Abstract
Sphingomonas paucimobilis SYK-6 is able to grow on various lignin-derived biaryls as the sole source of carbon and energy. These compounds are degraded to vanillate and syringate by the unique and specific enzymes in this strain. Vanillate and syringate are converted to protocatechuate (PCA) and 3-O-methylgallate (3MGA), respectively, by the tetrahydrofolate-dependent O-demethylases. Previous studies have suggested that these compounds are further degraded via the PCA 4,5-cleavage pathway. However, our subsequent analysis of the ligB insertion mutant, which encodes the β subunit of PCA 4,5-dioxygenase, suggested that at least one alternative route is involved in 3MGA degradation. In the present study, we isolated the desZ gene, which confers 3MGA degradation activity on Escherichia coli. The deduced amino acid sequence of desZ showed ca. 20 to 43% identity with the type II extradiol dioxygenases. Gas chromatography-mass spectrometry analysis suggested that DesZ catalyzes the 3,4-cleavage of 3MGA. Disruption of both desZ and ligB in SYK-6 resulted in loss of the dioxygen-dependent 3MGA transformation activity, but the resulting mutant retained the ability to grow on syringate. We found that the cell extract of the desZ ligB double mutant was able to convert 3MGA to gallate when tetrahydrofolate was added to the reaction mixture, and the cell extract of this mutant degraded gallate to the same degree as the wild type did. All these results suggest that syringate is degraded through multiple 3MGA degradation pathways in which ligAB, desZ, 3MGA O-demethylase, and gallate dioxygenase are participants.
The complex aromatic polymer lignin is the most abundant aromatic compound in nature, and its mineralization is an important step in the terrestrial carbon cycle. One of the most effective and expedient methods of converting lignin into useful materials is to use a bacterial enzyme system. In nature, it is thought that the degradation of native lignin is initiated by a reaction with lignin peroxidase, manganese peroxidase, and laccase, secreted by white rot fungi (10, 39). Bacteria contribute to the process of mineralization of abundant lignin-derived compounds found in soil (40). Sphingomonas paucimobilis SYK-6 is able to grow on various lignin-derived biaryls, including β-aryl ether (14-16), biphenyl (24, 25, 34), and diarylpropane, as the sole carbon and energy source. S. paucimobilis SYK-6 degrades these compounds with unique and specific enzymes, and thus the lignin degradation enzymes in SYK-6 would likely be an effective tool for the conversion of lignin to useful intermediate metabolites.
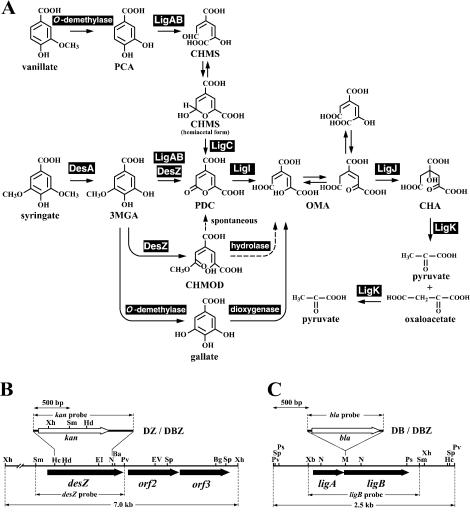
Lignin-derived compounds having syringyl (4-hydroxy-3,5-dimethoxyphenyl) and guaiacyl (4-hydroxy-3-methoxyphenyl) moieties are degraded to syringate and vanillate, respectively, by SYK-6. Syringate and vanillate are converted to 3-O-methylgallate (3MGA) and protocatechuate (PCA), respectively, by the tetrahydrofolate-dependent O-demethylases (21). It was previously assumed that 3MGA and PCA are further degraded via the PCA 4,5-cleavage pathway to generate oxaloacetate and pyruvate (Fig. 1). However, we found that disruptions of ligB, ligC, and ligI, which encode the β subunit of PCA 4,5-dioxygenase, 4-carboxy-2-hydroxymuconate-6-semialdehydedehydrogenase, and 2-pyrone-4,6-dicarboxylate (PDC) hydrolase, respectively, resulted in a growth defect on vanillate but not on syringate (18, 20; H. Aoshima, E. Masai, S. Nishikawa, Y. Katayama, and M. Fukuda, Abstr. 8th Int. Symp. Microb. Ecol., abstr. 93, 1998). In contrast, the mutant carrying an insertion of the 4-oxalomesaconate (OMA) hydratase gene (ligJ) was shown to have lost the ability to grow on both syringate and vanillate (9). These results suggested the presence of a pathway in which 3MGA is converted to OMA but not metabolized through PDC (Fig. 1).
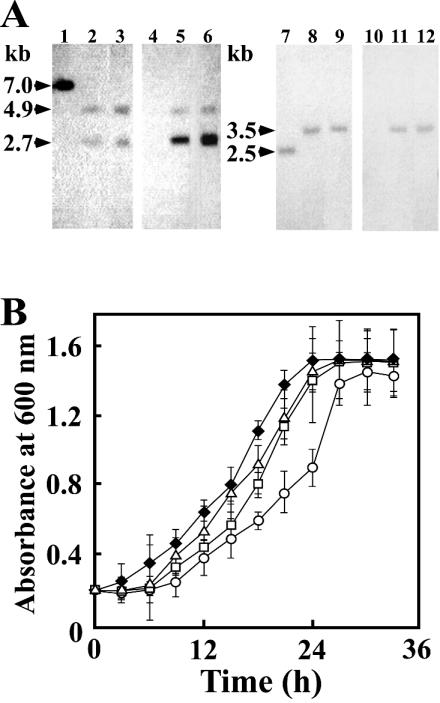
FIG. 1.
Proposed degradation pathways of syringate and vanillate by S. paucimobilis SYK-6 (A) and restriction maps of desZ (B) and ligAB (C). (A) Enzymes: DesA, syringate O-demethylase; DesZ, 3MGA 3,4-dioxygenase; LigA and LigB, small and large subunits, respectively, of PCA 4,5-dioxygenase; LigC, 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase; LigI, PDC hydrolase; LigJ, OMA hydratase; LigK, 4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetate decarboxylase. DesZ catalyzes the 3,4-cleavage of 3MGA to produce CHMOD. PDC was also produced from 3MGA catalyzed by DesZ. Both the direct production of PDC from 3MGA and the spontaneous conversion of CHMOD to PDC were suggested in this study. (B and C) Restriction maps of the 7.0-kb XhoI fragment and the 2.5-kb PvuII fragment carrying desZ and ligAB, respectively. Vertical bars above the restriction maps indicate the positions of kan and bla gene insertions in the desZ mutant (DZ), ligB mutant (DB), and desZ ligB double mutant (DBZ). Abbreviations: PCA, protocatechuate; CHMS, 4-carboxy-2-hydroxymuconate-6-semialdehyde; 3MGA, 3-O-methylgallate; PDC, 2-pyrone-4,6-dicarboxylate; CHMOD, 4-carboxy-2-hydroxy-6-methoxy-6-oxohexa-2,4-dienoate; OMA, 4-oxalomesaconate; CHA, 4-carboxy-4-hydroxy-2-oxoadipate. Abbreviations for restriction enzymes: Ba, BamHI; Bg, BglII; EI, EcoRI; EV, EcoRV; Hc, HincII; Hd, HindIII; M, MluI; N, NruI; Ps, PstI; Pv, PvuII; Sm, SmaI; Sp, SphI; Xb, XbaI; Xh, XhoI.
A specific ring cleavage dioxygenase for 3MGA has been reported only in Acinetobacter lwoffii (38) and Pseudomonas putida TMC (6), but the enzyme and gene have not yet been characterized. In the present study, we isolated and characterized the 3MGA dioxygenase gene, and the involvement of this gene in syringate degradation was examined. These studies revealed the presence of three different 3MGA degradation pathways in S. paucimobilis SYK-6.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. S. paucimobilis SYK-6 was grown on W minimal salt medium (24) containing 10 mM syringate or Luria-Bertani (LB) medium at 30°C. Pseudomonas putida PpY1100 was grown on LB medium. The SYK-6 mutants with insertions of desZ (DZ), ligB (DB), and desZ plus ligB (DBZ) were grown on LB medium containing 50 mg of kanamycin/liter, 300 mg of carbenicillin/liter, and 50 mg of kanamycin and 300 mg of carbenicillin/liter, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristica | Source or reference |
|---|---|---|
| S. paucimobilis | ||
| SYK-6 | Wild type; Nalr Smr | 11 |
| DZ | SYK-6 derivative; desZ::kan; Nalr Smr Kmr | This study |
| DB | SYK-6 derivative; ligB::bla; Nalr Smr Cbr | This study |
| DBZ | SYK-6 derivative; desZ::kan, ligB::bla; Nalr Smr Kmr Cbr | This study |
| P. putida PpY1100 | Nalr Smr | 11 |
| E. coli | ||
| HB101 | supE44 hsdS20 (rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 2 |
| MV1184 | ara Δ(lac-proAB) rspL thi (φ80 lacZΔM15) Δ(srl-recA)306::Tn10 F′ [traD36 proAB+lacIqlacZΔM15] | 41 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3); T7 RNA polymerase gene under the control of the lacUV5 promoter | 36 |
| Plasmids | ||
| pVK100 | Broad-host-range cosmid vector; Kmr Tcr | 5 |
| pUC19 | Cloning vector; Apr | 43 |
| pBluescript II KS(+) and SK(+) | Cloning vectors; Apr | 33 |
| pT7Blue | Cloning vector; Apr T7 promoter | Novagen |
| pET21a(+) | Expression vector; Apr T7 promoter | Novagen |
| pIK03 | KS(+) with a 1.3-kb EcoRV fragment carrying kan of pUC4K | 19 |
| pK19mobsacB | oriT sacB Kmr | 32 |
| pVK3-1 | pVK100 with approx 20-kb fragment carrying desZ | This study |
| pBX2F | KS(+) with a 7.0-kb XhoI fragment of pVK3-1 | This study |
| pBXSM1 | KS(+) with a 2.7-kb SmaI-XhoI fragment of pBX2F carrying desZ | This study |
| pTDZ | pT7Blue with a 1.0-kb PCR-amplified fragment carrying desZ | This study |
| pEDZA | pET21a(+) with a 1.0-kb NdeI fragment of pTDZ carrying desZ | This study |
| pBDDZ | pBXSMI with an insertion of the 1.3-kb EcoRV fragment carrying kan from pIK03 replacing the 0.8-kb HincII-NruI fragment | This study |
| pKDDZ | pK19mobsacB with a 2.8-kb XbaI-BglII fragment of pBDDZ | This study |
| pSSAB | SK(+) with a 1.5-kb XbaI-SmaI fragment carrying ligAB | This study |
| pKAB | pK19mobsacB with a 1.5-kb XbaI-SmaI fragment of pSSAB | This study |
| pAAB | pKAB with an insertion of the 1.0-kb fragment carrying bla from pUC19 into the MluI site | This study |
Abbreviations: Nalr, Smr, Kmr, Cbr, Apr, and Tcr, resistance to nalidixic acid, streptomycin, kanamycin, carbenicillin, ampicillin, and tetracycline, respectively.
Chemicals.
3MGA was synthesized from gallate methyl ester by the method of Scheline (31). Syringate, vanillate, protocatechuate, and gallate were purchased from Tokyo Kasei Kogyo Co. (Tokyo, Japan). Tetrahydrofolate was purchased from Sigma Chemical Co. (St. Louis, Mo.).
Cloning of the gene.
A partially SalI digested gene library of SYK-6 constructed with pVK100 as the vector was introduced into P. putida PpY1100 by triparental mating (5). The resulting transconjugants were grown on LB medium containing 50 mg of kanamycin/liter. When the turbidity of the culture at 600 nm reached 1.0, cells were harvested and washed with 50 mM Tris-HCl buffer (pH 7.5). Cells were resuspended in 1 ml of the same buffer. The 500-μl reaction mixture contained 495 μl of the cell suspension and 5 μl of 100 mM 3MGA (final concentration, 1 mM) and was incubated at 30°C for 20 h. The cells were removed by centrifugation (15,000 × g for 5 min), and the supernatant was filtered. The amount of 3MGA in the filtrates was analyzed by a high-pressure liquid chromatography (HPLC) system (HP1100 series LC-MSD; Agilent Technologies Co., Palo Alto, Calif.) with an ODS Hypersil C-18 column (4 by 125 mm; Agilent Technologies). The mobile phase was a mixture of water (89.5%), acetonitrile (9.5%), and acetic acid (1.0%), and the flow rate was 0.5 ml/min. Compounds were detected at 275 nm, and the retention time of 3MGA was 4.6 min.
A cosmid, pVK3-1, was obtained from a transconjugant that showed 3MGA degradation activity. The 7.0-kb XhoI fragment of pVK3-1, which conferred the 3MGA degradation activity on PpY1100 was cloned into pBluescript II KS(+) to generate pBX2F. pBXSM1 was obtained as the smallest plasmid that conferred the activity on E. coli.
DNA manipulations and nucleotide sequencing.
DNA manipulations were carried out essentially as described before (1, 28). A series of deletion derivatives of pBXSM1 were constructed with a Kilosequence kit (Takara Shuzo Co. Ltd., Kyoto, Japan). Nucleotide sequences were determined by the dideoxy termination method (29) with an ALFexpress DNA sequencer (Pharmacia Biotech, Milwaukee, Wis.). A Sanger reaction was performed with a Thermosequenase fluorescently labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). Analysis of the nucleotide sequence was performed with the GeneWorks program (IntelliGenetics, Inc., Mountain View, Calif.). Multiple sequence alignment was produced with the program Clustal W, and the phylogenetic tree was inferred from the alignments with the neighbor-joining method (27). Graphics for phylogenetic trees were produced with the TreeView program (23). The DDBJ databases were used for searching homologous proteins.
Expression of desZ in E. coli.
The coding region of desZ was amplified by PCR with Ex Taq polymerase (Takara Shuzo) with pBXSM1 as a template and the desZ-F3 primer (TGACATATGGCTGAGATCGTCC) and desZ-R3 primer (CATCAAGCTATCCTCTCACAGG). The 1.0-kb PCR product was cloned in pT7Blue and sequenced. The 1.0-kb NdeI fragment of the resulting plasmid was inserted into pET21a(+) to generate pEDZA. E. coli BL21(DE3) harboring pEDZA were grown in 5 liters of LB medium containing 100 mg of ampicillin/liter at 30°C. The expression of desZ was induced for 4 h by adding isopropyl-β-d-thiogalactopyranoside (final concentration, 1 mM) when the turbidity of the culture at 600 nm reached 0.5.
Preparation of cell extracts, protein determination, and PAGE.
Cells were harvested and resuspended in FE2 buffer consisting of 50 mM Tris-HCl buffer (pH 7.0), 10% glycerol, 0.1 mM ferrous ammonium sulfate, and 2 mM l-cysteine hydrochloride. Cells were ruptured by passage through a French pressure cell (Aminco, Urbana, Ill.), and centrifuged at 15,000 × g for 15 min. The supernatant was then used as a crude enzyme. The protein concentration was determined by the method of Bradford (3). The purity of the enzyme preparation was examined by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-PAGE).
Purification of DesZ and determination of the N-terminal amino acid sequence.
For purification of DesZ, streptomycin sulfate was added to the crude enzyme to a final concentration of 1%. The lysate was kept on ice for 10 min and centrifuged at 15,000 × g for 15 min to remove nucleic acids. The supernatant was recovered and then centrifuged again at 110,000 × g for 60 min. Enzyme purification was performed by the method described below with a BioCAD700E apparatus (PerSeptive Biosystems, Framingham, Mass.). The enzyme solution was applied to a POROS PI (polyethyleneimine) column (16 by 100 mm; PerSeptive Biosystems) previously equilibrated with FEA buffer, consisting of 50 mM Tris-HCl buffer (pH 8.0), 0.1 mM ferrous ammonium sulfate, and 2 mM l-cysteine hydrochloride containing 0.1 M NaCl. The enzyme was eluted with 402 ml of a linear gradient of 0.1 to 0.6 M NaCl. The fractions containing 3MGA dioxygenase activity, which eluted at approximately 0.39 M, were pooled, desalted, and concentrated by centrifugal filtration with a Centriplus YM-10 (Amicon, Beverly, Mass.). The resulting solution was applied to a POROS HQ (quaternized polyethyleneimine) column (4.6 by 100 mm; PerSeptive Biosystems) equilibrated with FEA buffer containing 0.2 M NaCl. The enzyme was eluted with 50 ml of a linear gradient of 0.2 to 0.7 M NaCl. The fractions containing 3MGA dioxygenase activity, which eluted at approximately 0.37 M, were pooled, desalted, and concentrated as described above.
To determine the N-terminal amino acid sequence of DesZ, purified DesZ was separated by SDS-12% PAGE and electroblotted onto a polyvinylidene difluoride membrane (Bio-Rad, Hercules, Calif.). The area on the membrane containing DesZ was cut out and analyzed on a PPSQ-21 protein sequencer (Shimadzu, Kyoto, Japan).
Identification of the reaction product.
The 1-ml assay mixture contained 50 mM Tris-HCl buffer (pH 7.0), 2 mM 3MGA, and the purified DesZ (100 μg of protein). Reactions were carried out at 30°C and stopped by the addition of methanol (final concentration, 25%). Precipitated protein was removed by centrifugation (15,000 × g for 10 min), and the supernatant was diluted 1:10, acidified, and extracted with ethyl acetate. The extract was trimethylsilylated (TMS), and then the derivatives were analyzed by gas chromatography-mass spectrometry (GC-MS). The substrate and reaction products were identified by GC-MS with a model 5971A with an Ultra-2 capillary column (50 m by 0.2 mm; Agilent Technologies). The column temperature was increased initially from 100 to 150°C and then from 150 to 300°C at rates of 20 and 3°C/min, respectively. The temperatures of injection and detection were 220 and 300°C, respectively. The mobile phase was a helium gas, and the flow rate was 1.0 ml/min.
Enzyme assay.
The dioxygenase activity of DesZ was assayed by measuring the substrate-dependent oxygen consumption rate. The 2-ml assay mixture contained GTA buffer consisting of 50 mM 3,3-dimethylglutarate, 50 mM Tris, and 50 mM 2-amino-2-methyl-1,3-propanediol (pH 7.0), purified DesZ (100 μg of protein), and 1 mM 3MGA, PCA, or gallate as a substrate. The reaction was carried out at 30°C, and the oxygen consumption rate was measured with an oxygen electrode (B-505; Iijima Electronics Manufacturing Co., Ltd., Aichi, Japan). One unit of enzyme activity was defined as the amount that consumed 1 μmol of O2 per min at 30°C. Specific activity was expressed as units per milligram of protein. Km and Vmax values were obtained from Hanes-Woolf plots and expressed as means of at least three independent experiments. For kinetic analysis of DesZ, the concentration of 3MGA was changed from 0.01 to 10 mM.
Construction of insertion mutants of S. paucimobilis SYK-6.
The 0.8-kb HincII-NruI fragment in the desZ gene of pBXSM1 was replaced with the 1.3-kb EcoRV fragment carrying the kanamycin resistance gene (kan) from pIK03. The 2.8-kb XbaI-BglII fragment of the resultant plasmid, pBDDZ, containing the inactivated desZ gene, was inserted into pK19mobsacB to generate pKDDZ. The 1.5-kb XbaI-SmaI fragment carrying ligAB of pSSAB was cloned into pK19mobsacB to generate pKAB. This plasmid was then digested with MluI in ligB, blunt ended with KOD DNA polymerase (Toyobo Co. Ltd., Osaka, Japan), and ligated with a blunt-ended 1.0-kb BspHI fragment containing the ampicillin resistance gene (bla) from pUC19. The resulting plasmid was designated pAAB.
pKDDZ and pAAB were introduced into SYK-6 cells by electroporation, and candidates for mutants were isolated as described previously (20). pKDDZ was further introduced into cells of the ligB mutant obtained (strain DB), and candidates for desZ ligB double mutants were isolated. To examine the disruption of each gene, Southern hybridization analysis was performed. Total DNAs of the candidates for desZ, ligB, and desZ ligB mutants were digested with XhoI, PvuII, or XhoI and PvuII, respectively. The 1.2-kb SmaI-PvuII fragment carrying desZ, the 1.5-kb XbaI-SmaI fragment carrying ligB, the 1.3-kb EcoRV fragment carrying kan, and the 1.0-kb BspHI fragment carrying bla were labeled with the DIG system (Roche Diagnostics, Indianapolis, Ind.) and used as probes.
Analysis of insertion mutants.
Degradation of 3MGA and gallate by SYK-6 and its mutants was assayed in a 2-ml mixture containing FE buffer, consisting of 50 mM Tris-HCl buffer (pH 7.0), 10% acetone, 10% glycerol, 1 mM FeSO4, 2 mM sodium ascorbate, 1 mM substrate, and cell extract. The assay mixtures for 3MGA and gallate contained 20 and 5 mg of protein, respectively. Reactions were performed at 30°C. A portion of the reaction mixture taken at various sampling points was diluted 1:10, acidified, and extracted with ethyl acetate. The extract was trimethylsilylated and then analyzed by GC-MS as described above.
Degradation of 3MGA in the presence of tetrahydrofolate was assayed in a 2-ml assay mixture containing 100 mM Tris-HCl (pH 8.0), 1 mM 3MGA, 1 mM tetrahydrofolate, and cell extracts of SYK-6 and the desZ ligB mutant (10 mg of protein). Reactions were carried out under anaerobic conditions at 30°C in an anaerobic box (Hirasawa Works Inc., Tokyo, Japan) that contained an atmosphere of 95% N2 and 5% H2. The reaction mixture was analyzed by GC-MS as described above.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper was deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession number AB110976.
RESULTS AND DISCUSSION
Cloning and nucleotide sequence of the 3MGA dioxygenase gene.
A gene library of SYK-6 constructed with the cosmid vector pVK100 was introduced into P. putida PpY1100, which was not able to degrade 3MGA. One thousand transconjugants were screened for 3MGA degradation activity by HPLC analysis, and two transconjugants that showed 3MGA degradation activity were found. One of the transconjugants harboring pVK3-1 degraded 3MGA within 20 h but showed no ability to degrade PCA. Because PCA 4,5-dioxygenase (LigAB) is known to catalyze the ring cleavage of PCA and 3MGA to generate 4-carboxy-2-hydroxymuconate-6-semialdehyde and PDC, respectively (17), the gene(s) included in pVK3-1 seemed to be different from ligAB.
A subcloning experiment with pVK3-1 revealed that pBX2F containing the 7.0-kb XhoI fragment conferred 3MGA degradation activity on E. coli MV1184. Further subcloning indicated that the 2.7-kb SmaI-XhoI fragment was necessary for the activity. The nucleotide sequence of the 2.7-kb SmaI-XhoI fragment was determined, and three open reading frames (ORFs) of 990, 660, and 651 bp were found. ORF1 encodes 330 amino acid residues with a molecular mass of 36,489 Da, and its deduced amino acid sequence showed 43% identity with 2,2′,3-trihydroxy-3′-methoxy-5,5′-dicarboxybiphenyl oxygenase (LigZ) of SYK-6, which is involved in lignin-related biphenyl degradation (24), and 17 to 21% identity with the β subunit of PCA 4,5-dioxygenase of SYK-6 (22), Sphingomonas sp. strain LB126 (42), and Comamonas testosteroni BR6020 (26). This result suggested that ORF1 encodes an extradiol dioxygenase for 3MGA, and this gene was designated desZ.
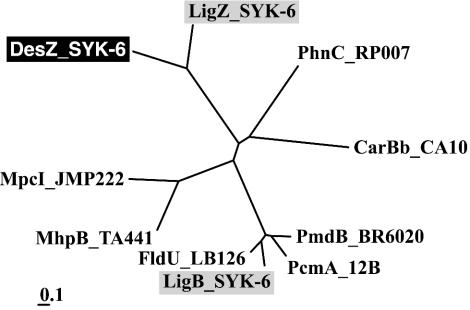
Extradiol dioxygenases are classified as type I or II on the basis of amino acid sequence similarity (8). Type I extradiol dioxygenases include a number of 2,3-dihydroxybiphenyl 1,2-dioxygenases and catechol 2,3-dioxygenases, and type II dioxygenases include the β subunit of PCA 4,5-dioxygenase (LigB) (22), LigZ (24), extradiol dioxygenase (PhnC) of Burkholderia sp. strain RP007 (13), and the β subunit of 2′-aminobiphenyl-2,3-diol 1,2-dioxygenase (CarBb) of Pseudomonas resinovorans CA10 (30). The phylogenetic tree of extradiol dioxygenases indicated that DesZ belongs to the family of type II extradiol dioxygenases (Fig. 2).
FIG. 2.
Phylogenetic tree of DesZ with type II extradiol dioxygenases. The scale corresponds to a genetic distance of 0.1 substitution per position (10% difference). Enzymes: MpcI_JMP222, catechol 2,3-dioxygenase I of Ralstonia eutropha JMP222 (S10154); MhpB_TA441, 3-(2,3-dihydroxyphenyl)propionate 1,2-dioxygenase of Comamonas testosteroni TA441 (BAA82879); CarBb_CA10, catalytic subunit of 2′-aminobiphenyl-2,3-diol 1,2-dioxygenase of Pseudomonas resinovorans CA10 (BAA21731); PhnC_RP007, extradiol dioxygenase of Burkholderia sp. strain RP007 (AAD09870); PmdB_BR6020, β subunit of PCA 4,5-dioxygenase of C. testosteroni BR6020 (AAM09637); LigB_SYK-6, β subunit of PCA 4,5-dioxygenase of SYK-6 (BAA97118); PcmA_12B, PCA 4,5-dioxygenase of Arthrobacter keyseri 12B (AAK16524); FldU_LB126, β subunit of putative PCA dioxygenase of Sphingomonas sp. strain LB126 (CAB87561); LigZ_SYK-6, OH-DDVA oxygenase of SYK-6 (BAA75884).
The crystallographic study revealed that the active site of LigB contains the Fe ion coordinated by His12, His61, and Glu242, and His195 is thought to act as the catalytic base (37). These amino acids were found to be conserved among almost all the type II enzymes. The residues His12, Glu287, and His250 (DesZ numbering), corresponding to His12, Glu242, and His195, respectively, of LigB, were well conserved in DesZ and LigZ, but the residue corresponding to His61 was not. However, His35 and His184 (DesZ numbering) were conserved between DesZ and LigZ. These residues might be involved in the coordination of the Fe ion.
Some type II dioxygenases, of which LigAB is one, contain two subunits, while others contain only a single subunit. Sugimoto et al. demonstrated that the α subunit of LigAB forms a lid that closes the open end of the binding pocket for PCA (37). In the case of the single-subunit-type enzymes, the insertion of ca. 40 amino acid residues is usually found (37). Such an insertion is thought to be folded and situated on top of the substrate-binding pocket to mimic the role of the α subunit of LigAB. On the other hand, the PCA 4,5-dioxygenase (PcmA) of Arthrobacter keyseri 12B is the enzyme corresponding to the α and β subunits of LigAB, which are joined to form a single polypeptide (7). DesZ might contain the region, which could assume the function of the α subunit of LigAB. Further research will be needed to determine whether such a region exists in DesZ.
The deduced amino acid sequences of ORF2 and ORF3 were similar to each other (34% identity) and showed 28 and 25% identity, respectively, with the putative 2-demethylmenaquinone 2-C-methyltransferase (MenG), involved in menaquinone synthesis in Methanococcus jannaschii (4). However, their actual functions remain to be determined.
Expression and purification of DesZ.
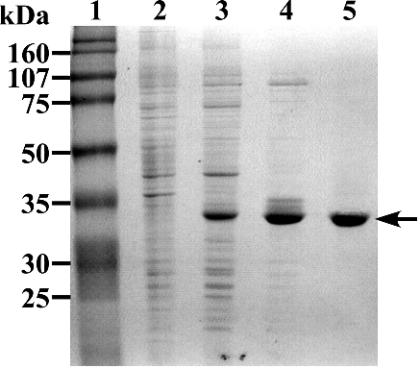
The desZ gene was expressed in E. coli BL21(DE3) harboring pEDZA under the control of the T7 promoter. Production of the 34-kDa protein was observed by SDS-PAGE (Fig. 3, lane 3). The size of this product is close to the value calculated from the deduced amino acid sequence of desZ (Mr 36,489).
FIG. 3.
SDS-PAGE analysis of protein fractions. Proteins were separated on an SDS-12% polyacrylamide gel and stained with Coomassie brilliant blue. Lanes; 1, molecular size markers; 2, crude extract of E. coli BL21(DE3) harboring pET21a(+) (10 μg of protein); 3, crude extract of E. coli BL21(DE3) harboring pEDZA (10 μg of protein); 4, polyethyleneimine (PI) fraction (5 μg of protein); 5, quaternized PI fraction (3 μg of protein). Molecular masses are given on the left.
In order to characterize the enzyme properties of the gene product of desZ, DesZ was purified to near homogeneity by a combination of column chromatography procedures with polyethyleneimine (PI) and quaternized PI (Table 2 and Fig. 3). However, the specific activity of the final preparation was almost the same as that of the PI fraction. DesZ seemed to be partially inactivated despite the fact that the purification was done in the presence of Fe2+ and cysteine. The N-terminal amino acid sequence of DesZ was determined, and the first 10 residues (AEIVLGIGTS) corresponded to the deduced amino acid sequence of desZ, with the exception of the first methionine.
TABLE 2.
Purification of DesZ from E. coli BL21(DE3) harboring pEDZA
| Fraction | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 158 | 250 | 1.58 | 100 | 1.0 |
| Polyethyleneimine (PI) | 13.0 | 44.0 | 3.38 | 18 | 2.1 |
| Quaternized PI | 4.50 | 15.8 | 3.51 | 6.3 | 2.2 |
Properties of DesZ.
The optimal temperature and pH for the dioxygenase activity of DesZ toward 3MGA were determined to be 30°C and 7.0, respectively. The Km and Vmax values for 3MGA were determined to be 210 ± 24 μM and 3.6 ± 0.2 U/mg, respectively. This Km was higher than those of the PCA 4,5-dioxygenase of Comamonas testosteroni for 3MGA (125 μM) and PCA (46 μM) (44). It is possible that the natural substrate of DesZ has not been identified.
The specific activities of DesZ toward 3MGA, PCA, and gallate were determined. DesZ showed relatively high dioxygenase activity for 3MGA (3.5 U/mg) and gallate (1.3 U/mg), but approximately 10 times lower activity than that for 3MGA was detected when PCA was used as a substrate. The kinetic values of DesZ for PCA could not be determined because of the low activity of DesZ toward PCA.
To examine the metal dependency of DesZ, 500 μM EDTA was added to the purified DesZ, and it was kept on ice for 20 h. No oxygen consumption activity toward 3MGA was detected in the reaction mixture containing the EDTA-treated enzyme, suggesting the requirement of a divalent cation by DesZ. The metal ions Fe2+, Fe3+, Co2+, Cu2+, Mg2+, Mn2+, and Zn2+ were added to the reaction mixture to a final concentration of 1 mM, and the resulting solutions were kept on ice for 1 h. The oxygen consumption activity toward 3MGA was recovered to 114% of the activity obtained with the purified DesZ only when Fe2+ was added to the reaction mixture. These results suggested that DesZ requires Fe2+ for its activity.
Identification of the reaction product.
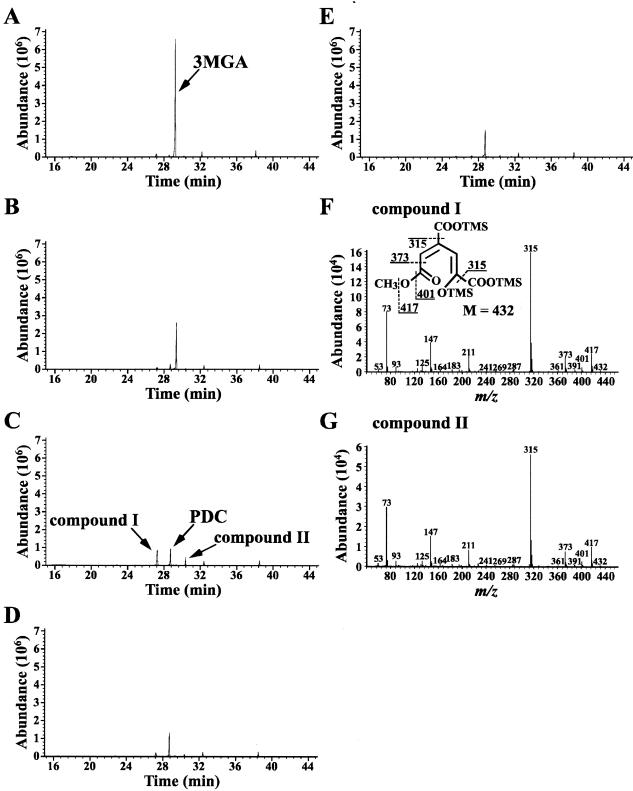
The reaction mixture containing 3MGA and purified DesZ was analyzed by GC-MS (Fig. 4). When the reaction mixture was analyzed immediately after the start of the reaction, the TMS derivative of 3MGA was detected with a retention time of 29.4 min (Fig. 4A). The abundance of this peak decreased significantly at 1 min, and the increase in the amount of peaks with retention times of 27.2 (compound I), 28.7, and 30.3 (compound II) min were observed (Fig. 4C). The mass spectra of compounds I and II were almost identical, suggesting that these are stereoisomers (Fig. 4F and G). The weight of the molecular ions, m/z 432 of compounds I and II, corresponded to the molecular weight of the TMS derivative of a ring cleavage form of 3MGA in which a molecular oxygen was incorporated (Fig. 4F). The major fragments at m/z 417, 401, 373, and 315 seemed to correspond to M-CH3, M-OCH3, M-COOCH3, and M-COOTMS, respectively (Fig. 4F). Generation of the fragment corresponding to M-COOCH3 strongly suggested that compounds I and II are 4-carboxy-2-hydroxy-6-methoxy-6-oxohexa-2,4-dienoate (CHMOD) produced by the cleavage of the C-3 and C-4 positions of 3MGA (Fig. 4F).
FIG. 4.
Identification of the reaction product from 3MGA catalyzed by DesZ. Panels A to E show gas chromatograms of TMS derivatives of the reaction products at the start and after 0.5, 1, 60, and 180 min of incubation, respectively. (F and G) Mass spectra of the TMS derivatives of compounds I and II, respectively.
GC-MS analysis revealed that the peak with a retention time of 28.7 min (Fig. 4B to E) corresponded to the TMS derivative of PDC. It was shown that PCA 4,5-dioxygenase catalyzes the direct conversion of 3MGA to PDC (12); DesZ, however, gave compounds I and II together with PDC as the reaction products from 3MGA. Until now, 3MGA dioxygenase activity has been detected only in a cell suspension of a 4-hydroxy-3-methoxymandelate degrader, Acinetobacter lwoffii (38). The 3,4,5-trimethoxycinnamate degrader Pseudomonas putida TMC has also been suggested to have this enzyme activity (6). In the case of A. lwoffii, CHMOD has been identified as a reaction product from 3MGA, and CHMOD has been shown to undergo spontaneous cyclization to PDC with the attack of an enolate oxygen on the ester carbonyl group and the release of methanol (38).
As shown in Fig. 4, 3MGA was completely degraded in the reaction mixture at 1 min of incubation with purified DesZ. The increase in the amount of PDC and the decrease in compounds I and II in the reaction mixture at 60 min of incubation suggested that PDC was produced from compounds I and II by a spontaneous reaction, similar to the case of A. lwoffii. However, the production of PDC was observed in the reaction mixture immediately after the start of the reaction (Fig. 4A) despite the fact that the half-life of CHMOD was estimated to be approximately 70 min (38). Therefore, PDC and CHMOD seem to have been directly produced from 3MGA by the reaction catalyzed by DesZ. It seems likely that CHMOD is transformed to 4-oxalomesaconate (OMA) by an unidentified hydrolase in SYK-6, as suggested in the case of P. putida TMC (6).
Disruption of desZ and ligB in S. paucimobilis SYK-6.
To investigate the roles of desZ and ligAB in syringate catabolism by SYK-6, ligB in SYK-6 was initially inactivated by the gene replacement technique with the ligB disruption plasmid pAAB, which was constructed by inserting the bla gene within ligB in pK19mobsacB. desZ in SYK-6 and the ligB mutant was also inactivated with the desZ disruption plasmid pKDDZ, which was constructed by replacing an internal segment of desZ in pK19mobsacB with the kan gene. These insertion mutations were confirmed by Southern hybridization analysis with the ligB, bla, desZ, and kan genes as probes (Fig. 1 and 5A). When the desZ and ligB mutants, strains DZ and DB, respectively, were grown in syringate, their growth rates were slightly decreased compared with that of SYK-6 (k = 0.12/h) (Fig. 5B). In the case of the desZ ligB double mutant (DBZ), the growth rate on syringate was decreased from 0.12 to 0.07/h. These results suggest that both desZ and ligAB are indeed involved in syringate degradation but are not essential to the growth of SYK-6 on syringate.
FIG. 5.
Disruption of desZ and ligB in SYK-6. (A) Southern blot analysis of the insertion mutants. Lanes: 1 and 4, total DNA of SYK-6 digested with XhoI; 2 and 5, total DNA of the desZ mutant (DZ) digested with XhoI; 3 and 6, total DNA of the desZ ligB double mutant (DBZ) digested with XhoI; 7 and 10, total DNA of SYK-6 digested with PvuII; 8 and 11, total DNA of the ligB mutant (DB) digested with PvuII; 9 and 12, total DNA of DBZ digested with PvuII. The 1.2-kb SmaI-PvuII fragment carrying desZ (lanes 1 to 3), the 1.3-kb EcoRV fragment carrying kan (lanes 4 to 6), the 1.5-kb XbaI-SmaI fragment carrying ligB (lanes 7 to 9), and the 1.0-kb BspHI fragment carrying bla (lanes 10 to 12) were used as probes. (B) Growth on syringate of SYK-6 (solid diamonds), DB (open triangles), DZ (open squares), and DBZ (open circles). These strains were grown in 10 ml of W medium containing 10 mM syringate. Each value is the average ± standard deviation of three independent experiments.
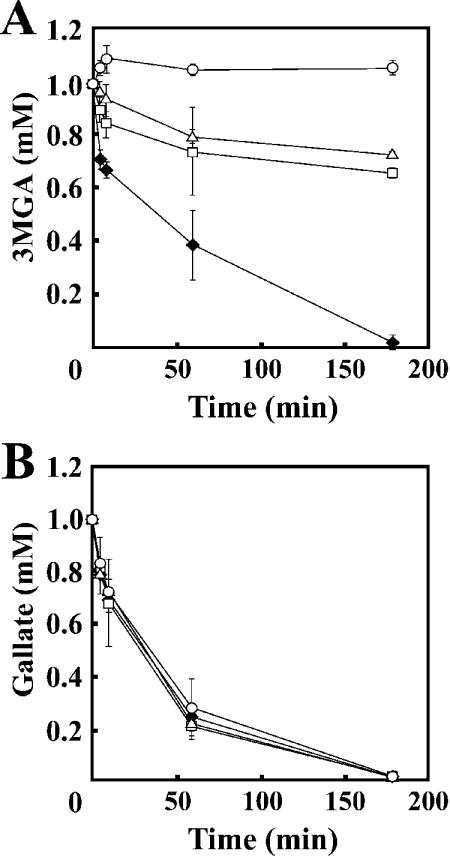
To estimate the level of participation of desZ and ligB in 3MGA degradation, the 3MGA degradation activities of cell extracts of DZ, DB, and DBZ grown on syringate were determined by GC-MS (Fig. 6A). Cell extracts of DZ and DB degraded only 30% of 3MGA over 3 h of incubation, while the cell extract of SYK-6 degraded all the 3MGA. Furthermore, DBZ completely lost its transformation activity toward 3MGA under the assay conditions used. Accordingly, it can be concluded that only DesZ and LigAB are fundamentally engaged in the ring cleavage of 3MGA. However, DBZ was able to grow on syringate, raising the possibility that a cofactor-dependent enzyme is involved in 3MGA degradation.
FIG. 6.
Degradation of 3MGA and gallate by insertion mutants. One millimolar 3MGA (A) and gallate (B) was incubated with the cell extracts (20 and 5 mg of protein for 3MGA and gallate degradation, respectively) of SYK-6 (solid diamonds), DB (open triangles), DZ (open squares), and DBZ (open circles). The degradation activities of the cell extracts toward these substrates were determined by GC-MS analysis. Each value is the average ± standard deviation of three independent experiments.
Identification of the alternative degradation pathway of 3MGA.
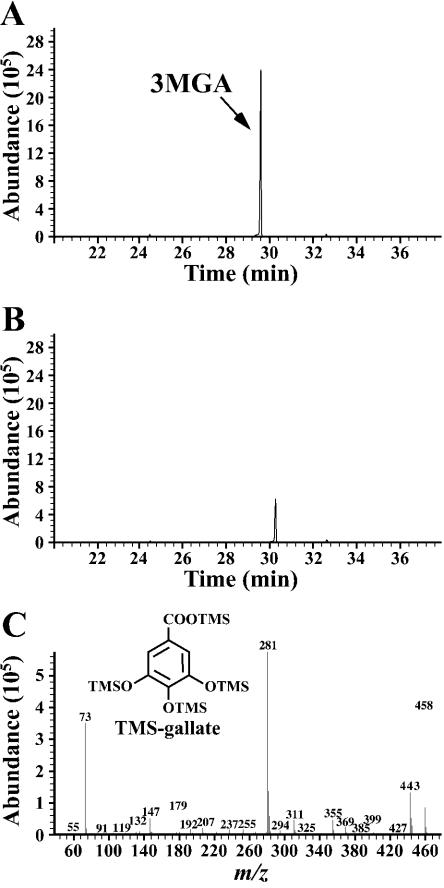
Recently, we identified and characterized the desA gene, which encodes the tetrahydrofolate-dependent O-demethylase for syringate (19). Because desA is essential for syringate degradation, it is apparent that syringate is degraded via 3MGA. We hypothesized that 3MGA is degraded to gallate by an unidentified O-demethylase that requires the presence of tetrahydrofolate in the cell extract of DBZ. To test this hypothesis, 3MGA was incubated with the cell extract of DBZ grown on syringate in the presence of tetrahydrofolate. This reaction was performed under anaerobic conditions to confirm the accumulation of gallate, as gallate degradation is thought to be catalyzed by a dioxygenase. GC-MS analysis of the reaction mixture incubated with the cell extract indicated complete transformation of 3MGA and significant accumulation of gallate (Fig. 7). The specific activity of DesA toward 3MGA was only 0.4% of that toward syringate; a distinct tetrahydrofolate-dependent O-demethylase therefore seems to be involved in the conversion of 3MGA to gallate.
FIG. 7.
Tetrahydrofolate-dependent transformation of 3MGA to gallate. 3MGA (1 mM) was incubated with the cell extract of DBZ (10 mg of protein) in the presence of 1 mM tetrahydrofolate. (A and B) Gas chromatograms of TMS derivatives of the reaction products at the start and after 3 h of incubation, respectively. (C) Mass spectrum of the compound with a retention time at 30.2 min in panel B.
The gallate degradation activities of the cell extracts of SYK-6, DB, DZ, and DBZ grown on syringate were examined. GC-MS analysis revealed that all these cell extracts completely degraded gallate within 3 h (Fig. 6B), indicating that disruption of both desZ and ligB did not affect the gallate degradation activity. Sparnins and Dagley reported that P. putida degrades gallate to oxaloacetate and pyruvate via OMA (35). They proposed that the ring cleavage dioxygenase is involved in the transformation of gallate to OMA. Similarly, SYK-6 seemed to contain gallate dioxygenase. The degradation activity of SYK-6 toward gallate measured at 1 min was 448% (153 mU/mg of protein) of the activity toward 3MGA (34.2 mU/mg of protein). This fact suggests that gallate dioxygenase plays a crucial role in syringate degradation.
In conclusion, we found three possible pathways for degradation of 3MGA in SYK-6, as follows (Fig. 1): (i) conversion of 3MGA to PDC by the reactions catalyzed by LigAB and DesZ; (ii) conversion of 3MGA to OMA by the reactions catalyzed by DesZ and a putative hydrolase; and (iii) conversion of 3MGA to OMA via gallate by the reactions catalyzed by 3MGA O-demethylase and gallate dioxygenase. However, the conversion of 3MGA to PDC by DesZ requires further experimental demonstration. Based on comparison of the growth rate between the desZ ligB double mutant and the wild type (Fig. 5B), the gallate degradation pathway seemed to be a main route of syringate degradation by SYK-6. The specific activity of LigAB toward 3MGA was only ca. 4% of that toward PCA (data not shown). In order to reinforce the ability to degrade syringate, these alternative degradation pathways might have been recruited in this strain. Isolation and characterization of the genes for the third pathway will enable us to estimate the actual contribution of each pathway to 3MGA degradation.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Encouragement of Young Scientists 13760062 from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Bolivar, F., and K. Backman. 1979. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 68:245-267. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 5.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnelly, M. I., and S. Dagley. 1981. Bacterial degradation of 3,4,5-trimethoxycinnamic acid with production of methanol. J. Bacteriol. 147:471-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton, R. W. 2001. Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J. Bacteriol. 183:3689-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eltis, L. D., and J. T. Bolin. 1996. Evolutionary relationships among extradiol dioxygenases. J. Bacteriol. 178:5930-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara, H., E. Masai, Y. Katayama, and M. Fukuda. 2000. The 4-oxalomesaconate hydratase gene, involved in the protocatechuate 4,5-cleavage pathway, is essential to vanillate and syringate degradation in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6950-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higuchi, T. 1971. Formation and biological degradation of lignins. Adv. Enzymol. Related Areas Mol. Biol. 34:207-283. [DOI] [PubMed] [Google Scholar]
- 11.Katayama, Y., S. Nishikawa, M. Nakamura, K. Yano, M. Yamasaki, N. Morohoshi, and T. Haraguchi. 1987. Cloning and expression of Pseudomonas paucimobilis SYK-6 genes involved in the degradation of vanillate and protocatechuate in P. putida. Mokuzai Gakkaishi 33:77-79. [Google Scholar]
- 12.Kersten, P. J., S. Dagley, J. W. Whittaker, D. M. Arciero, and J. D. Lipscomb. 1982. 2-Pyrone-4,6-dicarboxylic acid, a catabolite of gallic acids in Pseudomonas species. J. Bacteriol. 152:1154-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laurie, A. D., and G. Lloyd-Jones. 1999. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J. Bacteriol. 181:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masai, E., A. Ichimura, Y. Sato, K. Miyauchi, Y. Katayama, and M. Fukuda. 2003. Roles of the enantioselective glutathione S-transferases in cleavage of β-aryl ether. J. Bacteriol. 185:1768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masai, E., Y. Katayama, S. Kawai, S. Nishikawa, M. Yamasaki, and N. Morohoshi. 1991. Cloning and sequencing of the gene for a Pseudomonas paucimobilis enzyme that cleaves β-aryl ether. J. Bacteriol. 173:7950-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masai, E., Y. Katayama, S. Kubota, S. Kawai, M. Yamasaki, and N. Morohoshi. 1993. A bacterial enzyme degrading the model lignin compound β-etherase is a member of the glutathione-S-transferase superfamily. FEBS Lett. 323:135-140. [DOI] [PubMed] [Google Scholar]
- 17.Masai, E., Y. Katayama, S. Nishikawa, and M. Fukuda. 1999. Characterization of Sphingomonas paucimobilis SYK-6 genes involved in degradation of lignin-related compounds. J. Ind. Microbiol. Biotechnol. 23:364-373. [DOI] [PubMed] [Google Scholar]
- 18.Masai, E., K. Momose, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 2000. Genetic and biochemical characterization of 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase and its role in the protocatechuate 4,5-cleavage pathway in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 182:6651-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masai, E., M. Sasaki, Y. Minakawa, T. Abe, T. Sonoki, K. Miyauchi, Y. Katayama, and M. Fukuda. 2004. A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J. Bacteriol. 186:2757-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masai, E., S. Shinohara, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 1999. Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J. Bacteriol. 181:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishikawa, S., T. Sonoki, T. Kasahara, T. Obi, S. Kubota, S. Kawai, N. Morohoshi, and Y. Katayama. 1998. Cloning and sequencing of the Sphingomonas (Pseudomonas) paucimobilis gene essential for the O demethylation of vanillate and syringate. Appl. Environ. Microbiol. 64:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noda, Y., S. Nishikawa, K. Shiozuka, H. Kadokura, H. Nakajima, K. Yoda, Y. Katayama, N. Morohoshi, T. Haraguchi, and M. Yamasaki. 1990. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J. Bacteriol. 172:2704-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Page, R. D. M. 1996. Treeview: an application to display phylogenetic trees on personal computers. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 24.Peng, X., T. Egashira, K. Hanashiro, E. Masai, S. Nishikawa, Y. Katayama, K. Kimbara, and M. Fukuda. 1998. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl. Environ. Microbiol. 64:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng, X., E. Masai, Y. Katayama, and M. Fukuda. 1999. Characterization of the meta-cleavage compound hydrolase gene involved in degradation of the lignin-related biphenyl structure by Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 65:2789-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Providenti, M. A., J. Mampel, S. MacSween, A. M. Cook, and R. C. Wyndham. 2001. Comamonas testosteroni BR6020 possesses a single genetic locus for extradiol cleavage of protocatechuate. Microbiology 147:2157-2167. [DOI] [PubMed] [Google Scholar]
- 27.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato, S., N. Ouchiyama, T. Kimura, H. Nojiri, H. Yamane, and T. Omori. 1997. Cloning of genes involved in carbazole degradation of Pseudomonas sp. strain CA10: nucleotide sequences of genes and characterization of meta-cleavage enzymes and hydrolase. J. Bacteriol. 179:4841-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheline, R. R. 1966. Synthesis of 3-O-methylgallic acid. Acta Chem. Scand. 20:1182. [Google Scholar]
- 32.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 33.Short, J. M., J. M. Fernandez, J. A. Sorge, and W. D. Huse. 1988. λ ZAP: a bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res. 16:7583-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonoki, T., T. Obi, S. Kubota, M. Higashi, E. Masai, and Y. Katayama. 2000. Coexistence of two different O demethylation systems in lignin metabolism by Sphingomonas paucimobilis SYK-6: cloning and sequencing of the lignin biphenyl-specific O-demethylase (LigX) gene. Appl. Environ. Microbiol. 66:2125-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sparnins, V. L., and S. Dagley. 1975. Alternative routes of aromatic catabolism in Pseudomonas acidovorans and Pseudomonas putida: gallic acid as a substrate and inhibitor of dioxygenases. J. Bacteriol. 124:1374-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 37.Sugimoto, K., T. Senda, H. Aoshima, E. Masai, M. Fukuda, and Y. Mitsui. 1999. Crystal structure of an aromatic ring opening dioxygenase LigAB, a protocatechuate 4,5-dioxygenase, under aerobic conditions. Structure Fold. Des. 7:953-965. [DOI] [PubMed] [Google Scholar]
- 38.Sze, I. S., and S. Dagley. 1987. Degradation of substituted mandelic acids by meta fission reactions. J. Bacteriol. 169:3833-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ten Have, R., and P. J. M. Teunissen. 2001. Oxidative mechanisms involved in lignin degradation by white-rot fungi. Chem. Rev. 101:3397-3413. [DOI] [PubMed] [Google Scholar]
- 40.Vicuña, R. 1988. Bacterial degradation of lignin. Enzyme Microbiol. Technol. 10:646-655. [Google Scholar]
- 41.Vieira, J., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]
- 42.Wattiau, P., L. Bastiaens, R. van Herwijnen, L. Daal, J. R. Parsons, M. E. Renard, D. Springael, and G. R. Cornelis. 2001. Fluorene degradation by Sphingomonas sp. LB126 proceeds through protocatechuic acid: a genetic analysis. Res. Microbiol. 152:861-872. [DOI] [PubMed] [Google Scholar]
- 43.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 44.Zabinski, R., E. Münck, P. M. Champion, and J. M. Wood. 1972. Kinetic and Mössbauer studies on the mechanism of protocatechuic acid 4,5-oxygenase. Biochemistry 11:3212-3219. [DOI] [PubMed] [Google Scholar]