Abstract
Yersinia pestis has been historically divided into three biovars: antiqua, mediaevalis, and orientalis. On the basis of this study, strains from Microtus-related plague foci are proposed to constitute a new biovar, microtus. Based on the ability to ferment glycerol and arabinose and to reduce nitrate, Y. pestis strains can be assigned to one of four biovars: antiqua (glycerol positive, arabinose positive, and nitrate positive), mediaevalis (glycerol positive, arabinose positive, and nitrate negative), orientalis (glycerol negative, arabinose positive, and nitrate positive), and microtus (glycerol positive, arabinose negative, and nitrate negative). A 93-bp in-frame deletion in glpD gene results in the glycerol-negative characteristic of biovar orientalis strains. Two kinds of point mutations in the napA gene may cause the nitrate reduction-negative characteristic in biovars mediaevalis and microtus, respectively. A 122-bp frameshift deletion in the araC gene may lead to the arabinose-negative phenotype of biovar microtus strains. Biovar microtus strains have a unique genomic profile of gene loss and pseudogene distribution, which most likely accounts for the human attenuation of this new biovar. Focused, hypothesis-based investigations on these specific genes will help delineate the determinants that enable this deadly pathogen to be virulent to humans and give insight into the evolution of Y. pestis and plague pathogenesis. Moreover, there may be the implications for development of biovar microtus strains as a potential vaccine.
Plague, one of the most devastating diseases in human history, is caused by Yersinia pestis. Y. pestis has been responsible for three human plague pandemics: the Justinian plague (5th to 7th centuries), the Black Death (13th to 15th centuries), and modern plague (1870s to the present) (8). Y. pestis strains have been historically divided into three biovars, i.e., antiqua, mediaevalis, and orientalis, according to their ability to ferment glycerol and to reduce nitrate. Biovar antiqua is thought to be originally resident in Africa and is descended from bacteria that caused the Justinian Plague; biovar mediaevalis was originally resident in central Asia and is descended from bacteria that caused the Black Death; and biovar orientalis, which originated from southern China and is widespread at present, is associated with modern plague (11). This kind of biovar assignment is based on metabolic variations that do not seem to correlate with the virulence, while the correlation of each biovar with one of the pandemics is derived from epidemiological observations and historical records. In a DNA microarray-based genomic comparison of Y. pestis strains, we identified dozens of deletions of genomic regions (termed difference regions) in natural populations of Y. pestis (D. Zhou and R. Yang, unpublished data). The deletion or acquisition of difference regions under natural selection contributed greatly to the parallel microevolution of the Y. pestis genome. Furthermore, the biovar conversion was successfully integrated into the genome microevolution, and it was demonstrated that biovars mediaevalis and orientalis arose directly from antiqua in parallel, which is in agreement with previously published data (4, 6). In the present study, strains from Microtus-related plague foci, belonging to biovar mediaevalis according to the traditional biovar assignment, are proposed to constitute a new biovar, microtus (Microtus is the name of a rodent genus that belongs to order Rodentia and family Muridae), on the basis of their unique pathogenic, biochemical, and molecular features. We also established the probable molecular mechanisms for the metabolic variations between the four Y. pestis biovars, involving the inactivation of several metabolism-related genes.
Unique features of microtus strains: a new Y. pestis biovar.
Two independent groups have decoded the whole-genome sequences of two fully virulent Y. pestis strains, CO92 (biovar orientalis) and KIM (biovar mediaevalis) (1, 7). We completed the genome sequencing of a human-avirulent strain, 91001 (Y. Song and R. Yang, unpublished data). The genome of strain 91001 is composed of one chromosome and four plasmids (accession numbers AE017042, AE017043, AE017042, AE017045, and AE017046 for the chromosome, plasmid pCD1, plasmid pCRY, plasmid pMT1, and plasmid pPCP1, respectively). The genomic structure of strain 91001 differs dramatically from those of strains CO92 and KIM due to the rearrangements mediated by insertion sequence elements. In addition, strain 91001 has undergone a unique accumulation of gene loss and pseudogenes in its genome, which mostly likely contributes to its unique pathogenicity (Song and Yang, unpublished data).
Y. pestis 91001 was isolated from a Microtus-related plague focus (focus L [Table 1 ]). There are two Microtus-related foci in China, focus L and focus M (Table 1). Although the two foci are geographically distant, the phenotypic features of the Y. pestis isolates from these foci were almost identical but were divergent from those of Y. pestis isolates from the other plague foci in China (5). Biochemical assays demonstrated that microtus strains could not utilize arabinose, while all of the other types of Y. pestis strains could (5). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the water-soluble protein patterns revealed that microtus strains lost a 32-kDa band compared to all the other types of Y. pestis (5). In addition, microtus strains produced a unique random amplified polymorphic DNA profile (5).
TABLE 1.
The 260 natural isolates of Y. pestis used in PCR analysis
| Plague focus in Chinaa | Focus designation in this study | Biovar | No. of isolates tested |
|---|---|---|---|
| Marmota caudate plague focus of the Pamirs Plateau | A | antiqua | 11 |
| Marmota baibacina-Spermophilus undulates plague focus of the Tianshan Mountains | B | antiqua | 30 |
| Marmota Himalayana plague focus of the Qinghai-Gansu-Tibet Grassland | C | antiqua | 37 |
| Marmota himalayana plague focus of the Qilian Mountain | D | antiqua | 16 |
| Apodemus chevrieri-Eothenomys miletus plague focus of the highland of northwestern Yunnan Province | E | antiqua | 10 |
| Marmota himalayana plague focus of the Gangdisi Mountains | G | antiqua | 11 |
| Rattus flavipectus plague focus of Yunnan, Guangdong, and Fujian Provinces | F | orientalis | 22 |
| Marmota himalayana plague focus of the Kunlun Mountains | K | antiqua | 4 |
| mediaevalis | 9 | ||
| Spermophilus dauricus plague focus of the Song-Liao Plain | H | antiqua | 30 |
| Meriones unguiculatus plague focus of the Inner Mongolian Plateau | I | mediaevalis | 21 |
| Spermophilus dauricus alaschanicus plague focus of the Loess Plateau in Gansu and Ningxia Provinces | J | mediaevalis | 20 |
| Microtus brandti plague focus of the Xilin Gol Grassland | L | microtus | 19 |
| Microtus fuscus plague focus of the Qinghai-Tibet Plateau | M | microtus | 20 |
There are 11 natural plague foci in China (foci C, D, G, and K are the subfoci of the Marmota Himalayana plague focus of the Qinghai-Tibet Plateau). Focus N (Marmota sibirica plague focus of the Hulun Buir Plateau of Inner Mongolia) was discovered as early as 1911, but no strain of Y. pestis could be isolated from this focus since the 1950s, and there is no collection of bacterial strains from this focus in China at present. The Y. pestis strains used in this study are from all 10 foci, A to M.
In laboratories, microtus strains are lethal to Microtus and also are virulent to mouse and some other small rodents, but they are avirulent to larger mammals, such as guinea pig, rabbit, rhesus monkey, sheep, and human. There is no evidence that human plague can arise from microtus strains so far, although an epizootic of microtus plague occurs every year in these two foci. Furthermore, it has been demonstrated that strains from focus M are avirulent to humans by subcutaneous inoculation (3).
To determine the genomic variability between microtus strains and other types of Y. pestis strains, we performed DNA microarray-based comparative genomic hybridizations by use of whole-genome DNA microarrays of Y. pestis as described previously (12). A mixture of genomic DNAs of Y. pestis 91001 and 82009 (reference DNA) was coupled with genomic DNA from each of seven carefully selected Y. pestis strains (Table 2) (test DNA) by two-fluorescence hybridization. Cy3- or Cy5-labeled probes were generated by priming of the reference or test DNA with random hexamers and extension with Klenow polymerase. The coupled probes hybridized together with the microarrays. All hybridizations were performed in triplicate. An average intensity ratio (test DNA normalized intensity/reference DNA normalized intensity) was recorded for each gene and then was converted to log2.5 after a stringent procedure of filtering out data with low quality. Log values lower than −1 were taken as defining the absence of a gene in the seven strains. The microarray analysis identified five deletions of genomic regions specific for microtus strains. Each of these deletions was termed a region of deletion (RD) (see RD1 to RD5 in Table 3). In addition, the microarray analysis revealed that all the four strains from the two Microtus-related foci tested shared almost identical genomic contents. One or two genes were then arbitrarily chosen from each RD to represent the corresponding RD. Gene-specific primers for these selected genes were used to test a collection of 260 Y. pestis isolates (Table 1) for screening of the distribution of RDs in these strains. The PCR analysis confirmed the significant genomic differences between microtus strains and other types of Y. pestis strains revealed by microarray analysis. It can be safely concluded that there is a massive gene loss unique to microtus strains, e.g., the absence of five genomic fragments (RD1 to RD5) from the genomes of microtus strains.
TABLE 2.
Y. pestis strains used in microarray analysis
| Strain | Biovar | Features |
|---|---|---|
| 90001 | microtus | Natural isolate from focus L; human-avirulent strain |
| 91001 | microtus | Natural isolate from focus L; human-avirulent strain |
| N010001 | microtus | Natural isolate from focus M; human-avirulent strain |
| 18014 | microtus | Natural isolate from focus M; human-avirulent strain |
| 132002 | mediaevalis | Natural isolate from focus J; fully virulent strain |
| 49006 | antiqua | Natural isolate from focus A; fully virulent strain |
| 82009 | orientalis | Natural isolate from focus F; fully virulent strain |
TABLE 3.
Genomic features unique to microtus strainsa
| Gene in strain:
|
Annotated function | Feature in microtus strains | |
|---|---|---|---|
| CO92 | 91001 | ||
| YPPCP1.07 | pPCP08 | Pla, invasin/adhesin | Thr to Ile mutation at aab 273 |
| YPCD1.26c | pCD60 | YopM, effector of LCRc | 42 aa; deletion after aa 549 |
| YPCD1.31c | pCD52 | V antigen, LcrV | 16-bp deletions in 91001 at 969 bp after start codon |
| YPCD1.39c | pCD44 | YopN, effector of LCR | Leu-to-Phe mutation at aa 66 |
| YPMT1.23c | pMT024 | Unknown | Leu-to-Pro mutation at aa 496 |
| YPMT1.25c | pMT026 | Unknown | Gln-to-His mutation at aa 168 |
| YPMT1.43c | pMT056 | Unknown | Frameshift caused by 20-bp deletion at 169 bp after start codon |
| YPO1956 | YP1700 | Unknown | Disrupted by IS285 |
| YPO1973 | YP1715 | Putative GntR/family transcriptional regulatory protein | Disrupted by IS100 |
| YPO2258 | YP2054 | Arabinose operon regulatory protein, AraC | Frameshift caused by 112-bp deletion at 26 bp after start codon and a G insertion at 774 bp after start codon |
| YPO2729 | YP2435 | Putative membrane protein | Disrupted by IS285 |
| YPO2731 | YP2433 | Putative membrane protein | Frameshift caused by 2-bp deletion at 153 bp after start codon |
| YPO3049 | YP2671 | Putative binding protein-dependent transport system, inner membrane component | Frameshift caused by 7-bp deletion at 1,125 bp after start codon |
| YPO1986-1987 (RD1) | Exported protein | Absent | |
| YPO2096-2135 (RD2) | Prophage | Absent | |
| YPO2469 (RD3) | Unknown | Absent | |
| YPO2487-2489 (RD4) | Putative membrane protein | Absent | |
| YPO3046-3047 (RD5) | Putative sulfatase | Absent | |
All genes listed are intact in all of the Y. pestis strains except the microtus strains.
aa, amino acid.
LCR, low-calcium response.
The in silico comparison of the whole-genome sequences of Y. pestis CO92, KIM, and 91001 enables us to identify strain-specific point mutations that are not currently detectable with spotted DNA microarrays. In this study, our interest was focused on 12 genes (YPPCP1.07, YPCD1.26c, YPCD1.31c, YPCD1.39c, YPMT1.23c, YPMT1.25c, YPMT1.43c, YPO1956, YPO1973, YPO2729, YPO2731, and YPO3049 according to the CO92 genome annotation [Table 3]), each with a point mutation specific for the 91001 strain. The accumulation of these mutated genes in strain 91001 may contribute to its unique pathogenicity and host range (Song and Yang, unpublished data). Oligonucleotide primers designed from these mutated genes (Table 4) were used to screen for the distribution of the relevant mutations in the collection of 260 Y. pestis isolates. The PCR-based screening data revealed, for these 12 genes, that all of the microtus strains tested possessed the same mutations as strain 91001, while all of the other strains had the intact genes. It can be seen that microtus strains have accumulated many unique mutations resulting in the inactivation of the functional genes (Table 3).
TABLE 4.
Oligonucleotide primers used in this study
| Gene | Primer designation | Primer sequence (5′→3′) | Purpose |
|---|---|---|---|
| napA | NRF1 | CCCGGATGATGGTTATCTGT | DNA sequencing |
| NRR1 | CTGTGTTGGCTGTATCGCTG | DNA sequencing | |
| SN550 | TCCTTACCGTCAACCACC | DNA sequencing | |
| ASN645 | GAAGAGTACGCCAGCTTTGG | DNA sequencing | |
| SN1147 | GTGCTACCGCATGTAAACCC | DNA sequencing | |
| ASN1222 | CGGCAGATTGCTGAAACC | DNA sequencing | |
| SN1624 | GCCGTAAGCCATAGCCGATA | DNA sequencing | |
| ASN1688 | AGACTTCCTGGCTCAACACG | DNA sequencing | |
| SN2092 | CGGAGCCGAACATACCGAC | DNA sequencing | |
| ASN2304 | GGCTGAACTGCATCAAAGGC | DNA sequencing | |
| 613s | GTGCTACGATGATATTGAAG | Mutation screening | |
| 613m | GTGCTACGATGATATTGAAT | Mutation screening | |
| 1021s | ACACCGCTCATTTTGGCGGC | Mutation screening | |
| 1021m | ACACCGCTCATTTTGGCGGT | Mutation screening | |
| glpD | glpD-F1 | GGCTAGCCGCCTCAACAAAAACAT | Mutation screening |
| glpD-R1 | GGTCATACAAGAACAAGCCGGTGC | Mutation screening | |
| glpD-F2 | ATCGTGATCGGTGGCGGTA | Mutation screening | |
| glpD-R2 | GCTTCCAGCAGCAGTACCGA | Mutation screening | |
| araC | araC-F1 | CGGACGATATAAGGTTACG | DNA sequencing |
| araC-R1 | TCCGGTAATAAATTGACAG | DNA sequencing | |
| araC-F2 | ATTGTTGGTATCACCGCTG | DNA sequencing | |
| araC-R2 | TACTGGCATAACCGTAGC | DNA sequencing | |
| YPO2258F | AACCGCAACCCAATCCTTTG | Mutation screening | |
| YPO2258R | ATATAGCCCTTCATGCCGCC | Mutation screening | |
| YPPCP1.07 | pla-spe-M | CCACTATTCTTATCAATGA | Mutation screening |
| pla-spe-W | CCACTATTCTTATCAATGG | Mutation screening | |
| pla-chip | CCGGGAGTGCTAATGC | Mutation screening | |
| YPCD1.26c | yopM-F | AACTTGCCCTTCTTGACTGCG | Mutation screening |
| yopM-R | AGGGGGTAAATCGGGTAATG | Mutation screening | |
| yopM-R2 | ATAAATCGCAGTCAAGAAGGG | Mutation screening | |
| YPCD1.31c | TpCD1_lerV_RW | TTACCTCGTGTCATTTAC | Mutation screening |
| TpCD1_lerV_R | AGCTGGTATTCTTGAGTG | Mutation screening | |
| TpCD1_lerV_L | CCACCATTCAGGTGGATG | Mutation screening | |
| YPCD1.39c | TpCD1_LerE_R | CATGCTGACCAGAGCTTG | Mutation screening |
| TpCD1_LerE_LW | TCTCCGAGCGTAAGGAGC | Mutation screening | |
| TpCD1_LerE_LM | TCTCCGAGCGTAAGGAGT | Mutation screening | |
| YPMT1.23c | TpMT1_23CL | GTTAACAGTCTGCTGGTG | Mutation screening |
| TpMT1_23CRM | GTACTGACTTATTTTCAG | Mutation screening | |
| TpMT1_23CRW | GTACTGACTTATTTTCAA | Mutation screening | |
| YPMT1.25c | TpMT1_25CL | AACTCGCCTGACGCCAGC | Mutation screening |
| TpMT1_25CRM | TTACGCAGCTGCGCCACA | Mutation screening | |
| TpMT1_25CRW | TTACGCAGCTGCGCCACC | Mutation screening | |
| YPMT1.43c | TpMT1_43CRW | TACGAAATGCCAGCAGTG | Mutation screening |
| TpMT1_43CRM | GTTCAAGTGGATATGCAG | Mutation screening | |
| TpMT1_43CL | TTCTGTAGGATCTCTCTG | Mutation screening | |
| YPO1956 | pgm-middle-F | TGGCCAGTGTCATTATAC | Mutation screening |
| pgm-middle-R | TGTTTCACCTACCGTAGC | Mutation screening | |
| YPO1973 | hutC-F | AGCGCAGCAATGGACGAC | Mutation screening |
| hutC-R | GACCATAGAGCTGATAAC | Mutation screening | |
| YPO2729 | YPO2729F | GTCATTAAGCAGAAAAGCCGC | Mutation screening |
| YPO2729R | GTAGTGTCCATACCGCCAAC | Mutation screening | |
| YPO2731 | YPO2731F-W | CCTTATCCGTGCTTATGAACAC | Mutation screening |
| YPO2731F-M | CCTTATCCGTGCTTATGAACAT | Mutation screening | |
| YPO3049 | YPO3049RM1 | GCGATACCAATAAACAGATAACAG | Mutation screening |
| YPO3049RW1 | CAATAAACAGATAACAAATAAC | Mutation screening | |
| YPO3049R | AACCCTCTTTCAGCGAGTTATC | Mutation screening |
Plague is a typical zoonotic disease, primarily affecting rodent reservoirs in natural foci. Humans play no role in the long-term survival of this pathogen. However, typical natural isolates of Y. pestis possess the inherent ability to attack humans, and human plague cases occur with the transmission of the pathogen from reservoirs or flea vectors to humans. Table 3 lists the genomic features shown to be unique to microtus strains. These features provide a line of genomic evidences to support that the adaptation of microtus strains to Microtus-related niches in nature is a process of reductive evolution based on a massive loss or inactivation of genes. Some of the lost or defective genes, such as those encoding virulence determinants (e.g., Pla, LcrV, YopM, and YopN) or proteins related to host adaptation (e.g., metabolism pathway proteins, membrane proteins, and prophages), may be involved in plague pathogenesis. We believe that the inherent ability of microtus strains to attack humans is lost during this genome reduction course.
The microtus strains belong to biovar mediaevalis according to the traditional biovar assignment. However, microtus strains show great pathogenic, biochemical, and genetic differences from the typical mediaevalis strains. Biovar mediaevalis strains are believed to be linked with the Black Death pandemic, while microtus strains are thought to be avirulent to humans. It seems reasonable to classify these strains from Microtus-related plague foci as a new biovar, microtus. Here, based on their abilities to ferment glycerol and arabinose and to reduce nitrate, we assigned Y. pestis strains into four biovars: antiqua (glycerol positive, arabinose positive, and nitrate positive), mediaevalis (glycerol positive, arabinose positive, and nitrate negative), orientalis (glycerol negative, arabinose positive, and nitrate positive), and microtus (glycerol positive, arabinose negative, and nitrate negative). The first three biovars are linked to the first, second, and third human plague pandemics, respectively, while the last one is avirulent to humans, causing the microtus plague and its epizootics in the natural foci.
Glycerol fermentation defect in biovar orientalis: the glpD gene.
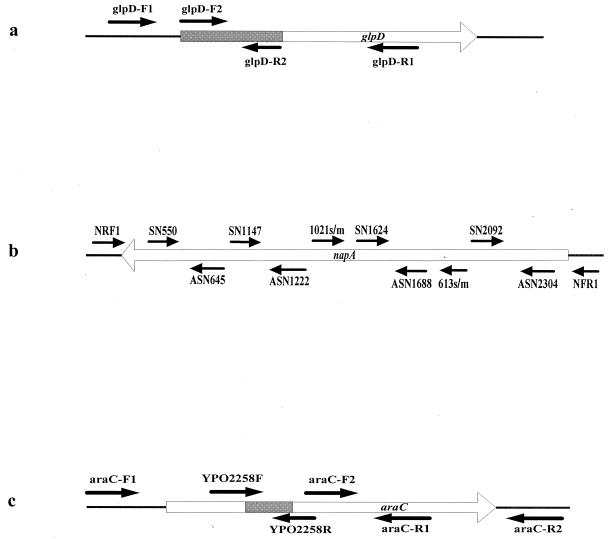
The CO92 strain (glycerol negative) has a defective glycerol-3-phosphophate dehydrogenase gene (glpD) with a 93-bp in-frame deletion (7). The loss of 93 bp results in a 31-amino-acid deletion within the N terminus of the GlpD protein; it most likely accounts for the glycerol-negative phenotype of the orientalis strains (6). We tested the collection of 260 Y. pestis isolates for the presence of the 93-bp deletion by using four primers, glpD-F1, glpD-F2, glpD-R1, and glpD-R2 (Fig. 1a). Each glycerol-positive strain (biovars antiqua, mediaevalis, and microtus) gave a 508-bp product for the primer pair glpD-F1-glpD-R1, a 281-bp product for glp-DF1-glpD-R2, and a 310-bp product for glpD-F2-glpD-R1, while each glycerol-negative strain (biovar orientalis) gave a 415-bp product for glpD-F1-glpD-R1 and no product for glpD-F1-glpD-R2 and glpD-F2-glpD-R1. The results of PCR analysis indicate that biovar orientalis has a truncated glpD gene, while the other three biovars each harbor an intact glpD gene. Eight of the 415-bp amplicons from the primer pair of glpD-F1 and glpD-R1 were then picked randomly for sequencing, which demonstrated that each amplicon contained the 93-bp deletion. The perfect consensus between the glycerol-negative phenotype and the presence of a 93-bp deletion within the glpD gene further confirms that it is this 93-bp deletion that results in the glycerol-negative phenotype of biovar orientalis strains.
FIG. 1.
Graphic representation of the primer locations in genes glpD (a), napA (b), and araC (c).
Nitrate reduction defect in biovars mediaevalis and microtus: the napA gene.
Nitrate reductases are widespread in bacteria and play a very important role in nitrogen assimilation and dissimilation. Three types of nitrate-reducing systems, i.e., cytoplasmic assimilatory nitrate reductase, membrane-bound respiratory nitrate reductase, and periplasmic nitrate reductase (NAP), were found in bacteria, in relation to active-site constitution, subunit structure, cell localization, etc. (9). Many of the pathogenic bacteria that may have to scavenge nitrate from the low levels present have the genetic information for NAP. Indeed in some of these bacteria, e.g., Y. pestis, NAP is the only nitrate reductase present.
In Y. pestis, the structural gene for NAP is named napA. The KIM strain (nitrate negative) has a mutation of napA with a GAA to TAA mutation at the 205th codon, while a GCC to ACC mutation occurs at the 341st codon within napA of the 91001 strain (nitrate negative). In our study, the napA genes from 11 mediaevalis strains and 7 microtus strains were sequenced by using the primers listed in Table 4. Each of the tested genes had a single nucleotide substitution. All 11 mediaevalis strains had the same mutated napA gene as KIM, while all 7 microtus strains had the same mutated gene as 91001. We further tested the 260 Y. pestis isolates for the presence of the two kinds of mutations by allele-specific PCR (Fig. 1b). All 50 mediaevalis strains analyzed were positive for the primer pair 613 M-SN1624 and negative for all of the other primer pairs, indicating that the mutation within napA is same as that of KIM. All 39 microtus strains tested were positive for the primer pair 1021 M-ASN1688 and negative for all of the other primer pairs, indicating that the mutation is same as in 91001.
The sequencing and PCR data demonstrate all nitrate-negative strains possess a mutated napA gene. The null mutation (GAA to TAA) within napA inactivates the expression of the NapA protein of biovar mediaevalis. The GCC-to-ACC mutation leads to the replacement of the hydrophobic Ala by the hydrophilic Thr at the 341st residue within the NapA protein of biovar microtus. The corresponding residue within the NapA protein of Desulfovibrio desulfuricans is a hydrophobic Val located in domain III of NAP (2). This domain is coated with clustered charged residues as well as hydrophobic residues; when binding with the cofactor of molybdenum, it provides a suitable environment for binding nitrate. Small changes in the vicinity of the molybdenum catalytic site, e.g., the hydrophobic conversion of the 341st residue within the NapA protein of Y. pestis biovar microtus, may be sufficient for the inactivation of the NAP enzyme.
The data presented here indicate that a point mutation at the 613th or 1,021th site within the napA gene most likely results in the nitrate-negative phenotype of Y. pestis biovars mediaevalis and microtus, respectively. This is the continued within-species microevolution after the speciation of Y. pestis. Indeed, the genotype and phenotype of Y. pestis itself have been continually adapting to the changes of niches under natural selection, with the resulting expansion of plague foci. Mutations at different positions of the napA gene led to the parallel evolution of the same nitrate-negative phenotype of the two biovars mediaevalis and microtus.
Arabinose utilization defect in biovar microtus: the araC gene.
The arabinose operon of Y. pestis harbors six genes, araA, araB, araF, araG, araH, and araC (7). In Escherichia coli, the ara operon codes for three enzymes that are required to catalyze the metabolism of arabinose: arabinose isomerase, encoded by araA; ribulokinase, encoded by araB; and ribulose-5-phosphate epimerase, encoded by araD (10). The araC gene encodes a regulatory protein. When arabinose is absent, AraC binds to two sites of the operon to transform the operon into a looped DNA, blocking the ParaBAD promoter. The presence of arabinose promotes the rearrangement of AraC from a state in which it represses transcription of the ParaBAD promoter to one in which it activates transcription of the ParaBAD promoter.
The 91001 strain (arabinose negative) has a mutation of araC with a 122-bp frameshift deletion from base 27 to 138 and a G insertion at base 773, which may account for the inactivation of the ara operon involved in arabinose utilization. The araC genes from 24 Y. pestis isolates were sequenced by use of the four primers araC-F1, araC-R1, araC-F2, and araC-R2 (Table 4). The sequencing data indicate that the araC genes of the 8 microtus strains tested are inactivated by the same mutation as in the 91001 strain, while those of the remaining 16 strains are intact. The 260 Y. pestis isolates were then analyzed for the presence of the 122-bp deletion by using the primer pair YPO2258F-YPO2258R (Fig. 1c). It was found that all 39 microtus strains contained the 122-bp deletion within araC, in contrast to the other strains tested. This 100% correlation between the arabinose-negative phenotype and the presence of an inactivated araC gene indicates that most likely the inability of biovar microtus strains to utilize arabinose is due to inactivation of this gene.
Concluding remarks.
On the basis of thorough microarray-, PCR-, and sequencing-based studies with 260 isolates of Y. pestis and the identification of an arabinose genetic and phenotypic marker, we propose a new Y. pestis biovar, microtus. Compared to other types of Y. pestis, biovar microtus strains have a unique genomic profile of gene loss and pseudogene distribution. The specific loss of genes or gene functions documented for this new group of strains is thought to be responsible for the human attenuation of biovar microtus strains. The results provide new avenues for focused hypothesis-based investigations to help delineate the determinants that enable this deadly pathogen to be virulent to humans, which will give insight into the evolution of Y. pestis and plague pathogenesis. Moreover, there may be implications for developing biovar microtus strains as a potential vaccine.
Acknowledgments
Financial support for this work came from the National High Technology Research and Development Program of China (Program 863, no. 2001-AA223061) and the National Natural Science Foundation of China (no. 30371284).
REFERENCES
- 1.Deng, W., V. Burland, G. Plunkett, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dias, J. M., M. E. Than, A. Humm, R. Huber, G. P. Bourenkov, and H. D. Bartunik. 1999. Crystal structure of the first dissimilatory nitrate reductase at 1.9 Å solved by MAD methods. Struct. Fold Des. 7:65-79. [DOI] [PubMed] [Google Scholar]
- 3.Fan, Z., Y. Luo, S. Wang, L. Jin, X. Zhou, J. Liu, Y. Zhang, and F. Li. 1995. Microtus brandti plague in the Xilin Gol Grassland was inoffensive to humans. Chin. J. Control Endem. Dis. 10:56-57. (In Chinese.) [Google Scholar]
- 4.Gonzalez, M. D., C. A. Lichtensteiger, R. Caughlan, and E. R. Vimr. 2002. Conserved filamentous prophage in Escherichia coli O18:K1:H7 and Yersinia pestis biovar Orientalis. J. Bacteriol. 184:6050-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu, Z., R. Hai, and F. Li. 2001. The discovery and study of Microtus fuscus natural plague foci in Qinghai-Tibet plateau. Chin. J. Control Endem. Dis. 16:321-327. (In Chinese.) [Google Scholar]
- 6.Motin, V. L., A. M. Georgescu, J. M. Elliott, P. Hu, P. L. Worsham, L. L. Ott, T. R. Slezak, B. A. Sokhansanj, W. M. Regala, R. R. Brubaker, and E. Garcia. 2002. Genetic variability of Yersinia pestis isolates as predicted by PCR-based IS100 genotyping and analysis of structural genes encoding glycerol-3-phosphate dehydrogenase (glpD). J. Bacteriol. 184:41019-41027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B., Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 8.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardson, D. J., B. C. Berks, D. A. Russell, S. Spiro, and C. J. Taylor. 2001. Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell Mol. Life Sci. 58:165-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheppard, D., and E. Englesberg. 1966. Positive control in the l-arabinose gene-enzyme complex of Escherichia coli B/r exhibited with stable merodiploids. Cold Spring Harbor Symp. Quant. Biol. 31:345-347. [DOI] [PubMed] [Google Scholar]
- 11.Wren, B. W. 2003. The Yersinia—a model genus to study the rapid evolution of bacterial pathogens. Nat. Rev. Microbiol. 1:55-64. [DOI] [PubMed] [Google Scholar]
- 12.Zhou, D., Y. Han, E. Dai, D. Pei, Y. Song, J. Zhai, Z. Du, J. Wang, Z. Guo, and R. Yang. 2004. Identification of signature genes for rapid and specific characterization of Yersinia pestis. Microbiol. Immunol. 48:263-269. [DOI] [PubMed]