Abstract
The symporter YhcL and two ATP binding cassette transporters, YtmJKLMN and YckKJI, were shown to mediate l-cystine uptake in Bacillus subtilis. A triple ΔyhcL ΔytmJKLMN ΔyckK mutant was unable to grow in the presence of l-cystine and to take up l-cystine. We propose that yhcL, ytmJKLMN, and yckKJI should be renamed tcyP, tcyJKLMN, and tcyABC, respectively. The l-cystine uptake by YhcL (Km = 0.6 μM) was strongly inhibited by seleno-dl-cystine, while the transport due to the YtmJKLMN system (Km = 2.5 μM) also drastically decreased in the presence of dl-cystathionine, l-djenkolic acid, or S-methyl-l-cysteine. Accordingly, a ΔytmJKLMN mutant did not grow in the presence of 100 μM dl-cystathionine, 100 μM l-djenkolic acid, or 100 μM S-methyl-l-cysteine. The expression of the ytmI operon and the yhcL gene was regulated in response to sulfur availability, while the level of expression of the yckK gene remained low under all the conditions tested.
Eukaryotic, bacterial, and archaeal cells contain a large number of integral membrane proteins and protein complexes involved in solute transport across the membrane. In Escherichia coli and Bacillus subtilis as many as 285 and 239 genes encode membrane transport proteins, respectively (25, 30), and 14% (E. coli) and 20% (B. subtilis) of these proteins are proposed to participate in amino acid transport (25). However, identification of permease function is often complex due to the existence of paralogues with similar or different substrate specificities and due to functional redundancy (14). Complete genome sequences and expression profiling experiments provide a powerful tool for analyzing global transcriptional patterns and for identifying gene function. DNA arrays have been used to investigate the changes in gene transcript levels during growth of the B. subtilis wild-type strain in the presence of sulfate or methionine as a sole sulfur source (2). Many sulfur-regulated genes encode transporters that are good candidates for the uptake of sulfur-containing compounds (2). B. subtilis can use methionine, homocysteine, cystathionine, cystine, sulfate, sulfite, thiosulfate, and sulfonates as sole sulfur sources, indicating that these sulfur compounds are efficiently taken up. A sulfate permease (CysP), a sulfonate ATP binding cassette (ABC) transporter (SsuABC), and a methionine ABC transporter (MetNPQ) have been experimentally characterized (15, 21, 43). In contrast, little is known about the transport of cysteine and its oxidized form cystine in this bacterium. Cysteine uptake has been investigated mainly in Saccharomyces cerevisiae and in enterobacteria (9, 17). In yeast, seven different permeases, which also transport other amino acids, participate in cysteine uptake. It therefore appears that cysteine is not taken up by one specific permease but rather is taken up by multiple transporters with broad specificity (9). In Salmonella enterica serovar Typhimurium, l-cystine is taken up by three different systems, CTS-1 (Km = 2 μM), CTS-2 (Km = 0.1 μM), and CTS-3 (nonsaturable) (3). In E. coli, two kinetically identifiable cystine transport systems are present; one is shared with diaminopimelic acid and several cystine analogues, and the other is more specific (4). The first system, which is sensitive to osmotic shock, corresponds to an ABC transporter. The bacterial ABC importers include one or two ATP binding proteins localized to the inner side of the cytoplasmic membrane, one or two transmembrane proteins, and a high-affinity solute binding protein external to the cytoplasmic membrane (5, 13, 39). The periplasmic cystine binding protein from E. coli has been characterized and was identified as FliY (6, 24). Its synthesis was increased during sulfate starvation (27). The fliY gene forms an operon with fliA encoding σF, which participates in transcription of class III genes involved in flagellar synthesis (24). FliY is not required for motility, and its possible role in flagellar synthesis remains to be established. The YecS and YecC proteins probably correspond to the E. coli permease and ATP binding protein of a cystine ABC transporter, but this has never been substantiated by experimental data (8, 14). The yecS and yecC genes are separated from fliY only by the yedO gene encoding a d-cysteine desulfhydrase. It has been proposed that these genes could form an operon (36, 41). In gram-positive bacteria, the solute binding protein BspA has been shown to be required for l-cystine uptake in Lactobacillus fermentum (42). In this work, we tested the possible involvement in l-cystine transport of different permeases whose synthesis was increased in the presence of methionine (2). We characterized the three transporters involved in l-cystine uptake in B. subtilis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The B. subtilis strains used in this work are listed in Table 1. E. coli cells were grown in Luria-Bertani broth (31). B. subtilis was grown in SP medium or in minimal medium (6 mM K2HPO4, 4.4 mM KH2PO4, 0.3 mM trisodium citrate, 4 mM MgCl2, 250 μM CaCl2, 10 μM MnCl2, 0.5% glucose, 50 mg of l-tryptophan liter−1, 11 mg of ferric ammonium citrate liter−1, 0.1% l-glutamine) supplemented with one of the following sulfur sources: 1 mM K2SO4, 1 mM l-methionine, 1 mM dl-homocysteine, 20 μM to 1 mM l-cystine, 0.1 to 1 mM dl-cystathionine, 0.1 to 1 mM l-djenkolic acid, or 0.1 to 1 mM S-methylcysteine. In this minimal medium, residual growth in the absence of any added sulfur source was observed. To avoid this problem, an exhausted minimal medium was obtained by growing B. subtilis 168 in a sulfur-free minimal medium and then centrifuging the culture and filtering the supernatant (15). Antibiotics were added at the following concentrations when required: ampicillin, 100 μg ml−1; chloramphenicol, 5 μg ml−1; spectinomycin, 100 μg ml−1; kanamycin, 5 μg ml−1; and erythromycin plus lincomycin, 1 and 25 μg ml−1, respectively. Solid media were prepared by addition of 20 g of Noble agar (Difco) liter−1. Standard procedures were used to transform E. coli and B. subtilis (19, 31).
TABLE 1.
B. subtilis strains used
| B. subtilis strain | Genotypea | Source or reference |
|---|---|---|
| 168 | trpC2 | Laboratory stock |
| BSIP1214 | trpC2 ytlI::aphA3 | 7 |
| BSIP1256 | trpC2 amyE::(PytmI′-lacZ cat) | pDIA5599 → 168 |
| BSIP1321 | trpC2 amyE::(PyhcL′-lacZ cat) | pDIA5629 → 168 |
| BSIP1389 | trpC2 ΔytmJKLMN::aphA3 | —b |
| BSIP1398 | trpC2 ΔytmJKLMN::aphA3 yhcL′::lacZ-erm ΔyhcL | BSIP1389 → BFS1605 |
| BSIP1534 | trpC2 ΔyhcL::spc | pDIA5669 → 168 |
| BSIP1570 | trpC2 ΔyxeMNO::cat | — |
| BSIP1572 | trpC2 ΔytmJKLMN::aphA3 ΔyxeMNO::cat | BSIP1389 → BSIP1570 |
| BSIP1575 | trpC2 ΔyxeMNO::cat ΔyhcL::spc | BSIP1570 → BSIP1534 |
| BSIP1576 | trpC2 ΔytmJKLMN::aphA3 ΔyxeMNO::cat ΔyhcL::spc | BSIP1534 → BSIP1572 |
| BSIP1582 | trpC2 ΔytmJKLMN::aphA3 ΔyhcL::spc | BSIP1389 → BSIP1534 |
| BSIP1601 | trpC2 ΔytmJKLMN::aphA3 ΔyxeMNO::cat ΔyhcL::lacZ-erm ΔyhcL | BSIP1570 → BSIP1398 |
| BSIP1603 | trpC2 ΔytmJKLMN::aphA3 ΔyxeMNO::cat yhcL′::lacZ-erm ΔyhcL yckK::Tn10 | Transposon mutagenesis |
| BSIP1643 | trpC2 ΔytmJKLMN::aphA3 yckK′::lacZ-erm ΔyckK | BSIP1389 → BFS4376 |
| BSIP1644 | trpC2 ΔyxeMNO::cat yckK′::lacZ-erm ΔyckK | BSIP1570 → BFS4376 |
| BSIP1645 | trpC2 ΔyhcL::spc yckK′::lacZ-erm ΔyckK | BSIP1534 → BFS4376 |
| BSIP1646 | trpC2 ΔytmJKLMN::aphA3 ΔyxeMNO::cat yckK′::lacZ-erm ΔyckK | BFS4376 → BSIP1572 |
| BSIP1647 | trpC2 ΔyxeMNO::cat ΔyhcL::spc yckK′::lacZ-erm ΔyckK | BFS4376 → BSIP1575 |
| BSIP1648 | trpC2 ΔytmJKLMN::aphA3 ΔyxeMNO::cat ΔyhcL::spc yckK′::lacZ-erm ΔyckK | BFS4376 → BSIP1576 |
| BSIP1649 | trpC2 ΔytmJKLMN::aphA3 ΔyhcL::spc yckK′::lacZ-erm ΔyckK | BFS4376 → BSIP1582 |
| BSIP1689 | trpC2 amyE::(PyhcL′-lacZ cat) ΔytmJKLMN::aphA3 | BSIP1389 → BSIP1321 |
| BSIP1690 | trpC2 amyE::(PytmI′-lacZ cat) ΔytmJKLMN::aphA3 | BSIP1389 → BSIP1256 |
| BSIP1691 | trpC2 amyE::(PyhcL′-lacZ cat) ΔyhcL::spc | BSIP1534 → BSIP1321 |
| BSIP1692 | trpC2 amyE::(PytmI′-lacZ cat) ΔyhcL::spc | BSIP1534 → BSIP1256 |
| BSIP1693 | trpC2 amyE::(PyhcL′-lacZ cat) ΔytmJKLMN::aphA3 ΔyhcL::spc | BSIP1534 → BSIP1689 |
| BSIP1694 | trpC2 amyE::(PytmI′-lacZ cat) ΔytmJKLMN::aphA3 ΔyhcL::spc | BSIP1534 → BSIP1690 |
| BSIP1701 | trpC2 amyE::(PyhcL′-lacZ cat) ΔytlI::aphA3 | BSIP1214 → BSIP1321 |
| BFS1605 | trpC2 yhcL′::lacZ-erm ΔyhcL | S. Bron |
| BFS4376 | trpC2 yckK′::lacZ-erm ΔyckK | K. Yamane |
cat, pC194 chloramphenicol acetyltransferase gene; aphA3, Enterococcus faecalis kanamycin resistance gene; spc, Staphylococcus aureus spectinomycin resistance gene; erm, pMUTIN erythromycin resistance gene.
—, see Materials and Methods.
The loss of amylase activity was detected as previously described (38). β-Galactosidase specific activity was measured as described by Miller (23) with cell extracts obtained by lysozyme treatment. Protein concentrations were determined by the method of Bradford. One unit of β-galactosidase activity was defined as the amount of enzyme that produced 1 nmol of o-nitrophenol min−1 at 28°C. The mean values for at least three independent experiments are presented below. The standard deviations were less than 15%.
The resistance of B. subtilis strains to a toxic analogue of l-cystine, seleno-dl-cystine, was tested as follows. Overnight cultures were grown in minimal medium containing l-methionine. The cells were diluted to an optical density at 600 nm of 1 and spread on plates of the corresponding agar medium. A paper disk (diameter, 6 mm) was laid on the agar and soaked with 10 μl of a 50 mM seleno-dl-cystine solution. The area of growth inhibition was measured.
DNA manipulations.
Plasmids from E. coli and chromosomal DNA from B. subtilis were prepared according to standard procedures. Restriction enzymes, Taq DNA polymerase, and phage T4 DNA ligase were used as recommended by the manufacturers. DNA fragments were purified from agarose gels with a Qiaquick kit (QIAGEN, Basel, Switzerland). DNA sequences were determined by using the dideoxy chain termination method with plasmid DNA as the template and a Thermo Sequenase kit (Amersham Pharmacia Biotech).
Plasmid and strain construction.
Plasmid pAC6 (38) allowed construction of transcriptional fusions between the yhcL and ytmI promoter regions and the promoterless lacZ gene. The DNA fragments corresponding to a region upstream from yhcL (nucleotides −206 to +59 relative to the yhcL translational start site) or a region upstream from ytmI (nucleotides −131 to −9 relative to the ytmI translational start site) were amplified by PCR with the creation of EcoRI and BamHI sites. The PCR products were inserted into pAC6, yielding pDIA5629 (PyhcL) and pDIA5599 (PytmI), respectively. These plasmids were linearized with ScaI, which allowed insertion of the transcriptional lacZ fusions as single copies at the amyE locus (Table 1).
A yhcL deletion mutant (BSIP1534) was constructed as follows. We cloned a 472-bp EcoRI/SmaI fragment, a SmaI/SmaI fragment carrying the spc spectinomycin cassette, and a 755-bp SmaI/BamHI DNA fragment between the EcoRI and BamHI sites of pJH101 (10) to obtain plasmid pDIA5669. The EcoRI/SmaI fragment and the SmaI/BamHI DNA fragment were generated by PCR by using oligonucleotides with the creation of restriction sites. These fragments corresponded to the 5′ part of yhcL until codon 64 and the 3′ part of yhcL with the 117 last codons. Plasmid pDIA5669 was linearized at the unique ScaI site and used to transform B. subtilis 168. Spectinomycin-resistant, chloramphenicol-sensitive integrants arose through a double-crossover event in which most of the yhcL coding sequence was deleted and replaced by the spectinomycin resistance gene (Table 1 and Fig. 1).
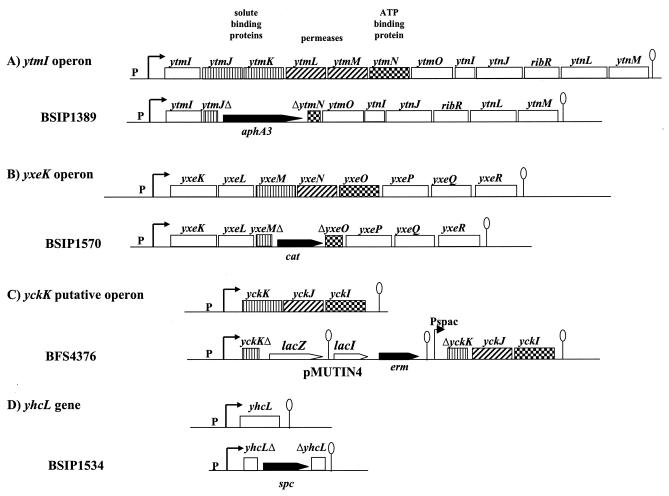
FIG. 1.
Genetic organization and disruption of the different transporters. The putative promoters of the ytmI, yxek, yckK, and yhcL genes or operons and the transcriptional terminators are indicated. The checkered boxes correspond to the ATP binding cassettes of the ABC transporters, the cross-hatched boxes correspond to the membrane permeases, and the striped boxes correspond to the solute binding proteins.
To construct strain BSIP1389, in which the ytmJKLMN genes were replaced by a kanamycin resistance cassette, a four-primer PCR procedure was used (44). The regions upstream from ytmJ (nucleotides −873 to +224 relative to the translational start site of ytmJ) and downstream from ytmN (nucleotides −614 to +420 relative to the translational stop site of ytmN) were amplified by PCR so that 21-bp fragments corresponding to the aphA3 gene were introduced at one of the ends. The ytmJ upstream region and the ytmN downstream region overlapping the aphA3 gene at one of the ends then served as long primers for PCR with aphA3 as the template. In this second PCR, two external primers were added. The final product, corresponding to the two regions flanking the ytmJ and ytmN genes with the inserted aphA3 cassette between them, was used to transform B. subtilis 168, giving strain BSIP1389 (Table 1 and Fig. 1). To limit the polar effect on the genes located downstream from ytmN, the terminator of the aphA3 gene was absent and the cassette was transcribed in the same orientation as the ytmI operon. To confirm transcription of the downstream genes, we showed that a ytnJ′-lacZ fusion (BFS70) was expressed in the ΔytmJKLMN mutant.
To construct strain BSIP1570, in which the yxeMNO genes were replaced by a chloramphenicol resistance cassette (cat gene), a four-primer PCR procedure was used as described above for deletion of the ytmJKLMN genes. The regions upstream from yxeM (nucleotides −983 to +42 relative to the translational start site of yxeM) and downstream from yxeO (nucleotides +317 to +1191 relative to the translational start site) were amplified by PCR. To limit the polar effect on the genes located downstream from yxeO, the terminator of the cat gene was absent and the cassette was transcribed in the same orientation as the yxeK operon (Fig. 1).
We used some strains disrupted by fusion with the lacZ reporter gene within the framework of European Union and Japanese projects for functional analysis of the B. subtilis genome (http://locus.jouy.inra.fr/cgi-bin/genmic/madbase/progs/madbase.operl and http://bacillus.genome.ad.jp) (16).
Transposon mutagenesis.
A transposon bank was constructed by introduction of the mini-Tn10 delivery vector pIC333 (37) into the B. subtilis 168 strain. Several thousand independent clones were pooled, and nine samples of chromosomal DNA were prepared for further use (33). To obtain selenocystine-resistant clones, B. subtilis BSIP1601 (ΔytmJKLMN::aphA3 ΔyxeMNO::cat yhcL′::lacZerm ΔyhcL) was transformed with chromosomal DNA corresponding to the prepared transposon bank (33). Clones were selected on SP plates containing spectinomycin (60 μg ml−1). By using velvet replicas, clones were transferred onto minimal medium plates containing methionine (250 μM), selenocystine (10 μM), and spectinomycin (60 μg ml−1). After 24 h of growth, selenocystine-resistant clones were isolated. The single-transposon insertion event was confirmed by backcrossing into strain BSIP1601, and the strain was checked for selenocystine resistance. To determine the location of the transposon insertion, chromosomal DNA was prepared, digested with EcoRI, and self-ligated. After transformation in E. coli, spectinomycin-resistant clones were selected, and the corresponding plasmids were sequenced. The following primers were used for sequencing of transposon insertions: Tn10 left (5′GGCCGATTCATTAATGCAGGG3′) and Tn10 right (5′CGATATTCACGG TTTACCCAC3′).
l-Cystine uptake and substrate specificity of transporters.
Cells were grown in minimal medium in the presence of l-methionine as the sole sulfur source to the middle of the exponential growth phase. They were harvested by centrifugation for 10 min and washed twice with medium A (6 mM K2HPO4, 4.4 mM KH2PO4, 0.3 mM trisodium citrate, 200 mM xylose, 0.5% glucose, 0.2 mM K2SO4, 60 μg of chloramphenicol ml−1). The substrate l-[14C]cystine was added at a concentration of 20 μM, and the reaction mixture was incubated at 37°C. Samples (200 μl) were withdrawn at intervals and filtered through 0.45-μm-pore-size Durapore membranes (HVLP02500; Millipore). The filters were washed with 5 ml of medium A, dried, and transferred to vials containing 15 ml of a counting scintillant (NBCS 104; Amersham Biosciences) for determination of the radioactivity.
To determine the apparent Km values of YhcL and YtmJKLMN, we used strains BSIP1646 (ΔytmJKLMN::aphA3 ΔyxeMNO::cat yckK′::lacZ erm ΔyckK) and BSIP1647 (ΔyxeMNO::cat ΔyhcL::spc yckK′::lacZ erm ΔyckK) lacking all the other possible l-cystine transport systems, respectively. To estimate the Km of YhcL, the rates of uptake were determined at 20°C to slow down the uptake and thus facilitate measurement of the initial rates. The experiments were performed at 37°C for YtmJKLMN.
The effects of addition of unlabeled amino acids or sulfur compounds at a concentration of 50 μM to the reaction mixture containing 5 μM l-[14C]cystine were determined. A 3-min transport kinetics analysis was performed to ensure linearity of the curves. The inhibition of l-cystine uptake at 2 min was determined.
RESULTS
B. subtilis transport systems whose synthesis is derepressed in the presence of methionine.
Transcriptome experiments revealed that several genes encoding transporters (yhcL, ytmJ, ytmK, ytmL, ytmM, ytmN, yxeM, and yxeO) were expressed more in the presence of methionine than in the presence of sulfate (2). The yhcL gene encodes a membrane protein belonging to the dicarboxylate amino acid:cation (Na+ and/or H+) symporter family (TC 2.A.23) (30). This type of transporter, which is found in eukaryotes, archaebacteria, and bacteria, is involved in the transport of dicarboxylic acids, of semipolar and neutral amino acids (Ala, Ser, Cys, and Thr), of both neutral and acidic amino acids, or of dibasic amino acids (29). The bacterial dicarboxylate amino acid:cation symporters are about 450 amino acids long (range, 420 to 491 amino acids) and have 10 to 12 putative transmembrane segments (35). In B. subtilis, four members of this family are present; they are two glutamate transporters (GltP and GltT), a C4-dicarboxylic acid transporter (DctP), and YhcL (1, 18, 40).
The YtmJKLMN and YxeMNO proteins belong to the polar amino acid uptake transporter family of the ABC transporters (TC 3.A.1.3) (14, 28). The YtmJ, YtmK, and YxeM polypeptides exhibit similarities to the l-cystine binding protein of E. coli, FliY (28, 28, and 35% identity, respectively) (6). The YtmL, YtmM, and YxeN proteins are similar to E. coli YecS (35, 32, and 48% identity, respectively), while YtmN and YxeO exhibit 50 and 52% identity to YecC. The ytmJKLMN genes belong to the large ytmIJKLMNO-ytnIJ-ribR-ytnLM operon (7, 34), while the yxeMNO genes are part of the yxeKLMNOPQR operon (Fig. 1). Remarkably, six of the eight products of the latter operon exhibit sequence similarities with the proteins encoded by the ytmI operon. These two operons could have evolved from the same ancestral operon by sequence duplications and rearrangements.
Due to their similarities with the l-cystine uptake system from E. coli, the YxeMNO and YtmJKLMN proteins are good candidates for l-cystine transporters in B. subtilis. The yhcL gene, which is expressed in the same conditions as the ytmI and yxeK operons, could also be involved in the uptake of this compound. To test the role of YxeMNO, YtmJKLMN, and YhcL in l-cystine transport, single and multiple mutants were constructed. We disrupted the coding regions of the yxeMNO, ytmJKLMN, and yhcL genes by double-crossover events that resulted in marker replacement (Fig. 1) (see Materials and Methods). In order to avoid major polar effects on downstream genes, the ytmJKLMN and yxeMNO genes were replaced by antibiotic resistance genes inserted in the same orientation with their transcription terminators deleted.
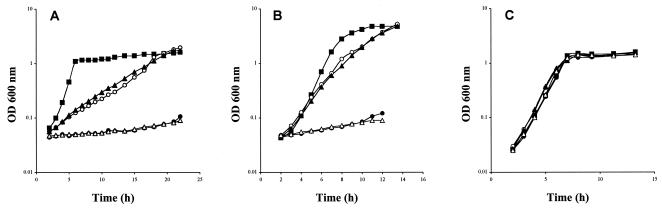
Strain BSIP1576 (ΔyxeMNO::cat ΔytmJKLMN::aphA3 ΔyhcL::spc), in which the three systems were inactivated, could still grow in the presence of 1 mM l-cystine (Fig. 2B). This indicates that at least one more l-cystine transporter exists in B. subtilis.
FIG. 2.
Growth of the wild-type strain and of different mutants with several transporters inactivated in the presence of l-cystine or sulfate. Growth curves for the following strains are shown: 168 (▪), BSIP1582 (ΔytmJKLMN::aphA3 ΔyhcL::spc) (○), BSIP1576 (ΔyxeMNO::cat ΔytmJKLMN::aphA3 ΔyhcL::spc) (▴), BSIP1649 (ΔytmJKLMN::aphA3 ΔyhcL::spc yckK′::lacZ-erm ΔyckK) (•), and BSIP1648 (ΔyxeMNO::cat ΔytmJKLMN::aphA3 ΔyhcL::spc yckK′::lacZ-erm ΔyckK) (▵). Cells were grown in exhausted minimal medium containing either 20 μM cystine (A), 1 mM cystine (B), or 40 μM sulfate (C). OD 600 nm, optical density at 600 nm.
Screening of a transposon library for selenocystine-resistant mutants in a ΔyhcL ΔytmJKLMN ΔyxeMNO strain.
Selenocystine is a toxic analogue of l-cystine. We therefore used this molecule to identify B. subtilis mutants with inactivated l-cystine uptake systems. The wild-type strain was sensitive to this toxic compound, and the growth inhibition area on plates was 3.8 cm wide in the presence of 10 μl of a 50 mM selenocystine solution. The growth inhibition of strain BSIP1576 was quite similar to that of the wild-type strain, but spontaneous selenocystine-resistant mutants appeared close to the paper disk containing selenocystine (data not shown). These spontaneous mutants showed no growth inhibition in the same conditions. This strongly suggested that a unique active cystine transporter was present in the triple mutant BSIP1576 (ΔyxeMNO::cat ΔytmJKLMN::aphA3 ΔyhcL::spc).
To identify this additional transporter, random transposition mutagenesis was performed. Both strain BSIP1576 and the mini-Tn10 transposon (37) carried a spectinomycin resistance marker. Next, we constructed strain BSIP1601 (ΔyxeMNO::cat ΔytmJKLMN::aphA3 yhcL′::lacZerm). In this strain the spectinomycin cassette disrupting the yhcL gene in strain BSIP1576 was replaced by a lacZ-erm gene obtained from strain BFS1605 (yhcL′::lacZ-erm ΔyhcL). Strain BSIP1601 was transformed with a random transposon library (see Materials and Methods). Mutants were selected for growth in the presence of 10 μM seleno-dl-cystine on plates containing l-methionine as the sole sulfur source. In order to ascertain that the selenocystine-resistant phenotype did not come from secondary mutations but was directly related to the transposon insertion, chromosomal DNA was extracted from each putative mutant and back-transformed into strain BSIP1601, with selection for the transposon antibiotic marker. The selenocystine resistance phenotype was subsequently checked, and a mutant that passed the test was retained. The insertion site of the mini-Tn10 transposon was then determined. Sequence analysis revealed that the transposon had been inserted into codon 182 of the yckK gene, which encodes the solute binding protein of a polar amino acid ABC transporter (http://genolist.pasteur.fr/SubtiList). Interestingly, the yckK gene seems to form an operon with the yckJ and the yckI downstream genes. YckJ corresponds to the permease, and YckI corresponds to the ATP binding protein of an ABC transporter.
Phenotypes of mutants with the four different uptake systems inactivated.
To investigate the relative roles of the yxeMNO, ytmJKLMN, yhcL, and yckKJI genes in l-cystine transport, several mutants carrying one to four mutations were constructed. To do this, a yckK mutant, BFS4376 (yckK′::lacZ-erm ΔyckK) obtained during the Bacillus Functional Analysis Program, was used (Table 1). We examined the effects of yxeMNO, ytmJKLMN, yhcL, and yckK gene disruptions on B. subtilis growth in sulfur-exhausted minimal medium containing either 20 μM l-cystine, 1 mM l-cystine, or 40 μM sulfate (Fig. 2). Single yxeMNO, ytmJKLMN, yhcL, and yckK mutants grew similar to the wild-type strain in the presence of sulfate or l-cystine at both concentrations (data not shown). Strains with two different transporters inactivated were therefore grown with 20 μM or 1 mM l-cystine. Strains BSIP1572, BSIP1575, BSIP1643, BSIP1644, and BSIP1645 grew similar to the wild-type strain at both concentrations (data not shown). In contrast, strain BSIP1582 (ΔytmJKLMN::aphA3 ΔyhcL::spc) displayed significant growth retardation with both L-cystine concentrations (Fig. 2A and B), while it grew as well as the wild-type strain with sulfate (Fig. 2C). The doubling times of this mutant were 220 and 60 min in the presence of 20 μM and 1 mM l-cystine, respectively, instead of the 40 min observed for the wild-type strain (Fig. 2A and B). To test the participation of each component of the ABC transporter encoded by the ytmI operon, we used the ytmJ, ytmK, ytmL, and ytmM mutants constructed during the Bacillus Functional Analysis Program (16). In these mutants, the downstream genes are expressed under control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter to avoid major polar effects (Fig. 1). Like strain BSIP1582 (ΔytmJKLMN::aphA3 ΔyhcL::spc), all the double mutants with mutations in yhcL and ytmJ, ytmK, ytmL, or ytmM poorly grew in the presence of 20 μM cystine and IPTG (data not shown). The disruption of the downstream genes ytmO and ytnJ (Fig. 1) in a ΔyhcL background led to normal growth in the same conditions. The results strongly suggest that the YtmJKLMN and YhcL systems are involved in l-cystine transport. In contrast, participation of YxeMNO seems unlikely since strains BSIP1582 (ΔytmJKLMN::aphA3 ΔyhcL::spc) and BSIP1576 (ΔyxeMNO::cat ΔytmJKLMN::aphA3 ΔyhcL::spc) grew similarly in the presence of 20 μM or 1 mM l-cystine (Fig. 2A and B).
To determine the involvement of the YckKJI ABC transporter in the residual growth observed for a ΔyhcL ΔytmJKLMN double mutant, we tested the effect of inactivation of yckK in a BSIP1582 mutant. In the presence of a low or high l-cystine concentration, the growth of strain BSIP1582 (ΔytmJKLMN::aphA3 ΔyhcL::spc) was completely abolished by introduction of a yckK gene disruption (Fig. 2A and B). As a control, we verified that all the strains grew similar to the wild-type strain with sulfate (Fig. 2C). These results strongly suggest that YckK participates in l-cystine transport. Inactivation of the transporters encoded by the ytmI operon, the yhcL gene, and the possible yckKJI operon led to a complete absence of growth of B. subtilis with l-cystine, indicating that three l-cystine uptake systems are present in this bacterium. Moreover, it seems that YtmJKLMN and YhcL are higher-affinity transporters than YckKJI. Indeed, the double mutant BSIP1582 (ΔytmJKLMN::aphA3 ΔyhcL::spc) displayed growth retardation with 20 μM cystine, while strains BSIP1643 (ΔytmJKLMN::aphA3 yckK′::lacZ-erm) and BSIP1645 (ΔyhcL::spc yckK′::lacZ-erm) did not (Fig. 2A and data not shown).
l-Cystine uptake by the YhcL, YtmJKLMN, and YckKJI transporters.
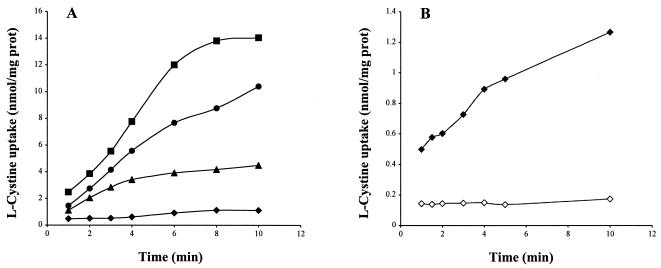
To confirm the role of the symporter and the two ABC transporters in l-cystine uptake, the abilities of B. subtilis strains 168, BSIP1534 (ΔyhcL::spc), BSIP1389 (ΔytmJKLMN::aphA3), BSIP1582 (ΔytmJKLMN::aphA3 ΔyhcL::spc), and BSIP1649 (ΔytmJKLMN::aphA3 ΔyhcL::spc yckK′::lacZ-erm) to take up this compound were tested. The data for uptake of l-[14C]cystine at a concentration of 20 μM were compared for the different backgrounds after growth of the strains with l-methionine, which allowed a high level of expression of the ytmI operon and the yhcL gene (2). A decrease in l- [14C]cystine uptake was observed in the ΔytmJKLMN or ΔyhcL mutant compared to the wild-type strain (Fig. 3A). The initial rates of l-cystine uptake were 1.9 nmol/min/mg of protein for the wild-type strain, 1.4 nmol/min/mg of protein for the ΔytmJKLMN mutant, and 0.85 nmol/min/mg of protein for the ΔyhcL mutant. The l-cystine uptake detected in the double mutant (ΔyhcL::spc ΔytmJKLMN::aphA3) was strongly reduced compared to the transport in single mutants (Fig. 3A). However, a low uptake rate (initial rate, 0.120 nmol/min/mg of protein) was obtained with strain BSIP1582, and uptake was completely abolished in strain BSIP1649 (ΔytmJKLMN::aphA3 ΔyhcL::spc yckK′::lacZerm) (Fig. 3B). This indicates that YckK was involved in the residual l-cystine transport observed in a ΔyhcL ΔytmJKLMN mutant. Three systems participated in l-cystine uptake in B. subtilis: the YhcL symporter and the two ABC transporters encoded by the ytmI and yckKJI operons. We then used strains BSIP1646 (ΔytmJKLMN::aphA3 ΔyxeMNO::cat yckK′::lacZ-erm) and BSIP1647 (ΔyxeMNO::cat ΔyhcL::spc yckK′::lacZ-erm) to estimate the Km values of the YhcL and YtmJKLMN transporters, respectively. The apparent Km values for l-cystine were 0.6 μM for the YhcL symporter and 2.5 μM for the YtmJKLMN ABC transporter.
FIG. 3.
Time course for l-[14C]cystine uptake into B. subtilis cells. (A) Comparison of l-cystine uptake in strains 168 (▪), BSIP1389 (ΔytmJKLMN::aphA3) (•), BSIP1534 (ΔyhcL::spc) (▴), and BSIP1582 (ΔytmJKLMN::aphA3 ΔyhcL::spc) (⧫). (B) Comparison of l-cystine uptake in strains BSIP1582 (ΔytmJKLMN::aphA3 ΔyhcL::spc) (⧫) and BSIP1649 (ΔytmJKLMN::aphA3 ΔyhcL::spc yckK′::lacZ-erm ΔyckK) (⋄). Cells were grown at 37°C in minimal medium containing 250 μM l-methionine. The cystine uptake analysis was performed in minimal medium in the presence of 20 μM l-[14C]cystine as described in Materials and Methods.
Substrate specificity of the YtmJKLMN and YhcL systems.
To estimate the substrate specificity of YhcL and YtmJKLMN, different sulfur-containing compounds and several amino acids were tested. To do this, we measured the inhibition of labeled l-cystine uptake in the presence of a 10-fold excess of nonradioactive compounds in strains BSIP1647 and BSIP1646 containing the YtmJKLMN and YhcL transporters, respectively. In the presence of 50 μM l-cystine, 50 μM l-djenkolic acid, 50 μM dl-cystathionine, 50 μM seleno-dl-cystine, or 50 μM S-methyl- l-cysteine, the YtmJKLMN-dependent l-[14C]cystine uptake was reduced by 88, 89, 80, 80, and 73%, respectively (Table 2). Additionally, the levels of inhibition for diaminopimelic acid, dl-lanthionine, l-methionine sulfoxide, l-methionine, l-arginine, l-glutamine, l-histidine, and dl-homocystine were 24 to 45%, while there was no significant inhibition by l-cysteine or oxidized glutathione (Table 2). These results suggest that the ABC transporter encoded by the ytmI operon participates in the uptake of l-cystine, djenkolic acid, cystathionine, selenocystine, and S-methylcysteine and that it can also recognize a much broader range of substrates.
TABLE 2.
Inhibition of YtmJKLMN-dependent l-[14C]cystine uptake by different compounds
| Competitora | % Inhibiton |
|---|---|
| l-Cystine | 88 ± 4 |
| l-Djenkolic acid | 89 ± 2 |
| dl-Cystathionine | 80 ± 6 |
| Seleno-dl-cystine | 80 ± 3 |
| S-Methyl-l-cysteine | 73 ± 8 |
| l-Histidine | 45 ± 2 |
| l-Methionine | 40 ± 1 |
| dl-Diaminopimelic acid | 39 ± 1 |
| dl-Lanthionine | 32 ± 3 |
| l-Glutamine | 32 ± 4 |
| l-Arginine | 28 ± 2 |
| dl-Homocystine | 24 ± 2 |
| l-Cysteine | <10 |
| Oxidized glutathione | <10 |
l-[14C]cystine was used at a concentration of 5 μM, and the unlabeled amino acids or sulfur compounds were present at a concentration of 50 μM. The initial l-[14C]-cystine uptake rate was measured.
For YhcL, a decrease in l-[14C]cystine uptake was observed when a 10-fold excess of nonradioactive l-cystine (92%) or seleno-dl-cystine (67%) was added (Table 3). The levels of inhibition for l-leucine, l-alanine, and l-valine were 48, 43, and 42%, respectively. Decreases in l-cystine uptake (18 to 40%) were also detected in the presence of S-methylcysteine, l-cysteine, l-dkenkolic acid, dl-cystathionine, l-methionine, and dl-homocystine (Table 3). The YhcL symporter seems to be more specific for l-cystine, but it could also transport a much broader range of amino acids and sulfur compounds.
TABLE 3.
Inhibition of YhcL-dependent l-[14C]cystine uptake in the presence of different compounds
| Competitora | % Inhibiton |
|---|---|
| l-Cystine | 92 ± 3 |
| Seleno-dl-cystine | 67 ± 4 |
| l-Leucine | 48 ± 6 |
| l-Valine | 43 ± 2 |
| l-Alanine | 42 ± 7 |
| S-Methyl-l-cysteine | 40 ± 1 |
| l-Cysteine | 38 ± 1 |
| l-Djenkolic acid | 20 ± 1 |
| dl-Cystathionine | 19 ± 1 |
| l-Methionine | 19 ± 1 |
| dl-Homocystine | 18 ± 2 |
| dl-Homocysteine | <10 |
| Oxidized glutathione | <10 |
| Reduced glutathione | <10 |
l-[14C]cystine was used at a concentration of 5 μM, and the unlabeled amino acids or sulfur compounds were present at a concentration of 50 μM. The initial l-[14C]cystine uptake rate was measured.
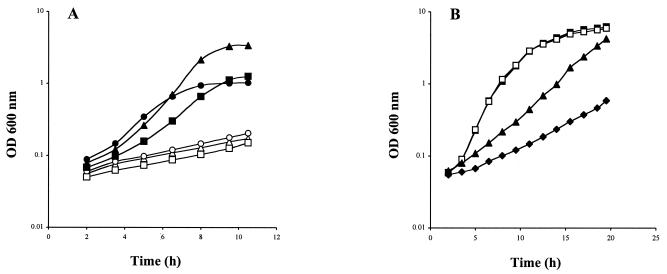
We then tested the effect of an ytmJKLMN gene disruption on utilization of djenkolic acid, cystathionine, and S-methylcysteine, a set of substrates more specific for this ABC transporter (Table 2). Growth of strain BSIP1389 (ΔytmJKLMN::aphA3) in the presence of 100 μM djenkolic acid, 100 μM cystathionine, or 100 μM S-methylcysteine was strongly reduced compared to growth of the wild-type strain (Fig. 4A). The ABC transporter encoded by the ytmI operon was therefore required for growth with these products at a concentration of 100 μM. We also tested the ability of this mutant to utilize these sulfur compounds at a higher concentration. The growth of the ΔytmJKLMN mutant with 1 mM cystathionine or 1 mM djenkolic acid remained reduced compared to the growth of the wild-type strain (data not shown). However, the residual growth observed was abolished in a ΔytmJKLMN ΔyhcL ΔyckK mutant (data not shown). In contrast, the growth of strain BSIP1389 was restored in the presence of 1 mM S-methylcysteine (Fig. 4B). Significant growth retardation was observed only in a ΔytmJKLMN ΔyhcL ΔyckK mutant. Introduction of a deletion of the yxeMNO genes in this triple mutant led to a further decrease in growth (Fig. 4B). This indicates that YhcL, YckK, and YxeMNO are probably involved to the import of S-methylcysteine together with YtmJKLMN. The residual growth of the quadruple mutant could have been due either to the existence of an additional, uncharacterized transporter or to the presence of traces of another sulfur compound in S-methylcysteine.
FIG. 4.
Growth of the wild-type strain and of different mutants in the presence of dl-cystathionine, l-djenkolic acid, or S-methyl-l-cysteine. (A) Growth curves for strain 168 (solid symbols) and BSIP1389 (ΔytmJKLMN::aphA3) (open symbols) in the presence of 100 μM dl-cystathionine (circles), 100 μM l-djenkolic acid (triangles), or 100 μM S-methyl-l-cysteine (squares). (B) Growth curves for strain 168 (▪), strain BSIP1389 (ΔytmJKLMN::aphA3) (□), strain BSIP1649 (ΔytmJKLMN::aphA3 ΔyhcL::spc yckK′::lacZ-erm ΔyckK) (▴), and strain BSIP1648 (ΔyxeMNO::cat ΔytmJKLMN::aphA3 ΔyhcL::spc yckK′::lacZ-erm ΔyckK) (⧫) in the presence of 1 mM S-methyl-l-cysteine. OD 600 nm, optical density at 600 nm.
Regulation of expression of the genes encoding l-cystine transporters.
The level of transcription of the ytmI gene is high in the presence of methionine, taurine, or glutathione and very low in the presence of sulfate, thiosulfate, and cysteine (7). To determine whether the yhcL, ytmI, and yckK genes are coregulated, expression of a yhcL′-lacZ, ytmI′-lacZ, or yckK′-lacZ transcriptional fusion was measured after growth with 1 mM l-methionine, 1 mM sulfate, or 20 μM or 1 mM l-cystine (Table 4). The expression of the yckK′-lacZ fusion was low in all the conditions tested. The β-galactosidase activities of the yhcL′-lacZ and ytmI′-lacZ fusions were 30- and 1,000-fold higher in the presence of methionine than in the presence of sulfate, respectively (Table 4) (2). The expression of these genes was also reduced in the presence of 1 mM cystine (Table 4). In the presence of 20 μM l-cystine, the expression of the yhcL′-lacZ fusion was quite similar during exponential growth and at the beginning of stationary phase, while ytmI expression increased when cells reached the stationary phase (Table 4). This increase in expression of the ytmI-lacZ fusion during the stationary phase was not observed in the presence of 1 mM l-cystine. Since l-cystine at a concentration of 20 μM is most probably the limiting growth factor, l-cystine depletion could lead to an increase in ytmI transcription.
TABLE 4.
Regulation of expression of a ytmI′-lacZ fusion, a yhcL′-lacZ fusion, and a yckK′-lacZ fusion in different backgrounds
| Strain | Relevant genotype | β-Galactosidase activity (nmol of o-nitrophenol min−1 mg of protein−1) witha:
|
|||
|---|---|---|---|---|---|
| l-Methionine (1 mM) | Sulfate (1 mM) |
l-Cystine
|
|||
| 20 μM | 1 mM | ||||
| BSIP1256 | amyE::(PytmI′-lacZ) | 975 | 1 | 5 (175) | 4 |
| BSIP1690 | amyE::(PytmI′-lacZ) ΔytmJKLMN::aphA3 | 400 | 1 | 5 | 9 |
| BSIP1692 | amyE::(PytmI′-lacZ) ΔyhcL::spc | 785 | 1 | 270 | 155 |
| BSIP1694 | amyE::(PytmI′-lacZ) ΔytmJKLMN::aphA3 ΔyhcL::spc | 415 | 1 | 1,075 | 940 |
| BSIP1321 | amyE::(PyhcL′-lacZ) | 215 | 7 | 15 (17) | 8 |
| BSIP1689 | amyE::(PyhcL′-lacZ) ΔytmJKLMN::aphA3 | 185 | 2 | 14 | 9 |
| BSIP1691 | amyE::(PyhcL′-lacZ) ΔyhcL::spc | 205 | 8 | 110 | 65 |
| BSIP1693 | amyE::(PyhcL′-lacZ) ΔytmJKLMN::aphA3 ΔyhcL::spc | 130 | 1 | 70 | 70 |
| BSIP1701 | amyE::(PyhcL′-lacZ) ΔytlI::aphA3 | 220 | 9 | 14 | 6 |
| BFS4376 | yckK′-lacZ | 38 | 38 | 33 | 25 |
| BSIP1645 | yckK′-lacZ ΔyhcL::spc | 40 | NDb | 40 | 31 |
Cells were grown in minimal medium containing different sulfur sources. Cells were harvested during exponential growth, and the β-galactosidase activity was determined as described in Materials and Methods. For 20 μM l-cystine, a second sample was harvested at the beginning of the stationary phase for strains BSIP1256 and BSIP1321; the β-galactosidase activities obtained in these conditions are indicated in parentheses.
ND, not determined.
We then tested the effect of inactivation of the two major l-cystine transport systems, YhcL and YtmJKLMN, on their own expression (Table 4). In the presence of 20 μM or 1 mM l-cystine, significant increases in expression of the ytmI′-lacZ and yhcL′-lacZ fusions were observed in a ΔyhcL mutant but not in a ΔytmJKLMN background. The upregulation of the ytmI′-lacZ fusion was more important in a ΔyhcL ΔytmJKLMN background. Deletion of the genes encoding the transporters had no effect on their expression in the presence of sulfate (Table 4). The increase in expression of the ytmI and yhcL genes in a yhcL mutant was specific for l-cystine.
The ytlI gene is located upstream of the ytmI operon and is transcribed divergently. YtlI is a LysR-type regulator that positively controls the expression of the ytmI operon (7). The role of YtlI in the regulation of expression of yhcL was investigated. Inactivation of the ytlI gene did not modify the expression pattern of the yhcL′-lacZ fusion (Table 4). YtlI is therefore not involved in the regulation of yhcL gene expression in response to sulfur availability.
DISCUSSION
Twenty-one families of secondary carriers, 11 families of ABC transporters, and three families of channel proteins mediate the transport of amino acids, peptides, and their derivatives into or out of living cells (29). In this work, we identified proteins involved in the import of l-cystine, the oxidized form of l-cysteine. In B. subtilis, four possible l-cystine transporters, YtmJKLMN, YckKJI, YxeMNO, and YhcL, were found by using differential expression in response to sulfur availability or screening for resistance to the toxic compound selenocystine (2; this study). The growth phenotypes of single and multiple mutants and the uptake of labeled l-cystine in these strains revealed the presence of three l-cystine transporters in this bacterium (Fig. 5). Surprisingly, the YxeMNO ABC transporter, which is the transporter that is most similar to the FliY/YecS/YecC system of E. coli (14), did not participate to the import of l-cystine. The YxeMNO system is probably involved in the uptake of another sulfur compound in B. subtilis. A ΔytmJKLMN ΔyhcL ΔyckK triple mutant is unable to grow in the presence of l-cystine (Fig. 2). This indicates that YhcL, YckK, and at least one protein of the YtmJKLMN system are required for l-cystine uptake in B. subtilis. The slow growth of double mutants with disruptions in yhcL and either ytmJ, ytmK, ytmL or ytmM suggests that YtmJ, YtmK, YtmL, and YtmM participate in l-cystine transport. Although direct experimental evidence for the participation of the ATP binding protein YtmN is lacking, it seems very likely that this protein is also required. For the YckKJI ABC transporter, the three different proteins also probably participate in l-cystine uptake. We propose that the symporter YhcL should be renamed TcyP (transporter of cystine). The ABC transporters YckKJI and YtmJKLMN are also renamed TcyABC and TcyJKLMN, respectively. A ΔtcyP ΔtcyJKLMN double mutant grows poorly in the presence of a low l-cystine concentration. The apparent Km values of TcyP (0.6 μM) and TcyJKLMN (2.5 μM) indicate that these two systems correspond to high-affinity l-cystine transporters. In contrast, several elements strongly suggest that TcyABC is a lower-affinity l-cystine transporter. Compared to the growth rate of the wild-type strain, the growth rate of the ΔtcyP ΔtcyJKLMN mutant is 5.5-fold lower with 20 μM l-cystine but only slightly reduced in the presence of 1 mM l-cystine (Fig. 2). The uptake of l-cystine at a concentration of 20 μM is also very low in a mutant containing only TcyABC, and the initial rate observed corresponds to less than 10% of the rate of the wild-type strain (Fig. 3).
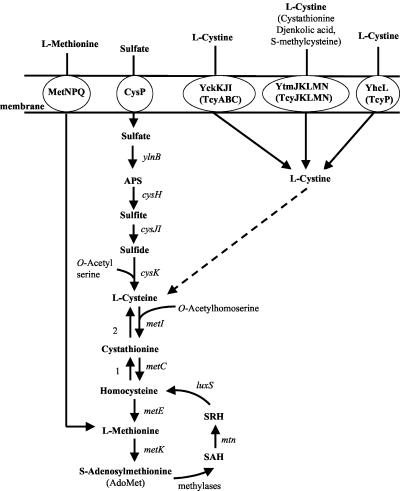
FIG. 5.
Transport and biosynthesis of sulfur-containing amino acids. The enzymes present in B. subtilis are indicated by the corresponding genes, as follows: ylnB, ATP sulfurylase; cysH, adenosine 5′-phosphosulfate reductase; cysJI, sulfite reductase; cysK, O-acetylserine sulfydrylase; metI, cystathionine γ-synthase/O-acetylhomoserine sulfydrylase; metC, cystathionine β-lyase; metE, methionine synthase; metK, S-adenosylmethionine synthase; mtnA, S-adenosyl homocysteine/methylthioadenosine nucleosidase; luxS, S-ribosylhomocysteine hydrolase; cysP, sulfate permease; metNPQ, methionine permease. SAH, S-adenosyl homocysteine; SRH, S-ribosylhomocysteine; MTA, methylthioadenosine; APS, adenosine 5′-phosphosulfate.
The ytmI operon and the tcyP gene are expressed differently in response to sulfur availability, while the level of expression of the tcyABC operon remains low in all the conditions tested. The expression of genes encoding amino acid permeases is often increased when the imported amino acid is depleted (3, 12, 15, 22, 26, 32). In particular, the synthesis of the high-affinity cystine ABC transporter from S. enterica serovar Typhimurium is decreased in the presence of cysteine as part of the CysB regulon (3). The expression of the genes encoding the two high-affinity transporters of B. subtilis is increased in conditions under which there is probable cysteine limitation; these conditions include the beginning of the stationary phase for the ytmI operon, the absence of the TcyP transporter, and maybe also the presence of methionine as the sole sulfur source. However, there are several differences in the regulation of expression of tcyP and ytmI: (i) the ytmI operon seems to be more sensitive to l-cystine starvation; (ii) the expression of the ytmI operon is repressed more than 200-fold in the presence of sulfate or l-cystine instead of the 30-fold observed for the tcyP gene (Table 4); (iii) the expression of some genes of the ytmI operon is induced by disulfide stress (20); (iv) a common target in the promoter region of tcyP and ytmI was not identified; and (v) the YtlI activator does not play a role in the regulation of tcyP expression. This indicates that YtlI is specifically involved in the control of the ytmI operon and that there is an uncharacterized regulator for the tcyP gene (2, 7; this study). It is also noteworthy that the high level of regulation of the ytmI operon could be due to the existence of a cascade of regulation and that the expression of ytlI is itself regulated (2).
In E. coli, the FliY/YecS/YecC ABC transporter displays a broader specificity for a variety of cystine analogues, while the second uncharacterized system is much more specific for l-cystine (4). This is reminiscent of the properties of the B. subtilis TcyJKLMN and TcyP transporters, respectively. Indeed, we found that the TcyJKLMN system is the major transporter of djenkolic acid, cystathionine, and, to a lesser extent, S-methylcysteine (Fig. 5). Two solute binding proteins exhibiting 57% identity, TcyJ and TcyK, are encoded by the ytmI operon. Characterization of the substrate specificities of TcyJ and TcyK, which might be different, requires further investigation. The FliY/YecS/YecC ABC transporter can also transport diaminopimelic acid (4). The l-cystine uptake by the TcyJKLMN transporter seems to be decreased in the presence of diaminopimelic acid (39% inhibition). A transporter specific for diaminopimelic acid, which is not involved in l-cystine uptake, is present in Bacillus megaterium (11). In contrast, it has been shown that B. subtilis is not able to take up diaminopimelic acid after growth in the presence of sulfate (11). This could be due to repression of the synthesis of the TcyJKLMN system in these conditions. Further work is needed to determine whether the TcyJKLMN transporter is able to efficiently take up diaminopimelic acid.
Probable l-cystine symporters, which are highly similar to TcyP, are present in Bacillus anthracis, Bacillus cereus, Staphylococcus aureus, Staphylococcus epidermidis, Oceanobacillus iheyensis, and Enterococcus faecalis. A TcyP-like protein is also found in several γ-proteobacteria, including E. coli, Salmonella enterica serovar Typhi, S. enterica serovar Typhimurium, Haemophilus influenzae, Photorhabdus luminescens, Yersinia pestis, Vibrio cholerae, and Pseudomonas putida. Interestingly, the YdjN polypeptide from E. coli, which exhibits 45% identity to TcyP, could correspond to the second uncharacterized l-cystine transporter (4). The ABC transporter encoded by the ytmI operon is present only in Listeria species (http://genolist.pasteur.fr). The TcyKLMN proteins exhibit 65 to 75% identity with the corresponding Listeria polypeptides. However, a unique solute binding protein is present in Listeria (Lmo2349 or Lin2443) instead of the two proteins present in B. subtilis, TcyJ and TcyK. A TcyABC-like ABC transporter is found in B. anthracis, B. cereus, Bacillus halodurans, S. aureus, S. epidermidis, Clostridium acetobutylicum, Neisseria meningitidis, H. influenzae, and probably several lactobacilli. The l-cystine binding protein BspA from L. fermentum (42) exhibits 36% identity to TcyA and less than 27% identity to TcyJ and TcyK. For the polar amino acid ABC transporter, identification of specific ligands is complex, and apparent extensive duplication and divergence of the sequence have occurred through evolution. The conservation of l-cystine binding proteins appears to be low (6, 42; this study). Further investigations are needed to analyze the sequence diversity of l-cystine binding proteins and to determine the substrate specificities of these different proteins.
Acknowledgments
We thank G. Rapoport and E. Dassa for helpful discussions and critical reading of the manuscript.
This research was supported by grants from the Ministère de l'Education Nationale de la Recherche et de la Technologie, the Centre National de la Recherche Scientifique (URA 2171), the Institut Pasteur, the Université Paris 7, and the European Biotech Program (contract QLG2 CT9901455).
REFERENCES
- 1.Asai, K., S. H. Baik, Y. Kasahara, S. Moriya, and N. Ogasawara. 2000. Regulation of the transport system for C4 dicarboxylic acids in Bacillus subtilis. Microbiology 146:263-271. [DOI] [PubMed] [Google Scholar]
- 2.Auger, S., A. Danchin, and I. Martin-Verstraete. 2002. Global expression profile of Bacillus subtilis grown in the presence of sulfate or methionine. J. Bacteriol. 184:5179-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baptist, E. W., and N. M. Kredich. 1977. Regulation of l-cystine transport in Salmonella typhimurium. J. Bacteriol. 131:111-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, E. A., and L. A. Heppel. 1972. A binding protein involved in the transport of cystine and diaminopimelic acid in Escherichia coli. J. Biol. Chem. 247:7684-7694. [PubMed] [Google Scholar]
- 5.Boos, W., and J. M. Lucht. 1996. Periplasmic binding protein-dependent ABC transporters, p. 1175-1209. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 6.Butler, J. D., S. W. Levin, A. Facchiano, L. Miele, and A. B. Mukherjee. 1993. Amino acid composition and N-terminal sequence of purified cystine binding protein of Escherichia coli. Life Sci. 52:1209-1215. [DOI] [PubMed] [Google Scholar]
- 7.Coppee, J. Y., S. Auger, E. Turlin, A. Sekowska, J. P. Le Caer, V. Labas, V. Vagner, A. Danchin, and I. Martin-Verstraete. 2001. Sulfur-limitation-regulated proteins in Bacillus subtilis: a two-dimensional gel electrophoresis study. Microbiology 147:1631-1640. [DOI] [PubMed] [Google Scholar]
- 8.Dassa, E., M. Hofnung, I. T. Paulsen, and M. H. Saier. 1999. The Escherichia coli ABC transporters: an update. Mol. Microbiol. 32:887-889. [DOI] [PubMed] [Google Scholar]
- 9.During-Olsen, L., B. Regenberg, C. Gjermansen, M. C. Kielland-Brandt, and J. Hansen. 1999. Cysteine uptake by Saccharomyces cerevisiae is accomplished by multiple permeases. Curr. Genet. 35:609-617. [DOI] [PubMed] [Google Scholar]
- 10.Ferrari, F. A., A. Nguyen, D. Lang, and J. A. Hoch. 1983. Construction and properties of an integrable plasmid for Bacillus subtilis. J. Bacteriol. 154:1513-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gally, D., C. R. Harwood, and A. R. Archibald. 1991. Diaminopimelic acid uptake by Bacillus megaterium: influence of growth conditions and other amino acids. Lett. Appl. Microbiol. 12:54-58. [Google Scholar]
- 12.Glansdorff, N. 1996. Biosynthesis of arginine and polyamines, p. 409-433. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 13.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67-113. [DOI] [PubMed] [Google Scholar]
- 14.Hosie, A. H. F., and P. S. Poole. 2001. Bacterial ABC transporters of amino acids. Res. Microbiol. 152:259-270. [DOI] [PubMed] [Google Scholar]
- 15.Hullo, M. F., S. Auger, E. Dassa, A. Danchin, and I. Martin-Verstraete. 2004. The metNPQ operon of Bacillus subtilis encodes an ABC permease transporting methionine sulfoxide, d- and l-methionine. Res. Microbiol. 155:80-86. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 18.Krom, B. P., J. B. Warner, W. N. Konings, and J. S. Lolkema. 2003. Transporters involved in uptake of di- and tricarboxylates in Bacillus subtilis. Antonie Leeuwenhoek 84:69-80. [DOI] [PubMed] [Google Scholar]
- 19.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leichert, L. I. O., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mansilla, M. C., and D. de Mendoza. 2000. The Bacillus subtilis cysP gene encodes a novel sulphate permease related to the inorganic phosphate transporter (Pit) family. Microbiology 146:815-821. [DOI] [PubMed] [Google Scholar]
- 22.Merlin, C., G. Gardiner, S. Durand, and M. Masters. 2002. The Escherichia coli metD locus encodes an ABC transporter which includes Abc (MetN), YaeE (MetI), and YaeC (MetQ). J. Bacteriol. 184:5513-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Mytelka, D. S., and M. J. Chamberlin. 1996. Escherichia coli fliAZY operon. J. Bacteriol. 178:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulsen, I. T., M. K. Sliwinski, and M. H. Saier. 1998. Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J. Mol. Biol. 277:573-592. [DOI] [PubMed] [Google Scholar]
- 26.Pittard, A. J. 1996. Biosynthesis of aromatic amino acids, p. 458-484. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 27.Quadroni, M., W. Staudenmann, M. Kertesz, and P. James. 1996. Analysis of global responses by protein and peptide fingerprinting of proteins isolated by two-dimensional gel electrophoresis. Application to the sulfate-starvation response of Escherichia coli. Eur. J. Biochem. 239:773-781. [DOI] [PubMed] [Google Scholar]
- 28.Quentin, Y., G. Fichant, and F. Denizot. 1999. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 287:467-484. [DOI] [PubMed] [Google Scholar]
- 29.Saier, M. H. 2000. Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology 146:1775-1795. [DOI] [PubMed] [Google Scholar]
- 30.Saier, M. H., S. R. Goldman, R. R. Maile, M. S. Moreno, W. Weyler, N. Yang, and I. T. Paulsen. 2002. Transport capabilities encoded within the Bacillus subtilis genome. J. Mol. Microbiol. Biotechnol. 4:37-67. [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 32.Sarsero, J. P., E. Merino, and C. Yanofsky. 2000. A Bacillus subtilis gene of previously unknown function, yhaG, is translationally regulated by tryptophan-activated TRAP and appears to be involved in tryptophan transport. J. Bacteriol. 182:2329-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekowska, A., and A. Danchin. 2002. The methionine salvage pathway in Bacillus subtilis. BMC Microbiol. 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekowska, A., S. Robin, J. J. Daudin, A. Henaut, and A. Danchin. 2001. Extracting biological information from DNA arrays: an unexpected link between arginine and methionine metabolism in Bacillus subtilis. Genome Biol. 2:0019. [DOI] [PMC free article] [PubMed]
- 35.Slotboom, D. J., W. N. Konings, and J. S. Lolkema. 1999. Structural features of the glutamate transporter family. Microbiol. Mol. Biol. Rev. 63:293-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soutourina, J., S. Blanquet, and P. Plateau. 2001. Role of d-cysteine desulfhydrase in the adaptation of Escherichia coli to d-cysteine. J. Biol. Chem. 276:40864-40872. [DOI] [PubMed] [Google Scholar]
- 37.Steinmetz, M., and R. Richter. 1994. Easy cloning of mini-Tn10 insertions from the Bacillus subtilis chromosome. J. Bacteriol. 176:1761-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stülke, J., I. Martin-Verstraete, M. Zagorec, M. Rose, A. Klier, and G. Rapoport. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 25:65-78. [DOI] [PubMed] [Google Scholar]
- 39.Tam, R., and M. H. Saier. 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57:320-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tolner, B., T. Ubbink-Kok, B. Poolman, and W. N. Konings. 1995. Characterization of the proton/glutamate symport protein of Bacillus subtilis and its functional expression in Escherichia coli. J. Bacteriol. 177:2863-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomii, K., and M. Kanehisa. 1998. A comparative analysis of ABC transporters in complete microbial genomes. Genome Res. 8:1048-1059. [DOI] [PubMed] [Google Scholar]
- 42.Turner, M. S., T. Woodberry, L. M. Hafner, and P. M. Giffard. 1999. The bspA locus of Lactobacillus fermentum BR11 encodes an l-cystine uptake system. J. Bacteriol. 181:2192-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Ploeg, J. R., N. J. Cummings, T. Leisinger, and I. F. Connerton. 1998. Bacillus subtilis genes for the utilization of sulfur from aliphatic sulfonates. Microbiology 144:2555-2561. [DOI] [PubMed] [Google Scholar]
- 44.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]