Abstract
The current study examined the effects of operant ethanol (EtOH) self-administration on gene expression in the nucleus accumbens (ACB) and amygdala (AMYG) of inbred alcohol-preferring (iP) rats. Rats self-trained on a standard two-lever operant paradigm to administer either water-water, EtOH (15% v/v)-water, or saccharin (SAC; 0.0125% g/v)-water. Animals were killed 24 hr after the last operant session, and the ACB and AMYG dissected; RNA was extracted and purified for microarray analysis. For the ACB, there were 513 significant differences at the p < 0.01 level in named genes: 55 between SAC and water; 215 between EtOH and water, and 243 between EtOH and SAC. In the case of the AMYG (p < 0.01), there were 48 between SAC and water, 23 between EtOH and water, and 63 between EtOH and SAC group. Gene Ontology (GO) analysis indicated that differences in the ACB between the EtOH and SAC groups could be grouped into 15 significant (p < 0.05) categories, which included major categories such as synaptic transmission, cell and ion homeostasis, and neurogenesis, whereas differences between the EtOH and water groups had only 4 categories, which also included homeostasis and synaptic transmission. Several genes were in common between the EtOH and both the SAC and water groups in the synaptic transmission (e.g., Cav2, Nrxn, Gabrb2, Gad1, Homer1) and homeostasis (S100b, Prkca, Ftl1) categories. Overall, the results suggest that changes in gene expression in the ACB of iP rats are associated with the reinforcing effects of EtOH.
Keywords: microarrays, gene expression, ethanol self-administration, alcohol-preferring rats, nucleus accumbens, amygdala
INTRODUCTION
Microarray analysis has emerged as a tool to study the multiple complex effects of pharmacological treatments on changes in gene expression. Examining innate differences and changes in gene expression in response to ethanol (EtOH) in lines or strains of mice and rats with divergent responses to ethanol could provide important clues toward identifying genes and gene networks involved in vulnerability to high alcohol drinking. Further, examining changes in gene expression resulting from chronic EtOH drinking could provide clues to identifying genes and gene networks involved in maintaining high alcohol drinking behavior. Thus far, changes in gene expression under operant EtOH self-administration conditions have not been conducted with rats that have been bred for high alcohol drinking behavior.
Animal models have been used to study the influence of genetic factors on the effects of alcohol and on alcohol drinking behavior (reviewed by Bell et al 2005; McBride and Li 1998; Murphy et al 2002). Selective breeding programs have developed lines of rats with divergent alcohol drinking behaviors. The results of these studies provide convincing data that genetics can markedly influence alcohol-drinking behavior. Many studies have been conducted with these rat lines and, thus far, the overall results suggest that differences in the complex interactions of a number of neurotransmitter systems and multiple intracellular events in several CNS regions may contribute to a predisposition for high alcohol drinking behavior (reviewed by Bell et al, 2005; McBride and Li 1998; Murphy et al, 2002).
Innate genetic expression differences between high and low alcohol consuming rodent lines have been indicated in several studies. Edenberg et al. (2005) examined differences in gene expression in the hippocampus (HIP) of inbred alcohol-preferring (iP) and inbred alcohol-non-preferring (iNP) rats, and reported differences in expression of genes involved in cell growth and adhesion, cellular stress reduction and anti-oxidation, protein trafficking, cellular signaling pathways, and synaptic function. Worst et al. (2005) reported on the transcriptome analysis in the frontal cortex of alcohol-naïve AA (Alko, alcohol) and ANA (Alko, non-alcohol) rats, and found differences between the AA and ANA rats in mRNA levels that could alter transmitter release (e.g., vesicle-associated membrane protein 2, syntaxin 1, syntaxin binding protein). In the whole brain analysis of inbred long-sleep and inbred short-sleep mice, expression of genes encoding for tyrosine protein kinase and ubiquitin carboxyl terminal hydrolase were higher in the brain of long-sleep mice (Xu et al., 2001). In a comprehensive transcriptome meta-analysis of different mice strains, Mulligan et al. (2006) identified several cis-regulated candidate genes for an alcohol preference QTL on chromosome 9.
Alterations in gene expression produced by exposure to alcohol have been reported in a few studies. Acute EtOH injections (6 g/kg; i.p.) produced changes in whole brain of C57BL/6J and DBA/2J mice (high and low alcohol drinkers, respectively) in expression of genes involved in regulating cell signaling, gene regulation, and homeostasis/stress response (Treadwell and Singh, 2004). Kerns et al. (2005) reported that acute i.p. ethanol injections altered, in the nucleus accumbens (ACB), prefrontal cortex and ventral tegmental area (VTA) of C57BL/6J and DBA/2J mice, expression of genes involved in glucocorticoid signaling, neurogenesis, myelination, neuropeptide signaling, and retinoic acid signaling. Differences were found in the dorsal HIP of Lewis rats given 12% EtOH or water for 15 months in expression of genes coding for oxidoreductases and ADP-ribosylation factors (Saito et al., 2002). In contrast, Saito et al. (2004) found no statistically significant effects of chronic free-choice alcohol drinking on gene expression in the striatum of C57BL/6By mice. The above studies were conducted using EtOH injections or 24-hr free-choice drinking. Moreover, other then the study of Kerns et al., (2005) using i.p. EtOH injections, none of the other studies reported data on limbic regions that are involved in mediating alcohol drinking. Therefore, it would be important to determine the effects of alcohol drinking on changes in gene expression in limbic regions that are involved in regulating alcohol drinking.
The nucleus accumbens (ACB) and amygdala (AMYG) are considered to be involved in mediating the reinforcing effects of EtOH and EtOH drinking (c.f., Koob et al., 1998; McBride and Li, 1998). Therefore, it would be important to determine changes in gene expression in these two limbic structures following EtOH self-administration. The objectives of the present study were to determine changes in gene expression associated with operant EtOH self-administration by inbred P rats. The use of operant procedures allowed determining the effects of the reinforcing effects of EtOH on gene expression under a controlled pattern of EtOH access and intake. Previous studies did not use operant techniques, nor did these studies use a controlled pattern of EtOH intake. Moreover, previous EtOH drinking studies did not examine changes in gene expression in the ACB and AMYG. In addition, a group self-administering saccharin (SAC) was used for comparison purposes to provide data on changes associated with learning the operant procedure, and motor activity related to lever responses. The present study was designed to test the hypothesis that EtOH self-administration would produce regional changes within the ACB and AMYG of iP rats in the expression of genes associated with intracellular signaling and synaptic transmission, and that these changes would be different from changes observed with SAC and water self-administration.
METHODS
To reduce genetic variability, inbred adult (90-100 days old) male rats from the iP (5C) strains were used in these experiments. Inbreeding by brother-sister mating was initiated after the S30 generation of mass selection; the inbred strain was in the F37 generation for these experiments. Rats were maintained on a 12-hr reversed light-dark cycle (lights off at 0900 hr). Food and water were available ad libitum throughout the experiment, except during operant testing. The animals used in these experiments were maintained in facilities fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC). All research protocols were approved by the institutional animal care and use committee and are in accordance with the guidelines of the Institutional Care and Use Committee of the National Institute on Drug Abuse, National Institutes of Health, and the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council 1996).
EtOH-naïve iP rats were self-trained on a standard two-lever operant paradigm using daily 1-hr sessions, as previously described for P rats (Rodd-Henricks et al., 2002a,b). Rats (n = 6/group) were allowed to self-administer either water-water, EtOH (15% v/v)-water, or SAC (0.0125% g/v)-water. The fixed-ratio (FR) requirement was increased on the EtOH and SAC levers, and on one of the levers in the water-water group, until a concurrent FR5-FR1 schedule of reinforcement was reached. Operant sessions were conducted over a 10-week period. A computer controlled the operant programs and recorded all data; the number of responses on both levers and the number of reinforcements obtained were recorded throughout all sessions. Sessions were 60 min in duration, occurring daily during the dark cycle. All operant sessions were conducted between 1100 and 1700. Previous research indicated that approximately 90-95% of the predicted fluid intake is consumed during the 60-min sessions (Rodd et al., 2003).
Animals were killed by decapitation approximately 24 hr after the last operant session. In this study, the 24-hr time point was chosen to allow (a) comparison of the EtOH group with the other two groups without EtOH being present; and (b) detection of changes in gene expression associated with self-administration behavior separated from a pharmacological response to EtOH.
Rats were killed within the same 2-hr time frame over 2 days with equal number of animals from each group being killed on each day to minimize differences in time of sacrifice and dissection, and maintain the experimental balance across groups. The head was immediately placed in a cold box maintained at −15°C, where the brain was rapidly removed and placed on a glass plate for dissection. All equipment used to obtain tissue was treated with RNAse Zap (Ambion, Inc. Austin, TX) to prevent RNA degradation. The ACB and AMYG were dissected according to the coordinates of Paxinos and Watson (1998). Briefly, the ACB was dissected from a 2-mm section generated by a coronal cut at 2 mm anterior to the optic chiasm (Bregma 1.70 mm) and a coronal cut at the optic chiasm (Bregma −0.26 mm). The AMYG was dissected by a cut at the lateral borders of the lateral hypothalamus (Bregma −2.12 mm) and ventral of the rhinal fissure, with cortical tissue then trimmed at the lateral edges of the dissected slice. Dissected tissues were immediately homogenized in Trizol reagent (Invitrogen, Carlsbad, CA) and processed according to the manufacturer's protocol, but with twice the suggested ratio of Trizol to tissue (Edenberg et al., 2005). Ethanol precipitated RNA was further purified through RNeasy® columns (Qiagen, Valencia, CA) according to the manufacturer's protocol. The yield, concentration and purity of the RNA were determined by running a spectrum from 210 to 350 nm, and analyzing the ratio of large and small ribosomal RNA bands using an Agilent Bioanalyzer. Yields and purity of the RNA were excellent.
Microarray procedures
Separate preparations of total RNA were made from individual CNS regions from each animal. Samples were not pooled. Standard Affymetrix protocols (GeneChip® Expression Analysis Technical Manual, Rev. 5 and updates) were used to synthesize biotinylated cRNA, starting with 5 ug total RNA from each region, using the Affymetrix kits for cDNA synthesis, in vitro transcription and sample cleanup. Fifteen μg of fragmented, biotinylated cRNA from each independent sample were mixed into 300 μl of hybridization cocktail, of which 200 μl was used for each hybridization. Hybridization was for 17 hr at 42°C. Samples were hybridized to the Affymetrix GeneChip® (Rat Genome 230 2.0 array GeneChips). Washing and scanning of the GeneChips were carried out according to standard protocols, as previously described (Edenberg et al, 2005; McClintick et al., 2003).
To minimize potential systematic errors, all stages of the experiment were balanced across experimental groups. That is, equal numbers of animals in each group were sacrificed within the same 2-hr time frame each day, and equal numbers of RNA preparations from the representative groups were processed through the labeling, hybridization, washing and scanning protocols on a given day, in a counterbalanced order, using premixes of reagents.
Statistical and neuroinformatics analysis of microarray data
Each GeneChip® was scanned using an Affymetrix Model 3000 scanner and underwent image analysis using Affymetrix GCOS software. Microarray data will be available from the National Center for Biotechnology Information's Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/, (Barrett et al. 2005; Edgar et al., 2002). Raw .cel files were then imported into the statistical programming environment R (R: A language and environment for statistical computing Ver 2.2.0; R Foundation for Statistical Computing, 2005) for further analysis with tools available from the Bioconductor Project (Gentleman et al. 2004), themselves further expanded by the authors using the R language. Expression data from the 18 arrays of each region were normalized within-region and converted to log(2) using the Robust Multi-chip Average (RMA) method (Irizarry et al. 2003) implemented in the Bioconductor package RMA. As a standardization step to facilitate later comparisons with other experiments, expression levels were scaled such that the mean expression of all arrays was log2(1000). As we were primarily concerned with identifying genes that could be subjected to further bioinformatic analysis, all probesets currently annotated by Affymetrix as “expressed sequence tags” or whose gene names contain the words “riken”, “predicted”, or “similar to” were filtered out. We next filtered out probe sets with a very low likelihood of actual expression in our samples, accomplished with the Bioconductor package “genefilter.” Probe sets that did not have at least 25% of samples with normalized scaled expression greater than 64 were filtered out. Linear modeling to calculate gene-wise p values for the contrasts of the EtOH group versus water group, SAC group versus water group, and EtOH group versus SAC group was performed using the package Limma (Smyth 2004); probe sets were considered to be statistically significant at p < 0.01, with a false discovery rate (FDR) less than 0.3.
Testing for over-representation of Gene Ontology (Harris et al. 2004; Ashburner et al. 2000) biologic process (GO) categories was performed using the Bioconductor package GOstats (Gentleman 2004). Briefly, for each gene set tested, a list of unique Entrez-Gene identifiers was constructed. This list was then compared to the list of all known Entrez-Gene identifiers that are represented on the Affymetrix chipset Rat Genome 230 2.0. Identification of over-represented GO categories was then accomplished within GOstats using the hypergeometric distribution. To filter out uninteresting categories, only those categories with greater than 9 and less than 300 genes represented on the chipset were included in the analysis, as were categories with less than 5 significant genes. GO categories were called significant at p < 0.05. Co-citation and network analyses were conducted with Ingenuity®.
Quantitative Real-Time PCR
Real-Time PCR was carried out using SybrGreen chemistry and the ABI Prism 7700 Sequence Detection System (Applied Biosystems). The amplification primers were designed using Primer Express software (Applied Biosystems). Total RNA, isolated for the microarray analyses, was employed for these analyses. Following reverse transcription of the RNA (TaqMan Reverse Transcription Reagents, Applied Biosystems), an aliquot of each reverse transcription reaction was amplified in triplicate. This reaction was repeated to generate 6 values for each test group. Two control reactions were run for each RNA preparation: 1) a reverse transcription and PCR reaction with no added RNA to control for contamination of the reagents; and 2) a PCR reaction without the reverse transcription reaction in the presence of RNA to detect DNA contamination of the RNA preparation. To correct for sample-to-sample variation, an endogenous control (GAPDH) was amplified with the target and served as an internal reference to normalize the data. Relative quantification of data from the ABI Prism 7700 Sequence Detection System was performed using the standard curve method (Applied Biosystems, User Bulletin #2; htpp://www.appliedbiosystems.com). Quantitative RT-PCR (qRT-PCR) measurements were conducted on genes to verify differences observed with microarray hybridization. Genes were selected on the basis of significant differential expression, relatively large fold changes, and the availability of primers.
RESULTS
Average responses on the FR5 lever indicated that there was a significant group effect (F2,15 values > 162.54, p values < 0.001); post-hoc comparisons indicated that the SAC group responded significantly more than the EtOH and water groups, and the EtOH group responded significantly more than the water group (Fig. 1). Responding by the SAC group was approximately 1.5-fold higher than the EtOH group and 25-fold higher than the water group. Responding on the alternate lever for water was low for all 3 groups and was comparable to responses on the FR5 lever by the water group (~20 responses/session).
Fig. 1.
Responses per session on the lever paired with ethanol, saccharin or water (FR5 lever) by the 3 groups of iP rats (n = 6/group). Data are the means ± SEM. Responding by the saccharin group was significantly higher than responding by other 2 groups; responding by the EtOH group was significantly higher than responding by the water group. Lever presses on the alternate lever for water (FR1 lever) are not shown but are comparable to the lever presses by the water group on the FR5 lever (~20 responses/session).
The average number of SAC reinforcements was 104, which would produce intakes of approximately 10 ml of 0.0125% SAC per session. The average number of EtOH reinforcements was 61, which would produce intakes of approximately 6 ml of 15% EtOH per session. Given that the average body weight was 410 g at the end of testing, the amount of EtOH consumed would be equivalent to approximately 1.7 g/kg/session. This level of EtOH self-administering was reached for at least 21 consecutive days. Previous research indicated that this level of intake would result in blood ethanol concentrations greater than 80 mg% in the P rat (c.f. Murphy et al., 2002, Rodd-Henricks et al., 2001).
Gene expression in the ACB
Comparing across the 3 groups, there were 513 differences in named gene expression in the ACB, with 55 differences between the SAC and water groups, 215 differences between the EtOH and water groups, and 243 differences between the EtOH and SAC groups. Most of the differences were in the range of 1.15 to 1.25-fold.
There were 55 differences (p < 0.01) in gene expression in the SAC versus the water group, with 31 genes having higher and 24 genes having lower expression in the SAC group (Table 1). However, with a FDR of 0.87, these differences could have occurred by chance alone.
TABLE 1.
Genes that were different in the nucleus accumbens of iP rats between the Saccharin and Water groups at P < 0.01 (FDR > 0.8)
| Gene Symbol | Name | Fold Change | Limma p-value |
|---|---|---|---|
| Nt5dc2 | 5′-nucleotidase domain containing 2 | −1.11 | 0.009 |
| Ar | androgen receptor | −1.15 | 0.005 |
| Aqp11 | aquaporin 11 | −1.14 | 0.006 |
| Bcl2l1 | Bcl2-like 1 | −1.15 | 0.001 |
| Clstn2 | calsyntenin 2 | −1.13 | 0.009 |
| Csnk1d | casein kinase 1, delta | −1.12 | 0.008 |
| C8b | complement component 8, beta polypeptide (mapped) | −1.11 | 0.004 |
| Cpne9 | copine family member IX | −1.13 | 0.005 |
| Cxxc4 | CXXC finger 4 | −1.17 | 0.006 |
| Doc2a | Double C2, alpha | −1.14 | 0.003 |
| Dusp1 | dual specificity phosphatase 1 | −1.33 | 0.009 |
| Gsk3b | glycogen synthase kinase 3 beta /// glycogen synthase kinase 3 beta | −1.14 | 0.007 |
| Gna11 | guanine nucleotide binding protein, alpha 11 /// guanine nucleotide binding protein, alpha 11 | −1.18 | 0.003 |
| Bat5 | HLA-B associated transcript 5 | −1.11 | 0.002 |
| Homer1 | homer homolog 1 (Drosophila) | −2.00 | 0.001 |
| Jun | Jun oncogene /// Jun oncogene | −1.13 | 0.009 |
| Numb | Numb gene homolog (Drosophila) | −1.17 | 0.005 |
| Col2a1 | procollagen, type II, alpha 1 | −1.15 | 0.002 |
| Pdcd8 | Programmed cell death 8 | −1.16 | 0.003 |
| Pcsk1 | proprotein convertase subtilisin/kexin type 1 | −1.13 | 0.002 |
| Scrg1 | scrapie responsive gene 1 | −1.14 | 0.008 |
| Scamp5 | secretory carrier membrane protein 5 | −1.17 | 0.004 |
| Tmed3 | transmembrane emp24 domain containing 3 /// transmembrane emp24 domain containing 3 | −1.13 | 0.009 |
| Tnfaip6 | tumor necrosis factor alpha induced protein 6 | −1.10 | 0.009 |
| Arpc1b | actin related protein 2/3 complex, subunit 1B | 1.16 | 0.004 |
| Adra2c | adrenergic receptor, alpha 2c | 1.15 | 0.005 |
| Cacnb1 | calcium channel, voltage-dependent, beta 1 subunit | 1.13 | 0.008 |
| Cast | calpastatin | 1.13 | 0.006 |
| Cnksr3 | Cnksr family member 3 | 1.17 | 0.006 |
| Coil | Coilin | 1.20 | 0.010 |
| Cfb | complement factor B /// complement factor B | 1.20 | 0.007 |
| Ddx27 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 | 1.17 | 0.006 |
| H2afx | dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) | 1.13 | 0.007 |
| Eef2k | eukaryotic elongation factor-2 kinase | 1.18 | 0.009 |
| Eif4a2 | Eukaryotic translation initiation factor 4A2 | 1.15 | 0.004 |
| Fkbp11 | FK506 binding protein 11 /// FK506 binding protein 11 | 1.13 | 0.005 |
| Gpnmb | glycoprotein (transmembrane) nmb /// glycoprotein (transmembrane) nmb | 1.14 | 0.004 |
| Gpm6b | Glycoprotein m6b | 1.26 | 0.006 |
| Gbp2 | guanylate nucleotide binding protein 2 | 1.19 | 0.001 |
| Ifitm3 | interferon induced transmembrane protein 3 | 1.32 | 0.002 |
| Neurod1 | Neurogenic differentiation 1 | 1.16 | 0.006 |
| Nexn | nexilin | 1.30 | 0.002 |
| Nexn | nexilin | 1.24 | 0.005 |
| Nfs1 | nitrogen fixation gene 1 (S. cerevisiae) | 1.12 | 0.005 |
| Ppig | Peptidylprolyl isomerase G | 1.20 | 0.008 |
| Pola2 | Polymerase (DNA directed), alpha 2 | 1.21 | 0.003 |
| Kcnd1 | potassium voltage-gated channel, Shal-related family, member 1 | 1.15 | 0.002 |
| Ptprc | protein tyrosine phosphatase, receptor type, C /// protein tyrosine phosphatase, receptor type, C | 1.21 | 0.003 |
| Rimbp2 | RIM binding protein 2 /// RIM binding protein 2 | 1.11 | 0.008 |
| RT1-Aw2 // | RT1 class Ib, locus Aw2 /// RT1 class Ia, locus A2 /// RT1 class I, A3 | 1.21 | 0.001 |
| Snrpb | Small nuclear ribonucleoprotein polypeptides B and B1 | 1.21 | 0.001 |
| Slc15a3 | solute carrier family 15, member 3 | 1.16 | 0.008 |
| Tada1l | transcriptional adaptor 1 (HFI1 homolog, yeast) like | 1.14 | 0.002 |
| Usf2 | upstream transcription factor 2 | 1.24 | 0.000 |
| Wwp1 /// A | WW domain containing E3 ubiquitin protein ligase 1 /// adipose differentiation related protein | 1.12 | 0.005 |
Table 2 lists the genes that were significantly different between the EtOH and water groups. Among the 215 named genes listed, 131 genes had higher and 84 genes lower expression levels in the EtOH compared to the water group. Several neurotransmitter receptors had lower expression levels in the EtOH group; these included the Htr2a, Htr5a, Gabrb1, Gabrb2, Grm1, and Sstr1, whereas only P2ry13 had higher expression in the EtOH group.
TABLE 2.
Genes that were significantly different in the nucleus accumbens of iP rats between the Ethanol and Water groups at p < 0.01 (FDR = 0.2-0.3)
| Gene Symbol | Name | Fold Change | Limma p-value |
|---|---|---|---|
| Pdpk1 | 3-phosphoinositide dependent protein kinase-1 | −1.45 | 0.003 |
| Htr2a | 5-hydroxytryptamine (serotonin) receptor 2A | −1.27 | 0.007 |
| Htr5a | 5-hydroxytryptamine (serotonin) receptor 5A | −1.18 | 0.009 |
| Ahi1 | Abelson helper integration site 1 | −1.31 | 0.006 |
| Adar | adenosine deaminase, RNA-specific | −1.14 | 0.006 |
| Atrx | alpha thalassemia/mental retardation syndrome X-linked homolog (human) | −1.22 | 0.006 |
| Appbp2 | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | −1.14 | 0.007 |
| Agtr1a | angiotensin II receptor, type 1 (AT1A) | −1.16 | 0.006 |
| Amh | anti-Mullerian hormone | −1.17 | 0.009 |
| Ap1gbp1 | AP1 gamma subunit binding protein 1 | −1.13 | 0.006 |
| Alg2 | asparagine-linked glycosylation 2 homolog (yeast, alpha-1,3-mannosyltransferase) | −1.22 | 0.004 |
| Alg2 | asparagine-linked glycosylation 2 homolog (yeast, alpha-1,3-mannosyltransferase) | −1.21 | 0.008 |
| Atxn3 | ataxin 3 | −1.17 | 0.000 |
| Atp2b4 | ATPase, Ca++ transporting, plasma membrane 4 | −1.27 | 0.001 |
| Blnk | B-cell linker | −1.12 | 0.005 |
| Bcl2l1 | Bcl2-like 1 | −1.22 | 0.000 |
| Bid | BH3 interacting domain death agonist /// BH3 interacting domain death agonist | −1.14 | 0.003 |
| Cacna2d1 | calcium channel, voltage-dependent, alpha2/delta subunit 1 | −1.29 | 0.002 |
| Cacnb4 | calcium channel, voltage-dependent, beta 4 subunit | −1.20 | 0.006 |
| Camk4 | calcium/calmodulin-dependent protein kinase IV | −1.38 | 0.000 |
| Clstn2 | calsyntenin 2 | −1.16 | 0.002 |
| Csnk1e | casein kinase 1, epsilon | −1.17 | 0.007 |
| Cstf1 | cleavage stimulation factor, 3′ pre-RNA, subunit 1 | −1.14 | 0.008 |
| Clock | clock homolog (mouse) | −1.17 | 0.006 |
| Cxxc4 | CXXC finger 4 | −1.26 | 0.000 |
| Ccnh | cyclin H | −1.21 | 0.002 |
| Cftr | cystic fibrosis transmembrane conductance regulator homolog | −1.13 | 0.005 |
| Cyp11b1 | cytochrome P450, subfamily 11B, polypeptide 1 /// cytochrome P450, subfamily 11B, polypeptide 1 | −1.21 | 0.005 |
| Dusp12 | dual specificity phosphatase 12 | −1.15 | 0.008 |
| Gabrb1 | gamma-aminobutyric acid (GABA-A) receptor, subunit beta 1 | −1.15 | 0.003 |
| Gabrb2 | gamma-aminobutyric acid (GABA-A) receptor, subunit beta 2 | −1.32 | 0.004 |
| Grm1 | glutamate receptor, metabotropic 1 | −1.19 | 0.000 |
| Gad1 | glutamic acid decarboxylase 1 | −1.25 | 0.003 |
| Gsk3b | glycogen synthase kinase 3 beta /// glycogen synthase kinase 3 beta | −1.13 | 0.009 |
| Gnaq | Guanine nucleotide binding protein, alpha q polypeptide | −1.27 | 0.000 |
| Gnaq | guanine nucleotide binding protein, alpha q polypeptide /// guanine nucleotide binding protein, alpha q polypeptide | −1.33 | 0.000 |
| Impact | imprinted and ancient | −1.18 | 0.005 |
| Kifc3 | Kinesin family member C3 | −1.19 | 0.001 |
| Mkks | McKusick-Kaufman syndrome protein | −1.15 | 0.004 |
| Map1b | microtubule-associated protein 1b | −1.34 | 0.000 |
| Mapk8ip3 | mitogen-activated protein kinase 8 interacting protein 3 | −1.23 | 0.006 |
| Mllt10 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 | −1.21 | 0.001 |
| Myh8 | myosin, heavy polypeptide 8, skeletal muscle, perinatal | −1.10 | 0.010 |
| Nmt1 | N-myristoyltransferase 1 | −1.14 | 0.005 |
| Nedd4a | neural precursor cell expressed, developmentally down-regulated gene 4A | −1.17 | 0.009 |
| 2610020o0 | nuclear NF-kappaB activating protein | −1.23 | 0.002 |
| Npap60 | nuclear pore associated protein | −1.15 | 0.001 |
| Npap60 | Nuclear pore associated protein | −1.14 | 0.003 |
| P34 | p34 protein | −1.14 | 0.004 |
| Pnma1 | paraneoplastic antigen MA1 | −1.16 | 0.008 |
| Pip5k2b | phosphatidylinositol-4-phosphate 5-kinase, type II, beta | −1.21 | 0.008 |
| Prps2 | phosphoribosyl pyrophosphate synthetase 2 | −1.13 | 0.007 |
| Kcnk9 | potassium channel, subfamily K, member 9 | −1.19 | 0.003 |
| Kcns2 | potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 | −1.12 | 0.010 |
| Kcnh2 | potassium voltage-gated channel, subfamily H (eag-related), member 2 | −1.14 | 0.009 |
| Kcnq3 | potassium voltage-gated channel, subfamily Q, member 3 | −1.17 | 0.004 |
| Col2a1 | procollagen, type II, alpha 1 | −1.13 | 0.004 |
| Pcsk1 | proprotein convertase subtilisin/kexin type 1 | −1.12 | 0.002 |
| Prkca | protein kinase C, alpha /// protein kinase C, alpha | −1.12 | 0.009 |
| Prkab2 | protein kinase, AMP-activated, beta 2 non-catalytic subunit | −1.17 | 0.007 |
| Prkacb | protein kinase, cAMP dependent, catalytic, beta | −1.29 | 0.000 |
| Ppp2r1a | protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), alpha isoform | −1.15 | 0.008 |
| Ramp3 | receptor (calcitonin) activity modifying protein 3 | −1.14 | 0.009 |
| Reln | reelin | −1.24 | 0.007 |
| Rnf12 | ring finger protein 12 | −1.18 | 0.003 |
| Styxl1 | Serine/threonine/tyrosine interacting-like 1 | −1.22 | 0.001 |
| Sgtb | small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta | −1.27 | 0.010 |
| Slc2a3 | solute carrier family 2 (facilitated glucose transporter), member 3 /// solute carrier family 2 (facilitated glucose transporter), member 3 | −1.20 | 0.007 |
| Slc22a4 | solute carrier family 22 (organic cation transporter), member 4 | −1.13 | 0.003 |
| Sstr1 | somatostatin receptor 1 | −1.24 | 0.001 |
| St8sia3 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 | −1.21 | 0.005 |
| Stch | stress 70 protein chaperone, microsome-associated, 60kD human homolog | −1.16 | 0.003 |
| Syt6 | synaptotagmin VI | −1.22 | 0.006 |
| Txndc13 | thioredoxin domain containing 13 | −1.23 | 0.008 |
| Tgfb1i4 | Transforming growth factor beta 1 induced transcript 4 | −1.15 | 0.002 |
| Tmod2 | tropomodulin 2 | −1.16 | 0.006 |
| Tpm3 | tropomyosin 3, gamma | −1.12 | 0.004 |
| Wars | tryptophanyl-tRNA synthetase | −1.13 | 0.006 |
| Flk | tyrosine protein kinase FLK | −1.10 | 0.007 |
| Usp11 | ubiquitin specific protease 11 | −1.24 | 0.006 |
| Ube4a | ubiquitination factor E4A, UFD2 homolog (S. cerevisiae) | −1.12 | 0.010 |
| Vti1a | vesicle transport through interaction with t-SNAREs homolog 1A (yeast) | −1.20 | 0.010 |
| Wdr47 | WD repeat domain 47 | −1.23 | 0.008 |
| Wbp4 | WW domain binding protein 4 | −1.12 | 0.004 |
| Zfp483 | zinc finger protein 483 | −1.25 | 0.002 |
| Zdhhc22 | zinc finger, DHHC-type containing 22 | −1.15 | 0.002 |
| Akap8l | A kinase (PRKA) anchor protein 8-like | 1.17 | 0.006 |
| Abhd1 | abhydrolase domain containing 1 | 1.21 | 0.006 |
| Aco2 | Aconitase 2, mitochondrial | 1.20 | 0.007 |
| Actn1 | actinin, alpha 1 | 1.16 | 0.010 |
| Alb | albumin /// albumin | 1.21 | 0.005 |
| As3mt | arsenic (+3 oxidation state) methyltransferase | 1.13 | 0.006 |
| Abcb1a | ATP-binding cassette, sub-family B (MDR/TAP), member 1A /// ATP-binding cassette, sub-family B (MDR/TAP), member 1A | 1.21 | 0.004 |
| Abcc4 | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 /// ATP-binding cassette, sub-family C (CFTR/MRP), member 4 | 1.19 | 0.005 |
| Atp2b1 | ATPase, Ca++ transporting, plasma membrane 1 | 1.22 | 0.001 |
| B2m | beta-2 microglobulin | 1.14 | 0.004 |
| B2m | Beta-2 microglobulin | 1.15 | 0.008 |
| Cdh11 | Cadherin 11 | 1.23 | 0.002 |
| Cib1 | calcium and integrin binding 1 (calmyrin) | 1.12 | 0.006 |
| Cacna2d3 | Calcium channel, voltage-dependent, alpha 2/delta 3 subunit | 1.33 | 0.004 |
| Camk2b | calcium/calmodulin-dependent protein kinase II, beta | 1.10 | 0.009 |
| Car6 | carbonic anhydrase 6 | 1.19 | 0.001 |
| Cflar | CASP8 and FADD-like apoptosis regulator | 1.27 | 0.001 |
| Cav2 | caveolin 2 | 1.17 | 0.003 |
| Cebpa | CCAAT/enhancer binding protein (C/EBP), alpha | 1.26 | 0.005 |
| Cd81 | CD 81 antigen | 1.10 | 0.006 |
| Cd99 | CD99 antigen | 1.12 | 0.002 |
| Cdca1 | cell division cycle associated 1 | 1.12 | 0.005 |
| Cxcl14 | chemokine (C-X-C motif) ligand 14 /// chemokine (C-X-C motif) ligand 14 | 1.13 | 0.009 |
| Chi3l1 | chitinase 3-like 1 | 1.14 | 0.007 |
| Clcn3 | chloride channel 3 | 1.18 | 0.002 |
| Ccdc5 | coiled-coil domain containing 5 | 1.15 | 0.008 |
| Ctdsp1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 | 1.14 | 0.005 |
| Cst3 | cystatin C | 1.14 | 0.010 |
| P22k15 | cystatin related protein 2 | 1.14 | 0.001 |
| Dhx57 | DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 | 1.11 | 0.007 |
| Ddn | dendrin | 1.16 | 0.009 |
| Dcir3 | dendritic cell inhibitory receptor 3 | 1.14 | 0.010 |
| Dscr1l1 | Down syndrome critical region gene 1-like 1 /// Down syndrome critical region gene 1-like 1 | 1.23 | 0.006 |
| Dullard | Dullard homolog (Xenopus laevis) | 1.13 | 0.005 |
| Dtnb | Dystrobrevin, beta | 1.14 | 0.009 |
| Efemp2 | EGF-containing fibulin-like extracellular matrix protein 2 | 1.13 | 0.007 |
| Emcn | endomucin | 1.23 | 0.001 |
| Ftl1 | ferritin light chain 1 /// ferritin light chain 1 | 1.13 | 0.006 |
| Gkap1 | G kinase anchoring protein 1 | 1.29 | 0.006 |
| Galm | galactose mutarotase | 1.13 | 0.003 |
| Gjb6 | gap junction membrane channel protein beta 6 | 1.15 | 0.005 |
| Glul | glutamate-ammonia ligase (glutamine synthase) /// glutamate-ammonia ligase (glutamine synthase) | 1.14 | 0.008 |
| Gpd1 | glycerol-3-phosphate dehydrogenase 1 (soluble) /// glycerol-3-phosphate dehydrogenase 1 (soluble) | 1.20 | 0.005 |
| Gpm6b | Glycoprotein m6b | 1.27 | 0.004 |
| Csf2ra | Granulocyte-macrophage colony stimulating receptor alpha | 1.15 | 0.010 |
| H2afy | H2A histone family, member Y | 1.14 | 0.001 |
| Bat1a | HLA-B-associated transcript 1A | 1.13 | 0.009 |
| Homer1 | homer homolog 1 (Drosophila) | 1.75 | 0.005 |
| Hyal3 | Hyaluronoglucosaminidase 3 | 1.12 | 0.003 |
| Id4 | inhibitor of DNA binding 4 /// inhibitor of DNA binding 4 | 1.15 | 0.001 |
| Itgb1 | integrin beta 1 (fibronectin receptor beta) | 1.17 | 0.001 |
| Itgb1 | integrin beta 1 (fibronectin receptor beta) /// integrin beta 1 (fibronectin receptor beta) | 1.18 | 0.001 |
| Klhl5 | kelch-like 5 (Drosophila) | 1.24 | 0.009 |
| Kif1a | kinesin family member 1A | 1.16 | 0.003 |
| Klf15 | Kruppel-like factor 15 | 1.14 | 0.006 |
| Letm2 | Leucine zipper-EF-hand containing transmembrane protein 2 | 1.17 | 0.002 |
| Lig3 | Ligase III, DNA, ATP-dependent | 1.14 | 0.010 |
| Man2c1 | mannosidase, alpha, class 2C, member 1 | 1.14 | 0.005 |
| 39148 | Membrane-associated ring finger (C3HC4) 7 | 1.16 | 0.006 |
| Mag | myelin-associated glycoprotein | 1.15 | 0.008 |
| Mcl1 | myeloid cell leukemia sequence 1 | 1.12 | 0.009 |
| Mrlcb | myosin light chain, regulatory B | 1.16 | 0.010 |
| Nrd1 | Nardilysin, N-arginine dibasic convertase 1 | 1.17 | 0.009 |
| --- | Nclone10 mRNA | 1.21 | 0.002 |
| Ndnl2 | Necdin-like 2 | 1.20 | 0.004 |
| Nrxn3 | neurexin 3 | 1.25 | 0.003 |
| Nfia | nuclear factor I/A | 1.19 | 0.007 |
| Nfib | nuclear factor I/B | 1.12 | 0.009 |
| Nfib | nuclear factor I/B | 1.15 | 0.005 |
| Nfkbia | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha /// nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | 1.19 | 0.006 |
| Numa1 | Nuclear mitotic apparatus protein 1 | 1.14 | 0.002 |
| Nr1h3 | nuclear receptor subfamily 1, group H, member 3 /// nuclear receptor subfamily 1, group H, member 3 | 1.15 | 0.007 |
| Odz2 | Odd Oz/ten-m homolog 2 (Drosophila) | 1.27 | 0.010 |
| Pctk1 | PCTAIRE-motif protein kinase 1 | 1.16 | 0.001 |
| Ppig | Peptidylprolyl isomerase G | 1.22 | 0.004 |
| Prdx6 | Peroxiredoxin 6 | 1.19 | 0.001 |
| Ppap2b | phosphatidic acid phosphatase type 2B | 1.15 | 0.002 |
| Pik4ca | Phosphatidylinositol 4-kinase, catalytic, alpha polypeptide | 1.21 | 0.009 |
| Pla2g6 | phospholipase A2, group VI | 1.14 | 0.004 |
| Plscr3 | phospholipid scramblase 3 | 1.10 | 0.009 |
| Plag1 | pleiomorphic adenoma gene 1 | 1.18 | 0.004 |
| Pola2 | Polymerase (DNA directed), alpha 2 | 1.20 | 0.003 |
| Polb | Polymerase (DNA directed), beta | 1.21 | 0.010 |
| Psg4 | pregnancy specific beta-1-glycoprotein 4 | 1.16 | 0.008 |
| Col11a2 | procollagen, type XI, alpha 2 (mapped) | 1.19 | 0.003 |
| Pkig | protein kinase inhibitor, gamma | 1.09 | 0.007 |
| Prkwnk1 | Protein kinase, lysine deficient 1 | 1.13 | 0.004 |
| Ptp4a2 | protein tyrosine phosphatase 4a2 | 1.11 | 0.007 |
| Ptpn2 | protein tyrosine phosphatase, non-receptor type 2 | 1.12 | 0.009 |
| Ptprf | protein tyrosine phosphatase, receptor type, F | 1.17 | 0.003 |
| Plp | proteolipid protein | 1.15 | 0.006 |
| Ptk2 | PTK2 protein tyrosine kinase 2 /// PTK2 protein tyrosine kinase 2 | 1.12 | 0.007 |
| P2ry13 | Purinergic receptor P2Y, G-protein coupled, 13 | 1.24 | 0.005 |
| Ua20 | Putative UA20 protein | 1.14 | 0.007 |
| Rims1 | regulating synaptic membrane exocytosis 1 | 1.20 | 0.000 |
| Rgc32 | response gene to complement 32 | 1.15 | 0.005 |
| Rgc32 | Response gene to complement 32 | 1.19 | 0.003 |
| Rpe65 | retinal pigment epithelium 65 | 1.17 | 0.001 |
| Arhgef1 | Rho guanine nucleotide exchange factor (GEF) 1 | 1.19 | 0.007 |
| Rnasen | ribonuclease III, nuclear | 1.14 | 0.005 |
| Rpl13a | ribosomal protein L13A /// ribosomal protein L13A | 1.14 | 0.005 |
| Rps29 | ribosomal protein S29 | 1.14 | 0.006 |
| Rps3a | ribosomal protein S3a | 1.11 | 0.010 |
| Rps6ka2 | Ribosomal protein S6 kinase polypeptide 2 | 1.22 | 0.009 |
| Rnf44 | Ring finger protein 44 | 1.12 | 0.007 |
| S100b | S100 protein, beta polypeptide | 1.14 | 0.008 |
| Scamp1 | Secretory carrier membrane protein 1 | 1.18 | 0.002 |
| Sepw1 | selenoprotein W, muscle 1 | 1.13 | 0.008 |
| Sdccag1 | serologically defined colon cancer antigen 1 | 1.14 | 0.005 |
| Shank1 | SH3 and multiple ankyrin repeat domains 1 | 1.19 | 0.003 |
| Shank2 | SH3/ankyrin domain gene 2 /// SH3/ankyrin domain gene 2 | 1.17 | 0.004 |
| Slc2a1 | solute carrier family 2 (facilitated glucose transporter), member 1 /// solute carrier family 2 (facilitated glucose transporter), member 1 | 1.15 | 0.004 |
| Slc22a17 | solute carrier family 22 (organic cation transporter), member 17 | 1.11 | 0.005 |
| Slc23a2 | Solute carrier family 23 (nucleobase transporters), member 2 | 1.19 | 0.003 |
| Slc25a25 | solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 /// solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 | 1.15 | 0.003 |
| Slc33a1 | Solute carrier family 33 (acetyl-CoA transporter), member 1 | 1.23 | 0.008 |
| Slc34a1 | solute carrier family 34 (sodium phosphate), member 1 | 1.18 | 0.005 |
| Slc4a4 | Solute carrier family 4, member 4 | 1.22 | 0.001 |
| Scd2 | stearoyl-Coenzyme A desaturase 2 | 1.15 | 0.009 |
| Sc5d | sterol-C5-desaturase (fungal ERG3, delta-5-desaturase) homolog (S. cerevisae) /// sterol-C5-desaturase (fungal ERG3, delta-5-desaturase) homolog (S. cerevisiae) | 1.21 | 0.000 |
| Sympk | symplekin | 1.12 | 0.004 |
| Sv2a | synaptic vesicle glycoprotein 2a | 1.22 | 0.001 |
| Sdc4 | syndecan 4 | 1.17 | 0.009 |
| Tbkbp1 | TBK1 binding protein 1 | 1.22 | 0.001 |
| Thap7 | THAP domain containing 7 | 1.16 | 0.001 |
| pur-beta | Transcription factor Pur-beta /// Transcription factor Pur-beta | 1.17 | 0.001 |
| Tmem10 | transmembrane protein 10 /// transmembrane protein 10 | 1.14 | 0.010 |
| Ets1 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 1.22 | 0.000 |
| Vcam1 | vascular cell adhesion molecule 1 /// vascular cell adhesion molecule 1 | 1.14 | 0.008 |
| Zfp212 | Zinc finger protein 212 | 1.11 | 0.004 |
| Zfp36l2 | zinc finger protein 36, C3H type-like 2 | 1.15 | 0.002 |
| Zfp423 | zinc finger protein 423 | 1.20 | 0.002 |
| Zfand3 | zinc finger, AN1-type domain 3 | 1.10 | 0.007 |
| Zswim6 | Zinc finger, SWIM domain containing 6 | 1.16 | 0.004 |
There were approximately 243 significant differences in named genes (P < 0.01) between the EtOH and SAC groups (Table 3), with 148 genes having higher and 95 genes having lower expression in the EtOH versus the SAC group. Genes for several transmitter receptors had lower expression in the EtOH group than the SAC group; these included Gabrb2. Gabrb3, Gria2, Gria3 and Oprk1; only the expression of the Tacr3 gene was higher in the EtOH than SAC group.
TABLE 3.
Genes that were significantly different in the nucleus accumbens of iP rats between the ethanol and saccharin groups at p < 0.01 (FDR = 0.2-0.3)
| Gene Symbol | Name | Fold Change | Limma p-value |
|---|---|---|---|
| Pdpk1 | 3-phosphoinositide dependent protein kinase-1 | −1.47 | 0.002 |
| Ap3m2 | adaptor-related protein complex 3, mu 2 subunit | −1.11 | 0.004 |
| Adar | adenosine deaminase, RNA-specific | −1.16 | 0.003 |
| Atrx | alpha thalassemia/mental retardation syndrome X-linked homolog (human) | −1.29 | 0.001 |
| Atrx | alpha thalassemia/mental retardation syndrome X-linked homolog (human) | −1.26 | 0.001 |
| Aplp2 | amyloid beta (A4) precursor-like protein 2 | −1.30 | 0.003 |
| App | Amyloid beta (A4) precursor protein | −1.13 | 0.004 |
| App | amyloid beta (A4) precursor protein /// amyloid beta (A4) precursor protein | −1.27 | 0.003 |
| Appbp2 | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | −1.14 | 0.007 |
| Arih1 | ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila) | −1.14 | 0.001 |
| Actr3 | ARP3 actin-related protein 3 homolog (yeast) | −1.20 | 0.008 |
| Atxn3 | ataxin 3 | −1.15 | 0.001 |
| Atp2b4 | ATPase, Ca++ transporting, plasma membrane 4 | −1.25 | 0.002 |
| Atp6v1b2 | ATPase, H transporting, lysosomal V1 subunit B2 | −1.16 | 0.009 |
| Birc4 | baculoviral IAP repeat-containing 4 | −1.30 | 0.007 |
| Bag4 | BCL2-associated athanogene 4 | −1.16 | 0.003 |
| Bfar | bifunctional apoptosis regulator | −1.20 | 0.009 |
| Blcap | bladder cancer associated protein homolog (human) | −1.25 | 0.001 |
| Bmp3 | bone morphogenetic protein 3 | −1.11 | 0.009 |
| Cacnb4 | calcium channel, voltage-dependent, beta 4 subunit | −1.31 | 0.004 |
| Cacnb4 | calcium channel, voltage-dependent, beta 4 subunit | −1.20 | 0.006 |
| Camk4 | calcium/calmodulin-dependent protein kinase IV | −1.23 | 0.002 |
| Camk4 | calcium/calmodulin-dependent protein kinase IV | −1.32 | 0.001 |
| Csen | calsenilin, presenilin binding protein, EF hand transcription factor | −1.23 | 0.008 |
| Csnk1e | casein kinase 1, epsilon | −1.15 | 0.007 |
| Cp | ceruloplasmin /// ceruloplasmin | −1.30 | 0.004 |
| Cct3 | Chaperonin subunit 3 (gamma) | −1.14 | 0.004 |
| Cldn1 | claudin 1 /// claudin 1 | −1.15 | 0.005 |
| Ccnh | cyclin H | −1.21 | 0.002 |
| Dcbld2 | discoidin, CUB and LCCL domain containing 2 | −1.12 | 0.009 |
| Dlgh2 | discs, large homolog 2 (Drosophila) | −1.17 | 0.010 |
| Ddit4l | DNA-damage-inducible transcript 4-like /// DNA-damage-inducible transcript 4-like | −1.18 | 0.006 |
| Dnm3 | dynamin 3 | −1.17 | 0.010 |
| Elavl2 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) | −1.19 | 0.008 |
| Enah | enabled homolog (Drosophila) /// enabled homolog (Drosophila) | −1.13 | 0.009 |
| Extl3 | exostoses (multiple)-like 3 | −1.14 | 0.007 |
| Fgl2 | fibrinogen-like 2 | −1.19 | 0.010 |
| Gabrb2 | gamma-aminobutyric acid (GABA-A) receptor, subunit beta 2 | −1.32 | 0.004 |
| Gabrb3 | gamma-aminobutyric acid (GABA-A) receptor, subunit beta 3 | −1.33 | 0.002 |
| Gria2 | glutamate receptor, ionotropic, AMPA2 | −1.16 | 0.001 |
| Gria3 | glutamate receptor, ionotropic, AMPA3 (alpha 3) /// glutamate receptor, ionotropic, AMPA3 (alpha 3) | −1.19 | 0.009 |
| Gad1 | glutamic acid decarboxylase 1 | −1.23 | 0.006 |
| Gad2 | glutamic acid decarboxylase 2 | −1.32 | 0.007 |
| Gpiap1 | GPI-anchored membrane protein 1 | −1.31 | 0.002 |
| Grb2 | growth factor receptor bound protein 2 | −1.14 | 0.006 |
| Gnaq | Guanine nucleotide binding protein, alpha q polypeptide | −1.20 | 0.001 |
| Gnaq | guanine nucleotide binding protein, alpha q polypeptide /// guanine nucleotide binding protein, alpha q polypeptide | −1.30 | 0.001 |
| Hnrpm | heterogeneous nuclear ribonucleoprotein M | −1.13 | 0.008 |
| Hk1 | hexokinase 1 | −1.22 | 0.006 |
| Igf2r | insulin-like growth factor 2 receptor /// insulin-like growth factor 2 receptor | −1.14 | 0.004 |
| Ifitm3 | interferon induced transmembrane protein 3 | −1.30 | 0.002 |
| Kifc3 | Kinesin family member C3 | −1.15 | 0.007 |
| Lmo4 | LIM domain only 4 | −1.28 | 0.007 |
| Mak10 | MAK10 homolog, amino-acid N-acetyltransferase subunit, (S. cerevisiae) | −1.11 | 0.009 |
| Map1b | microtubule-associated protein 1b | −1.37 | 0.000 |
| Mllt10 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 | −1.23 | 0.000 |
| Ncam2 | neural cell adhesion molecule 2 | −1.23 | 0.005 |
| Nedd4a | neural precursor cell expressed, developmentally down-regulated gene 4A | −1.18 | 0.006 |
| Nxph3 | neurexophilin 3 | −1.14 | 0.009 |
| Neurod1 | Neurogenic differentiation 1 | −1.15 | 0.008 |
| Nln | neurolysin (metallopeptidase M3 family) | −1.15 | 0.003 |
| 2610020o0 | nuclear NF-kappaB activating protein | −1.21 | 0.006 |
| Npap60 | nuclear pore associated protein | −1.18 | 0.000 |
| Npap60 | Nuclear pore associated protein | −1.14 | 0.002 |
| Nupl1 | nucleoporin like 1 | −1.16 | 0.008 |
| Oprk1 | opioid receptor, kappa 1 | −1.20 | 0.007 |
| Otud4 | OTU domain containing 4 | −1.20 | 0.003 |
| Osbpl2 | oxysterol binding protein-like 2 | −1.21 | 0.005 |
| P34 | p34 protein | −1.14 | 0.003 |
| Pik3r3 | phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 | −1.16 | 0.009 |
| Pafah1b1 | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | −1.19 | 0.007 |
| Kcnj9 | potassium inwardly-rectifying channel, subfamily J, member 9 | −1.18 | 0.008 |
| Pja2 | praja 2, RING-H2 motif containing | −1.16 | 0.001 |
| Prkca | protein kinase C, alpha /// protein kinase C, alpha | −1.17 | 0.001 |
| Prkacb | protein kinase, cAMP dependent, catalytic, beta | −1.29 | 0.000 |
| Ppp3r1 | protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I) | −1.29 | 0.004 |
| Clcn4-2 | putative chloride channel 4-2 | −1.15 | 0.005 |
| Rasgrp1 | RAS guanyl releasing protein 1 | −1.31 | 0.004 |
| Ramp3 | receptor (calcitonin) activity modifying protein 3 | −1.14 | 0.008 |
| Rp1h | retinitis pigmentosa 1 homolog (human) | −1.16 | 0.005 |
| Scamp1 | secretory carrier membrane protein 1 | −1.14 | 0.009 |
| Sel1h | Sel1 (suppressor of lin-12) 1 homolog (C. elegans) | −1.19 | 0.001 |
| Styxl1 | Serine/threonine/tyrosine interacting-like 1 | −1.17 | 0.007 |
| Sgtb | small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta | −1.28 | 0.009 |
| Snrpb | Small nuclear ribonucleoprotein polypeptides B and B1 | −1.22 | 0.001 |
| Scn2b | sodium channel, voltage-gated, type II, beta | −1.45 | 0.002 |
| Slc2a3 | solute carrier family 2 (facilitated glucose transporter), member 3 /// solute carrier family 2 (facilitated glucose transporter), member 3 | −1.22 | 0.005 |
| Slc23a2 | solute carrier family 23 (nucleobase transporters), member 2 | −1.16 | 0.005 |
| Stc2 | Stanniocalcin 2 | −1.09 | 0.008 |
| Stch | stress 70 protein chaperone, microsome-associated, 60kD human homolog | −1.20 | 0.000 |
| Strn | striatin, calmodulin binding protein | −1.17 | 0.004 |
| Syt6 | synaptotagmin VI | −1.21 | 0.009 |
| Txndc13 | thioredoxin domain containing 13 | −1.23 | 0.008 |
| Tef | thyrotroph embryonic factor | −1.20 | 0.009 |
| Tmed5 | transmembrane emp24 protein transport domain containing 5 | −1.19 | 0.008 |
| Uhmk1 | U2AF homology motif (UHM) kinase 1 | −1.25 | 0.005 |
| Ube4a | ubiquitination factor E4A, UFD2 homolog (S. cerevisiae) | −1.13 | 0.006 |
| Vti1a | vesicle transport through interaction with t-SNAREs homolog 1A (yeast) | −1.21 | 0.007 |
| Wwp1 /// A | WW domain containing E3 ubiquitin protein ligase 1 /// adipose differentiation related protein | −1.12 | 0.004 |
| Zfp161 | zinc finger protein 161 | −1.11 | 0.009 |
| Zfp260 | zinc finger protein 260 | −1.13 | 0.002 |
| Zfp483 | zinc finger protein 483 | −1.30 | 0.000 |
| Nt5c3l | 5′-nucleotidase, cytosolic III-like | 1.14 | 0.008 |
| Arbp | acidic ribosomal phosphoprotein P0 | 1.16 | 0.004 |
| Alb | albumin | 1.22 | 0.001 |
| Alb | albumin /// albumin | 1.35 | 0.000 |
| Aspa | aspartoacylase | 1.12 | 0.010 |
| Arid1b | AT rich interactive domain 1B (Swi1 like) | 1.23 | 0.006 |
| Abcb10 | ATP-binding cassette, sub-family B (MDR/TAP), member 10 | 1.12 | 0.010 |
| Abcc4 | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 /// ATP-binding cassette, sub-family C (CFTR/MRP), member 4 | 1.24 | 0.001 |
| Blnk | B-cell linker | 1.16 | 0.002 |
| B2m | beta-2 microglobulin | 1.12 | 0.009 |
| Bckdha | branched chain ketoacid dehydrogenase E1, alpha polypeptide | 1.14 | 0.010 |
| Bckdk | branched chain ketoacid dehydrogenase kinase | 1.16 | 0.003 |
| Cdh11 | Cadherin 11 | 1.23 | 0.003 |
| Cflar | CASP8 and FADD-like apoptosis regulator | 1.29 | 0.001 |
| Ctnnb1 | Catenin (cadherin associated protein), beta 1 | 1.25 | 0.009 |
| Cav2 | caveolin 2 | 1.21 | 0.001 |
| Cav2 | caveolin 2 | 1.16 | 0.005 |
| Cd99 | CD99 antigen | 1.10 | 0.006 |
| Ctbs | chitobiase, di-N-acetyl- | 1.12 | 0.006 |
| Clcn3 | chloride channel 3 | 1.15 | 0.008 |
| Ccdc23 | coiled-coil domain containing 23 | 1.10 | 0.005 |
| Ckb | creatine kinase, brain /// creatine kinase, brain | 1.13 | 0.005 |
| Ctdsp1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 | 1.12 | 0.009 |
| Cugbp2 | CUG triplet repeat, RNA binding protein 2 | 1.20 | 0.008 |
| P22k15 | cystatin related protein 2 | 1.15 | 0.001 |
| Cox6a1 | cytochrome c oxidase, subunit VIa, polypeptide 1 /// cytochrome c oxidase, subunit VIa, polypeptide 1 | 1.13 | 0.002 |
| Cyp4f2 | cytochrome P450, family 4, subfamily F, polypeptide 2 /// cytochrome P450, family 4, subfamily F, polypeptide 2 | 1.16 | 0.003 |
| Ddt | D-dopachrome tautomerase | 1.14 | 0.007 |
| Dedd | Death effector domain-containing | 1.19 | 0.002 |
| Dlgh1 | Discs, large homolog 1 (Drosophila) | 1.28 | 0.000 |
| Dlgh2 | Discs, large homolog 2 (Drosophila) | 1.24 | 0.002 |
| Dscam | Down syndrome cell adhesion molecule | 1.15 | 0.010 |
| Dusp6 | Dual specificity phosphatase 6 | 1.13 | 0.006 |
| E2f5 | E2F transcription factor 5 /// E2F transcription factor 5 | 1.19 | 0.004 |
| Egr2 | early growth response 2 /// early growth response 2 | 1.43 | 0.004 |
| Emcn | endomucin | 1.23 | 0.001 |
| Edg2 | endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor, 2 | 1.16 | 0.005 |
| Fads1 | fatty acid desaturase 1 | 1.19 | 0.002 |
| Fasn | fatty acid synthase /// fatty acid synthase | 1.13 | 0.008 |
| Fos | FBJ murine osteosarcoma viral oncogene homolog /// FBJ murine osteosarcoma viral oncogene homolog | 1.52 | 0.003 |
| Fcgrt | Fc receptor, IgG, alpha chain transporter | 1.15 | 0.008 |
| Ftl1 | ferritin light chain 1 /// ferritin light chain 1 | 1.15 | 0.003 |
| Fau | Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) protein | 1.13 | 0.006 |
| Fzd2 | frizzled homolog 2 (Drosophila) | 1.13 | 0.003 |
| Gtf3a | general transcription factor III A | 1.15 | 0.009 |
| Gpx4 | glutathione peroxidase 4 /// glutathione peroxidase 4 | 1.15 | 0.004 |
| Gpd1 | Glycerol-3-phosphate dehydrogenase 1 (soluble) | 1.44 | 0.009 |
| Gpd1 | glycerol-3-phosphate dehydrogenase 1 (soluble) /// glycerol-3-phosphate dehydrogenase 1 (soluble) | 1.23 | 0.002 |
| Gp1bb /// Sept5 | glycoprotein Ib, beta polypeptide /// septin 5 | 1.16 | 0.009 |
| Gm2a | GM2 ganglioside activator protein | 1.13 | 0.009 |
| Gramd3 | GRAM domain containing 3 | 1.17 | 0.001 |
| Gamt | guanidinoacetate methyltransferase | 1.14 | 0.004 |
| Hesl | hairy and enhancer of split 1 (Drosophila) | 1.19 | 0.006 |
| Hhex | hematopoietically expressed homeobox | 1.17 | 0.008 |
| Hist1h4b | histone cluster 1, H4b /// histone cluster 1, H4b | 1.13 | 0.004 |
| Bat5 | HLA-B associated transcript 5 | 1.09 | 0.009 |
| Homer1 | homer homolog 1 (Drosophila) | 3.49 | 0.000 |
| Hyal3 | Hyaluronoglucosaminidase 3 | 1.13 | 0.005 |
| Hadh2 | hydroxyacyl-Coenzyme A dehydrogenase type II /// hydroxyacyl-Coenzyme A dehydrogenase type II | 1.13 | 0.002 |
| Hsd11b1 | hydroxysteroid 11-beta dehydrogenase 1 /// hydroxysteroid 11-beta dehydrogenase 1 | 1.17 | 0.004 |
| Hcn1 | Hyperpolarization-activated cyclic nucleotide-gated potassium channel 1 | 1.23 | 0.003 |
| Impa2 | inositol (myo)-1(or 4)-monophosphatase 2 | 1.17 | 0.005 |
| Ifngr | interferon gamma receptor 1 | 1.14 | 0.005 |
| Il12a | interleukin 12a /// interleukin 12a | 1.16 | 0.002 |
| Klk6 | kallikrein 6 | 1.20 | 0.004 |
| Klf15 | Kruppel-like factor 15 | 1.15 | 0.004 |
| Klf4 | Kruppel-like factor 4 (gut) | 1.38 | 0.001 |
| Ldhd | lactate dehydrogenase D | 1.13 | 0.005 |
| Ldhd | lactate dehydrogenase D | 1.15 | 0.001 |
| Matr3 | matrin 3 | 1.21 | 0.002 |
| Mkks /// Cldn1 | McKusick-Kaufman syndrome protein /// Claudin 1 | 1.13 | 0.007 |
| 39143 | membrane-associated ring finger (C3HC4) 2 | 1.15 | 0.001 |
| Mt3 | metallothionein 3 /// metallothionein 3 | 1.14 | 0.003 |
| MAST1 | microtubule associated serine/threonine kinase 1 | 1.15 | 0.001 |
| Mfge8 | milk fat globule-EGF factor 8 protein | 1.15 | 0.005 |
| Map2k3 | mitogen activated protein kinase kinase 3 | 1.16 | 0.002 |
| Mag | myelin-associated glycoprotein | 1.16 | 0.004 |
| Mal | myelin and lymphocyte protein, T-cell differentiation protein | 1.12 | 0.007 |
| Mog | myelin oligodendrocyte glycoprotein | 1.16 | 0.002 |
| Mcl1 | myeloid cell leukemia sequence 1 | 1.12 | 0.010 |
| --- | Nclone10 mRNA | 1.24 | 0.000 |
| Necap2 | NECAP endocytosis associated 2 | 1.14 | 0.008 |
| Nedd9 | neural precursor cell expressed, developmentally down-regulated gene 9 | 1.17 | 0.002 |
| Nrxn3 | neurexin 3 | 1.31 | 0.001 |
| Ntrk2 | neurotrophic tyrosine kinase, receptor, type 2 | 1.51 | 0.000 |
| Nfia | nuclear factor I/A | 1.20 | 0.004 |
| Nfib | nuclear factor I/B | 1.15 | 0.010 |
| Nfkbia | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha /// nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | 1.23 | 0.002 |
| Nr4a3 | Nuclear receptor subfamily 4, group A, member 3 | 1.28 | 0.003 |
| Nr4a3 | nuclear receptor subfamily 4, group A, member 3 /// nuclear receptor subfamily 4, group A, member 3 | 1.56 | 0.000 |
| Numb | Numb gene homolog (Drosophila) | 1.18 | 0.002 |
| Olig1 | oligodendrocyte transcription factor 1 | 1.16 | 0.009 |
| Por | P450 (cytochrome) oxidoreductase /// P450 (cytochrome) oxidoreductase | 1.11 | 0.010 |
| Pnlip | pancreatic lipase /// pancreatic lipase | 1.18 | 0.001 |
| Prdx6 | Peroxiredoxin 6 | 1.15 | 0.005 |
| Ppan | peter pan homolog (Drosophila) | 1.10 | 0.006 |
| Pik4ca | Phosphatidylinositol 4-kinase, catalytic, alpha polypeptide | 1.13 | 0.009 |
| Pitpnm1 | phosphatidylinositol transfer protein, membrane-associated 1 | 1.13 | 0.008 |
| Pea15 | phosphoprotein enriched in astrocytes 15 | 1.13 | 0.004 |
| Pttg1ip | pituitary tumor-transforming 1 interacting protein | 1.11 | 0.006 |
| Pllp | plasma membrane proteolipid | 1.13 | 0.007 |
| Plekhc1 | pleckstrin homology domain containing, family C (with FERM domain) member 1 | 1.16 | 0.004 |
| Plag1 | pleiomorphic adenoma gene 1 | 1.19 | 0.003 |
| Pnkp | polynucleotide kinase 3′-phosphatase | 1.14 | 0.001 |
| Kcnn2 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 /// potassium intermediate/small conductance calcium-activated channel, sub-family N, member 2 | 1.17 | 0.007 |
| Kcnh3 | potassium voltage-gated channel, subfamily H (eag-related), member 3 | 1.13 | 0.008 |
| Kcnd3 | potassium voltage gated channel, Shal-related family, member 3 | 1.16 | 0.007 |
| Pias4 | protein inhibitor of activated STAT, 4 | 1.13 | 0.003 |
| Prkwnk1 | Protein kinase, lysine deficient 1 | 1.21 | 0.008 |
| Plp | proteolipid protein | 1.14 | 0.009 |
| Ua20 | Putative UA20 protein | 1.14 | 0.007 |
| Qscn6 | Quiescin Q6 | 1.13 | 0.004 |
| Rab34 | RAB34, member of RAS oncogene family | 1.13 | 0.009 |
| Rad23a | RAD23a homolog (S. cerevisiae) | 1.14 | 0.009 |
| Rassf4 | Ras association (RalGDS/AF-6) domain family 4 | 1.15 | 0.010 |
| Rgc32 | response gene to complement 32 | 1.23 | 0.000 |
| Rpe65 | retinal pigment epithelium 65 | 1.14 | 0.005 |
| Rpl10a | ribosomal protein L10A | 1.18 | 0.004 |
| Rpl28 | ribosomal protein L28 | 1.14 | 0.002 |
| Rpl29 | ribosomal protein L29 | 1.15 | 0.004 |
| Rpl32 | ribosomal protein L32 | 1.19 | 0.001 |
| Rps15 | ribosomal protein S15 | 1.20 | 0.003 |
| Rps5 | ribosomal protein S5 | 1.13 | 0.005 |
| Rnf167 | ring finger protein 167 | 1.11 | 0.004 |
| --- | RM2 mRNA, partial sequence | 1.47 | 0.001 |
| S100a1 | S100 calcium binding protein A1 | 1.10 | 0.006 |
| S100b | S100 protein, beta polypeptide | 1.14 | 0.008 |
| Scrg1 | scrapie responsive gene 1 | 1.18 | 0.002 |
| Sepw1 | selenoprotein W, muscle 1 | 1.13 | 0.006 |
| Sgk | serum/glucocorticoid regulated kinase | 1.29 | 0.006 |
| Sh3glb1 | SH3-domain GRB2-like B1 (endophilin) | 1.16 | 0.008 |
| Sirt2 | sirtuin (silent mating type information regulation 2 homolog) 2 (S. cerevisiae) | 1.16 | 0.002 |
| Slc23a2 | Solute carrier family 23 (nucleobase transporters), member 2 | 1.22 | 0.001 |
| Spata6 | spermatogenesis associated 6 | 1.11 | 0.008 |
| Sc5d | sterol-C5-desaturase (fungal ERG3, delta-5-desaturase) homolog (S. cerevisae) /// sterol-C5-desaturase (fungal ERG3, delta-5-desaturase) homolog (S. cerevisiae) | 1.19 | 0.001 |
| Srebf1 | sterol regulatory element binding factor 1 /// sterol regulatory element binding factor 1 | 1.14 | 0.003 |
| Strn3 | Striatin, calmodulin binding protein 3 | 1.13 | 0.008 |
| Sv2a | synaptic vesicle glycoprotein 2a | 1.18 | 0.003 |
| Stx5a | syntaxin 5a | 1.10 | 0.008 |
| Snta1 | syntrophin, acidic 1 | 1.12 | 0.007 |
| Tacr3 | tachykinin receptor 3 | 1.13 | 0.005 |
| Tbkbp1 | TBK1 binding protein 1 | 1.23 | 0.001 |
| Tspan2 | tetraspanin 2 | 1.12 | 0.006 |
| Thap7 | THAP domain containing 7 | 1.19 | 0.000 |
| Tst | thiosulfate sulfurtransferase | 1.20 | 0.001 |
| Tmed3 | transmembrane emp24 domain containing 3 /// transmembrane emp24 domain containing 3 | 1.15 | 0.004 |
| Uba52 | ubiquitin A-52 residue ribosomal protein fusion product 1 | 1.19 | 0.002 |
| Unc13c | unc-13 homolog C (C. elegans) | 1.19 | 0.005 |
| Ets1 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 1.15 | 0.007 |
| Vat1 | vesicle amine transport protein 1 homolog (T californica) | 1.18 | 0.002 |
| Zfp335 | zinc finger protein 335 | 1.13 | 0.006 |
There were 4 significant GO categories that differed between the EtOH and water groups, and 15 GO categories that differed between the EtOH and SAC groups (Table 4). General categories such as cell and ion transport and homeostasis, and synaptic transmission appeared in both lists of GO categories. Additional major GO categories in the EtOH versus SAC contrast included endocytosis, neurogenesis and ensheathment of neurons. Several genes listed in the synaptic transmission category for both EtOH contrasts included Grm1, Rims1, Htr2a, Htr5a, Gria2, Gria3, Sv2a, Scn2b, Gad1, Gad2, Camk4, Gabrb1, Gabrb2, Gabrb3, Cav2, Nrxn3, S100b and Oprk1 (Tables 1 and 2).
TABLE 4.
Significant GO categories for EtOH versus Water and EtOH versus SAC comparisons
| Term | P-value | No. sig genes | Total genes |
|---|---|---|---|
| I. EtOH versus water significant categories | |||
| anion transport | 0.0367 | 5 | 65 |
| calcium ion transport | 0.016 | 6 | 72 |
| chemical homeostasis | 0.0104 | 10 | 151 |
| synaptic transmission | 0.01618 | 15 | 288 |
| II. EtOH versus SAC significant categories | |||
| calcium ion homeostasis | 0.01 | 9 | 92 |
| cell ion homeostasis | 0.00 | 17 | 132 |
| cell maturation | 0.01 | 6 | 50 |
| chemical homeostasis | 0.00 | 19 | 178 |
| endocytosis | 0.02 | 5 | 47 |
| ensheathment of neurons | 0.00 | 7 | 33 |
| forebrain development | 0.00 | 7 | 35 |
| membrane organization and biogenesis | 0.02 | 9 | 116 |
| myelination | 0.00 | 5 | 27 |
| negative regulation of transcription from RNA polymerase II promoter | 0.04 | 6 | 73 |
| neurogenesis | 0.05 | 15 | 265 |
| neurological process | 0.00 | 24 | 272 |
| nucleocytoplasmic transport | 0.05 | 5 | 56 |
| potassium ion transport | 0.02 | 7 | 80 |
| synaptic transmission | 0.00 | 17 | 233 |
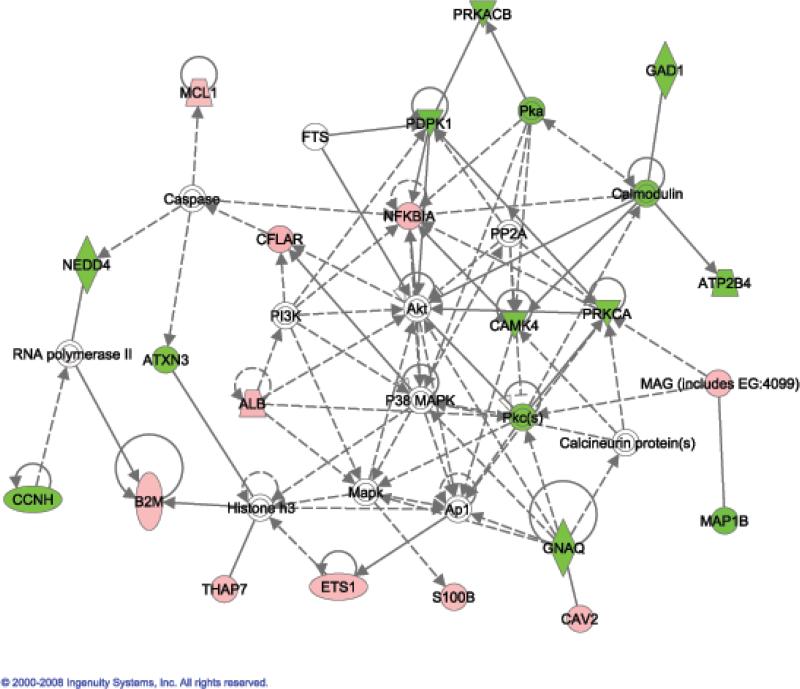
There were 73 genes that were significantly changed in the same direction in the EtOH group versus both the water and SAC groups, with 40 genes having higher and 33 genes lower expression in the EtOH group (Table 5). There were 11 genes within the synaptic transmission category that were in common in both contrasts, with 7 genes (Cav2, Homer1, Nrxn3, Pik4ca, Plp, S100b and Sv2a) having higher, and 4 genes (Camk4, Gabrb2, Gad1 and Syt6) having lower expression in the EtOH group. There were 7 genes within a combined homeostasis/transport category that were in common in the EtOH group versus the SAC and water groups, with 5 genes (S100b, Sv2a, Clcn3, Ftl1 and Alb) having higher and only 2 genes (Prkca and Atp2b4) having lower expression in the EtOH group.
TABLE 5.
Genes that were significantly different and changed in the same direction in the nucleus accumbens of iP rats for the Ethanol group versus both the Saccharin and Water groups
| Symbol | Gene Description | Higher (+) or Lower (-) with EtOH | GO category |
|---|---|---|---|
| Pdpk1 | 3-phosphoinositide dependent protein kinase-1 | - | |
| Adar | adenosine deaminase, RNA-specific | - | |
| Alb | albumin | + | h/t |
| Atrx | alpha thalassemia/mental retardation syndrome X-linked homolog (human) | - | |
| Appbp2 | amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | - | |
| Atxn3 | ataxin 3 | - | |
| Atp2b4 | ATPase, Ca++ transporting, plasma membrane 4 | - | h/t |
| Abcc4 | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 | + | |
| Blnk | B-cell linker | - | |
| B2m | beta-2 microglobulin | + | |
| Cdh11 | Cadherin 11 | + | |
| Cacnb4 | calcium channel, voltage-dependent, beta 4 subunit | - | |
| Camk4 | calcium/calmodulin-dependent protein kinase IV | - | st |
| Csnk1e | casein kinase 1, epsilon | - | |
| Cflar | CASP8 and FADD-like apoptosis regulator | + | |
| Cav2 | caveolin 2 | + | st |
| Cd99 | CD99 antigen | + | |
| Clcn3 | chloride channel 3 | + | h/t |
| Ctdsp1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 | + | |
| Ccnh | cyclin H | - | |
| P22k15 | cystatin related protein 2 | + | |
| Emcn | endomucin | + | |
| Ftl1 | ferritin light chain 1 | + | h/t |
| Gabrb2 | gamma-aminobutyric acid (GABA-A) receptor, subunit beta 2 | - | st |
| Gad1 | glutamic acid decarboxylase 1 | - | st |
| Gpd1 | glycerol-3-phosphate dehydrogenase 1 (soluble) | + | |
| Gnaq | Guanine nucleotide binding protein, alpha q polypeptide | - | |
| Homer1 | homer homolog 1 (Drosophila) | + | st |
| Hyal3 | Hyaluronoglucosaminidase 3 | + | |
| Kifc3 | Kinesin family member C3 | - | |
| Klf15 | Kruppel-like factor 15 | + | |
| Map1b | microtubule-associated protein 1b | - | |
| Mag | myelin-associated glycoprotein | + | |
| Mcl1 | myeloid cell leukemia sequence 1 | + | |
| Mllt10 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 | - | |
| --- | Nclone10 mRNA | + | |
| Nedd4a | neural precursor cell expressed, developmentally down-regulated gene 4A | - | |
| Nrxn3 | neurexin 3 | + | st |
| Nfia | nuclear factor I/A | + | |
| Nfib | nuclear factor I/B | + | |
| Nfkbia | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | + | |
| 2610020o08rik | nuclear NF-kappaB activating protein | - | |
| Npap60 | nuclear pore associated protein | - | |
| P34 | p34 protein | - | |
| Prdx6 | Peroxiredoxin 6 | + | |
| Pik4ca | Phosphatidylinositol 4-kinase, catalytic, alpha polypeptide | + | st |
| Plag1 | pleiomorphic adenoma gene 1 | + | |
| Prkca | protein kinase C, alpha | - | h/t |
| Prkacb | protein kinase, cAMP dependent, catalytic, beta | - | |
| Prkwnk1 | Protein kinase, lysine deficient 1 | + | |
| Plp | proteolipid protein | + | st |
| Ua20 | Putative UA20 protein | + | |
| Ramp3 | receptor (calcitonin) activity modifying protein 3 | - | |
| Rgc32 | response gene to complement 32 | + | |
| Rpe65 | retinal pigment epithelium 65 | + | |
| S100b | S100 protein, beta polypeptide | + | st, h/t |
| Scamp1 | Secretory carrier membrane protein 1 | + | |
| Sepw1 | selenoprotein W, muscle 1 | + | |
| Styxl1 | Serine/threonine/tyrosine interacting-like 1 | - | |
| Sgtb | small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta | - | |
| Slc2a3 | solute carrier family 2 (facilitated glucose transporter), member 3 | - | |
| Slc23a2 | Solute carrier family 23 (nucleobase transporters), member 2 | + | |
| Sc5d | sterol-C5-desaturase (fungal ERG3, delta-5-desaturase) homolog (S. cerevisae) | + | |
| Stch | stress 70 protein chaperone, microsome-associated, 60kD human homolog | - | |
| Sv2a | synaptic vesicle glycoprotein 2a | + | st, h/t |
| Syt6 | synaptotagmin VI | - | st |
| Tbkbp1 | TBK1 binding protein 1 | + | |
| Thap7 | THAP domain containing 7 | + | |
| Txndc13 | thioredoxin domain containing 13 | - | |
| Ube4a | ubiquitination factor E4A, UFD2 homolog (S. cerevisiae) | - | |
| Vti1a | vesicle transport through interaction with t-SNAREs homolog 1A (yeast) | - | |
| Ets1 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | + | |
| Zfp483 | zinc finger protein 483 | - |
Abbreviation: st = synaptic transmission; h/t = homeostasis/transport
Gene expression in the AMYG
In the AMYG, comparing across the 3 groups, there were 134 differences (p < 0.01) in the expression of named genes, with 48 differences between the SAC and water groups, 23 differences between the EtOH and water groups, and 63 differences between the EtOH and SAC groups (Table 6). However, because of the high FDR, these differences could have occurred by chance alone.
TABLE 6.
Genes that were different in the amygdala of iP rats between the Ethanol, Saccharin and Water groups at p < 0.01 (FDR > 0.5)
| Gene Symbol | Name | Fold Change | Limma p-value |
|---|---|---|---|
| I. Saccharin versus water (FDR = 1.0) | |||
| Adcy3 | adenylate cyclase 3 | −1.13 | 0.008 |
| Anxa4 | annexin A4 | −1.17 | 0.004 |
| Abca1 | ATP-binding cassette, sub-family A (ABC1), member 1 | −1.20 | 0.009 |
| Atg7 | Autophagy-related 7 (yeast) | −1.12 | 0.006 |
| Dusp1 | dual specificity phosphatase 1 | −1.32 | 0.000 |
| Dusp1 | dual specificity phosphatase 1 | −1.23 | 0.005 |
| Dusp5 | dual specificity phosphatase 5 | −1.23 | 0.009 |
| Dusp9 | dual specificity phosphatase 9 | −1.18 | 0.005 |
| Ephb6 | Eph receptor B6 | −1.20 | 0.009 |
| Foxp1 | forkhead box P1 | −1.20 | 0.005 |
| Hs3st2 | heparan sulfate (glucosamine) 3-O-sulfotransferase 2 | −1.21 | 0.001 |
| Hpca | hippocalcin | −1.26 | 0.002 |
| Homer1 | homer homolog 1 (Drosophila) | −1.97 | 0.001 |
| Klf10 | Kruppel-like factor 10 /// Kruppel-like factor 10 | −1.28 | 0.000 |
| Lgr4 | leucine-rich repeat-containing G protein-coupled receptor 4 | −1.16 | 0.002 |
| Map1lc3b | Microtubule-associated protein 1 light chain 3 beta | −1.28 | 0.000 |
| Nr4a3 | nuclear receptor subfamily 4, group A, member 3 /// nuclear receptor subfamily 4, group A, member 3 | −1.39 | 0.003 |
| Pvalb | parvalbumin | −1.18 | 0.003 |
| Kcnab1 | potassium voltage-gated channel, shaker-related subfamily, beta member 1 | −1.19 | 0.007 |
| Kcnab1 | potassium voltage-gated channel, shaker-related subfamily, beta member 1 | −1.25 | 0.006 |
| Prkcb1 | protein kinase C, beta 1 | −1.14 | 0.006 |
| Pprf18 | PRP18 pre-mRNA processing factor 18 homolog (yeast) | −1.16 | 0.003 |
| Ramp3 | receptor (calcitonin) activity modifying protein 3 | −1.29 | 0.000 |
| --- | RM2 mRNA, partial sequence | −1.21 | 0.010 |
| Slit2 | slit homolog 2 (Drosophila) | −1.28 | 0.003 |
| Trpv6 | Transient receptor potential cation channel, subfamily V, member 6 | −1.36 | 0.001 |
| Zfand2a | zinc finger, AN1-type domain 2A /// zinc finger, AN1-type domain 2A | −1.12 | 0.009 |
| A2m | alpha-2-macroglobulin /// alpha-2-macroglobulin | 1.26 | 0.004 |
| Cd44 | CD44 antigen | 1.21 | 0.004 |
| Ceacam1 | CEA-related cell adhesion molecule 1 | 1.17 | 0.006 |
| Cybrd1 | cytochrome b reductase 1 /// cytochrome b reductase 1 | 1.20 | 0.006 |
| Dspp | dentin sialophosphoprotein | 1.13 | 0.008 |
| Dpp6 | Dipeptidylpeptidase 6 | 1.17 | 0.005 |
| Doc2g | double C2, gamma | 1.14 | 0.007 |
| DLP2 | Dynein-like protein 2 | 1.15 | 0.007 |
| Fcgr2b | Fc receptor, IgG, low affinity IIb | 1.29 | 0.004 |
| Gja4 | gap junction membrane channel protein alpha 4 | 1.14 | 0.006 |
| Gipr | gastric inhibitory polypeptide receptor | 1.14 | 0.007 |
| Igh-1a | immunoglobulin heavy chain 1a (serum IgG2a) | 1.94 | 0.008 |
| Irf3 | interferon regulatory factor 3 | 1.12 | 0.010 |
| Kazald1 | Kazal-type serine peptidase inhibitor domain 1 | 1.14 | 0.008 |
| LMO7 | LIM domain only protein 7 | 1.16 | 0.007 |
| Phactr2 | Phosphatase and actin regulator 2 | 1.23 | 0.001 |
| Pik4cb | Phosphatidylinositol 4-kinase, catalytic, beta polypeptide | 1.14 | 0.010 |
| Arhgdib | Rho, GDP dissociation inhibitor (GDI) beta | 1.13 | 0.009 |
| RT1-Bb | RT1 class II, locus Bb | 1.25 | 0.010 |
| Srpk3 | serine/arginine-rich protein specific kinase 3 | 1.32 | 0.000 |
| Smyd2 | SET and MYND domain containing 2 | 1.14 | 0.007 |
| Stx4a | Syntaxin 4A (placental) | 1.16 | 0.009 |
| Vwf | von Willebrand factor /// von Willebrand factor | 1.24 | 0.007 |
| II. Ethanol versus water (FDR = 1.0) | |||
| Bfar | bifunctional apoptosis regulator | −1.24 | 0.007 |
| Cast | Calpastatin | −1.16 | 0.004 |
| Eif4g2 | Eukaryotic translation initiation factor 4 gamma, 2 | −1.22 | 0.003 |
| Gabbr1 | gamma-aminobutyric acid (GABA) B receptor 1 | −1.33 | 0.005 |
| Homer2 | homer homolog 2 (Drosophila) | −1.16 | 0.008 |
| Igf2r | insulin-like growth factor 2 receptor /// insulin-like growth factor 2 receptor | −1.19 | 0.005 |
| Il1rap | interleukin 1 receptor accessory protein | −1.15 | 0.009 |
| Rab27a | RAB27A, member RAS oncogene family | −1.23 | 0.007 |
| Slc30a7 | solute carrier family 30 (zinc transporter), member 7 | −1.18 | 0.009 |
| Tef | thyrotroph embryonic factor | −1.20 | 0.006 |
| Tgfbr1 | transforming growth factor, beta receptor 1 /// transforming growth factor, beta receptor 1 | −1.18 | 0.004 |
| Acvr1 | activin A receptor, type 1 | 1.14 | 0.009 |
| Aldh5a1 | Aldehyde dehydrogenase family 5, subfamily A1 | 1.19 | 0.004 |
| Amt | aminomethyltransferase (glycine cleavage system protein T) /// aminomethyltransferase (glycine cleavage system protein T) | 1.14 | 0.009 |
| Acly | ATP citrate lyase /// ATP citrate lyase | 1.16 | 0.005 |
| Bhlhb2 | Basic helix-loop-helix domain containing, class B2 | 1.25 | 0.006 |
| Dtnbp1 | distrobrevin binding protein 1 | 1.16 | 0.007 |
| Ifngr | interferon gamma receptor 1 | 1.14 | 0.005 |
| Lpxn | leupaxin | 1.13 | 0.007 |
| Rpo1-4 | RNA polymerase 1-4 | 1.16 | 0.005 |
| Serpinc1 | Serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 | 1.17 | 0.009 |
| Stom | stomatin | 1.21 | 0.008 |
| Ttc23 | tetratricopeptide repeat domain 23 | 1.18 | 0.004 |
| III. Ethanol versus saccharin (FDR = 0.5 - 0.8) | |||
| A2m | alpha-2-macroglobulin /// alpha-2-macroglobulin | −1.23 | 0.008 |
| Atp5i | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit e /// ATP synthase, H+ transporting, mitochondrial F0 complex, subunit e | −1.19 | 0.002 |
| Bckdha | branched chain ketoacid dehydrogenase E1, alpha polypeptide | −1.17 | 0.007 |
| Cacna1c | Calcium channel, voltage-dependent, L type, alpha 1C subunit | −1.20 | 0.003 |
| Camk2d | Calcium/calmodulin-dependent protein kinase II, delta | −1.13 | 0.008 |
| Ceacam1 | CEA-related cell adhesion molecule 1 | −1.19 | 0.004 |
| Cops3 | COP9 (constitutive photomorphogenic) homolog, subunit 3 (Arabidopsis thaliana) | −1.14 | 0.008 |
| Ckap5 | Cytoskeleton associated protein 5 | −1.15 | 0.009 |
| Dgki | Diacylglycerol kinase, iota | −1.19 | 0.006 |
| Dscr1l1 | Down syndrome critical region gene 1-like 1 | −1.25 | 0.005 |
| Fmo2 | flavin containing monooxygenase 2 | −1.21 | 0.007 |
| Gspt1 | G1 to S phase transition 1 | −1.15 | 0.001 |
| Gipr | gastric inhibitory polypeptide receptor | −1.15 | 0.003 |
| Glt8d1 | Glycosyltransferase 8 domain containing 1 | −1.17 | 0.002 |
| Hcr | HCR (a-helix coiled-coil rod homolog) | −1.17 | 0.006 |
| Igh-1a | immunoglobulin heavy chain 1a (serum IgG2a) | −1.92 | 0.009 |
| Maea | Macrophage erythroblast attacher | −1.18 | 0.005 |
| Hnt | Neurotrimin | −1.21 | 0.009 |
| Ntrk1 | Neurotrophic tyrosine kinase, receptor, type 1 | −1.17 | 0.004 |
| Pctp | phosphatidylcholine transfer protein | −1.14 | 0.003 |
| Pabpn1 | poly(A) binding protein, nuclear 1 | −1.16 | 0.003 |
| Psmb8 | proteosome (prosome, macropain) subunit, beta type 8 /// proteosome (prosome, macropain) subunit, beta type 8 | −1.33 | 0.002 |
| Pycard | PYD and CARD domain containing | −1.17 | 0.007 |
| Rasgrp4 | RAS guanyl releasing protein 4 | −1.14 | 0.009 |
| Reep4 | receptor accessory protein 4 | −1.17 | 0.004 |
| Serinc3 | Serine incorporator 3 | −1.21 | 0.002 |
| Vps54 | Vacuolar protein sorting 54 (yeast) | −1.15 | 0.008 |
| Wdr46 | WD repeat domain 46 | −1.20 | 0.006 |
| Nt5c3l | 5′-nucleotidase, cytosolic III-like | 1.22 | 0.002 |
| Acvr1 | activin A receptor, type 1 | 1.20 | 0.001 |
| Adcy3 | adenylate cyclase 3 | 1.14 | 0.005 |
| Acly | ATP citrate lyase /// ATP citrate lyase | 1.14 | 0.008 |
| B3gat2 | beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) | 1.14 | 0.006 |
| Cacna2d3 | calcium channel, voltage-dependent, alpha 2/delta 3 subunit /// calcium channel, voltage-dependent, alpha 2/delta 3 subunit | 1.13 | 0.009 |
| Crem | cAMP responsive element modulator | 1.15 | 0.001 |
| Ckb | creatine kinase, brain /// creatine kinase, brain | 1.13 | 0.007 |
| Dlgap2 | discs, large (Drosophila) homolog-associated protein 2 | 1.15 | 0.002 |
| Dusp1 | dual specificity phosphatase 1 | 1.36 | 0.000 |
| Dusp1 | dual specificity phosphatase 1 | 1.28 | 0.001 |
| Egr2 | early growth response 2 /// early growth response 2 | 1.40 | 0.001 |
| Fpgt | Fucose-1-phosphate guanylyltransferase | 1.18 | 0.006 |
| Gcnt2 | glucosaminyl (N-acetyl) transferase 2, I-branching enzyme | 1.13 | 0.006 |
| Gpd1 | glycerol-3-phosphate dehydrogenase 1 (soluble) /// glycerol-3-phosphate dehydrogenase 1 (soluble) | 1.22 | 0.004 |
| Hs3st2 | heparan sulfate (glucosamine) 3-O-sulfotransferase 2 | 1.27 | 0.000 |
| Homer1 | homer homolog 1 (Drosophila) | 2.19 | 0.000 |
| Klf10 | Kruppel-like factor 10 /// Kruppel-like factor 10 | 1.27 | 0.000 |
| Masp1 | mannan-binding lectin serine peptidase 1 | 1.21 | 0.009 |
| Nedd9 | neural precursor cell expressed, developmentally down-regulated gene 9 | 1.14 | 0.005 |
| Nedd9 | neural precursor cell expressed, developmentally down-regulated gene 9 | 1.18 | 0.005 |
| Nr4a3 | Nuclear receptor subfamily 4, group A, member 3 | 1.30 | 0.007 |
| Nr4a3 | nuclear receptor subfamily 4, group A, member 3 /// nuclear receptor subfamily 4, group A, member 3 | 1.56 | 0.000 |
| Pvalb | parvalbumin | 1.26 | 0.000 |
| Kcnab1 | potassium voltage-gated channel, shaker-related subfamily, beta member 1 | 1.26 | 0.005 |
| Pnoc | prepronociceptin | 1.23 | 0.008 |
| P2ry12 | purinergic receptor P2Y, G-protein coupled 12 | 1.12 | 0.010 |
| Ramp3 | receptor (calcitonin) activity modifying protein 3 | 1.30 | 0.000 |
| Rgs18 | Regulator of G-protein signaling 18 | 1.35 | 0.003 |
| Rnf138 | ring finger protein 138 | 1.14 | 0.001 |
| --- | RM2 mRNA, partial sequence | 1.36 | 0.000 |
| Slc33a1 | Solute carrier family 33 (acetyl-CoA transporter), member 1 | 1.14 | 0.008 |
| St3gal5 | ST3 beta-galactoside alpha-2,3-sialyltransferase 5 | 1.16 | 0.002 |
| Txnl4b | thioredoxin-like 4B | 1.15 | 0.007 |
| Tle4 | transducin-like enhancer of split 4, E(spl) homolog (Drosophila) | 1.14 | 0.009 |
| Trpv6 | Transient receptor potential cation channel, subfamily V, member 6 | 1.27 | 0.006 |
| Tpbg | trophoblast glycoprotein | 1.30 | 0.006 |
| Tsc22d3 | TSC22 domain family 3 /// TSC22 domain family 3 | 1.15 | 0.002 |
Quantitative RT-PCR confirmation
Because there were more significant differences and more significant GO categories between the EtOH versus SAC group than between the EtOH versus water group, genes selected for qRT-PCR confirmation (Table 7) were chosen from the EtOH-SAC comparison (Table 3). Among the 12 genes tested, 9 were confirmed as changing significantly in the same direction as the microarray values (Table 7). Of the remaining 3 genes, Map1b changed in the same direction with both measures (however, the RT-PCR values were not statistically different), Camk4 was not changed in the RT-PCR measure, and Nrxn3 changed significantly in both measures, but in opposite directions (Table 7). Similar to previous studies from our lab (Edenberg et al., 2005; Kimpel et al., 2007), there was a high degree of concordance between the microarray and RT-PCR results. However, the lack of agreement between the two measures for Camk4 and Nrxn3 suggests the results for these two genes are inconclusive.
TABLE 7.
Quantitative RT-PCR confirmation of differences observed in the nucleus accumbens between EtOH and SAC groups
| Gene Symbol | Gene Name | Microarray Fold change | qRT-PCR fold change | Microarray p-value | qRT-PCR p-value |
|---|---|---|---|---|---|
| Cacnb4 | Calcium channel, voltage dependent, beta 4 subunit | −1.31 | −1.28 | 0.004 | 0.003 |
| Camk4 | Calcium/calmodulin-dependent protein kinase IV | −1.23 | 1.01 | 0.002 | 0.42 |
| Cflar | CASP8 and FADD-like apoptosis regulator - intron | 1.29 | 1.04 | 0.001 | 0.036 |
| Cflar | CASP8 and FADD-like apoptosis regulator - exon | 1.29 | 1.05 | 0.001 | 0.001 |
| Gabrb2 | GABA-A receptor, beta 2 subunit | −1.31 | −1.05 | 0.004 | 0.069 |
| Gnaq | Guanine nucleotide binding protein, alpha q polypeptide | −1.30 | −1.04 | 0.001 | 0.063 |
| Homer1 | Homer homolog 1 (Drosophila) - exon | −1.15 | −1.33 | 0.089 | 0.075 |
| Homer1 | Homer homolog 1 (Drosophila) - intron | 3.49 | 2.52 | 0.001 | 0.001 |
| Map1b | Microtubule-associated protein 1b | −1.37 | −1.04 | 0.001 | 0.12 |
| Nrxn3 | Neurexin 3 | 1.31 | −1.31 | 0.001 | 0.001 |
| Pdpk1 | 3-phosphoinositide dependent protein kinase-1 | −1.47 | −1.15 | 0.002 | 0.007 |
| Prkacb | Protein kinase, cAMP-dependent, catalytic, beta | −1.29 | −1.08 | 0.001 | 0.030 |
Negative values indicate that EtOH values are lower than SAC values; positive values indicate that EtOH values are higher than SAC values.
Supplemental tables
See Supplemental tables A and B for more complete information on data for differences in the ACB between the EtOH and water groups, and between the EtOH and SAC groups.
DISCUSSION
The major findings of this study are that, compared to the water control group, EtOH self-administration, but not SAC self-administration, produced changes in named gene expression in the ACB of iP rats (Tables 1 and 2), whereas significant changes in named gene expression were not observed in the AMYG (Table 6). The effects of EtOH self-administration on gene expression in the ACB is not due to the presence of EtOH in the tissue at the time of killing, because animals were killed 24 hr after the last operant session. Also, the differences between the EtOH and water groups do not appear to be due to motor activity, learning or conditioning factors associated with the operant task, because the SAC group learned the task as well as the EtOH group and responded more on the active lever than the water lever (Fig. 1), but there were no significant differences in gene expression in the ACB between the SAC and water groups (Table 1). Changes associated with the operant task may have occurred in the ACB of EtOH and SAC groups, but these changes were not detectable after 24 hr, as suggested by the SAC versus water contrast (Table 1). The changes that persisted for 24 hr in the ACB of the EtOH group may be due to the chronic effects of EtOH exposure and changes associated with the CNS reinforcing effects of EtOH. More robust differences between the EtOH and the other groups may have been observed with the present experimental conditions, if the ACB shell had been analyzed separately from the core, and if shorter time points had been analyzed.
The apparent lack of finding significant changes in gene expression in the AMYG between any of the groups may be due to the combination of factors, i.e., (a) changes are occurring but they do not persist for 24 hr, and (b) measuring the whole AMYG may mask changes occurring within distinct amygdaloid nuclei. It is also possible in the AMYG, and to a lesser extent in the ACB, only small changes in mRNA may be needed to maintain larger changes in protein levels that may have developed with chronic drinking. Therefore, many changes may have occurred in the AMYG and ACB that are not detected with microarray analyses, but may be detected with sensitive proteomics methods.
Common differences in the EtOH group compared to both the SAC and water groups could indicate differences in the CNS reinforcing effects of EtOH, the chronic general pharmacological actions of EtOH, and conditioning factors associated with the operant EtOH sessions. In the ACB, there were 73 genes that were significantly different in the EtOH group versus both the water and SAC groups (Table 5). GO analysis indicated two general overlapping categories in the contrasts of EtOH versus water and EtOH versus SAC (Table 4), i.e., synaptic transmission and homeostasis/transport. Seven of the 11 genes that were changed in the same direction in the ACB had higher expression in the EtOH group (Table 5), suggesting increased transmission at certain synapses in the ACB. In contrast, the lower expression of Gad1 and Gabrb2 may indicate reduced transmission at certain GABAA receptors. If reduced transmission is occurring at certain GABA synapses and increased transmission is occurring at non-inhibitory synapses, the net results could indicate increased excitatory synaptic function within the ACB of the EtOH group. In addition, 5 of the 7 genes in common between the EtOH and both the other two groups in the homeostasis/transport category had higher expression in the EtOH group (Table 5), suggesting that the ACB may have reached a different homeostatic state as a result of chronic EtOH self-administration.
Ingenuity® analysis indicated a network of genes, involved in intracellular signaling pathways (e.g., Prkca, Gnaq, Prkacb), that mainly had reduced expression in the EtOH group compared to the other groups (Fig. 2). These results could suggest that chronic EtOH may be reducing general cellular functions, some of which are calcium-dependent. In contrast, other genes involved in pro-inflammatory responses (e.g., Cflar, Mcl1) and histone regulation (e.g., Thap7, Est1) appear mainly to have higher expression in the ACB of the EtOH group (Fig. 2). Overall, these results suggest that chronic EtOH self-administration may be producing effects on multiple intracellular systems that could alter cellular function and the response of these cells to environmental alterations.
Fig. 2.
Ingenuity® analysis showing co-citation and networks for genes that were significantly different between the ethanol group and the saccharin group. Green indicates genes that had reduced expression in the ethanol group, and red indicates genes that had higher expression in the ethanol group. Open symbols indicate that these genes were not statistically different between the ethanol group and the other two groups, but these genes were highly linked to multiple genes that were significantly changed. See tables 2 and 3 for abbreviations of genes that changed significantly. Reduced expression of genes involved in intracellular signaling networks is depicted in green on the right hand part of the figure. Increased expression of genes involved in pro-inflammatory responses and histone regulation is shown in red on the left side.
In the ACB, the two main GO categories represented were synaptic transmission and homeostasis/transport for the EtOH group versus the other two groups. In the synaptic transmission category, Homer1, Sv2a and Cav2 had higher expression levels in the EtOH group than in the SAC and water groups (Table 5). The Homer 1 genes are part of a family of synaptic scaffolding proteins that are involved in regulating the insertion of metabotropic glutamate (mGlu) receptors into the synaptic plasma membrane (Kammermeier, 2006; Tappe and Kuner, 2006). The protein for Cav2 can also function as a scaffolding protein and interact with mGlu receptors (Burgueno et al., 2004), as well as other receptors, e.g., dopamine D1 (Yu et al., 2004) and muscarinic (Perez-Rosello et al., 2005) receptors. The synaptic vesicle glycoprotein 2a (Sv2a) is involved in regulating exocytosis (Xu and Bajjalieh, 2001; Crowder et al., 1999). Overall, these changes suggest that complex neuronal alterations may be occurring to increase neuronal function at certain synapses.
Expression of Gpd1 was elevated in the ACB of the alcohol group in the present study (Table 5); similar findings were reported for Gpd1 in the hippocampus of C57 mice exposed to EtOH in a vapor chamber (Daniels and Buck 2002), although opposite effects were observed for Gpd1 in the hippocampus of rats that had been on a forced liquid diet for several months (Saito et al 2002). An increased expression of Kruppel-like factors (Klf), transcription factors possibly involved in controlling neuronal morphogenesis (Laub et al., 2005), was observed in the present study in the ACB (Table 5), and in the study of Daniels and Buck (2002). The increased expression of Klf might reflect alterations in neuronal structure.
Some of the changes observed with EtOH self-administration in the present study have also been reported for human alcoholics. Lewohl et al. (2000) examined differences in gene expression in the frontal cortex of human alcoholics and controls, and reported reduced expression of Gabrb2 and microtubule-associated protein 4. In the present study (Table 5), lower expression levels of Gabrb2 and Map1b were observed in the ACB of the alcohol group. Flatscher-Bader et al. (2005) reported reduced expression of synaptogamin 1 (involved in exocytosis) in the ACB of human alcoholics, whereas, in the present, lower expression levels of Syt6 were observed in the ACB of the EtOH group (Table 5). The study of Lewohl et al. (2000) reported lower expression levels of genes for many myelin proteins in the frontal cortex of alcoholics. However, in the present study, lower expression levels of genes for myelin-associated proteins were not observed, suggesting that similar signs of neuronal damage were not evident in the ACB of the iP rats self-administering EtOH, as were found for human alcoholics (Lewohl et al. 2000).
Acute EtOH administration increased expression of Klf15 and Nfkbia in the whole brain of C57 and DBA mice (Treadwell and Singh 2004), a finding also observed in the ACB of the EtOH group in the present study (Table 5), suggesting that acute EtOH administration can increase expression of genes for transcription factors and that these effects persist with chronic EtOH exposure. In contrast to the decreased expression of Gabrb2 in the ACB of the chronic EtOH group (Table 5), acute EtOH administration increased Gabrb1 gene expression in the ACB of mice (Kerns et al., 2005).
If there were innate differences in certain CNS regions that predispose certain individuals to high alcohol drinking behavior, then one hypothesis could be that expression of these genes is altered by EtOH. Kimpel et al (2007) reported that there were innate differences in gene expression in 5 CNS regions, i.e., ACB, AMYG, frontal cortex, hippocampus, striatum, between the iP and iNP rats. Comparison of the expression of genes that changed in the ACB of the EtOH group versus the other 2 groups, with innate differences in gene expression between iP and iNP rats indicated a number of overlapping genes (summarized in Table 8). Sixteen named genes that differed between the iP and iNP rats also differed in the EtOH group versus both the SAC and water groups. A change in the opposite direction between innate and EtOH self-administration values might suggest that alcohol drinking is attempting to bring the expression of these genes toward a normal value. On the other hand, the expression of genes that changed in the same direction between the innate and EtOH self-administration studies might indicate that these genes are involved in vulnerability to high alcohol drinking and maintaining high alcohol drinking after it has begun. Genes that were changed in the same direction with alcohol drinking as were found between the iP versus the iNP rats (Table 8) included several genes coding for proteins involved in neurotransmission/synaptic function (e.g., Gnaq, Syt6, Sv2a, Plp). Compared to changes observed between iP and iNP rats (Kimpel et al., 2007), alcohol self-administration produced changes in the opposite direction for several of genes coding for proteins involved in synaptic transmission (e.g., Homer1, Gabrb2) or intracellular signaling (Prkca), suggesting that alcohol drinking may be attempting to re-establish ‘normal’ levels of the proteins produced by these genes.
Table 8.
Comparison of innate differences in gene expression between iP and iNP rats and effects of EtOH self-administration by iP rats on gene expression in the nucleus accumbens
| Gene description | iP vs iNP | EtOH vs SAC & water |
|---|---|---|
| Proteolipid protein | Plp (+) | Plp (+) |
| Adenosine monophosphate deaminase/adenosine deaminase | Ampd3 (+) | Adar (−) |
| 3-phosphoglycerate dehydrogenase/glycerol-3-phosphate dehydrogenase | Phgdh (−) | Gdp1 (+) |
| Beta-2 microglobulin | B2m (−) | B2m (+) |
| ATPase, Ca++ transporting, plasma membrane | Atp2a2 (−) | Atp2b4 (−) |
| Guanine nucleotide binding protein alpha | Gnao (−) | Gnaq (−) |
| Homer homolog 1, 2 (Drosophila) | Homer2 (−) | Homer1 (+) |
| Microtubule-associated proteins tau, 1A/1B light chain 3, 1b | Mapt (−); Map1lc3b (+) | Map1b (−) |
| Casein kinase 1 delta/epsilon | Csnk1d (−) | Csnk1e (−) |
| Synaptogamin 6 | Syt6 (−) | Syt6 (−) |
| Albumin | Alb (+) | Alb (+) |
| Ferritin heavy/light chain 1 | Fth1 (+) | Ftl1 (+) |
| Gamma-aminobutyric acid receptor subunit beta 1, 2 | Gabrb1 (+) | Gabrb2 (−) |
| Response gene to complement 32 | Rgc32 (+) | Rgc32 (+) |
| Synaptic vesicle glycoprotein 2b, 2a | Sv2b (+) | Sv2a (+) |
| Protein kinase C, alpha, delta, gamma | Prkcd (+) Prkcg (+) |
Prkca (−) |
Plus (+) symbol indicates higher expression in iP compared to iNP or higher expression in EtOH group versus SAC and Water groups; minus (−) symbol indicates the opposite.
In conclusion, the current study indicates that the ACB may be an important limbic structure regulating the reinforcing effects of EtOH in iP rats, and that changes in the expression of genes involved in synaptic transmission, homeostasis and intracellular signaling may contribute to this regulation. The study has some shortcomings, i.e., there may be a number of false positives in our analysis, and only a limited number of genes were confirmed. Future studies should be directed at analyzing more discrete sub-regions and nuclei within the ACB and AMYG at shorter time points after the operant sessions.
Supplementary Material
ACKNOWLEDGEMENTS
The present study was supported in by AA07611, AA07462, AA13521 [INIA Project], AA13522 [INIA Project], AA016652 [INIA Project] and INGEN® (which is partially funded by Lilly Endowment Inc.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles--database and tools. Nucleic Acids Res. 2005;33(Database issue):D562–566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RL, Rodd ZA, Murphy JM, McBride WJ. Use of selectively bred alcohol-preferring rats to study alcohol abuse, relapse and craving. In: Preedy VR, Watson RR, editors. Comprehensive Handbook of Alcohol Related Pathology. Vol. 3. Academic Press, Elsevier Science; New York: 2005. pp. 1515–1533. [Google Scholar]
- Burgueno J, Canela EI, Mallol J, Franco R, Ciruela F. Mutual regulation between metabotropic glutamate type 1 alpha receptor and caveolin proteins: from traffic to constitutive activity. Exp Cell Res. 2004;300:23–34. doi: 10.1016/j.yexcr.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Crowder KM, Gunther JM, Jones TA, Hale BD, Zhang HZ, Peterson MR, Scheller RH, Chavkin C, Bajjalieh SM. Abnormal neurotransmission in mice lacking synaptic vesicle protein 2A (SV2A). Proc Natl Acad Sci USA. 1999;96:15268–152673. doi: 10.1073/pnas.96.26.15268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels GM, Buck KJ. Expression profiling identifies strain-specific changes associated with ethanol withdrawal in mice. Genes Brain Behav. 2002;1:35–45. doi: 10.1046/j.1601-1848.2001.00008.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Strother WN, McClintick JN, Tian H, Stephans M, Jerome RE, Lumeng L, Li T-K, McBride WJ. Gene expression in the hippocampus of inbred alcohol-preferring and -nonpreferring rats. Genes Brain Behav. 2005;4:20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, Wilce PA. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem. 2005;93:359–370. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- Gentleman RC. Using GO for statistical analysis. Proc COMPSTAT. 2004;2004:171–180. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 32 Database issue. 2004:D258–261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kammermeier PJ. Surface clustering of metabotropic glutamate receptor 1 induced by long Homer proteins. BMC Neurosci. 2006;7:1. doi: 10.1186/1471-2202-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MF. Ethanol-responsive brain region expression networks: Implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpel MW, Strother WN, McClintick JN, Carr LG, Edenberg HJ, McBride WJ. Functional gene expression differences between inbred alcohol-preferring (iP) and –non-preferring (iNP) rats in five brain regions. Alcohol. 2007;41:95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Roberts AJ, Schulteis G, Parsons LF, Heyser CJ, Hyytia P, Merlopich E, Weiss F. Neurocircuitry targets in ethanol reward and dependence. Alcohol Clin Exp Res. 1998;22:3–9. [PubMed] [Google Scholar]
- Laub F, Lei L, Sumiyoshi H, Kajimura D, Dragomir C, Smaldone S, Puche AC, Petros TJ, Mason C, Parada LF, Ramirez F. Transcription factor KLF7 is important for neuronal morphogenesis in selected regions of the nervous system. Mol Cell Biol. 2005;25:5699–5711. doi: 10.1128/MCB.25.13.5699-5711.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortes. Alcohol Clin Exp Res. 2000;24:1873–1882. [PubMed] [Google Scholar]
- McBride WJ, Li T- K. Animal models of alcoholism: neurobiology of high alcohol-drinking behavior in rodents. Crit Rev Neurobiol. 1998;12:339–369. doi: 10.1615/critrevneurobiol.v12.i4.40. [DOI] [PubMed] [Google Scholar]
- McClintick JN, Jerome RE, Nicholson CR, Crabb DW, Edenberg HJ. Reproducibility of oligonucleotide arrays using small samples. BMC Genomics. 2003;4:1–15. doi: 10.1186/1471-2164-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomerav I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GJ, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci (USA) 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JM, Stewart RB, Bell RL, Badia-Elder NE, Carr LG, McBride WJ, Lumeng L, Li T- K. Phenotypic and genotypic characterization of the Indiana University rat lines selectively bred for high and low alcohol preference. Behav Genet. 2002;32:363–388. doi: 10.1023/a:1020266306135. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. Academic Press; New York: 1998. [Google Scholar]
- Perez-Rosello T, Figueroa A, Salgado H, Vilchis C, Tecuapetia F, Guzman JN, Galarraga E, Bargas J. Cholinergic control of firing pattern and neurotransmission in rat neostriatal projection neurons: role of Cav2.1 and Cav2.2 Ca+2 channels. J Neurophysiol. 2005;93:2507–2519. doi: 10.1152/jn.00853.2004. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bell RL, Kuc KA, Murphy JM, Lumeng L, Li T- K, McBride WJ. Effects of repeated alcohol deprivations on operant ethanol self-administration by alcohol-preferring (P) rats. Neuropsychopharmacology. 2003;28:1614–1621. doi: 10.1038/sj.npp.1300214. [DOI] [PubMed] [Google Scholar]
- Rodd-Henricks ZA, Bell RL, Kuc KA, Murphy JM, McBride WJ, Lumeng L, Li T- K. Effects of concurrent access to multiple ethanol concentrations and repeated deprivations on alcohol intake of alcohol-preferring rats. Alcohol Clin Exp Res. 2001;25:1140–1150. [PubMed] [Google Scholar]
- Rodd-Henricks ZA, Bell RL, Kuc KA, Murphy JM, McBride WJ, Lumeng L, Li T- K. Effects of ethanol exposure on subsequent acquisition and extinction of ethanol self-administration and expression of alcohol-seeking behavior in adult alcohol-preferring (P) rats. I. Periadolescent exposure. Alcohol Clin Exp Res. 2002a;26:1632–1641. doi: 10.1097/01.ALC.0000036301.36192.BC. [DOI] [PubMed] [Google Scholar]
- Rodd-Henricks ZA, Bell RL, Kuc KA, Murphy JM, McBride WJ, Lumeng L, Li T- K. Effects of ethanol exposure on subsequent acquisition and extinction of ethanol self-administration and expression of alcohol-seeking behavior in adult alcohol-preferring (P) rats. II. Adult exposure. Alcohol Clin Exp Res. 2002b;26:1642–1652. doi: 10.1097/01.ALC.0000036302.73712.9D. [DOI] [PubMed] [Google Scholar]
- Saito M, Smiley J, Toth R, Vadasz C. Microarray analysis of gene expression in rat hippocampus after chronic ethanol treatment. Neurochem Res. 2002;27:1221–1229. doi: 10.1023/a:1020937728506. [DOI] [PubMed] [Google Scholar]
- Saito M, Szakall I, Toth R, Kovacs KM, Oros M, Prasad VV, Blumenberg M, Vadasz C. Mouse striatal transcriptome analysis: effects of oral self-administration of alcohol. Alcohol. 2004;32:223–241. doi: 10.1016/j.alcohol.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differntial expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3(1) doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Tappe A, Kuner R. Regulation of motor performance and striatal function by synaptic scaffolding proteins of the Homer 1 family. Proc Natl Acad Sci USA. 2006;103:774–779. doi: 10.1073/pnas.0505900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treadwell JA, Singh SM. Microarray analysis of mouse brain gene expression following acute ethanol treatment. Neurochem Res. 2004;29:357–369. doi: 10.1023/b:nere.0000013738.06437.a6. [DOI] [PubMed] [Google Scholar]
- Worst TJ, Tan JC, Robertson DJ, Freeman WM, Hyytia P, Kiianmaa K, Vrana KE. Transcriptome analysis of frontal cortex in alcohol-preferring and nonpreferring rats. J Neurosci Res. 2005;80:529–538. doi: 10.1002/jnr.20496. [DOI] [PubMed] [Google Scholar]
- Xu T, Bajjalieh SM. SV2 modulates the size of the readily releasable pool of secretory vesicles. Nature Cell Biol. 2001;3:691–698. doi: 10.1038/35087000. [DOI] [PubMed] [Google Scholar]
- Xu Y, Ehringer M, Yang F, Sikela JM. Comparison of global brain gene expression profiles between inbred long-sleep and inbred short-sleep mice by high-density gene array hybridization. Alcohol Clin Exp Res. 2001;25:810–818. [PubMed] [Google Scholar]
- Yu P, Yang Z, Jones JE, Wang Z, Owens SA, Mueller SC, Felder RA, Jose PA. D1 dopamine receptor signaling involves caveolin-2 in HEK-293 cells. Kidney Intl. 2004;66:2167–2180. doi: 10.1111/j.1523-1755.2004.66007.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.