Abstract
The biocontrol activity of various fluorescent pseudomonads towards plant-pathogenic fungi is dependent upon the GacA/GacS-type two-component system of global regulators and the RpoS transcription sigma factor. In particular, these components are required for the production of antifungal antibiotics and exoenzymes. To investigate the effects of these global regulators on the expression of biocontrol factors by plant-associated bacteria other than Pseudomonas spp., gacA/gacS and rpoS homologues were cloned from biocontrol strain IC1270 of Serratia plymuthica, which produces a set of antifungal compounds, including chitinolytic enzymes and the antibiotic pyrrolnitrin. The nucleotide and deduced protein sequence alignments of the cloned gacA/gacS-like genes—tentatively designated grrA (global response regulation activator) and grrS (global response regulation sensor) and of the cloned rpoS gene revealed 64 to 93% identity with matching genes and proteins of the enteric bacteria Escherichia coli, Pectobacterium carotovora subsp. carotovora, and Serratia marcescens. grrA, grrS, and rpoS gene replacement mutants of strain IC1270 were deficient in the production of pyrrolnitrin, an exoprotease, and N-acylhomoserine lactone quorum-sensing signal molecules. However, neither mutant appeared to differ from the parental strain in the production of siderophores, and only grrA and grrS mutants were deficient in the production of a 58-kDa endochitinase, representing the involvement of other sigma factors in the regulation of strain IC1270's chitinolytic activity. Compared to the parental strain, the grrA, grrS, and rpoS mutants were markedly less capable of suppressing Rhizoctonia solani and Pythium aphanidermatum under greenhouse conditions, indicating the dependence of strain IC1270's biocontrol property on the GrrA/GrrS and RpoS global regulators.
Several global regulators of gene expression are currently known in gram-negative bacteria. The GacA/GacS regulatory cascade has been shown to control exoenzyme and secondary-metabolite production in Pseudomonas species, including plant growth-promoting and biocontrol strains of fluorescent pseudo-monads (5, 9, 16, 23, 45), and similar two-component systems have been detected in Escherichia coli (32, 33), plant-pathogenic strains of Pectobacterium (Erwinia) carotovora subsp. carotovora (14), and other gram-negative bacteria (reviewed in references 17 and 19). The GacA response regulator protein described in various pseudomonads (17, 19, 12) is similar to UvrY in E. coli (33) and ExpA in P. carotovora subsp. carotovora (14), while the GacS protein of pseudomonads (5, 9, 45), a transmembrane sensor kinase responsible for transferring a phosphoryl group to the receiver domain of the cognate response regulator GacA, is similar to BarA in E. coli (32), ExpS in P. carotovora subsp. carotovora (14), and many others (19). Recent evidence indicates that GacS/GacA and similar networks have a major impact on target gene expression at the posttranscriptional level via positive control of transcription of small regulatory RNAs able to titrate translation repressors like mRNA-binding proteins (10, 18, 34).
Regulation by the quorum-sensing signal molecules N-acylhomoserine lactones (acyl-HSLs) has been found to be important in the production of secondary metabolites and exoenzymes in many gram-negative bacteria (29, 35). Acyl-HSLs serve as global regulatory signals in nature, but in many bacteria, including Pseudomonas aureofaciens and P. aeruginosa species, their production is under the control of a GacA/GacS-type two-component signal transduction system (5, 36).
Eubacterial RNA polymerase uses the sigma subunit for recognition of and transcription initiation from promoter DNA sequences. The sigma factors belonging to members of the σ70 family are classified as either primary or alternative, based on functional differences (25). Primary sigma factors such as σ70 (RpoD) are usually essential, as they are responsible for the transcription of housekeeping genes in vegetative cells. The alternative sigma factor σ38 (RpoS) plays a key role in the survival of bacteria during starvation or exposure to stress conditions and is required for the expression of many genes in the stationary growth phase but may be nonessential for cell viability (20). Regulation specific for gene expression in the stationary or exponential growth phase is provided by relative changes in the concentrations of the stationary and housekeeping sigma factors (38, 39) and by anti-sigma factors, like RssB in E. coli, which can inhibit the expression of σs-dependent genes in the presence of high σs levels (20). In turn, the GacA/GacS system in P. fluorescens strain Pf-5 (45) and BarA regulator in E. coli (31) are most likely directly required for rpoS transcription and σs accumulation.
Manipulation of the GacA/GacS and sigma regulators has proven to be an innovative approach to improving the biocontrol activity of fluorescent pseudomonads strains (30). More knowledge about these global regulators in other nonfluorescent pseudomonad species could open the way to extending the range of plant-associated bacteria able to serve as superior biocontrol agents of plant pathogens. We chose strain IC1270, previously described as Enterobacter agglomerans (6-8) and recently reidentified based on 16S rRNA gene partial sequences analysis as Serratia plymuthica (GenBank accession no. AY551332), as a model to investigate whether the GacA/GacS and sigma regulators are responsible for antifungal activity in bacterial biocontrol agents other than P. fluorescens. Strain IC1270 was isolated from the rhizosphere of grape and described as a wide-range antagonist of many microorganisms, including plant-pathogenic fungi, producing two N-acetyl-β-d-glucosaminidases of 89 and 67 kDa, the 58-kDa endochitinase ChiA, and the antibiotic pyrrolnitrin (3-chloro-4-[2′-nitro-3′-chlorophenyl]-pyrrole) (6-8). Strain IC1270's homologues of genes encoding the GacA/GacS-type two-component global regulation systems and σ38 (RpoS) transcription factors were cloned and characterized. The grrA, grrS, and rpoS mutants of strain IC1270 were found to be deficient in the production of antifungal compounds, including pyrrolnitrin, exoprotease, and the ChiA endochitinase (only the grrA and grrS mutants), as well as in antifungal activity against Rhizoctonia solani and Pythium aphanidermatum. In addition, production of quorum-sensing acyl-HSL signals by strain IC1270 was also found to be affected by GrrA/GrrS and RpoS regulators.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. Liquid or solid (1.5% [wt/vol] agar) Luria broth and agar (LB/A) and potato-dextrose broth and agar (PDB/A) were used as growth media. V8 vegetable juice (Campbell Soup Co., Camden, N.J.) supplemented with calcium carbonate (2.5 g/liter) was used as the growth medium for Pythium aphanidermatum. To induce chitinolytic activity, bacteria were grown in liquid synthetic medium (SM) with 0.2% (wt/vol) colloidal chitin as described previously (8). To detect pyrrolnitrin production, strain IC1270 was grown in M9 minimal medium (3) supplemented with 2% (wt/vol) glycerol as the sole carbon source. M9 medium supplemented with 0.4% (wt/vol) glucose was used for growth of the Agrobacterium tumefaciens NTL4/pZLR4 reporter strain. Antibiotic supplements were used at the following concentratios: ampicillin, 100 μg/ml; carbenicillin, 50 μg/ml; rifampin, 40 μg/ml; tetracycline, 20 μg/ml; and gentamicin, 20 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Agrobacterium tumefaciens NTL4/pZLR4 | Gmr Cbr; β-galactosidase induction-based acyl-HSL bioreporters | 41 |
| Chromobacterium violaceum CV026 | Violacein production-based acyl-HSL bioreporter | 26 |
| Escherichia coli | ||
| DH5α | F−endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 φ80dlacZΔM15 λ− | 3 |
| S17-1(λ-pir) | thi pro hsdR hsdM recA rpsL RP4-2 (Tcr::Mu) (Kmr::Tn7) | 42 |
| Pseudomonas fluorescens 2-79 | Phz+ | 43 |
| Serratia plymuthica (previously Enterobacter agglomerans) | ||
| IC1270 | Rifr, isolated from rhizosphere of grape in Samarkand region, Republic of Uzbekistan | 8 |
| IC1270#20 | grrA (=gacA) Rifr Gmr gene replacement mutant of strain IC1270 | This study |
| IC1270#93 | grrS (=gacS), Rifr Gmr gene replacement mutant of strain IC1270 | This study |
| IC1270#823 | rpoS Rifr Gmr gene replacement mutant of strain IC1270 | This study |
| Plasmids | ||
| pSB401 | Tcr, lux-based acyl-HSL bioreporter | 47 |
| pSB536 | Ampr, lux-based acyl-HSL bioreporter | 47 |
| pSB1075 | Ampr, lux-based acyl-HSL bioreporter | 47 |
| pGEM-T Easy | Ampr | Promega Corp. |
| p34S-Gm | Source of Gmr cassette | 11 |
| pEX18Tc gene replacement vector | TcroriT+sacB | 21 |
Ampr, ampicillin resistance; Gmr, gentamicin resistance; Tcr, tetracycline resistance; Rifr, rifampicin resistance; Cbr, carbenicillin resistance; sacB, sucrose-inducible gene of Bacillus subtilis, the expression of which is lethal in the presence of 5% sucrose; Phz+, production of phenazine-1-carboxylic acid.
DNA manipulations.
Total genomic DNA isolation, restriction enzyme digestions, agarose gel electrophoresis, electroporation, PCR, and Southern hybridization were generally performed according to standard procedures (3). Enzymes were purchased from MBI Fermentas (Vilnius, Lithuania) or Promega (Madison, Wis.) and used according to the manufacturers' directions. Inverse PCR was performed as described previously (44). The general strategy for cloning all three regulatory genes was the following. For each gene, we first designed two primers to the conserved regions of the corresponding gene of P. carotovora subsp. carotovora, E. coli, or P. fluorescens (see Table S1, primers A1 to A6, in the supplemental material). The PCR products obtained with strain IC1270 genomic DNA as the template were ligated into a pGEM-T Easy (Promega) vector, the ligation mixtures were electroporated into E. coli strain DH5α, and corresponding selected clones were subjected to sequencing. Then the inverse PCR technique was used to extend the obtained partial sequences in both directions.
Briefly, genomic DNA of strain IC1270 was digested with restriction endonucleases which did not cut the obtained sequences and subjected to self-ligation. These self-ligation products were used as templates in a PCR with primers designed to the partial sequences in opposite orientations (see Table S1, primers B1 to B6, in the supplemental material). The corresponding products of the inverse PCR were cloned into pGEM-T Easy and sequenced. For final cloning of the complete sequences of each gene, primers C1 to C6 (see Table S1 in the supplemental material) were designed to the 5′ and 3′ regions of the corresponding sequences obtained by inverse PCR. The PCR products obtained with this third pair of primers, representing the complete sequence of the desired S. plymuthica strain IC1270 regulatory gene, were in turn cloned into the pGEM-T Easy vector and sequenced.
PCR was performed in a total volume of 25 μl, containing 1.5 mM MgCl2, 200 μM concentrations of each of the four deoxynucleoside triphosphates, 10 pmol of each PCR primer, 1 U of error-proof Taq DNA polymerase (Boehringer, Mannheim, Germany), and 100 ng of strain IC1270's total DNA. The following program was used for thermal cycling: 94°C for 3 min, 55°C for 2 min, and 72°C for 2 min, then 30 cycles at 94°C for 30 s, 55°C for 1 min, and 72°C for 1.5 min, and a final extension at 72°C for 10 min. PCR products were resolved on a 1% agarose gel in Tris-acetate-EDTA buffer and purified with a QIAquick gel extraction kit (Qiagen Inc., Hilden, Germany). PCR products used as DNA probes for Southern blot analysis of genomic DNA of IC1270 and its derivatives were labeled by [32P]dCTP (3,000 Ci mmol−1) with the Rediprime II DNA labeling system (Amersham). The membranes incubated with the labeled probes at 65°C were washed twice (10 min each time) in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% (wt/vol) sodium dodecyl sulfate (SDS) at the same temperature and subjected to autoradiography.
The complete nucleotide sequence of the PCR product was determined with the method of automated DNA sequencing, utilizing the BigDye Terminator cycle sequencing chemistry from Applied Biosystems (ABI, Perkin-Elmer, Foster City, Calif.), ABI 3700 DNA analyzer, and the ABI's data collection and sequence analysis software. Standard T7 and SP6 primers and a set of specific internal primers designed to overlap and match the end parts of the sequences determined in each previous step (see above) were used for sequencing from both ends of the PCR product inserted into the pGEM-T vector in order to obtain the entire sequence of the inserted DNA. Comparative sequence analysis was performed with the ClustalW multiple sequence alignment program.
Gene replacement mutagenesis.
To investigate their specific functions, the grrA, grrS, and rpoS genes of strain IC1270 were each inactivated by inserting a suicide vector into the bacterial chromosome by homologous recombination with the sacB-based strategy (40). Briefly, a 931-bp fragment bearing the gentamicin resistance gene aac1, encoding gentamicin acetyltransferase 3-1, was cut by SmaI or BamHI from the cloning vector p34S-Gm (11) and inserted into grrA, grrS, and rpoS gene sequences cloned into the pGEM-T Easy vector as described above. The sites for insertion of the gentamicin cassette were EcoR47III, EheI, and BamHI for grrA, grrS, and rpoS, respectively. The interrupted genes were recloned at the EcoRI site into the pEX18Tc gene replacement vector (21) and introduced by electroporation into the E. coli S17-1 strain, used then as a donor to transfer the constructs into strain IC1270 by mating. The double-crossover progeny were selected on LB plates supplemented with 5% (wt/vol) sucrose, gentamicin (21 μg/ml), and rifampin (40 μg/ml). The gentamicin-resistant, rifampin-resistant, sucrose-resistant, tetracycline-sensitive clones were initially assessed by performing colony PCR on gentamicin-resistant isolates with two aac1 (gentamicin resistance)-specific primers (Gm-F332, 5′-GGCTCAAGTATGGGCATCATT-3′ and Gm-R781, 5′-GGCGGTACTTGGGTCGATA-3′) as described (21), and the gene replacements were further verified by Southern analysis.
The genomic DNAs of strain IC1270 and the selected mutants were digested with EcoRV, which cut the gentamicin resistance cassette only at site 781 (GenBank accession no. AF062079), separated electrophoretically, and transferred to Hybond-N+ membranes. Full 32P-labeled sequences of grrA, grrS, and rpoS, obtained by EcoRI restriction of the corresponding pGEM-T Easy-based constructs purified from an agarose gel, were used as probes for these EcoRV digests. A 32P-labeled 449-bp aac1 fragment that did not include the EcoRV site was obtained with the gentamicin-specific primers described above and used as probe 2 for reprobing.
Pyrrolnitrin purification and assay.
A crude extract of the antibiotic from cells grown on plates with M9 agar medium supplemented with 2% glycerol was prepared by chloroform extraction as described previously (6). The amount of pyrrolnitrin obtained after evaporation of chloroform and dissolution in acetonitrile was measured by high-pressure liquid chromatography (HPLC) (Spectrum Chromatography, Houston, Tex.) with a Luna (Phenomenex, Torrance, Calif.) reverse-phase C18 column (5 μm, 250 by 4.6 mm) and eluted isocratically (45% H2O, 30% acetonitrile, 25% methanol) at a flow rate of 1 ml/min. Antibiotic absorption was monitored with a diode array detector (UV6000) at 225 nm, the wavelength commonly used to monitor pyrrolnitrin (9). Purified pyrrolnitrin used as a standard was kindly supplied by Karl-Heinz van Pee (Institute for Biochemie, Technical University Dresden, Dresden, Germany).
Assays of chitinolytic, proteolytic, and siderophore activities.
Detection of chitinolytic enzymes in extracellular proteins following gel electrophoresis was generally performed as described previously (8). Secreted proteins, first concentrated and dialyzed in an ultrafiltration system with a cutoff of 10 kDa (Vivascience, Lincoln, United Kingdom), were separated by SDS-10% polyacrylamide gel electrophoresis (PAGE) and then reactivated by removing SDS with the casein-EDTA procedure (27). Enzyme activity was detected on gels with the fluorescent substrate 4-methylumbelliferyl-β-d-N,N′-diacetylchitobioside (Sigma, St. Louis, Mo.), which can detect both exo- and endochitinolytic enzyme types produced by strain IC1270 (9). Proteolytic activity was assayed on Bacto-litmus milk (Difco) and skim milk (Difco) agar plates by monitoring the appearance of halos of casein degradation around the bacterial colonies. For detection of siderophores, the plate assay (2) was performed with the ternary complex chrome azurol S-iron(III)-hexacyltrimethylammonium bromide (CAS) as an indicator. Siderophore production was revealed by the appearance of yellow halos surrounding bacterial growth on the blue background. In all plate tests, the plates were incubated at 30°C until halos appeared around the colonies.
Assays for acyl-HSL production.
Ethyl acetate extracts of the tested strains, prepared as described (4, 41), were evaporated and dissolved in acetonitrile. Bioluminescence and violacein production assays for acyl-HSLs with lux-based reporter plasmids and the strain Chromobacterium violaceum CV026, respectively, were performed essentially as described by Camara et al. (4). Light emission was monitored with a CL-BIS luminometer (DNR Imaging Systems Ltd.). Violacein production was indicated by dark blue-purple pigmentation of the bacterial lawn surrounding the wells made in the solidified top Luria agar layer.
β-Galactosidase induction with the A. tumefaciens NTL4/pZLR4 reporter strain was performed as described previously (41). Formation of a diffuse blue zone surrounding the tested colony or the extract spot indicated production of acyl-HSLs. Synthetic acyl-HSL standards (kindly supplied by Paul Williams, Nottingham University, Nottingham, United Kingdom) were spotted on plates to confirm reporter strain activity. For detection of the acyl-HSLs by thin-layer chromatography (TLC) analysis, the procedure described by Shaw et al. (41) with the NTL4/pZLR4 reporter strain was used. Ethyl acetate extracts and synthetic acyl-HSL standards were spotted onto glass-backed RP18 F254S reverse-phase TLC plates (Merck), and samples were separated with 60% (vol/vol) methanol in water as the solvent. Then TLC plates were overlaid with the reporter strain seeded in water-soft agar supplemented with glucose (0.2%) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (40 μg/ml). After overnight incubation at 30°C, acyl-HSLs were located visually as blue spots. TLC analysis was repeated at least twice with samples obtained independently for each assay.
Inhibition of Rhizoctonia solani and Pythium aphanidermatum.
Assays of fungal growth suppression on plates were performed as described (8). Under greenhouse conditions, R. solani infection of beans (Faseolus vulgaris L., cv. wax yellow) and Pythium aphanidermatum infection of cucumber (Cucumis sativus L. cv. Cfir 413) were used as the model systems. Briefly, in the case of infection with R. solani, polypropylene boxes were two-thirds filled with sandy loam soil (pH 7.2). A seed cover layer (one-third of the pot's depth) was infected with a preparation of R. solani mycelium blended and mixed with soil. Bacteria in tap water (ca. 109 cells/ml) were applied by drenching immediately after infection and on the fourth day from sowing. In the case of Pythium aphanidermatum infection, the soil was inoculated by oospore suspension (≈5 × 105 spores/ml). Before sowing, the seeds were soaked for 10 min in the bacterial suspension (≈109 cells/ml in tap water) being tested, then the seeds were sown in the soil and additionally inoculated by dripping 100 μl of the same suspension of bacterial cells onto the seeds before covering them with the infected soil. In both schemes of experiments, 10 seeds were placed in each box. In the disease control, tap water was used instead of the bacterial suspension. In the seed germination control, the soil was not infected with fungal pathogen and the pots were irrigated only with tap water. Disease incidence was determined after 10 to 14 days as the percentage of seedlings with R. solani root rot or Pythium aphanidermatum pre- and postemergence damping-off disease.
Nucleotide sequence accession numbers.
The GenBank accession numbers for regulatory genes grrA, grrS, and rpoS from IC1270 are AY057388, AY057389, and AY057391, respectively.
RESULTS
Cloning and characterization of strain IC1270 genes encoding GacA/GacS-type global regulators.
The homologues of the two-component regulator genes were cloned from strain IC1270 as described in Materials and Methods with a combination of PCR and inverse PCR with corresponding pairs of primers and sequenced. The sequences were compared with the corresponding genes expA, expS, uvrY, and barA from the taxonomically similar enteric bacteria P. carotovora subsp. carotovora and E. coli (GenBank accession nos. X95564, Y13670, AE000284, and D10888, respectively), with Serratia marcescens genome sequences available at www.sanger.ac.uk/Projects/S_marcescens/, and with genes gacA and gacS (= apdA) from P. fluorescens strain Pf5 (GenBank accession nos. AF065156 and U30858, respectively). The last were chosen for comparison because they are from a biocontrol strain that produces several antifungal compounds similar to those produced by strain IC1270, including pyrrolnitrin, protease, and siderophore (9, 45).
The 1,484-bp sequence included the open reading frame of the 657-bp-long gacA-like gene of S. plymuthica IC1270 (tentatively designated grrA for global response regulation activator), and the deduced amino acid sequence of a tentative GrrA protein with a calculated molecular mass of 25 kDa was determined. Computer analysis revealed 87, 78, 76, and 47% identity between grrA and sequence sm733b12 p1k of S. marcescens genes grrA, uvrY, expA, and gacA, respectively. A database search revealed that the deduced GrrA protein amino acid sequence was 88, 84, and 57% identical to response regulators UvrY, ExpA, and GacA, respectively. The N-terminal phosphate acceptor domain containing the phosphorylated aspartate residue (Asp-8, -9, and -54 and lysine-114 in GrrA) and the C-terminal DNA-binding domain, including the helix-turn-helix motif described in similar proteins of other gram-negative bacteria (19, 23), appear to be conserved. (see Fig. S1-A in the supplemental material).
The strain IC1270 homologue of the sensor kinase GacS was characterized by cloning the 3,311-bp nucleotide sequence, including a 2,727-bp-long open reading frame, with 89, 68, 65, and 50% identity to sequence sm670e08.q1k of S. marcescens and to gacS family genes expS, barA, and gacS (= apdA) of P. carotovora subsp. carotovora, E. coli, and P. fluorescens strain Pf5, respectively. Based on this identity, the cloned gacS homologue was tentatively designated grrS (for global response regulation sensor). The sequence of the deduced GrrS protein (907 amino acids, with a molecular mass of 101.6 kDa) was found to be 66, 64, and 37% identical to those of the ExpA, BarA, and GacS (=ApdA) sensor kinases, respectively. Similar to previously described GacS-type sensor kinases (9, 14, 19, 32), the GrrS contains linker, transmitter, receiver, and carboxy-terminal histidine phosphotransfer (Hpt) output domains with conserved H-N-G-F boxes associated with the complex phosphorelay sensor kinases, which may serve to modulate the phosphorylation of GacA-type response regulators (19) (see Fig. S1-B in the supplemental material).
Cloning and characterization of strain IC1270 homologues of RpoS sigma factor gene.
The S. plymuthica strain IC1270 rpoS homologue was characterized by cloning the 1,888-bp sequence, including the 996-bp-long open reading frame. The nucleotide sequence alignment of the cloned rpoS gene with sequence sm556b09.q1k from genome of S. marcescens and corresponding genes from P. carotovora (GenBank accession no. U66542), E. coli (GenBank accession no. D13548), and P. fluorescens (GenBank accession no. U34203) revealed 89, 81, 81, and 66% identity, respectively. The alignment of the deduced 331-amino-acid-long 38-kDa RpoS protein of S. plymuthica with corresponding sequences of proteins from these three bacterial species demonstrated the presence of highly conserved residues. Within the most highly conserved regions 1 through 4 of the σ70 family of sigma factors described in other bacteria (25), the identity to the RpoS proteins of E. coli, P. carotovora, and P. fluorescens was 93, 91, and 63%, respectively (see Fig. S1-C in the supplemental material).
Southern blot analysis revealed that each of the three tested genes is present in strain IC1270's genomic DNA in a single copy. Hybridization of digests with EcoRI, which did not cut any of the three gene coding sequences, and with the specific DNA probe led to the appearance of only one band. At the same time, hybridization with genomic DNA digested with enzymes cutting the corresponding coding sequence at only one site led to the formation of two bands instead of one (data not shown).
Isolation and characterization of strain IC1270 gene replacement mutants.
To demonstrate the dependence of strain IC1270 antifungal activity on the GacA/GacS- and RpoS-type global regulators, grrA, grrS, and rpoS mutants were constructed by homologous recombination with the pEX18Tc suicide vector carrying copies of these genes disrupted by insertion of the gentamicin resistance cassette. Double recombinants sensitive to tetracycline due to loss of part of the vector DNA were verified by Southern hybridization analysis (data not shown). Three mutant isolates, IC1270#20 (grrA), IC1270#93 (grrS), and IC1270#823 (rpoS), were compared with the wild-type strain for their ability to produce pyrrolnitrin, chitinolytic enzymes, exoprotease, siderophore, and N-acyl-HSL quorum-sensing signal molecules and their ability to suppress the fungal pathogens R. solani and Pythium aphanidermatum on plates and under greenhouse conditions.
(i) Chitinolytic activity.
Strain IC1270, producing a set of chitinolytic enzymes, hydrolyzed colloidal chitin when grown on solid medium supplemented with colloidal chitin as the sole carbon source. Clear zones of chitin degradation around the growing colonies were observed (8). Three fluorescent bands, corresponding to two exochitinases with apparent molecular masses of 89 and 67 kDa and one 58-kDa endochitinase (ChiA) were detected in the extracellular proteins of strains IC1270 and mutant IC1270#823 (rpoS) (Fig. 1, lanes 1 and 4) indicating that other sigma factors are required for the expression of genes encoding these chitinases. However, the 58-kDa band was not visible in the extracellular proteins of mutants IC1270#20 and IC1270#93 (Fig. 1, lanes 2 and 3), demonstrating the GrrA/GrrS dependence of ChiA endochitinase production by strain IC1270.
FIG. 1.
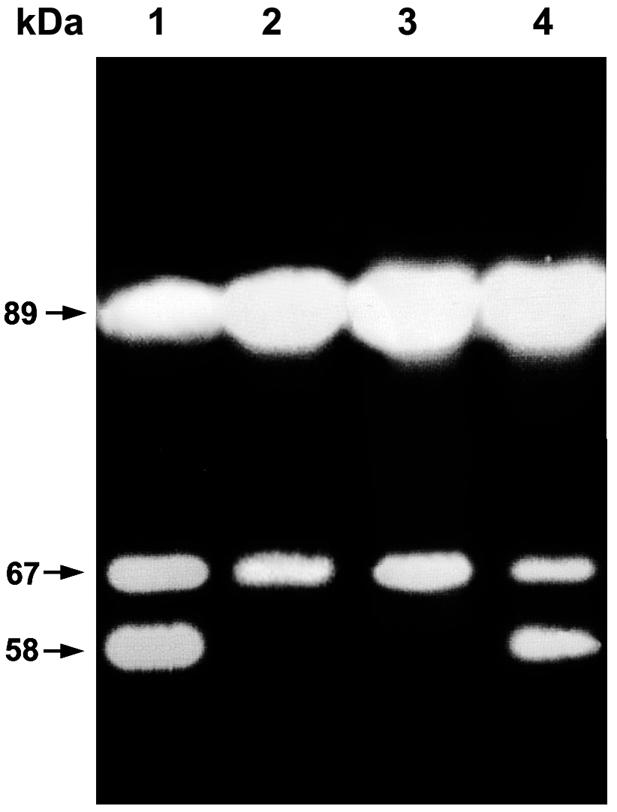
Detection of chitinolytic activity of extracellular proteins produced by S. plymuthica strain IC1270 and its derivatives grown on SM with chitin after separation by SDS-PAGE. Chitinolytic activity was detected with 4-methylumbelliferyl-β-d-N,N′-diacetylchitobioside as the substrate. Lanes 1 to 4, extracellular proteins from strains IC1270, IC1270#20 (grrA), IC1270#93 (grrS), and IC1270#823 (rpoS), respectively. Low-range prestained SDS-PAGE standards (Bio-Rad) were used as size markers.
(ii) Pyrrolnitrin, exoprotease, and siderophore production.
HPLC analysis demonstrated drastically decreased production of pyrrolnitrin by the IC1270#20 (grrA) and especially by the IC1270#93 (grrS) and IC1270#823 (rpoS) mutants relative to the parental strain, indicating that pyrrolnitrin production in IC1270 is under the control of the GrrA/GrrS and RpoS regulators (Table 2). The same appears to be true for the control of exoprotease production, since no zones of casein hydrolysis around the colonies of grrA, grrS, and rpoS mutants were observed on Bacto-litmus milk or skim milk agar plates. In contrast, IC1270 and all three mutants were equally proficient at siderophore production (Table 2).
TABLE 2.
Comparative characteristics of S. plymuthica IC1270 and its grrA, grrS, and rpoS mutants
| Property | Strain or mutanta
|
|||
|---|---|---|---|---|
| IC1270 | IC1270#20 (grrA) | IC1270#93 (grrS) | IC1270#823 (rpoS) | |
| Suppression of R. solani (halo size, mm)b | 14 ± 2.0 | 10 ± 1.4 | − | − |
| Suppression of P. aphanidermatum (halo size, mm)b | 13 ± 1.5 | 9 ± 1.5 | − | − |
| Pyrrolnitrin production (μg/ml)c | 108 ± 14 | 13.2 ± 3.1 | 2.8 ± 0.2 | 3.6 ± 0.4 |
| Exoprotease productionb | + | − | − | − |
| Siderophore production (mm)b | 19 ± 4 | 14.3 ± 3.2 | 12.6 ± 2.3 | 21 ± 4.1 |
| Biocontrol of R. solani in beans (IDR, %)d | 48.5a | 8.8b | 11.1b | 7.9b |
| Biocontrol of P. aphanidermatum in cucumber (seedling emergence)e | 95.3a | 74.6b | 60.8b | 76.4b |
| Biocontrol of P. aphanidermatum in cucumber (seedling survival)f | 84.8a | 58.5b | 56.0b | 53.6b |
| Biocontrol of P. aphanidermatum in cucumber (IDR, %)g | 68.5a | 37.5b | 24.4b | 29.7b |
+, detected; −, not detected.
Mean ± SD for four independent plates.
Data from HPLC analysis. Values are means ± SD from three replicate preparations.
Index of disease reduction (IDR) in treatments was calculated according to the formula 1 − [(percent of root rot-affected seedlings out of the total number of apparently healthy seedlings)/66.8] × 100, where percent disease in disease control, 66.8% ± 4.9%, was estimated as the number of root rot-affected seedlings divided by the total number of apparently healthy seedlings and multiplied by 100. The data from four independent experiments with five replicates for each treatment variant are summarized.
Percent seedling emergence was estimated as the percentage of germinated seedlings divided by the percentage of seedlings that emerged in a germination control (89.2 ± 3) multiplied by 100. In the disease control (treatment with water), percent seedling emergence was 33.6. Here and below, values not followed by the same letter were significantly different, as determined by the all-pairs Tukey-Kramer test at P = 0.05 with the JMP 5.0.1a (SAS Institute Inc., Cary, N.C.) program.
Percent postemergence seedling survival was calculated by dividing the number of seedlings that were alive 12 days after planting by the number of emerged seedlings and multiplying by 100. For the disease control, it was 29.0.
Calculated by the formula 1 − [(percentage of total disease in treatment)/(percent of total disease in disease control)] × 100, where the percentage of total (pre- and postgermination) disease in the disease control was calculated as [(number of nongerminated seedlings plus number of germinated seedlings affected by damping-off)/(average number of seedlings germinated in the germination control)] × 100. Data from three independent experiments with five replicates in each treatment are summarized.
(iii) Pattern of acyl-HSL signals.
Five reporter strains, C. violaceum CV026, A. tumefaciens NTL4/pZLR4, and three derivatives of strain E. coli S17-1 carrying pSB401, pSB536, or pSB1075 lux-based reporter plasmids (Table 1) were used to detect strain IC1270's ability to produce acyl-HSL signal molecules. With the CV026 reporter, specific for acyl-HSLs with side chains ranging from four to eight carbons in length (4, 26), pSB536, highly specific for the C4-HSL (N-butyryl-l-homoserine lactone [BHL]) signal molecule, and pSB1075, preferentially used for the detection of long, 10- to 14-carbon acyl-HSLs (47), we did not detect acyl-HSLs in petri dish or TLC assays with either the culture supernatants or the ethyl acetate extracts, indicating that strain IC1270 is unable to produce acyl-HSLs, produces them in small amounts, or produces acyl-HSLs that are undetectable by these reporters. However, the pSB401 reporter, which responds preferentially to C6 and C8 unsubstituted, 3-oxo- and 3-hydroxy-substituted molecules (47), exhibited bioluminescence when the ethyl acetate extract of strain IC1270 was applied to the petri dish in a rather large amount (data not shown).
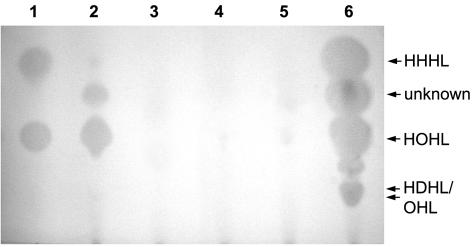
Although the A. tumefaciens NTL4/pZLR4 reporter was shown to be specific only for certain acyl-HSLs, being, for example, unable to detect BHL (41), the presence of several acyl-HSL molecules in ethyl acetate extracts of the strain IC1270 supernatant was observed by the appearance of blue spots in tests on petri dishes (data not shown) and by TLC analysis. Three blue spots appeared on TLC plates overlaid with this reporter (Fig. 2, lane 2). The Rf of the observed spots was similar to those in extracts of P. fluorescens strain 2-79 (Fig. 2, lane 6), whose pattern of acyl-HSL molecules was described previously (41).
FIG. 2.
Effect of global regulators on the production of acyl-HSLs by S. plymuthica IC1270. Acyl-HSLs were detected by TLC on RP-18 plates with A. tumefaciens NTL4/pZLR4 overlay. Lanes: 1, mixture of HHHL (10 μg ml−1, 8 μl) and HOHL (1 μg ml−1, 5 μl) standards; 2, 3, 4, and 5, ethyl acetate extracts of strains IC1270, IC1270#20, IC1270#93, and IC1270#823, respectively, 20 μl of each; 6, ethyl acetate extract of strain 2-79, 5 μl.
By comparing the Rf values of the three compounds from extracts of IC1270 and 2-79 and of chemical standards 3-hydroxy-C6-HSL (HHHL) and 3-hydroxy-C8-HSL (HOHL), we tentatively identified the acyl-HSLs produced by strain IC1270 as HHHL (Rf = 0.56), HOHL (Rf = 0.34), and an unidentified compound (Rf = 0.47) also visible in ethyl acetate extracts of strain 2-79 (41). However, it appears that strain IC1270 produced these acyl-HSLs, particularly HHHL, in relatively small amounts, since a fourfold higher volume of extract from IC1270 needed to be applied to see the spots in comparison with the volume of extract from strain 2-79 grown to the same optical density (Fig. 2). Mutants defective in grrA, grrS, or rpoS downregulated production of these acyl-HSLs by strain IC1270 (Fig. 2, lanes 3, 4, and 5), perhaps indicating indirect involvement of the GrrA/GrrS and RpoS global regulators in strain IC1270's quorum-sensing network.
Antifungal activity.
Strain IC1270 suppresses the growth of various fungal phytopathogens on plates, including R. solani and Pythium aphanidermatum (8). Large inhibition zones between these pathogenic fungi and the bacterium were observed, while grrS and rpoS mutants were deficient in this activity (Table 2). Under greenhouse conditions, IC1270 efficiently protected bean seedlings from R. solani root rot and cucumber seedlings from Pythium aphanidermatum damping-off diseases. We did not find differences in on-plate antifungal activity between the IC1279#20 (grrA) mutant and the parental strain (Table 2), probably because the residual level of pyrrolnitrin produced by this mutant is sufficient to suppress fungal growth in vitro. However, while the number of seedlings with root rot symptoms caused by R. solani decreased significantly when the strain IC1270 suspension was applied to the infested soil, all the mutants, including IC1279#20 (grrA), were deficient in biocontrol activity (Table 2). In the Pythium aphanidermatum-cucumber model, strain IC1270 suppressed preemergence damping-off and increased seedling survival (Table 2). As in the case of R. solani infection, all three mutants tested differed markedly from the parental strain in their biocontrol activity, being significantly less proficient in the ability to suppress both pre- and postemergence damping-off disease.
DISCUSSION
Three regulatory genes of S. plymuthica strain IC1270, cloned and characterized in this work, are homologues of the gacA/gacS two-component global regulation genes and transcription sigma factor rpoS gene described in many species of gram-negative bacteria (17, 19, 20, 25). The corresponding gacA/gacS homologues in strain IC1270 were tentatively designated grrA (for global response regulation activator) and grrS (for global response regulation sensor) in order to avoid any confusion that may arise from using the Pseudomonas-specific (and mnemonic) designation gac for the corresponding genes in unrelated S. plymuthica species. Both of these genes were found to be highly similar to sequences sm733b12.p1k and sm670e08.q1k from the genome of S. marcescens as well as to the corresponding genes of the GasA/GaS family described in other enteric bacteria (14, 19, 32, 33).
The relatively low identity between the grrA and grrS genes of S. plymuthica and their gacA and gacS homologues in fluorescent pseudomonads (47 and 50%, respectively) not only reflects the taxonomic distance between these bacteria but also fits with recent data showing the inability of a gacA probe developed from conserved sequences within the gacA genes of various Pseudomonas species to recognize similar genes in many other bacteria, including S. plymuthica, Pantoea agglomerans, and E. coli. Moreover, the gacA probe specific for Pseudomonas spp. was unable to detect plant-beneficial bacteria producing several antibiotics, chitinases, or siderophores other than pseudomonads from the rhizosphere (12).
The deduced GrrA and GrrS proteins were more similar to matching ExpA and ExpS proteins from P. carotovora subsp. carotovora (84 and 64% identity, respectively) and UvrY and BarA from E. coli (88 and 66% identity, respectively) than to GacA and GacS from P. fluorescens (57 and 37% identity, respectively). Similarly, database analysis revealed high (up to 94%) identity of S. plymuthica RpoS within regions 1 through 4, conserved among σ70 family sigma factors (25) with RpoS in E. coli and P. carotovora, whereas only 67% identity was found with RpoS from P. fluorescens. Likewise, the RpoD protein of strain IC1270 (GenBank accession number AY057390) was 93, 98, and 64% identical to the σ70 factors of E. coli, Salmonella enterica serovar Typhimurium, and P. fluorescens, respectively (M. Ovadis and L. Chernin, unpublished results).
Mutations in the grrA, grrS, and rpoS genes of S. plymuthica IC1270 significantly reduced the parental strain's ability to produce pyrrolnitrin (Table 2). Likewise, the ability of the GacA/GacS-type two-component systems and transcription sigma factors to regulate the formation of many extracellular compounds, including antibiotics contributing to biological control, has been described mainly in plant-associated fluorescent pseudomonads (9, 16, 38, 45). RpoS has been shown to be necessary for the expression of the pyrrolnitrin biosynthesis gene in biocontrol strains P. fluorescens Pf-5 (38, 45) and CHA0 (17). However, neither GrrA/GrrS nor RpoS affect siderophore production by strain IC1270, in agreement with the data of others showing that neither of these global regulators is involved in siderophore production by plant growth-promoting strains of Enterobacter cloacae (37) or Pseudomonas putida (22).
The pattern of acyl-HSLs detected in ethyl acetate extracts from strain IC1270 culture medium was very similar to that of strain P. fluorescens 2-79, known to produce mainly 3-hydroxyl-substituted derivatives of acyl-HSLs, including HHHL and HOHL (41). The reason for the similar acyl-HSL patterns in these unrelated bacterial species is unclear, but we observed similar acyl-HSL patterns between 2-79, several strains of diazotrophic bacteria of the genus Herbaspirillum, and the closely related genus Collimonas, both of which belong to the β-proteobacteria (X. Liu, W. de Boer, and L. Chernin, unpublished results).
Acyl-HSL production by strain IC1270 depends on GrrA/GrrS global regulation. This observation corresponds to other data showing that, in several strains of Pseudomonas, regulation by quorum sensing is typically under the control of a GacS/GacA system (5, 36). Furthermore, in P. carotovora subsp. carotovora and P. aeruginosa, quorum sensing is downregulated posttranscriptionally by the RsmA/RsmB and RsmA/RsmZ systems, respectively (10, 34). The inability of the rpoS mutant of strain IC1270 to produce acyl-HSLs demonstrates a relationship between stationary-phase sigma factor RpoS and quorum sensing in this bacterium. The reduction of acyl-HSL production was the most dramatic phenotypic change identified for rpoS mutants of the plant-pathogenic β-proteobacterium Ralstonia solanacearum (15). However, other data have shown that RpoS is not involved in the regulation of acyl-HSL production in P. putida WCS358 (22) or in the control of luminescence in Vibrio harveyi (24). Moreover, the rpoS null mutants displayed no difference in the accumulation of acyl-HSL signal molecules in Burkholderia cepacia (1), whereas they showed an elevated level of quorum-sensing gene transcription in Pseudomonas aeruginosa (46).
Positive control of chitinase production by the GacS/GacA system has been detected in plant-beneficial, root-colonizing strains of P. fluorescens BL915 (16) and Pseudomonas chlororaphis PCL1391 (Chin-A-Woeng, 2000, cited in reference 19), but the exact enzyme was not identified. In the grrA and grrS mutants of IC1270, we observed deficiency only in the 58-kDa ChiA chitinase but not the 89-kDa and 67-kDa exochitinases. Since rpoS mutant IC1270#823 was proficient at ChiA enzyme production, the grrA or gacS mutation is not likely to be exerting its effect via the repression of RpoS, the sigma factor known to be regulated by the GacA/GacS system (31, 45). In P. carotovora subsp. carotovora and P. fluorescens, the RsmA protein, proposed to be a downstream element in the GacS/GasA-type regulatory cascade promoting specific mRNA decay, needs to be scavenged by small regulatory RNAs such as RsmB, RsmZ, and RsmY to allow the translation of target mRNA (10, 18). Based on the taxonomic resemblance between S. plymuthica and P. carotovora, it can be assumed that similar posttranscriptional regulators in strain IC1270 are responsible for translation of chiA mRNAs as well as those of genes encoding exoprotease and pyrrolnitrin.
In plant growth-promoting fluorescent pseudomonads, global control by the GacA/GacS system has been shown to mediate the suppression of several fungal diseases, including Thielaviopsis basicola black root rot on tobacco (23), R. solani root rot on cotton (16), and Pseudomonas ultimum damping-off of cucumber (13). Our data showing that the IC1270#20 (grrA) and IC1270#93 (grrS) mutants deficient in ChiA chitinase and pyrrolnitrin production are also impaired in the suppression of R. solani and Pythium aphanidermatum demonstrates the involvement of two-component regulatory systems in biocontrol activity of other non-Pseudomonas plant-associated bacteria. The IC1270#823 (rpoS) mutant was deficient in both biocontrol activity and pyrrolnitrin production. In contrast, the RpoS mutant of strain P. fluorescens Pf-5 suppressed P. ultimum damping-off on cucumber as well as or even better than the wild-type strain (38).
Strain Pf-5 produces several antifungal antibiotics, including pyrrolnitrin, 2,4-diacetylphloroglucinol, and pyoluterin, but only pyrrolnitrin was not detected in the strain Pf-5 rpoS mutant, while production of two others increased significantly. Contrary to Pf-5, in strain IC1270 pyrrolnitrin appears to be the main antibiotic compound, and therefore the deficiency of grrA, grrS, and rpoS mutants in pyrrolnitrin production leads to a decline in the suppression of Pythium aphanidermatum, an oomycete fungus whose cell wall is composed mainly of β-glucan and cellulose and no appreciable chitin. This could explain why the rpoS mutant, which has the same pattern of chitinolytic enzymes as IC1270, and the grrA and grrS mutants, which are deficient in ChiA production, were similarly impaired in the biocontrol of this fungus.
The data presented here demonstrate that the ability to produce pyrrolnitrin is probably more important for strain IC1270's antifungal action than its chitinolytic activity. Nevertheless, other mechanisms, including competition for nutrients and induced resistance, recently shown to be involved in strain IC1270's biocontrol capability (28) could also be responsible for the observed residual biocontrol activity of its grrA, grrS, and rpoS mutants against both Pythium aphanidermatum and R. solani.
Supplementary Material
Acknowledgments
We thank S. Farrand, D. Mavrodi, S. Swift, L. Thomashow, P. Williams, and G. Zylistra for kind provision of strains and plasmids. We acknowledge S. Farrand, S. Swift, and P. Williams for helpful suggestions on the acyl-HSL assays and W. De Boer for help in 16S rRNA gene partial sequence analysis. Appreciation also goes to P. Williams and K.-H. van Pee for generous gifts of acyl-HSLs and pyrrolnitrin standards, respectively.
This research was supported in part by grant no. CA16-012, U.S.-Israel Cooperative Development Research Program, Economic Growth, U.S. Agency for International Development.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aguilar, C., I. Bertani, and V. Venturi. 2003. Quorum-sensing system and stationary-phase sigma factor (rpoS) of the onion pathogen Burkholderia cepacia genomovar I type strain, ATCC 25416. Appl. Environ. Microbiol. 69:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, D. B., and D. A. Zuberer. 1991. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 12:39-45. [Google Scholar]
- 3.Ausubel, F. M., R. Brendt, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. John Wiley & Sons Inc., New York, N.Y.
- 4.Camara, M., M. Daykin, and S. R. Chhabra. 1998. Detection, purification, and synthesis of N-acylhomoserine lactone quorum sensing signal molecules. Methods Microbiol. 27:319-330. [Google Scholar]
- 5.Chancey, S. T., D. W. Wood, and L. S. Pierson, III. 1999. Two component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 65:2294-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chernin, L. S., A. Brandis, Z. Ismailov, and I. Chet. 1996. Pyrrolnitrin production by an Enterobacter agglomerans strain with a broad spectrum of antagonistic activity towards fungal and bacterial phytopathogens. Curr. Microbiol. 32:208-212. [Google Scholar]
- 7.Chernin, L. S., L. De La Fuente, V. Sobolev, S. Haran, C. E. Vorgias, A. B. Oppenheim, and I. Chet. 1997. Molecular cloning, structural analysis, and expression in Escherichia coli of a chitinase gene from Enterobacter agglomerans. Appl. Environ. Microbiol. 63:834-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chernin, L. S., Z. Ismailov, S. Haran, and I. Chet. 1995. Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Appl. Environ. Microbiol. 61:1720-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbell, N. A., and J. E. Loper. 1995. A global regulator of second metabolite production in Pseudomonas fluorescens Pf-5. J. Bacteriol. 177:6230-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui, Y., A. Chatterjee, and A. K. Chatterjee. 2001. Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and harpin Ecc. Mol. Plant-Microbe Interact. 14:516-526. [DOI] [PubMed] [Google Scholar]
- 11.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Souza, J. T., M. Mazzola, and J. M. Raaijmakers. 2003. Conservation of the response regulator gene gacA in Pseudomonas species. Environ. Microbiol. 5:1328-1340. [DOI] [PubMed] [Google Scholar]
- 13.Duffy, B. K., and G. Defago. 2000. Controlling instability in gacS-gacA regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. J. Bacteriol. 66:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eriksson, A. R., R. A. Andersson, M. Pirhonen, and E. T. Palva. 1998. Two-component regulators involved in the global control of virulence in Erwinia carotovora subsp. carotovora. Mol. Plant-Microbe Interact. 11:743-752. [DOI] [PubMed] [Google Scholar]
- 15.Flavier, A. B., M. A. Schell, and T. P. Denny. 1998. An RpoS (σs) homologue regulates acylhomoserine lactone-dependent autoinduction in Ralstonia solanacearum. Mol. Microbiol. 28:475-486. [DOI] [PubMed] [Google Scholar]
- 16.Gaffney, T. D., S. T. Lam, J. Ligon, K. Gates, A. Frazelle, J. Di Maio, S. Hill, S. Goodwin, N. Torkewitz, A. M. Allshouse, H. J. Kempt, and J. O. Becker. 1994. Global regulation of expression of antifungal factors by a Pseudomonas fluorescens biological control strain. Mol. Plant-Microbe Interact. 7:455-463. [DOI] [PubMed] [Google Scholar]
- 17.Haas, D., and C. Keel. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41:117-153. [DOI] [PubMed] [Google Scholar]
- 18.Heeb, S., C. Blumer, and D. Haas. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184:1046-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol. Plant-Microbe Interact. 14:1351-1363. [DOI] [PubMed] [Google Scholar]
- 20.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 22.Kojic, M., G. Degrassi, and V. Venturi. 1999. Cloning and characterization of the rpoS gene from plant growth-promoting Pseudomonas putida WCS358: RpoS in not involved in siderophore and homoserine lactone production. Biochim. Biophys. Acta 1489:413-420. [DOI] [PubMed] [Google Scholar]
- 23.Laville, J., C. Voisard, C. Keel, M. Maurhofer, G. Defago, and D. Haas. 1992. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root of tobacco. Proc. Natl. Acad. Sci. USA 89:1562-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, Y. H., C. Miyamoto, and E. A. Meighen. 2002. Cloning, sequencing, and functional studies of the rpoS gene from Vibrio harveyi. Biochem. Biophys. Res. Commun. 293:456-462. [DOI] [PubMed] [Google Scholar]
- 25.Lonetto, M., M. Gribskov, and C. A. Gross. 1992. The σ70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 174:3843-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chabra, M. Camara, M. Daykin, J. H Lamb, S. Swift, B. W. Bycroft, G. S. A. B. Stewart, and P. Williams. 1997. Quorum sensing in Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 27.McGrew, B. R., and D. M. Green. 1990. Enhanced removal of detergent and recovery of enzymatic activity following sodium dodecyl sulfate-polyacrylamide gel electrophoresis: use of casein in gel wash buffer. Anal. Biochem. 189:68-74. [DOI] [PubMed] [Google Scholar]
- 28.Meziane, H., L. Chernin, and M. Höfte. 2003. Biological control of green mould on citrus fruits and the induction of resistance on bean by Pantoea agglomerans strain IC1270, p. 515-520. In J. W. Kloepper (ed.), Plant growth-promoting rhizobacteria. Indian Institute of Spices Research, Calicut, Kerala, India.
- 29.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 30.Morrissey, J. P., U. F. Walsh, A. O'Donnell, Y. Moenne-Loccoz, and F. O'Gara. 2002. Exploitation of genetically modified inoculants for industrial ecology applications. Antonie van Leeuwenhoek 81:599-606. [DOI] [PubMed] [Google Scholar]
- 31.Mukhopadhyay, S., J. P. Audia, R. N. Roy, and H. E. Schellhorn. 2000. Transcriptional induction of the conserved alternative sigma factor RpoS in Escherichia coli is dependent on BarA, a probable two-component regulator. Mol. Microbiol. 37:371-381. [DOI] [PubMed] [Google Scholar]
- 32.Nagasawa, S., S. Tokishita, H. Aiba, and T. Mizuno. 1992. A novel sensor-regulator protein that belongs to the homologous family of signal-transduction proteins involved in adaptive responses in Escherichia coli. Mol. Microbiol. 6:799-807. [DOI] [PubMed] [Google Scholar]
- 33.Pernestig, A. K., O. Melefors, and D. Georgellis. 2001. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J. Biol. Chem. 276:225-231. [DOI] [PubMed] [Google Scholar]
- 34.Pessi, G., F. Williams, Z. Hindle, K. Heurlier, M. T. G. Holden, M. Cámara, D. Haas, and P. Williams. 2001. The global post-transcriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183:6676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierson, L. S., III, D. W. Wood, and E. A. Pierson. 1998. Homoserine lactone-mediated gene regulation in plant-associated bacteria. Annu. Rev. Phytopathol. 36:207-225. [DOI] [PubMed] [Google Scholar]
- 36.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas.1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 37.Saleh, S. S., and B. R. Glick. 2001. Involvement of gacS and rpoS in enhancement of the plant growth-promoting capabilities of Enterobacter cloacae CAL2 and UW4. Can. J. Microbiol. 47:698-705. [DOI] [PubMed] [Google Scholar]
- 38.Sarniguet, A., J. Kraus, M. D. Henkels, A. M. Muehlchen, and J. E. Loper. 1995. The sigma factor σS affects antibiotic production and biological control activity of Pseudomonas fluorescens Pf-5. Proc. Natl. Acad. Sci. USA 92:12255-12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnider, U., C. Keel, C. Blumer, J. Troxler, G. Defago, and D. Haas. 1995. Amplification of the housekeeping sigma factor in Pseudomonas fluorescens CHA0 enhances antibiotic production and improves biocontrol abilities. J. Bacteriol. 177:5387-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 41.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 43.Thomashow, L. S., and D. M. Weller. 1988. Role of phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 170:3499-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trigilia, T., M. G. Peterson, and D. J. Kemp. 1988. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 16:8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whistler, C. A., N. A. Corbell, A. Sarniguet, W. Ream, and J. E. Loper. 1998. The two-component regulators GacA and GacS influence accumulation of the stationary-phase sigma factor σS and the stress response in Pseudomonas fluorescens Pf-5. J. Bacteriol. 180:6635-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winson, M. K., S. Swift, L. Fish, J. P. Throup, F. J.ørgensen, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. A. B. Stewart. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163:185-192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.