15.Motin, V. L., A. M. Georgescu, J. M. Elliott, P. Hu, P. L. Worsham, L. L. Ott, T. R. Slezak, B. A. Sokhansanj, W. M. Regala, R. R. Brubaker, and E. Garcia. 2002. Genetic variability of Yersinia pestis isolates as predicted by PCR-based IS100 genotyping and analysis of structural genes encoding glycerol-3-phosphate dehydrogenase (glpD). J. Bacteriol. 184:41019-41027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oyston, P. C., J. L. Prior, S. Kiljunen, M. Skurnik, J. Hill, and R. W. Titball. 2003. Expression of heterologous O-antigen in Yersinia pestis KIM does not affect virulence by the intravenous route. J. Med. Microbiol. 52:289-294. [DOI] [PubMed] [Google Scholar]
- 17.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 18.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porat, R., W. R. McCabe, and R. R. Burbaker. 1995. Lipopolysaccharide associated resistance to killing of yersinia by complement. J. Endotoxin Res. 2:91-97. [Google Scholar]
- 20.Radnedge, L., P. G. Agron, P. L. Worsham, and G. L. Andersen. 2002. Genome plasticity in Yersinia pestis. Microbiology 148:1687-1698. [DOI] [PubMed] [Google Scholar]
- 21.Rosqvist, R., M. Skurnik, and H. Wolf-Watz. 1988. Increased virulence of Yersinia pseudotuberculosis by two independent mutations. Nature 334:522-525. [DOI] [PubMed] [Google Scholar]
- 22.Rudolph, A. E., J. A. Stuckey, Y. Zhao, H. R. Matthews, W. A. Patton, J. Moss, and J. E. Dixon. 1999. Expression, characterization, and mutagenesis of the Yersinia pestis murine toxin, a phospholipase D superfamily member. J. Biol. Chem. 274:11824-11831. [DOI] [PubMed] [Google Scholar]
- 23.Simonet, M., B. D. Riot, N. Fortineau, and P. Berche. 1996. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect. Immun. 64:375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skurnik, M., and H. Wolf-Watz. 1989. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol. Microbiol. 3:517-529. [DOI] [PubMed] [Google Scholar]
- 25.Wren, B. W. 2003. The Yersinia—a model genus to study the rapid evolution of bacterial pathogens. Nat. Rev. Microbiol. 1:55-56. [DOI] [PubMed] [Google Scholar]