Abstract
In Methanothermobacter thermautotrophicus, oxaloacetate synthesis is a major and essential CO2-fixation reaction. This methanogenic archaeon possesses two oxaloacetate-synthesizing enzymes, pyruvate carboxylase and phosphoenolpyruvate carboxylase. The phosphoenolpyruvate carboxylase from this organism was purified to homogeneity. The subunit size of this homotetrameric protein was 55 kDa, which is about half that of all known bacterial and eukaryotic phosphoenolpyruvate carboxylases (PPCs). The NH2-terminal sequence identified this enzyme as the product of MTH943, an open reading frame with no assigned function in the genome sequence. A BLAST search did not show an obvious sequence similarity between MTH943 and known PPCs, which are generally well conserved. This is the first report of a new type of phosphoenolpyruvate carboxylase that we call PpcA (“A” for “archaeal”). Homologs to PpcA were present in most archaeal genomic sequences, but only in three bacterial (Clostridium perfringens, Oenococcus oeni, and Leuconostoc mesenteroides) and no eukaryotic genomes. PpcA was the only recognizable oxaloacetate-producing enzyme in Methanopyrus kandleri, a hydrothermal vent organism. Each PpcA-containing organism lacked a PPC homolog. The activity of M. thermautotrophicus PpcA was not influenced by acetyl coenzyme A and was about 50 times less sensitive to aspartate than the Escherichia coli PPC. The catalytic core (including His138, Arg587, and Gly883) of the E. coli PPC was partly conserved in PpcA, but three of four aspartate-binding residues (Lys773, Arg832, and Asn881) were not. PPCs probably evolved from PpcA through a process that added allosteric sites to the enzyme. The reverse is also equally possible.
The synthesis of oxaloacetate (OAA) is a major and essential CO2-fixation reaction in the methanarchaea (10, 11, 15, 16, 50, 52, 58, 60). These organisms possess an incomplete tricarboxylic acid (TCA) cycle which is used to generate intermediates (OAA and α-ketoglutarate [α-KG]) and a carrier (succinate) for the biosynthesis of amino acids and tetrapyrroles (10, 11, 15, 16, 50, 52, 58, 60). The organisms belonging to the orders of Methanococcales, Methanobacteriales, and Methanomicrobiales, which primarily use hydrogen as an energy source (2), employ a reductive sequence starting at OAA and terminating at α-KG (10, 11, 15, 16, 50, 52, 60).
Methanosarcina species, which predominantly depend on acetotrophic or methylotrophic methanogenesis for energy generation (2), use an oxidative branch of the TCA cycle that initiates with OAA and acetate and terminates at α-KG (52, 58). Hence, OAA synthesis is a central anabolic process in methanarchaea. Thus far, pyruvate carboxylase (PYC) (39, 41, 42, 50) and phosphoenolpyruvate carboxylase (PPC) (14, 31, 47, 60) have been found to be capable of fulfilling this requirement, as follows:
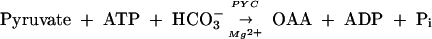 |
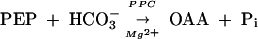 |
PYC is ubiquitous in the methanogens (39, 41, 42, 50), and the primary structure, kinetic characteristics, and expression patterns of the methanogen PYCs have been investigated (39, 41, 42, 50). Methanothermobacter thermautotrophicus (formerly known as Methanobacterium thermoautotrophicum strain ΔH) (2, 57, 61) and Methanothermus sociabilis also possess phosphoenolpyruvate carboxylase (14, 31, 47, 60), but very little of such information is available on these enzymes. Although the Methanothermus sociabilis PPC was purified to homogeneity, the primary structure and the encoding gene of this protein were not identified (31, 47). A search of the genome databases with the amino acid sequences of known PPCs as queries did not detect a homolog of this enzyme in the archaea, even though such homologs were readily found in bacteria and eukaryotes. For these reasons and the probability that the methanarchaeal PPCs have a novel structure, we purified the enzyme from M. thermautotrophicus. Our results show that the PPC from M. thermautotrophicus has a novel structure and that most archaea possess a homolog of this enzyme.
MATERIALS AND METHODS
Purification of phosphoenolpyruvate carboxylase from M. thermautotrophicus.
The enzyme purification was carried out aerobically at 4°C. Frozen pellets (87 g) of autotrophically grown M. thermautotrophicus (42) were thawed and suspended in 90 ml of lysis solution (100 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 20% glycerol, 1 mM dithiothreitol). The cells in this suspension were lysed as described previously (42). The resulting slurry was clarified by three centrifugation steps, the first of which was performed at 15,000 × g for 20 min, with the other two performed at 100,000 × g for 60 and 30 min. The supernatant from the final ultracentrifugation step was fractionated by two consecutive ammonium sulfate precipitation steps. First, the solution was brought to 30% saturation and then the pellet was recovered by centrifugation at 15,000 × g for 30 min and dissolved in 100 ml of buffer A (50 mM Tris-HCl [pH 7.5] in 20% glycerol). The supernatant was then brought to 60% saturation and the pellet was recovered as described above. The percent saturation values were for a temperature of 0°C.
After resuspension, the pellet from the 30% ammonium sulfate saturation was fractionated by anion-exchange chromatography on a 45-ml bed of DEAE-Sephadex A-25 (particle size, 40 to 120 μm; Amersham Biosciences Corp., Piscataway, N.J.) in a 1.5- by 30-cm Econo-column (Bio-Rad Laboratories, Richmond, Calif.). After equilibration of the bed with buffer A, the sample was loaded. The column was then washed with two column bed volumes of buffer A and eluted with a 200-ml linear gradient of 0 to 1 M NaCl in buffer A. The flow rates were 300 ml h−1 for packing and equilibration and 100 ml h−1 for all subsequent steps. The active fractions eluted at about 0.5 M NaCl, and these were pooled and concentrated on an Amicon YM-100 filter (100-kDa cutoff; Amicon, Beverly, Mass.). The retentate was desalted by dilution with buffer B (10 mM potassium phosphate buffer [pH 6.8], 20% glycerol) and further filtration. The desalted enzyme preparation was diluted with 20 ml of buffer B and fractionated by hydroxyapatite chromatography. For this purpose, a 0.75- by 15-cm column packed with Bio-Gel HTP (Bio-Rad Laboratories) to a bed volume of 4.6 ml was used. After the column bed was equilibrated with buffer B, the desalted enzyme preparation was loaded. The column was then washed with two column bed volumes of buffer B and eluted with a 25-ml gradient of 10 to 400 mM potassium phosphate buffer (pH 6.8) in 20% glycerol. The flow rates were 11 ml h−1 for packing and equilibration and 10 ml h−1 for all subsequent steps. The active fractions eluted at about 200 mM potassium phosphate buffer and were pooled.
Solid (NH4)2SO4 was added to a final concentration of 1 M to the pooled fractions, and the resulting preparation was fractionated by octyl Sepharose chromatography, for which a 0.75- by 25-cm column packed with octyl Sepharose CL-4B (particle size, 40 to 190 μm; Amersham Biosciences Corp.) to a bed volume of 8 ml was used. The column bed was equilibrated with 1 M ammonium sulfate in buffer C (25 mM Tris-HCl [pH 7.0], 20% glycerol). After loading of the active fractions from the hydroxyapatite chromatography step, the column was washed with two column bed volumes each of 1 and 0.7 M ammonium sulfate in buffer C and then eluted with a 40-ml reversed gradient of 0.7 to 0 M ammonium sulfate in buffer C. The flow rates were 70 ml h−1 for packing and equilibration and 50 ml h−1 for all subsequent steps. The phosphoenolpyruvate carboxylase activity was eluted at about 200 mM ammonium sulfate, and the active fractions were pooled.
Assays and data analysis.
The protein was assayed according to the Bradford method (3), using Pierce dye reagent (Pierce Biotechnology, Inc., Rockford, Ill.) and a bovine serum albumin standard. Phosphoenolpyruvate carboxylase activity was determined by coupling the reaction with malate dehydrogenase (MDH) and monitoring the oxidation of NADH spectrophotometrically at 340 nm (55). Unless otherwise mentioned, the assay was conducted aerobically at 50°C with a reaction mixture of the following composition: 50 mM Tris-HCl (pH 8.0), 0.2 mM NADH + H+, 5 mM KHCO3, 2.5 mM phosphoenolpyruvate (PEP), 5 mM MgCl2, and 1 U of thermophilic MDH from Thermus flavus (Sigma, St. Louis, Mo.) per ml. In the amount used, MDH did not limit the reaction rate in any assay. In most cases, the reaction was initiated by the addition of PPC. The assays that were performed to monitor the recoveries during the enzyme purification were initiated with PEP. To avoid interference from NADH oxidase activities, we performed assays with cell extracts and preparations from the ammonium sulfate precipitations anaerobically (38); other preparations were free of NADH oxidase activity. All pH values reported here were determined at 25°C. The initial velocity data were analyzed with the KinetAsyst program, version 1.01 (Intellikinetics, State College, Pa.).
Gel electrophoresis, determination of NH2-terminal sequence, and gel filtration chromatography.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with a slab gel according to the method of Laemmli (33). The NH2-terminal sequence was determined via automated Edman degradation in which a membrane blot of a denatured sample of the purified protein was processed (40). This experiment was performed by use of a Procise 494 HT sequencer (Applied Biosystems, Foster City, Calif.) in pulse-liquid mode at the Protein Sciences Facility, Biotechnology Center, University of Illinois, Urbana-Champaign. The analysis was done for 10 cycles. For gel filtration analysis, a HiPrep 16/60 Sephacryl S-300 HR column (bed volume, 120 ml) and a fast protein liquid chromatography system (Amersham Pharmacia Biotech, Inc.) were used. The development was isocratic, at a flow rate of 30 ml h−1, with a mobile phase of 50 mM sodium phosphate buffer (pH 7.2) containing 150 mM NaCl. For calibration of the column, the following molecular size standards (Bio-Rad Laboratories) were employed (mass, Stokes radius) (40): bovine thyroglobulin (670 kDa, 85 Å), bovine gammaglobulin (158 kDa, 52.5 Å), chicken ovalbumin (44 kDa, 30.5 Å), horse myoglobin (17 kDa, 19 Å), and vitamin B12 (1.357 kDa). For each application, a 0.5-ml solution containing these components, in the amounts of 2.5, 2.5, 2.5, 1.25, and 0.25 mg, respectively, was used. The phosphoenolpyruvate carboxylase sample analyzed was a partially purified preparation. It was obtained by precipitating proteins from a cell extract with ammonium sulfate to 60% saturation as described above and resuspending the resulting pellet in 50 mM Tris-HCl (pH 7.5) containing 20% glycerol. A 0.5-ml aliquot of this preparation containing 21.7 mg of protein was analyzed. The enzyme peak in the elution profile was identified by assaying the column fractions for activity.
Amino acid sequence alignment and phylogenetic inference.
Amino acid sequences were aligned automatically with the ClustalW program (version 1.82) (56) and then were analyzed by protein maximum likelihood methods with the ProML program (PHYLIP, version 3.6a2.1) (12) using the JTT amino acid substitution model. Bootstrap proportions were calculated with the Seqboot, ProML, and Consense programs from the PHYLIP package. A similar phylogenetic tree was inferred by the neighbor-joining method with the Protdist and Neighbor programs (12).
RESULTS
Purification of phosphoenolpyruvate carboxylase from M. thermautotrophicus.
The enzyme from M. thermautotrophicus was purified to homogeneity as described in Table 1. In the ammonium sulfate precipitation step, the 30% saturation pellet retained approximately 60% of the total recovered activity and the 60% saturation pellet had the remaining activity. Starting with the 30% ammonium sulfate saturation pellet, some purification experiments yielded homogeneous fractionations. Such homogeneous preparations were rare, and only a limited amount of the enzyme was available for characterization. It was also not possible to obtain a homogeneous preparation by fractionation of the 60% ammonium sulfate saturation pellet.
TABLE 1.
Purification of PPC from M. thermautotrophicus
| Purification step | Volume (ml) | Protein (mg) | Sp act (U/mg) | Total act (U) | Purification (fold) | % Recovery |
|---|---|---|---|---|---|---|
| Ultracentrifugation | 112 | 4,200 | 0.015 | 63 | ||
| Ammonium sulfate precipitation | 100 | 43.5 | 0.391a | 17a | 26 | 27 |
| DEAE-Sephadex chromatography | 32.5 | 5.68 | 5.5a | 31.3a | 367 | 50 |
| Concentration | 20 | 4.8 | 7.37 | 35.4 | 492 | 56 |
| Hydroxypatite chromatography | 10.5 | 1.05 | 23.2 | 24.3 | 1,543 | 39 |
| Octyl Sepharose chromatography | 14 | 0.07 | 260 | 18.2 | 17,333 | 29 |
The reported value is an underestimate, because ammonium sulfate and sodium chloride inhibited the enzyme.
Molecular and catalytic properties of phosphoenolpyruvate carboxylase.
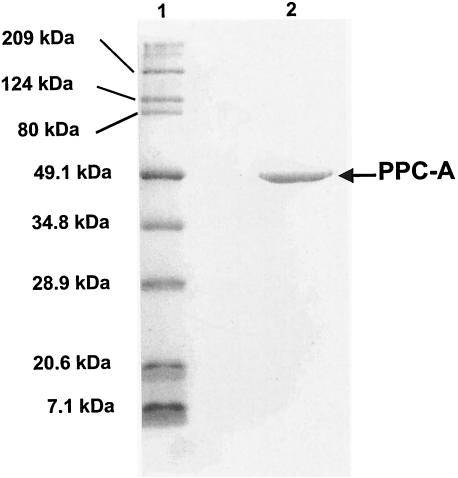
Based on SDS-PAGE data, we concluded that the purified enzyme consisted of only one type of polypeptide, with an apparent molecular mass of 50 kDa (Fig. 1). Due to the difficulty in obtaining an adequate amount of purified enzyme, we estimated the native molecular mass by using a partially purified preparation. From the gel filtration chromatography data, the Stokes radius and apparent native molecular mass were estimated to be 61.8 Å and 277 kDa, respectively. Therefore, the enzyme was homotetrameric. The NH2-terminal amino acid sequence for the polypeptide was MKVPRXMSTQ, where “X” was an unidentified residue. In the range of 25 to 70°C, the enzyme exhibited maximum specific activity at 62°C. From the linear segment (between 25 and 62°C) of the corresponding Arrhenius plot, the activation energy for OAA formation was calculated to be 53.8 kJ mol−1. The loss of HCO3− at lower pH values did not allow for an accurate determination of the optimum pH. The apparent pH optimum for the enzyme was 6.8, and the activities at pHs 6.3 and 7.2 were 5 and 92% of the maximum value, respectively. The activity was also highly inhibited by either NaCl or KCl (0.05 to 1.0 M). At a concentration of 0.25 M, each of these salts inhibited the enzyme by 60%; at 0.1 M, the inhibition was 27%.
FIG. 1.
SDS-PAGE with purified PPC from M. thermautotrophicus. Lane 1, molecular mass standards; lane 2, 2.5 μg of homogeneous enzyme from octyl Sepharose chromatography. The gel was stained with Coomassie blue R-250.
Mg2+ was required for activity and was not significantly inhibitory at concentrations up to 50 mM when the PEP concentration in the assay was 5 mM. The responses of the initial velocity to the variation in KHCO3 concentration in the 0.1 to 10 mM range when the PEP concentration in the assays was held constant at 5 mM followed the Henri-Michaelis-Menten equation well, and from a fit of the velocity data to this equation, the apparent Km value for KHCO3 was determined to be 1.8 ± 0.2 mM. A similar response was observed with 0.025 to 3.5 mM PEP when the KHCO3 concentration was fixed at 10 mM, and the Km value for PEP was 0.46 ± 0.03 mM. At PEP, KHCO3, and Mg2+ concentrations of 2.5, 5, and 5 mM, respectively, the specific activity of the homogenous enzyme was 260 U mg−1.
The effects of several potential modulators on enzyme activity were examined under standard assay conditions, except that the PEP and KHCO3 concentrations were 5 and 20 mM, respectively. Acetyl-CoA at the concentrations of 0.2 and 2 mM neither stimulated nor inhibited the activity of the enzyme. At a concentration of 10 mM, ATP, ADP, AMP, GTP, and aspartate reduced the activity by 92, 57, 16, 58, and 41%, respectively; the activity observed in the absence of any one of these compounds was taken as 100%. If the concentration of Mg2+ was increased to 15 mM, ATP, ADP, AMP, or GTP (each at a level of 10 mM) reduced the activity of the enzyme by 65, 46, 12, and 15%, respectively. The inhibition by aspartate was not influenced by such an increase in the Mg2+ level. At a concentration of 10 mM, glutamate and α-KG did not inhibit the enzyme significantly.
Phosphoenolpyruvate carboxylase gene of M. thermautotrophicus.
The NH2-terminal sequence of the purified enzyme was identical to residues 40 to 49 of MTH943, an ORF with no assigned function in the M. thermautotrophicus genome sequence (54). We named this ORF ppcA (“A” for “archaeal”). An analysis with the SignalP program (http://www.cbs.dtu.dk/services/SignalP/) (44) did not identify the first 39 residues of MTH943 as a potential signal sequence. The codon for the 40th residue (ATG) was preceded by a potential ribosome-binding site (GGAGTG, 7 to 12 bp upstream of ATG), but a recognizable ribosome-binding site was not apparent near the previously described start site for MTH943 (54). Therefore, we propose that the ATG codon for the Met residue at the 40th position of this ORF is the actual initiation codon for the ppcA gene. With this change, the calculated subunit molecular mass for the protein would be 55,042 Da, which was close to the experimentally determined value of 50 kDa (Fig. 1).
A BLAST search (1) with the amino acid sequence for a known PPC did not identify MTH943 as a PPC homolog. In contrast, bacterial and plant PPCs are readily identified through these searches. A pairwise comparison at the Pôle Bio-InformatiqueLyonnais site on the World Wide Web showed that PpcA had very little sequence similarity to the maize and E. coli PPCs (17, 22) and provided the following values (percentages of identity and percentages of strong similarity): for maize PPC and E. coli PPC, 38.45 and 21.44%; for E. coli PPC and PpcA, 12.12 and 12%; and for maize PPC and PpcA, 9.78 and 12.26%. PpcA is composed of 483 amino acid residues, whereas the E. coli and maize PPC polypeptides contain 883 and 970 residues, respectively (17, 22).
Distribution of archaeal phosphoenolpyruvate carboxylase homologs in other organisms.
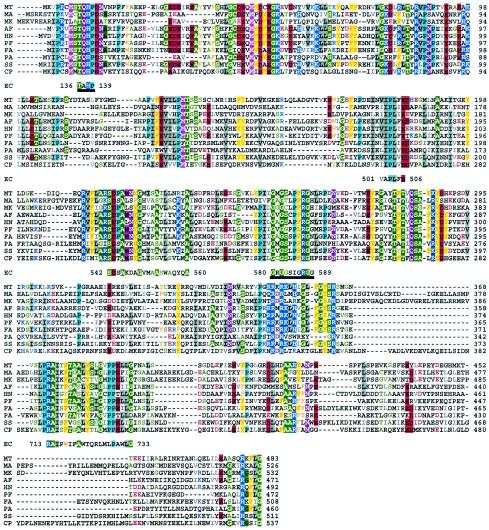
Interestingly, PpcA homologs were present in most archaea for which whole genome sequences are available (Table 2). A search of bacterial genomes identified only the following PpcA homologs (bacterium, open reading frame [ORF], accession number): Clostridium perfringens, CPE1094, NP_562010 (51); Oenococcus oeni MCW, Ooen1256, ZP_00070236; and Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293A, Lmes0541, ZP_00063059. None of these bacterial homologs have been assigned a function. A primary structure alignment indicated that the PpcA homologs were highly similar to each other (Fig. 2). A phylogenetic analysis clearly supported this relationship and did not reveal specific groupings within the PpcAs. The same analysis showed that the previously described PPCs have no discernible evolutionary relationships to the archaeal enzymes.
TABLE 2.
Distribution of OAA-synthesizing enzymes in archaeaa
| Organism (reference for experimental and genome sequence data)b | ORF no. or absence of ORFc
|
||
|---|---|---|---|
| PpcA | PYCA/PYCBd | PCKe | |
| Methanogens | |||
| Methanocaldococcus jannaschii (4, 39) | Absent | MJ1229/MJ1231 | Absent |
| Methanococcus maripaludis (50) (BX950229)f | Absent | MMP0341/MMP0340 | Absent |
| Methanopyrus kandleri (20, 53) | MK0190 | Absent | Absent |
| Methanothermobacter thermautotrophicus (14, 31, 42, 54) | MTH943 | MTH1917/MTH1107 | Absent |
| Methanosarcina barkeri (41) | METH1932 | METH3573/METH3572 | Absenth |
| Methanosarcina mazei (9) | MM3212 | MM1828/MM1827 | Absent |
| Methanosarcina acetivorans (18) | MA2690 | MA0675/MA0674 | Absent |
| Methanococcoides burtonii (48) | Absenth | Mbur125601/Mbur125501 | Absenth |
| Nonmethanogens | |||
| Halobacterium sp. NRC-1 (43) | VNG2259c | Absent | Absent |
| Archaeoglobus fulgidus (32) | AF1486 | Absent | Absent |
| Pyrococcus furiosus (45) | PF1975 | Absent | PF0289 |
| Pyrococcus horikoshii (28) | PH0016 | Absent | PH0312 |
| Pyrococcus abyssi (8) | PAB2342 | Absent | PAB1253 |
| Thermoplasma acidophilum (46) | Absent | Absent | Ta0123 |
| Thermoplasma volcanium (29) | Absent | Absent | TVN0200 |
| Ferroplasma acidarmanus | FACI0253 | Absenth | FACI0139 |
| Pyrobaculum aerophilum (13) | PAE3416 | Absent | Absent |
| Sulfolobus acidocaldarius (47a) | Present | —i | — |
| Sulfolobus solfataricus (49) | SSO2256 | Absent | SSO2537 |
| Sulfolobus tokadaii (26) | ST2101 | Absent | ST1058 |
| Aeropyrum pernix (27) | Absent | Absent | APE0033g |
The evidence for PpcA in M. thermautotrophicus and PYCA and PYCB in M. jannaschii, M. thermautotrophicus, and M. barkeri are experimental (31, 39, 41, 42; this work); M. maripaludis and S. acidocaldarius data are partly experimental (47a, 50). The rest of the data were inferred from the sequence data in the National Center for Biotechnology Information database.
Organisms in bold are Crenarchaea; all other organisms are Euryarchaea.
Retrieved from the National Center for Biotechnology Information database from similarity searches by use of MTH943 (for PpcA), M. thermautotrophicus PYC subunits (for PYCA and PYCB) (42), E. coli PCK (for ATP-dependent PCK) (35), and Myocbacterium smegmatis PCK (for GTP-dependent PCK) (37) as queries.
α4β4-type enzyme composed of PYCA and PYCB subunits.
GTP-dependent enzyme, unless otherwise indicated.
An accession number was used to refer to the unpublished genome data.
ATP-dependent enzyme.
Unfinished genome.
—, No genome sequence.
FIG. 2.
Primary structure alignment for PpcA from M. thermautotrophicus and its homologs. The segments of E. coli PPC that were successfully aligned with the PpcAs are shown with the position numbers for their termini. The sources for the sequences are as follows (abbreviation, organism, ORF number, accession number [reference]): MT, M. thermautotrophicus, MTH943, D69226 (54); MA, Methanosarcina acetivorans, MA2690, NP_617588 (18); MK, Methanopyrus kandleri, MK0190, NP_613477 (53); AF, Archaeoglobus fulgidus, AF1486, NP_070315 (32); HN, Halobacterium sp. NRC-1, VNG2259c, NP_280898 (43); PF, Pyrococcus furiosus, PF1975, NP_579704 (45); FA, Ferroplasma acidarmanus, FACI0253, ZP_00000247; PA, Pyrobaculum aerophilum PAE3416, NP_560717 (13); SS, Sulfolobus solfataricus, SSO2256, NP_343633 (49); CP, Clostridium perfringens CPE1094, NP_562010 (51). Shading with color shows sequence identity and a presentation of the residues in color indicates functional conservation, and in both cases the following color codes were used: blue, H, K, and R (basic); green, A, G, C, S, and T (small); gray, I, L, and V (aliphatic); pink, M; red, D and E (acidic); turquoise, P; violet, Q and N (neutral); orange (with or without yellow shading), F, Y, and W (aromatic). Two catalytically important and characteristic features for the E. coli PPC (24, 25) are marked underneath with black bars; these were found to be conserved in the PpcAs (Fig. 3).
Comparison of primary structures of M. thermautotrophicus PpcA and PPCs.
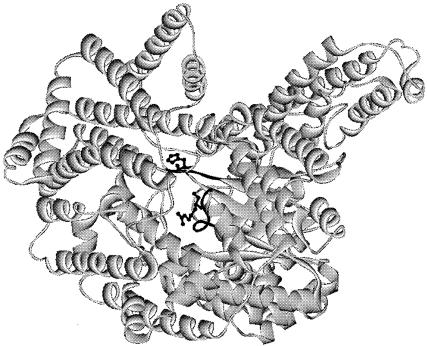
As mentioned above, a BLAST search (1) did not identify MTH943 as a PPC. PSI- and PHI-BLAST searches (1) revealed weak similarities between PpcA and several bacterial PPCs. An analysis with 3D-PSSM, a web-based program for protein fold recognition (http://www.sbg.bio.ic.ac.uk/servers/3dpssm/) (30), revealed the following PPC characteristics (24, 25) in the archaeal enzyme: class, alpha and beta proteins; fold, TIM beta/alpha-barrel; and superfamily, phosphoenolpyruvate/pyruvate domain. These clues, automated alignments (1, 56), and manual inspections helped us to find certain key PPC features in M. thermautotrophicus PpcA which are presented in Fig. 2. Figure 3 shows the three-dimensional structure of an Escherichia coli PPC subunit in which the highlighted sections represent two catalytically important sequence elements that were conserved in M. thermautotrophicus PpcA and its homologs.
FIG. 3.
Structural similarities between E. coli PPC and M. thermautotrophicus PpcA. The structure shown is for the E. coli enzyme (accession no. 1FIY) (24). Catalytically important 136TAHP139 and 580GRGGSIGRGGQ589 elements are shown in black, and the homologous sequences in M. thermautotrophicus PpcA were 9TQHP12 and 252GMGSAPFRGN261, respectively. The side chains shown correspond to His138 and Arg587 of E. coli PPC and were conserved in M. thermautotrophicus PpcA as His11 and Arg259.
DISCUSSION
We have purified and characterized the PPC from M. thermautotrophicus to homogeneity and determined that the subunit of this enzyme corresponds to MTH943 (54), an ORF that has no assigned function and is considered distinctive to the archaea (19). The molecular mass of the subunit of this enzyme was similar to that of the Methanothermus sociabilis PPC (60 kDa) (47) but was half that for all known bacterial and eukaryotic PPCs (96 to 116 kDa) (25). Also, the overall amino acid sequence of the M. thermautotrophicus enzyme was very dissimilar to that of the known PPCs. A careful inspection revealed that a few of the essential and characteristic PPC sequence features were partially conserved in the archaeal enzyme (Fig. 2 and 3). As described below, these conserved elements represented only a small part of the PPC protein and were located either at or near the catalytic core (24, 25). Also, the sequence features that allow for allosteric regulation of E. coli and maize PPCs (24, 25) were apparently absent from MTH943. Thus, the M. thermautotrophicus enzyme was clearly distinct from all previously described bacterial and eukaryotic PPCs, and we called this archaeal enzyme PpcA. Similar to the Methanothermus sociabilis PpcA (native mass, 240 kDa) (47) and all PPCs (24, 25), the M. thermautotrophicus enzyme (277 kDa) was homotetrameric. However, a final conclusion on the quaternary structure of the PpcAs must await an accurate determination of the native molecular mass for these proteins by use of a more appropriate method. The present values were derived solely from gel filtration data (this work; 47) and therefore are less reliable than values from analytical ultracentrifugation (7).
PPCs are present in all photosynthetic organisms, which include higher plants, green algae, and photosynthetic bacteria (25). They are also found in most nonphotosynthetic bacteria and protozoa, but not in molds, yeasts, and animals (25). In all of these organisms, PPC serves an anaplerotic role. The structure of this very widely distributed enzyme is highly conserved across phylogenetic boundaries (24, 25). Therefore, the observed lack of obvious similarity between the PPCs and M. thermautotrophicus PpcA came as a great surprise. An equally surprising observation was that PPCs were absent from the archaea, whereas PpcA was almost universally present in this domain. Interestingly, outside of the archaeal domain, PpcA homologs were found in only three bacteria, one of which (C. perfringens [51]) does not possess a PPC; the genome sequence data for the other two (O. oeni and L. mesenteroides) are not complete.
The alignment in Fig. 2 shows that some of the catalytically important sequence features of the bacterial and eukaryotic PPCs were partially conserved in the M. thermautotrophicus enzyme (Fig. 2). The PPCs possess a unique Gly-Arg loop sequence (GRGGXXGRGG, where XX = TV, SI, or SV) which appears in the E. coli and maize proteins as GRGGSIGR587GG (positions 580 to 589) (Fig. 3) and GRGGTVGR647GG (positions 640 to 649), respectively (24, 25). Arg587 of the E. coli PPC and Arg647 of the maize PPC are PEP-binding-site residues and are essential for catalysis (24, 25). This Gly-Arg loop forms a lid for the active site and thereby protects the reaction intermediates from attack by water (24). A sequence element homologous to this loop was present in each PpcA, and in the M. thermautotrophicus enzyme it was 252GMGSAPFR259GN261 (Fig. 2). However, the putative Gly-Arg element of the archaeal enzymes had only three Gly and one Arg residue (compared to six conserved Gly and two Arg residues in a PPC) and also contained a Pro and a Phe residue (Fig. 2). It will be interesting to study how the Pro and Phe residues affect the function of the Gly-Arg loop. The His177 residue of the maize PPC directly participates in catalysis (24, 25) and lies within a TXHP element that is fully conserved in all PPCs (residues 135 to 138 in E. coli PPC) (Fig. 2 and 3). The PpcA homologs also contained a TXHP sequence which belonged to a larger and fully conserved element, with a consensus of PXXM(M/A/C/S)TQHPD (residues 4 to 13 of the M. thermautotrophicus enzyme) (Fig. 2). In summary, His11and Arg259 of M. thermautotrophicus PpcA might represent functional equivalents of the catalytically active His138 and Arg587 of E. coli PPC (Fig. 2 and 3). The following sequence elements were found to be conserved in both PPCs and PpcAs (consensus, sequence numbers in M. thermautotrophicus PpcA, and sequence numbers in E. coli PPC): (V/I/L)(A/I)PL(F/V/I)E, 178 to 183, 501 to 506; SDXAX3GX3(S/A)X6A, 214 to 232, 542 to 560; RAIX(F/W/Y)X9-10P X3G, 373 to 392, 713 to 733. Leu504, Glu506, and Arg713 of E. coli PPC belong to these elements and have been implicated in the fixation of CO2 onto the enolate form of pyruvate, in binding Mn2+, and in catalysis, respectively (25).
E. coli PPC is activated by acetyl-CoA (6), whereas this compound did not have any effect on the activity of M. thermautotrophicus PpcA. The archaeal enzyme was relatively insensitive to aspartate, an allosteric inhibitor of the PPCs (5, 21). At a concentration of 10 mM, aspartate lowered the activity of PpcA by 41%. For the E. coli enzyme, the same degree of inhibition is caused by about 0.2 mM aspartate (55). Thus, the M. thermautotrophicus enzyme was about 50 times less sensitive to aspartate than the E. coli enzyme. The small PPC from Methanothermus sociabilis is also insensitive to aspartate (47). This difference between the two classes of enzymes fits the observation that of four aspartate-binding residues of E. coli PPCs (Arg587, Lys773, Arg832, and Asn881) (25), three are not conserved in the PpcAs. Arg587, which also has a catalytic role (24, 25), is conserved in the PpcAs (Fig. 2). Asn881 of E. coli PPC belongs to a carboxy-terminal NTG element (25). The terminal Gly residue is catalytically active and forms an ion pair with Arg587 (25). Aspartate binds to the Asn of the NTG element and prevents the terminal Gly from moving towards the ion-pairing Arg (25). While the PpcA homologs possess the carboxyl-terminal Gly residue (Fig. 2), the Asn881 of the E. coli enzyme was not conserved in these proteins. Lys773 and Arg832 of E. coli PPC are also not conserved in the PpcAs.
It is rare to find both a PPC and PYC in an organism, and wherever they coexist their activities are regulated differently (reviewed in reference 42). The kinetic properties of the PpcA and PYC from M. thermautotrophicus suggest that these OAA-synthesizing enzymes might be active in the cell under two different sets of conditions. The PYC from this methanogen shows a strong substrate inhibition with ATP and a negative cooperativity towards bicarbonate (42). Therefore, this enzyme will be more active at low ATP and high bicarbonate concentrations, a state that represents hydrogen limitation, among other things. Because the apparent Km value for bicarbonate of PpcA is 1.8 mM and that of PYC is 6.8 mM (42), PpcA will be the favored activity at low bicarbonate concentrations. It is possible that under biotin limitation a cell will fully or primarily depend on PpcA activity for OAA synthesis, because this organism does not produce holo-PYC if an exogenous source of biotin is not available (42). Although the PpcA of M. thermautotrophicus was inhibited by NaCl and KCl, the PYC of this organism is greatly stimulated by these salts and shows maximal activity if these are present in the assay at 0.2 and 0.4 M concentrations, respectively (42). When it is present at a level higher than that of ATP, Mg2+ inhibits PYC (42), whereas the PpcA activity was not influenced if this ion was present in a large excess over PEP. More precise comparisons of the physiological roles of the PpcA and PYC in a methanogen will require a genetic analysis. Since the Methanosarcina species possess both a PYC (41) and a PpcA homolog (Fig. 2 and Table 2) and are amenable to genetic manipulations (36), they offer a better system for such studies.
Table 2 summarizes our current knowledge of the OAA-synthesizing activities in the archaea, and the data show a very interesting pattern. A PpcA homolog was found in every methanogen, except Methanocaldococcus jannaschii (4) and Methanococcus maripaludis (accession number BX950229) (Table 2); the genome sequence information for Methanococcoides burtonii is incomplete (48). It has been shown that cell extracts of Methanococcus maripaludis lack PPC activity (50). The other archaeal genome sequences that lack PpcA homologs are those of Thermoplasma volcanium (29), Thermoplasma acidophilum (46), and Aeropyrum pernix (27) (Table 2), and these organisms probably rely solely on phosphoenolpyruvate carboxykinase (PCK) for OAA synthesis.
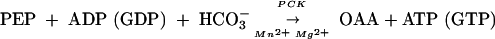 |
Thus, PpcA was widespread in the archael domain (Table 2). In contrast, PYC is very much a methanogen enzyme and PCK is probably restricted to the crenarchaeota (Table 2). A PCK can either be ATP dependent or GTP dependent (34), and most archaeal enzymes are expected to be GTP dependent (Table 2). The putative ATP-dependent PCK of Aeropyrum pernix (27) is an exception. Methanopyrus kandleri differs from other methanogens in that it lacks a PYC homolog (Table 2). It also does not possess a PCK (Table 2). It has been recently proposed that Methanopyrus kandleri uses a carboligase for OAA synthesis (53). Our analysis suggested that this extremely thermophilic archaeon probably uses a PpcA for this purpose, and a similar situation exists for Archaeoglobus fulgidus and Halobacterium sp. NRC-1 (Table 2). Among the crenarchaeota, Pyrobaculum aerophilum (13) presents such an example (Table 2).
The unusual catalytic and regulatory properties and structure of PpcA bring new thoughts on the evolution of PPCs (59). As shown in Fig. 3, the areas of clear similarities between PpcA and PPCs concern only the catalytic core. It is possible that PPCs evolved from a PpcA through a process that increased the subunit size and generated the structural attributes that are required for the allosteric regulation of activity (5, 21, 24, 25). It is equally possible that PpcA was derived from a complex PPC through the loss of certain features. A similar benefit could be realized in research aimed towards improving photosynthesis productivities in C3 plants in which the focus has been the introduction of an unregulated PPC (24, 25). PpcA forms a new topic in archaeal biochemistry and CO2 sequestration, as well as the primary productivity in the hydrothermal vent environment, where organisms such as Methanopyrus kandleri and Methanocaldococcus jannaschii live (20, 23) and employ two different OAA synthesis routes (Table 2) (39).
Acknowledgments
A part of the reported work was completed during B.M.'s postdoctoral research in the laboratory of Ralph S. Wolfe at the University of Illinois, and we thank him for encouragement and support. We thank David E. Graham for help in phylogenetic analysis, John A. Leigh for access to the M. maripaludis genome database before submission, and Endang Purwantini for discussions and comments on the manuscript.
Hiten Patel received a Colgate-Palmolive Undergraduate Research Award from the Biotechnology Center at the University of Illinois at Urbana-Champaign. This work was supported, in part, by a start-up fund to B.M. from the Virginia Bioinformatics Institute, Virginia Tech.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boone, D. R., W. B. Whitman, and P. Rouviere. 1993. Diversity and taxonomy of methanogens. In J. G. Ferry (ed.), Methanogenesis: ecology, physiology, biochemistry and genetics. Chapman and Hall, New York, N.Y.
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon Methanococcus janaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 5.Canovas, J. L., and H. L. Kornberg. 1966. Properties and regulation of phosphopyruvate carboxylase activity in Escherichia coli. Proc. R. Soc. Lond. B 165:189-205. [DOI] [PubMed] [Google Scholar]
- 6.Canovas, J. L., and H. L. Kornberg. 1965. Fine control of phosphopyruvate carboxylase activity in Escherichia coli. Biochim. Biophys. Acta 96:169-172. [DOI] [PubMed] [Google Scholar]
- 7.Cantor, C. R., and P. R. Schimmel. 1980. Techniques for the study of biological structure and function: biophysical chemistry, vol. 2. W. H. Freeman Co., New York, N.Y.
- 8.Chinen, A., I. Uchiyama, and I. Kobayashi. 2000. Comparison between Pyrococcus horikoshii and Pyrococcus abyssi genome sequences reveals linkage of restriction-modification genes with large genome polymorphisms. Gene 259:109-121. [DOI] [PubMed] [Google Scholar]
- 9.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, R. Martinez-Arias, A. Henne, A. Wiezer, S. Baumer, C. Jacobi, H. Bruggemann, T. Lienard, A. Christmann, M. Bomeke, S. Steckel, A. Bhattacharyya, A. Lykidis, R. Overbeek, H. P. Klenk, R. P. Gunsalus, H. J. Fritz, and G. Gottschalk. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453-461. [PubMed] [Google Scholar]
- 10.Ekiel, I., I. C. Smith, and G. D. Sprott. 1983. Biosynthetic pathways in Methanospirillum hungatei as determined by 13C nuclear magnetic resonance. J. Bacteriol. 156:316-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ekiel, I., G. D. Sprott, and G. B. Patel. 1985. Acetate and CO2 assimilation by Methanothrix concilii. J. Bacteriol. 162:905-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 2001. PHYLIP, Phylogeny Inference Package, version 3.6a2.1. Department of Genetics, University of Washington, Seattle.
- 13.Fitz-Gibbon, S. T., H. Ladner, U. J. Kim, K. O. Stetter, M. I. Simon, and J. H. Miller. 2002. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. USA 99:984-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs, G., and E. Stupperich. 1982. Autotrophic CO2 fixation pathway in Methanobacterium thermoautotrophicum. Zentbl. Bakteriol. Hyg. Abt. 1 Orig. C3 177:277-288. [Google Scholar]
- 15.Fuchs, G., and E. Stupperich. 1978. Evidence for an incomplete reductive carboxylic acid cycle in Methanobacterium thermoautotrophicum. Arch. Microbiol. 118:121-125. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs, G., E. Stupperich, and R. K. Thauer. 1978. Acetate assimilation and the synthesis of alanine, aspartate and glutamate in Methanobacterium thermoautotrophicum. Arch. Microbiol. 117:61-66. [DOI] [PubMed] [Google Scholar]
- 17.Fujita, N., T. Miwa, S. Ishijima, K. Izui, and H. Katsuki. 1984. The primary structure of phosphoenolpyruvate carboxylase of Escherichia coli. Nucleotide sequence of the ppc gene and deduced amino acid sequence. J. Biochem. (Tokyo) 95:909-916. [DOI] [PubMed] [Google Scholar]
- 18.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham, D. E., R. Overbeek, G. J. Olsen, and C. R. Woese. 2000. An archaeal genomic signature. Proc. Natl. Acad. Sci. USA 97:3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huber, R., M. Kurr, H. W. Jannasch, and K. O. Stetter. 1989. A novel group of abyssal methanogenic archaebacteria (Methanopyrus) growing at 110°C. Nature (London) 342:833-834. [Google Scholar]
- 21.Izui, K. 1970. Kinetic studies on the allosteric nature of phosphoenolpyruvate carboxylase from Escherichia coli. J. Biochem. (Tokyo) 68:227-238. [DOI] [PubMed] [Google Scholar]
- 22.Izui, K., S. Ishijima, Y. Yamaguchi, F. Katagiri, T. Murata, K. Shigesada, T. Sugiyama, and H. Katsuki. 1986. Cloning and sequence analysis of cDNA encoding active phosphoenolpyruvate carboxylase of the C4-pathway from maize. Nucleic Acids Res. 14:1615-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, W. J., J. A. Leigh, F. Mayer, C. R. Woese, and R. S. Wolfe. 1983. Methanococcus jannaschii sp. nov., an extreme thermophilic methanogen from a submarine hydrothermal vent. Arch. Microbiol. 136:254-261. [Google Scholar]
- 24.Kai, Y., H. Matsumura, T. Inoue, K. Terada, Y. Nagara, T. Yoshinaga, A. Kihara, K. Tsumura, and K. Izui. 1999. Three-dimensional structure of phosphoenolpyruvate carboxylase: a proposed mechanism for allosteric inhibition. Proc. Natl. Acad. Sci. USA 96:823-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kai, Y., H. Matsumura, and K. Izui. 2003. Phosphoenolpyruvate carboxylase: three-dimensional structure and molecular mechanisms. Arch. Biochem. Biophys. 414:170-179. [DOI] [PubMed] [Google Scholar]
- 26.Kawarabayasi, Y., Y. Hino, H. Horikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Kato, T. Yoshizawa, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, S. Masuda, M. Yanagii, M. Nishimura, A. Yamagishi, T. Oshima, and H. Kikuchi. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. DNA Res. 8:123-140. [DOI] [PubMed] [Google Scholar]
- 27.Kawarabayasi, Y., Y. Hino, H. Horikawa, S. Yamazaki, Y. Haikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, H. Nakazawa, M. Takamiya, S. Masuda, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, H. Kikuchi, et al. 1999. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 6:83-101, 145-152. [DOI] [PubMed]
- 28.Kawarabayasi, Y., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, S. Yamamoto, M. Sekine, S. Baba, H. Kosugi, A. Hosoyama, Y. Nagai, M. Sakai, K. Ogura, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Ohfuku, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, and H. Kikuchi. 1998. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5:55-76. [DOI] [PubMed] [Google Scholar]
- 29.Kawashima, T., N. Amano, H. Koike, S. Makino, S. Higuchi, Y. Kawashima-Ohya, K. Watanabe, M. Yamazaki, K. Kanehori, T. Kawamoto, T. Nunoshiba, Y. Yamamoto, H. Aramaki, K. Makino, and M. Suzuki. 2000. Archaeal adaptation to higher temperatures revealed by genomic sequence of Themoplasma volcanium. Proc. Natl. Acad. Sci. USA 97:14257-14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 31.Kenealy, W. R., and J. G. Zeikus. 1982. Characterization and function of phosphoenolpyruvate carboxylase in Methanobacterium thermoautotrophicum. FEMS Microbiol. 14:7-10.
- 32.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, J. C. Venter, et al. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 34.Matte, A., L. W. Tari, H. Goldie, and E. L. T. Delbaer. 1997. Structure and mechanism of phosphoenolpyruvate carboxylkinase. J. Biochem. 272:8105-8108. [DOI] [PubMed] [Google Scholar]
- 35.Medina, V., R. Pontarollo, D. Glaeske, H. Tabel, and H. Goldie. 1990. Sequence of the pckA gene of Escherichia coli K-12: relevance to genetic and allosteric regulation and homology of E. coli phosphoenolpyruvate carboxykinase with the enzymes from Trypanosoma brucei and Saccharomyces cerevisiae. J. Bacteriol. 172:7151-7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metcalf, W. W., J. K. Zhang, E. Apolinario, K. R. Sowers, and R. S. Wolfe. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. USA 94:2626-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukhopadhyay, B., E. M. Concar, and R. S. Wolfe. 2001. A GTP-dependent vertebrate-type phosphoenolpyruvate carboxykinase from Mycobacterium smegmatis. J. Biol. Chem. 276:16137-16145. [DOI] [PubMed] [Google Scholar]
- 38.Mukhopadhyay, B., and L. Daniels. 1989. Aerobic purification of N5,N10-methylenetetrahydromethanopterin dehydrogenase, separated from N5,N10-methylenetetrahydromethanopterin cyclohydrolase, from Methanobacterium thermoautotrophicum strain Marburg. Can J. Microbiol. 35:499-507. [DOI] [PubMed] [Google Scholar]
- 39.Mukhopadhyay, B., V. J. Patel, and R. S. Wolfe. 2000. A stable archaeal pyruvate carboxylase from the hyperthermophile Methanococcus jannaschii. Arch. Microbiol. 174:406-414. [DOI] [PubMed] [Google Scholar]
- 40.Mukhopadhyay, B., and E. Purwantini. 2000. Pyruvate carboxylase from Mycobacterium smegmatis: stabilization, rapid purification, molecular and biochemical characterization and regulation of the cellular level. Biochim. Biophys. Acta 1475:191-206. [DOI] [PubMed] [Google Scholar]
- 41.Mukhopadhyay, B., E. Purwantini, C. Kreder, and R. Wolfe. 2001. Oxaloacetate synthesis in the methanarchaeon Methanosarcina barkeri: pyruvate carboxylase genes and a putative Escherichia coli-type bifunctional biotin protein ligase gene (bpl/birA) exhibit a unique organization. J. Bacteriol. 183:3804-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukhopadhyay, B., S. F. Stoddard, and R. S. Wolfe. 1998. Purification, regulation, and molecular and biochemical characterization of pyruvate carboxylase from Methanobacterium thermoautotrophicum strain ΔH. J. Biol. Chem. 273:5155-5166. [DOI] [PubMed] [Google Scholar]
- 43.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nielsen, H., J. Engelbrecht, S. Brunak, and G. V. Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 45.Robb, F. T., D. L. Maeder, J. R. Brown, J. DiRuggiero, M. D. Stump, R. K. Yeh, R. B. Weiss, and D. M. Dunn. 2001. Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 330:134-157. [DOI] [PubMed] [Google Scholar]
- 46.Ruepp, A., W. Graml, M. L. Santos-Martinez, K. K. Koretke, C. Volker, H. W. Mewes, D. Frishman, S. Stocker, A. N. Lupas, and W. Baumeister. 2000. The genomic sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407:508-513. [DOI] [PubMed] [Google Scholar]
- 47.Sako, Y., K. Takai, A. Uchida, and Y. Ishida. 1996. Purification and characterization of phosphoenolpyruvate carboxylase from the hyperthermophilic archaeon Methanothermus sociabilis. FEBS Lett. 392:148-152. [DOI] [PubMed] [Google Scholar]
- 47a.Sako, Y., K. Takai, T. Nishizaka, and Y. Ishido. 1997. Biochemical relationship of phosphoenol pyruvate carboxylases (PEPCs) from thermophilic archaea. FEMS Michobiol. 153:159-165.
- 48.Saunders, N. F., T. Thomas, P. M. Curmi, J. S. Mattick, E. Kuczek, R. Slade, J. Davis, P. D. Franzmann, D. Boone, K. Rusterholtz, R. Feldman, C. Gates, S. Bench, K. Sowers, K. Kadner, A. Aerts, P. Dehal, C. Detter, T. Glavina, S. Lucas, P. Richardson, F. Larimer, L. Hauser, M. Land, and R. Cavicchioli. 2003. Mechanisms of thermal adaptation revealed from the genomes of the Antarctic Archaea Methanogenium frigidum and Methanococcoides burtonii. Genome Res. 13:1580-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shieh, J. S., and W. B. Whitman. 1987. Pathway of acetate assimilation in autotrophic and heterotrophic methanococci. J. Bacteriol. 169:5327-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simpson, P. G., and W. B. Whitman. 1993. Anabolic pathways in methanogens, p. 445-472. In J. G. Ferry (ed.), Methanogenesis: ecology, physiology, biochemistry, and genetics. Chapman and Hall, New York, N.Y.
- 53.Slesarev, A. I., K. V. Mezhevaya, K. S. Makarova, N. N. Polushin, O. V. Shcherbinina, V. V. Shakhova, G. I. Belova, L. Aravind, D. A. Natale, I. B. Rogozin, R. L. Tatusov, Y. I. Wolf, K. O. Stetter, A. G. Malykh, E. V. Koonin, and S. A. Kozyavkin. 2002. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. USA 99:4644-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teraoka, H., K. Izui, and H. Katsuki. 1974. Phosphoenolpyruvate carboxylase of Escherichia coli. Multiple conformational states elicited by allosteric effectors. Biochemistry 13:5121-5128. [DOI] [PubMed] [Google Scholar]
- 56.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wasserfallen, A., J. Nolling, P. Pfister, J. Reeve, and E. Conway de Macario. 2000. Phylogenetic analysis of 18 thermophilic Methanobacterium isolates supports the proposals to create a new genus, Methanothermobacter gen. nov., and to reclassify several isolates in three species, Methanothermobacter thermautotrophicus comb. nov., Methanothermobacter wolfeii comb. nov., and Methanothermobacter marburgensis sp. nov. Int. J. Syst. Evol. Microbiol. 50:43-53. [DOI] [PubMed] [Google Scholar]
- 58.Weimer, P. J., and J. G. Zeikus. 1979. Acetate assimilation pathway of Methanosarcina barkeri. J. Bacteriol. 137:332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Westhoff, P., and U. Gowik. 2004. Evolution of c4 phosphoenolpyruvate carboxylase. Genes and proteins: a case study with the genus Flaveria. Ann. Bot. (London) 93:13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeikus, J. G., G. Fuchs, W. Kenealy, and R. K. Thauer. 1977. Oxidoreductases involved in cell carbon synthesis of Methanobacterium thermoautotrophicum. J. Bacteriol. 132:604-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeikus, J. G., and R. S. Wolfe. 1972. Methanobacterium thermoautotrophicus sp. nov., an anaerobic, autotrophic, extreme thermophile. J. Bacteriol. 109:707-715. [DOI] [PMC free article] [PubMed] [Google Scholar]