Abstract
Salmonella strains utilize a type III secretion system for their successful survival and replications inside host cells. SseF is one of the several effector proteins that are required for conferring this survival ability by altering the trafficking of the Salmonella-containing vacuoles. These effector proteins often require appropriate chaperones to maintain their stabilities inside the bacteria. These chaperones are also known to assist the subsequent secretion and translocation of their substrates. We report here that SscB acts as the chaperone for SseF, an effector for the Salmonella pathogenicity island 2 (SPI-2). We found that the sscB gene is required for the formation of Salmonella sp.-induced continuous filaments in epithelial cells. Efficient Salmonella replication in macrophages requires SscB function. Intracellular and secretion levels of SseF are greatly reduced in an sscB mutant strain compared to the wild-type strain. A protein stability assay demonstrated that the half-life of SseF is significantly shortened in the absence of SscB. Transcriptional analysis of the sseF gene showed that the effect of SscB on the SseF level is not at the transcriptional level. A coprecipitation experiment indicated that SscB interacts with SseF. In summary, our results indicate that SscB is a chaperone for SPI-2 effector SseF to facilitate its secretion and function inside the host cells.
Salmonella species cause a number of food-borne diseases in humans and other warm-blooded animals; these diseases range from mild intestinal diarrhea to the more severe typhoid fever (1, 42). Virulent Salmonella strains possess the ability to successfully enter, survive, and replicate inside mammalian cells including professional macrophages. Salmonella species stay inside a closed membrane compartment after entry. Previous studies have demonstrated that Salmonella play a key role in subverting the cellular process for the biogenesis of the Salmonella-containing vacuoles (SCVs) (4, 12, 15, 17, 22, 26, 37, 44, 45, 47, 50). Salmonella enterica serovar Typhimurium encodes two type III secretion systems (TTSS) within the Salmonella pathogenicity island 1 (SPI-1) and SPI-2 (14, 40). These type III protein secretion and translocation systems function to inject a number of bacterial proteins (effectors) into the host cells to promote bacterial entry and subsequent survival inside the SCV (14, 40). These two TTSS have distinct functions during Salmonella-host interactions. Whereas SPI-1-encoded TTSS injects effector proteins into host cells to trigger invasion (51), the SPI-2-encoded TTSS injects effector proteins to facilitate the biogenesis of SCV inside the cells (25). Effector proteins contain secretion and translocation signals that are often located in their N termini, presumably recognized by the TTSS (30).
Genes located in SPI-2 encode proteins that make up the type III secretion apparatus, serve as transcriptional regulators, and function as effector proteins inside the host cells (22). A number of effector proteins are also encoded outside the SPI-2 locus. Most genes encoding components and effectors of the SPI-2 type III secretion are clustered and appear to be in the same transcriptional operon (19-21). This operon includes sseA-G and sscA-B. It was recently determined that SseB, SseC, and SseD are secreted proteins and are required for the translocation of other effector proteins (32). SseF and SseG, are putative effector proteins implicated in the formation of Salmonella-induced aggregation of host endosomes (17). More-recent studies have suggested that SseG is responsible for targeting Salmonella to the host Golgi network through poorly understood mechanisms (38).
Effector proteins often require appropriate chaperones to maintain their stabilities and subsequent secretion and translocation across the TTSS into mammalian cells (33). Several studies have described chaperones for SPI-1 secretion apparatus and effectors, including InvB, SicA, and SicP which are encoded within SPI-1 (3, 8, 13, 46). SseA is the only reported chaperone for the SPI-2-encoded secretion apparatus proteins SseB and SseD (7, 36, 53). No chaperones have been reported for any of the SPI-2 effector proteins, although SscA and SscB have been proposed to act as chaperones based on sequence analysis (6, 22). However, there is no experimental evidence to support this hypothesis.
Some of the hallmarks for putative type III effector chaperones are (i) that they are small proteins with a low molecular mass of <15 kDa; (ii) that genes encoding the chaperones are often situated in the vicinity of genes encoding their corresponding substrates; (iii) that these chaperones possess amphipathic helices at their C termini; and (iv) that these chaperones often have acidic pIs. SscB has been proposed as a chaperone based on its 23% identity and 36% similarity over 98 amino acid residues to IppI, a chaperone for Shigella flexneri invasion proteins (2, 22). Consistent with being a chaperone, SscB has a predicted molecular mass of 16.3 kDa and an acidic pI of 4.92 and is also predicted to contain a mostly alpha-helical secondary structure that extends throughout its entire length. In addition, the sscB gene is located immediately upstream of the sseF gene in the effector/chaperone region of SPI-2. We present evidence here that SscB is a chaperone for SseF.
MATERIALS AND METHODS
Bacterial strains.
Bacterial strains and plasmids used in the present study are listed in Table 1. Escherichia coli and serovar Typhimurium strains are routinely cultured in Luria-Bertani (LB) broth. Salmonella trains were grown under SPI-1 TTSS-inducing conditions (LB broth with 0.3 M NaCl) for all of the invasion experiments. When SPI-2 TTSS-inducing conditions were desired, the strains were grown in MgM minimal medium adjusted to either pH 7.0 (MgM7) or pH 5.0 (MgM5) (10). Antibiotics were used at the indicated concentrations: ampicillin at 120 μg ml−1, streptomycin at 25 μg ml−1, kanamycin at 40 μg ml−1, and tetracycline at 12 μg ml−1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| S. enterica serovar Typhimurium | ||
| SL1344 | Wild-type serovar Typhimurium; Strr | 23 |
| ZP18 | ΔssaV | This study |
| ZP19 | ΔsseF | This study |
| ZP22 | sscB::aphT | This study |
| ZP31 | ΔsseF::lacZ | This study |
| ZP33 | ΔsseF::lacZ sscB::aphT | This study |
| ZP39 | ΔssaV::lacZ | This study |
| ZP40 | ΔssaV::lacZ sscB::aphT | This study |
| ZP36 | ΔssaV ΔsseF sscB::aphT | This study |
| ZP41 | ΔssaV sscB::aphT | This study |
| E. coli | ||
| DH5αMCR | F−mcrA Δ(mrr-hsdRMS-mcrBC) | Gibco-BRL |
| SM10λpir | thi thr leu tonA Δacy supE recA::RP4-2-Tc::Mu(Kmr)λpir F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 | 31 |
| Top10 | deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pSB890 | R6K-derived ori; Tcr | 24 |
| pWSK29 | pSC101 ori; Apr | 48 |
| pZP226 | ΔssaV | This study |
| pZP227 | ΔsseF | This study |
| pZP259 | sscB::aphT | This study |
| pSB1040 | Promoterless lacZ gene | 34 |
| pZP369 | ΔsseF::lacZ | This study |
| pZP428 | ΔssaV::lacZ | This study |
| pZP281 | Arabinose-inducible sseF-M45 | This study |
| pZP284 | Arabinose-inducible sscBsseF-M45 | This study |
| pZP430 | Arabinose-inducible sifB-M45 | This study |
| pZP434 | Arabinose-inducible bicistronic sseF-M45sscB-his | This study |
| pZP445 | Arabinose-inducible bicistronic sseF-M45 ADF-his | This study |
| pZP492 | Arabinose-inducible sseJ-M45 | This study |
| pZP545 | ProsseA in pWSK29 | This study |
| pZP546 | sseF-M45 under ProsseA | This study |
| pZP549 | sscBsseF-M45 under ProsseA | This study |
| pZP548 | sscB-M45 under ProsseA | This study |
Strr, streptomycin resistance; Kmr, karamycin resistance; Tcr, tetracycline resistance; Apr, ampicillin resistance.
Mammalian cell lines and bacterial infection assay.
The murine macrophage RAW264.7 (TIB-71, American Type Culture Collection [ATCC]) and the human epithelial cell line HeLa (CCL-2 [ATCC]) were purchased from the ATCC cell biology stock center (Manassas, Va.). Both were maintained in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum. Salmonella infection of mammalian cells was conducted as previously described (52) at a multiplicity of infection (MOI) of 10. Bacterial infection of RAW264.7 macrophages and survival assays were carried out as described before (9, 22). Briefly, macrophages were seeded in 24-well plates at a density of 5 × 105 cells/well 24 h before infection. Bacteria were cultured to early stationary phase in LB medium at 37°C and diluted to an optical density at 600 nm of 0.1. The bacteria were opsonized for 20 min in DMEM containing 10% normal mouse serum (Gemini Bio-Products, Woodland, Calif.) at 37°C. Opsonized bacteria were added to RAW264.7 macrophage monolayer at an MOI of 10. Bacterial attachment was facilitated by centrifugation at 500 × g for 5 min at room temperature. The bacteria-cell mixtures were incubated for 30 min at 37°C in 5% CO2. After infection, the macrophages were washed twice with phosphate-buffered saline (PBS) to remove extracellular bacteria and incubated further in DMEM containing 10% fetal bovine serum and 16 μg of gentamicin ml−1. At 2 and 24 h after gentamicin treatment, infected macrophages were washed three times in PBS and lysed with 1% Triton X-100 and 0.1% sodium dodecyl sulfate (SDS). Samples were then serially diluted and plated on selective medium to enumerate the intracellular bacteria. The extent of replication was then determined by dividing the number of intracellular bacteria at 24 h by the number at 2 h.
Strain and plasmid constructions.
In-frame chromosomal deletions of genes in Salmonella strains were generated by using an allelic-exchange suicide vector as previously reported (24). Briefly, the DNA fragment with the in-frame deletion was cloned into the suicide vector pSB890. The resulting plasmid constructs (pZP226 for ssaV deletion and pZP227 for sseF) were transferred into serovar Typhimurium via conjugation, where they were integrated into the chromosome by homologous recombination. In order to construct the sscB mutant, a HincII promoterless kanamycin resistance gene cassette from pSB1046 was introduced into the SmaI site of the sscB gene. The inactivated sscB gene was then cloned into the suicide vector pSB890 to generate pZP259. Plasmid pZP259 was subsequently transferred into Salmonella by conjugation and integrated into the chromosome by homologous recombination as described above. To generate expression plasmids from the SseA promoter, a 450-bp DNA fragment containing the SPI-2 promoter ProsseA was obtained as described before (18) and cloned into the HindIII/EcoRI-digested pWSK29 to obtain pZP545. Various SPI-2 genes with the M45 tag were subcloned into the EcoRI/XbaI-digested pZP545.
To generate M45-tagged or His-tagged fusion proteins, the entire gene encoding the corresponding full-length protein was amplified by PCR and cloned into pBAD24 by using standard molecular biology techniques (29). The sseF gene was amplified by PCR with the primers 5′-CGGAATTCGCATGAAAATTCATATT-3′ and 5′CGCCATGGATGGTTCTCCCCGAGAT-3′, and the sifB gene was amplified with the primers 5′-GCGAATTCCCATGCCAATTACTATCGGGAGAG-3′ and 5′-GCGGATCCACTCTGGTGATGAGCCTCATT-3′. The sseJ gene was amplified with the primers 5′-GGAATTCCCATGCCATTGAGTGTTGG-3′ and 5′-GCGGATCCTTCAGTGGAATAATG-3′, and the sscB gene was amplified with the primers 5′-CGGAATTCGTATGATGATGAAAGA-3′ and 5′-GCGATATCAGCAATAAGAGTATCAA-3′.
Intracellular protein level and secretion.
The desired serovar Typhimurium strains were grown under SPI-2-inducing conditions described previously (10). Briefly, to check the intracellular level of different proteins, ZP18 (ΔssaV) and ZP41 (ΔssaV ΔsscB) containing pZP281 and pZP546 (encoding SseF-M45), pZP284 and pZP549 (encoding SscB and SseF-M45), pZP430 and pZP556 (SifB-M45), or pZP492 (SseJ-M45) were grown at 37°C for 8 h in LB medium. Bacterial cultures were washed twice and diluted 1:100 in MgM5 or MgM7. The cultures were grown with agitation at 200 rpm for 16 additional hours. Production of fusion proteins was induced by addition of 1 mM arabinose during the last 2 h of growth when expressed from the pBAD promoter. Bacteria were then pelleted by centrifugation at 10,000 × g for 20 min. Cells were lysed with SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer and subjected to SDS-PAGE analysis and immunoblotting. For protein secretion assays, wild type and ZP22 (sscB) containing pZP281 (encoding SseF-M45), pZP284 (encoding SscB and SseF-M45), pZP430 (SifB-M45), or pZP492 (SseJ-M45) were cultured as described above. Bacteria were pelleted by centrifugation at 10,000 × g, and the supernatants passed through a 0.2-μm-pore-size filter. Secreted proteins were collected by precipitation with 10% trichloroacetic acid and resuspended in SDS-PAGE sample buffer, followed by SDS-PAGE and Western blot analysis with anti-M45 antibody.
Transcriptional fusions and β-galactosidase assay.
A promoterless lacZ gene was introduced into the same XbaI site of pZP226 (in-frame deletion of ssaV in the suicide vector pSB890) and pZP227 (in-frame deletion of sseF in the suicide vector pSB890), respectively. The resulting plasmids pZP428 (ΔssaV::lacZ) and pZP369 (ΔsseF::lacZ) were then introduced into wild-type Salmonella or into the sscB mutant strain via conjugation and integrated into the chromosome by homologous recombination. The resulting strains—ZP31 (ΔsseF::lacZ), ZP33 (sscB ΔsseF::lacZ), ZP39 (ΔssaV::lacZ), and ZP40 (sscB ΔssaV::lacZ)—were grown overnight in LB medium, washed twice and diluted 1:50 in MgM5 or MgM7, and grown for 6 h. β-Galactosidase assays were performed as previously described (39).
Protein stability assay by gentamicin treatment.
The desired serovar Typhimurium strains, ZP18 (ΔssaV) and ZP41 (ΔssaV sscB::aphT) containing pZP281 (encoding SseF-M45) or pZP430 (SifB-M45) were grown under SPI-2-inducing conditions. Fusion proteins were induced by addition of 1 mM arabinose for 2 h. Bacteria were collected by centrifugation, and the pellets were washed three times with PBS to remove the residual arabinose. Gentamicin was added to 100 μg ml−1. Samples were taken at different times after the addition of gentamicin. Bacterial lysates were analyzed by SDS-PAGE and Western blotting as described above.
Protein copurification.
Serovar Typhimurium strain ZP36(ΔssaV ΔsseF sscB::aphT) carrying plasmid pZP434 (encoding SseF-M45 and His-SscB), pZP284 (encoding SseF-M45 and SscB), or pZP445 (encoding SseF-M45 and His-ADF) were grown in SPI-2-inducing minimal medium as described above and the expression of SseF-M45 and His-SscB were induced by addition of 1 mM arabinose. Cells were collected by centrifugation and lysed by sonication in lysis buffer containing 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, and 1 mM phenylmethylsulfonyl fluoride (pH 8.0). Slurry of Ni-nitrilotriacetic acid (NTA) agarose beads (Qiagen, Valencia, Calif.) was then added to the lysates, followed by incubation for 1 h at 4°C with rotation. The Ni-NTA beads were pelleted by centrifugation, washed five times with 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, and 1 mM phenylmethylsulfonyl fluoride (pH 8.0). Copelleted proteins were dissolved by adding SDS-PAGE sample buffer, followed by boiling for 5 min. Samples were analyzed by SDS-PAGE and Western blotting with anti-M45 antibodies.
Immunofluorescence.
For immunofluorescence analyses, HeLa cells were infected for 30 min with Salmonella at an MOI of 10 as described before. Extracellular bacteria were removed by washing with PBS and incubated with DMEM containing 10% fetal calf serum supplemented with gentamicin (100 μg ml−1) for 1 h. The medium was subsequently replaced with DMEM containing 10% fetal calf serum and 16 μg of gentamicin ml−1 for the remainder of the experiment. After infection, HeLa cells were washed three times with PBS and fixed with 3% paraformaldehyde for 15 min at room 22°C temperature before being permeabilized with 0.2% Triton X-100 in PBS. Cells were incubated with the primary antibody for 30 min after being blocked with 5% skim milk, washed three times with PBS, and incubated with the secondary antibody for 30 min. Serovar Typhimurium were identified by using rabbit anti-Salmonella O-antigen group B (Difco) and a secondary anti-rabbit antibody-Texas red conjugant (Molecular Probes, Eugene, Oreg.). LAMP-2 was detected with mouse anti-human LAMP-2 (H4B4; Developmental Studies Hybridoma Bank, The University of Iowa) and a secondary anti-mouse AF488 conjugant (Molecular Probes).
RESULTS
The sscB gene is required for the stability of SseF.
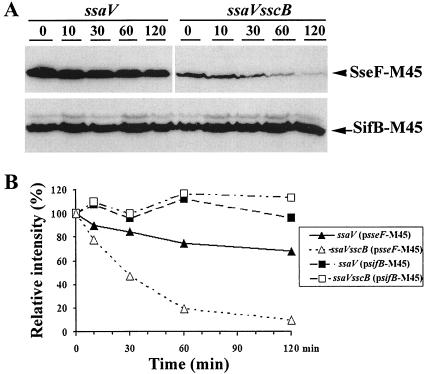
Chaperones for the type III secretion apparatus and related effectors often affect the stability and/or secretion of their cognate substrates (33). To investigate the possible role of SscB as a chaperone for SseF, the intracellular level of SseF was examined in the ssaV mutant and the sscB ssaV double-mutant strains. Examining the intracellular level of SseF in an ssaV mutant background eliminated the potential secretion variation because this strain is unable to secrete any known SPI-2 effectors (21). To facilitate the detection of SseF, a plasmid expressing a M45-tagged SseF from its native promoter (ProsseA) was introduced into the ssaV mutant and the sscB ssaV double-mutant strains. Levels of SseF-M45 were determined by Western blotting with a monoclonal antibody to the M45 epitope when grown in SPI-2-inducing MgM5 or MgM7 minimal medium. As shown in Fig. 1A, the intracellular levels of SseF in whole-cell lysates were not detected in the absence of SscB but were restored by coexpressing the sscB gene with sseF-M45 in the sscB ssaV mutant strain (Fig. 1A). This result suggests that SscB is required for the stability of cytoplasmic SseF.
FIG. 1.
Effect of SscB on the intracellular level of SseF. The secretion-deficient ssaV mutant strain (ZP18) and the ssaV sscB double-mutant strain (ZP41) carrying plasmids expressing either SseF-M45 (pZP281 and pZP546), SscBSseF-M45 (pZP284 and pZP549), SifB-M45 (pZP430), or SseJ-M45 (pZP492) were grown in the SPI-2-inducing MgM5 or MgM7 minimal medium, and protein expressions were either driven by the sseA promoter (A) or were induced by the addition of 1 mM arabinose (panel B). Bacteria were collected and lysed, and protein levels were determined as described in Materials and Methods.
To investigate whether the sscB mutant has a general defect in protein stability, we determined the protein levels of SifB and SseJ, two other SPI-2-secreted effectors. Under similar conditions as described above, we failed to detect SifB and SseJ from the ProsseA promoter in the presence or absence of SscB (data not shown). Therefore, we sought out to express SifB and SseJ from an arabinose-inducible promoter (pBAD24) promoter. To aid the detection of SseF, SifB, and SseJ, plasmids expressing M45-tagged SseF, SifB, and SseJ from an arabinose-inducible promoter (pBAD24) were constructed. These expression plasmids were then introduced into the ssaV mutant and the sscB ssaV double-mutant strains as described above. Production of the fusion proteins was induced with arabinose in SPI-2-inducing minimal medium, and the levels of SseF-M45, SifB-M45, and SseJ-M45 were determined by Western blotting. Consistent with data shown above (Fig. 1A), intracellular SseF in the whole-cell lysates was considerably less in the absence of SscB (Fig. 1B). This decrease was complemented by coexpressing sscB with sseF-M45 in the sscB ssaV mutant strain (Fig. 1). In contrast, the levels of SifB and SseJ did not change significantly in the presence or absence of SscB (Fig. 1B). This result suggests that SscB functions for the stability of intracellular SseF but not for SifB and SseJ.
To further demonstrate that SscB affects the level of SseF by increasing its stability, we examined the half-life of SseF in the presence or absence of SscB. Levels of SseF-M45 and SifB-M45 in the whole bacterial lysates were monitored over time by Western blotting after the addition of gentamicin, an antibiotic that stops protein synthesis in bacteria. Intracellular levels of SseF-M45 were monitored in the ssaV mutant and the ssaV sscB double-mutant strains. As shown in Fig. 2, although >70% of SseF-M45 was still detectable in the ssaV mutant strain, <10% remained in the ssaV sscB double-mutant strain 90 min after treatment with gentamicin. In addition, the level of SseF-M45 in the ssaV mutant strain was significantly higher than that in the ssaV sscB double-mutant strain at time zero. This is consistent with our previous results demonstrating that SscB affects the intracellular level of SseF-M45. In contrast, the levels of SifB-M45 in both the ssaV and the ssaV sscB mutant strains did not show any significant change over the time course of the experiments. These results suggested that SscB specifically affects the level of SseF by influencing its stability.
FIG. 2.
Effect of SscB on the stability of SseF. The secretion-deficient strains ZP18 (ΔssaV) and ZP41 (ΔssaV sscB::aphT), harboring pZP281(encoding SseF-M45) or pZP430 (encoding SifB-M45), were grown in MgM7, and protein expression was induced with 1 mM arabinose. Gentamicin (100 μg ml−1) was then added to inhibit protein synthesis. (A) The levels of SseF-M45 and SifB-M45 were examined at different times after gentamicin treatment by Western blotting with anti-M45 antibody. (B) The protein amounts were quantified by densitometry scanning. Experiments were repeated three times.
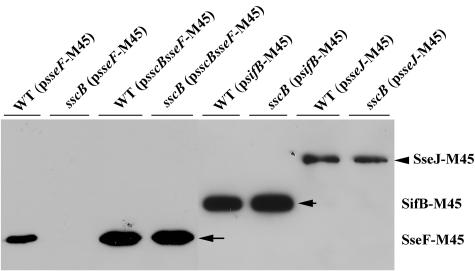
The sscB gene is required for SseF secretion.
Chaperones of the TTSS are not only required for the stability of their corresponding substrate but are also necessary for their secretion. We thus examined the effect of SscB on the secretion of SseF. A plasmid expressing the M45-tagged SseF was introduced into the wild type and into the sscB mutant strain. After growth in acidic SPI-2-inducing minimal medium, the secreted proteins were collected by trichloroacetic acid precipitation, and the amounts of SseF were examined by Western blotting with the monoclonal anti-M45 antibody. The sscB mutation completely abolished extracellular SseF, whereas it had no obvious effect on the extracellular level of SifB and SseJ (Fig. 3), indicating that the SPI-2 secretion pathway is not impeded in the sscB mutant strain. SseF secretion can be rescued by coexpressing sscB with sseF-M45 in the sscB mutant strain (Fig. 3). This result indicates that SscB influences not only the intracellular levels of SseF but also its secretion.
FIG. 3.
Effect of SscB on the secretion of SseF. Secretion of SseF-M45 (pZP281), SifB-M45 (pZP430), and SseJ-M45 (pZP492) were detected in either wild type (SL1344) or an sscB mutant strain (ZP22). Bacteria were grown in MgM5, and the culture supernatants were collected after centrifugation. Secreted proteins were precipitated and analyzed as described in Materials and Methods.
SscB does not significantly affect sseF transcription.
It has been reported that some chaperones affect the transcription and even the translation of their cognate substrates (8, 46). The selective effect of SscB on SseF could be partly explained if SscB alters the transcription of the sseF gene. To investigate this possibility, we constructed a transcriptional fusion of lacZ to the intact sseF gene and to ssaV, an SPI-2 TTSS apparatus gene, in the chromosome of the wild type or the sscB mutant strain. β-Galactosidase activities were determined in bacteria grown in SPI-2-inducible minimal medium MgM5 or MgM7. As shown in Fig. 4, there was no significant difference in the expression of lacZ fused to sseF or ssaV in the presence or absence of SscB. This result suggests that SscB exerts its effect on the level of SseF posttranscriptionally.
FIG. 4.
Effect of the sscB mutation on sseF transcription. Transcription of sseF-lacZ (A) and ssaV-lacZ (B [control]) were measured in the wild type or in the sscB mutant strain containing a chromosomal fusion of lacZ with sseF (ZP31 and ZP33, respectively) or ssaV (ZP39 and ZP40, respectively). Stationary-phase bacteria grown in LB medium were washed twice, diluted in MgM5 or MgM7, and grown for an additional 6 h. Samples were then collected and assayed for β-galactosidase activities (Miller units). The experiments were carried out three times with similar results.
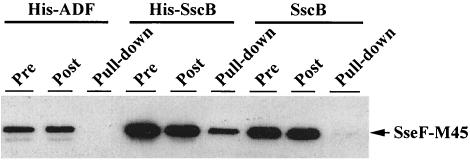
SscB interacts with SseF.
Chaperones often exert their function by associating specifically with their cognate substrates (41, 49). To examine the interaction between SscB and SseF, we coexpressed the M45-tagged SseF with His-tagged SscB from a plasmid with an arabinose-inducible promoter in the secretion-deficient Salmonella ssaV sseF sscB triple-mutant strain. Strains harboring the plasmid were grown in SPI-2-inducing minimal medium or LB medium in the presence or absence of arabinose. His-SscB was precipitated by the addition of Ni-NTA agarose beads, and His-SscB-associated proteins were then examined by Western blotting with an anti-M45 antibody. As a control, an untagged SscB and His-ADF (for actin depolymerization factor in eukaryotes) were produced in the same plasmid, and “pull-down” experiments carried out in an identical manner. As shown in Fig. 5, SseF-M45 was detected in the precipitated His-SscB fraction when His-SscB was used. In contrast, SseF-M45 was not detected when an untagged SscB or His-ADF was used in place of His-SscB (Fig. 5). These data showed that SscB specifically interacts with SseF, supporting the hypothesis that SscB serves as the chaperone for SseF.
FIG. 5.
SseF interacts with SscB in a coprecipitation assay. Serovar Typhimurium mutant strain ZP36 (ΔssaV ΔsseF sscB::aphT) containing plasmids expressing SseF-M45 and His-SscB (pZP434), SseF-M45 and SscB (pZP284), or SseF-M45 and His-ADF (pZP445) was grown in MgM7, and protein expression was induced with 1 mM arabinose. His-tagged proteins were precipitated from lysates by Ni-NTA agarose beads. Samples were taken before the addition of the beads (Pre) and after bead binding (Post). Coprecipitated proteins (Pull-down), Pre, and Post samples were all examined by Western blotting with the anti-M45 antibody.
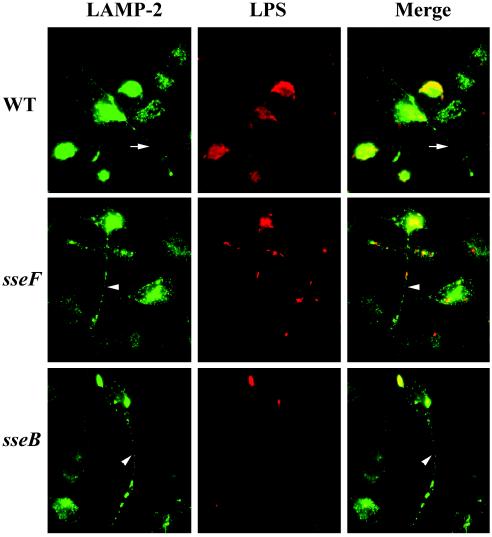
The sscB gene is required for the formation of continuous Salmonella-induced filaments.
Previous studies have shown that Salmonella infection induces the formation of filamentous structures (Sif) that are rich in lysosomal membrane glycoproteins (16). Salmonella-induced Sifs are connected to the bacterium-containing vacuoles (16). The formation of these filaments is dependent on the function of SPI-2 (26). Interestingly, strains that are deficient in SseF were still capable of forming aggregated endosomes, but the distribution of lysosomal membrane glycoproteins appeared to be punctate rather than continuous (26).
The ability of the sscB mutant strain to form filamentous structures in HeLa cells was analyzed by immunofluorescence microscopy. Consistent with previous studies, wild-type Salmonella sp. induced aggregation of LAMP-2-containing filamentous structures (Fig. 6). In contrast to the continuous distribution of LAMP-2 along Sifs induced by the wild-type Salmonella, the sscB mutant strain induced Sifs displaying punctate distribution of LAMP-2 (Fig. 6). This discontinuous distribution pattern of LAMP-2 is similar to the “pseudo-Sif” induced by the sseF mutant strain as reported previously (26). This result is consistent with the hypothesis that SscB functions as a chaperone for SseF.
FIG. 6.
The sscB mutation induced discontinuous Sifs. HeLa cells were infected with wild-type (SL1344), the sseF mutant (ZP19), or the sscB mutant (ZP22) strains. Sifs and vacuolar membranes were labeled with mouse anti-LAMP-2 H4B4 antibody and secondary Alexa Fluor 488 (green). Bacteria were visualized by staining with rabbit anti-O antigen and secondary antibody-Texas red (red). Arrows indicates Sifs, and arrowheads indicate the “pseudo-Sifs.”
SscB is required for efficient Salmonella replication in macrophages.
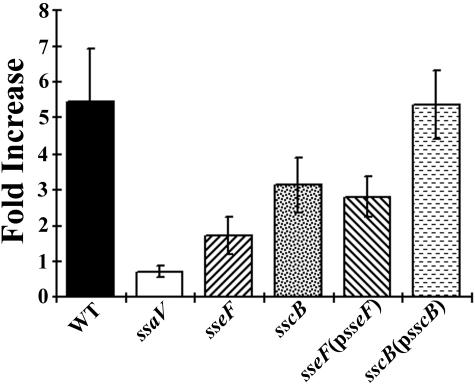
Previous studies have shown that SseF contributes to intracellular replication of serovar Typhimurium in macrophages (22). If SscB functions as a chaperone for SseF, mutations in sscB should affect Salmonella intracellular proliferation due to a reduced level and/or translocation of SseF. As shown in Fig. 7, the sscB mutant strain replicated less efficiently inside macrophage and was similar to that of the sseF mutant strain. This defect was complemented by introducing a plasmid (pZP283) encoding the full-length SscB into the sscB mutant strain. This complementation indicates that the slower replication of the sscB mutant strain was not due to a polar effect on a downstream sseF gene. This result further supports our hypothesis that SscB functions as a chaperone for SseF.
FIG. 7.
SscB is required for efficient intracellular survival of serovar Typhimurium in macrophages. RAW264.7 macrophages were infected with wild-type serovar Typhimurium and various mutant strains at an MOI of 10. Extracellular bacteria were removed by washing and gentamicin treatment. At 2 and 24 h after bacterial invasion, cells were lysed and the number of intracellular bacteria was determined by plating on LB medium-streptomycin plates. The data shown were obtained from three independent experiments, each performed in duplicate.
DISCUSSION
Salmonella strains utilize the SPI-2 for their successful survival and replications inside host cells by translocating SPI-2 effectors, including SseF, into the host cell. It has not been clear whether these SPI-2 effector proteins require any chaperones for their functions. We report here that SscB acts as the chaperone for SseF. Our data show that the SscB is required for the stability of cytoplasmic SseF and for its secretion into the culture medium. This is further corroborated by the protein stability assay demonstrated that the half-life of SseF is significantly shortened in the absence of SscB. The effect of SscB on SseF was shown to be posttranscriptional. In addition, SscB interacted with SseF in a coprecipitation experiment. Taken together, our results indicate that SscB functions as a chaperone for SseF.
It is becoming evident that many type III secretion components require cognate chaperones for their secretion into the extracellular medium and/or translocation into the host cells. In addition, complex functions have been described for chaperones SicA, SicP, InvB, and SseA involving the Salmonella TTSS apparatus and effectors. Although their substrates range from the type III secretion apparatus and effectors, they all have the ability to stabilize their substrates in the bacterial cytoplasm (3, 7, 8, 11, 13, 28, 36, 46, 53). Interestingly, SicA has been shown to regulate sopE gene transcription, possibly through InvF (8, 46). In addition, the Yersinia SycH chaperone was shown to relieve the posttranscriptional repression of Yop effector synthesis by binding to YscM1 and YscM2 (5). No chaperones have been reported for SPI-2 effectors despite speculation based on sequence homologies. Our data indicate that chaperones are also required for SPI-2 effector protein, SseF. Although SscB is essential for both the stability and the secretion of SseF, it appears to have a greater influence on SseF secretion than on its intracellular level, suggesting that the effect on SseF secretion may be independent of its effect on stability (Fig. 1B and 3). These data are consistent with previously published results, suggesting a role for chaperones in partially unfolding the substrates in order to maintain secretion-competent state for the type III secretion apparatus (43).
The phenotype of the sscB mutant can be complemented by coexpressing sscB with sseF in a bicistronic vector. Interestingly, the interaction between SscB and SseF can be detected only if SscB and SseF are coexpressed from the bicistronic vector (Fig. 5). No interaction was observed when SscB and SseF were expressed separately and bacteria lysates were mixed later. A similar requirement of a 40-kDa chaperone for the crystallization of the 34-kDa protein and formation of the inclusion bodies was reported in Bacillus thuringiensis subsp. thompsoni (35). It is tempting to speculate that proper folding of SseF requires the presence of SscB immediately after or during SseF translation. This hypothesis is consistent with the genetic arrangement of the sscB gene upstream of sseF in the Salmonella chromosome and apparently in the same operon (32). Further studies are required to explore this possibility.
The sseG gene, which encodes another SPI-2 effector of the TTSS, is located immediately downstream of the sseF gene and appears to be in the same transcriptional operon as sseA-G and sscA-B (19-21). SseG is also a putative effector protein implicated in the formation of Salmonella-induced aggregation of host endosomes (17). Recent studies have suggested that SseG is responsible for targeting Salmonella to the host Golgi network through an undefined mechanism (38). It has been suggested that SscB acts as a chaperone for both SseF and SseG (6, 22). One reason hampering the functional study of SPI-2 determinants is the difficulty in expressing these proteins in vitro for biochemical analysis. For example, we have not been able to detect any SseF when it is expressed from the chromosomal gene under its native promoter even though the sseF gene is transcribed (Fig. 4 and data not shown). Our data suggest that expressing these SPI-2 effector proteins from a medium-copy-number plasmid would facilitate their functional characterization. Our preliminary data have shown that the intracellular level of SseG is significantly decreased in the absence of SscB (data not shown). However, numerous attempts to express SseG to a detectable secretion level have failed (data not shown). Further studies are needed to explore whether SscB acts as a chaperone for SseG.
It is known that SPI-1 functions to promote invasion (51) and that SPI-2 functions to facilitate the biogenesis of SCV inside cells (25). These two TTSS apparently function in very different environments. Recent studies have shown that the chaperone-binding domain of SopE and SptP might, in addition to their stabilizing activities, confer secretion specificity. These two effectors are secreted through the flagellum-associated export system instead of SPI-1 TTSS when their chaperone-binding domains are deleted (27). Our data indicate that SscB is required both for stability and for secretion of SseF, suggesting that SscB may function in a mechanism similar to that of the SPI-1 effector chaperones. Analysis of how SscB functions may help us understand whether SPI-2 effector chaperones function differently from the SPI-1 effector chaperones, perhaps when salmonellae encounter different surrounding environments.
Acknowledgments
This research was supported by NIH grant AI49978 and AHA grant 0230286N to D.Z.
We thank Ferric Fang for providing technical help with the macrophage survival assay and Patrick Hearing for providing monoclonal anti-M45 antibodies. We thank Arthur Aronson for critically reviewing the manuscript.
REFERENCES
- 1.Anonymous. 1999. Summary of notifiable diseases—United States, 1998. Morb. Mortal. Wkly. Rep. 47:ii-92. [PubMed]
- 2.Baudry, B., M. Kaczorek, and P. J. Sansonetti. 1988. Nucleotide sequence of the invasion plasmid antigen B and C genes (ipaB and ipaC) of Shigella flexneri. Microb. Pathog. 4:345-357. [DOI] [PubMed] [Google Scholar]
- 3.Bronstein, P. A., E. A. Miao, and S. I. Miller. 2000. InvB is a type III secretion chaperone specific for SspA. J. Bacteriol. 182:6638-6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brumell, J. H., S. Kujat-Choy, N. F. Brown, B. A. Vallance, L. A. Knodler, and B. B. Finlay. 2003. SopD2 is a novel type III secreted effector of Salmonella typhimurium that targets late endocytic compartments upon delivery into host cells. Traffic 4:36-48. [DOI] [PubMed] [Google Scholar]
- 5.Cambronne, E. D., J. A. Sorg, and O. Schneewind. 2004. Binding of SycH chaperone to YscM1 and YscM2 activates effector yop expression in Yersinia enterocolitica. J. Bacteriol. 186:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 7.Coombes, B. K., N. F. Brown, S. Kujat-Choy, B. A. Vallance, and B. B. Finlay. 2003. SseA is required for translocation of Salmonella pathogenicity island-2 effectors into host cells. Microbes Infect. 5:561-570. [DOI] [PubMed] [Google Scholar]
- 8.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Groote, M. A., U. A. Ochsner, M. U. Shiloh, C. Nathan, J. M. McCord, M. C. Dinauer, S. J. Libby, A. Vazquez-Torres, Y. Xu, and F. C. Fang. 1997. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. USA 94:13997-14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 11.Ehrbar, K., A. Friebel, S. I. Miller, and W. D. Hardt. 2003. Role of the Salmonella pathogenicity island 1 (SPI-1) protein InvB in type III secretion of SopE and SopE2, two Salmonella effector proteins encoded outside of SPI-1. J. Bacteriol. 185:6950-6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freeman, J. A., M. E. Ohl, and S. I. Miller. 2003. The Salmonella enterica serovar Typhimurium translocated effectors SseJ and SifB are targeted to the Salmonella-containing vacuole. Infect. Immun. 71:418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu, Y., and J. E. Galán. 1998. Identification of a specific chaperone for SptP, a substrate of the centisome 63 type III secretion system of Salmonella typhimurium. J. Bacteriol. 180:3393-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galán, J. E., and R. D. Curtiss. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-del Portillo, F., and B. Finlay. 1995. Targeting of Salmonella typhimurium to vesicles containing lysosomal membrane glycoproteins bypasses compartments with mannose 6-phosphate receptors. J. Cell Biol. 129:81-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-del Portillo, F., M. B. Zwick, K. Y. Leung, and B. B. Finlay. 1993. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc. Natl. Acad. Sci. USA 90:10544-10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guy, R. L., L. A. Gonias, and M. A. Stein. 2000. Aggregation of host endosomes by Salmonella requires SPI2 translocation of SseFG and involves SpvR and the fms-aroE intragenic region. Mol. Microbiol. 37:1417-1435. [DOI] [PubMed] [Google Scholar]
- 18.Hansen-Wester, I., B. Stecher, and M. Hensel. 2002. Type III secretion of Salmonella enterica serovar Typhimurium translocated effectors and SseFG. Infect. Immun. 70:1403-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hensel, M., J. E. Shea, A. J. Baumler, C. Gleeson, F. Blattner, and D. W. Holden. 1997. Analysis of the boundaries of Salmonella pathogenicity island 2 and the corresponding chromosomal region of Escherichia coli K-12. J. Bacteriol. 179:1105-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 21.Hensel, M., J. E. Shea, B. Raupach, D. Monack, S. Falkow, C. Gleeson, T. Kubo, and D. W. Holden. 1997. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol. Microbiol. 24:155-167. [DOI] [PubMed] [Google Scholar]
- 22.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 23.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 24.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 25.Knodler, L. A., and O. Steele-Mortimer. 2003. Taking Possession: biogenesis of the Salmonella-containing vacuole. Traffic 4:587-599. [DOI] [PubMed] [Google Scholar]
- 26.Kuhle, V., and M. Hensel. 2002. SseF and SseG are translocated effectors of the type III secretion system of Salmonella pathogenicity island 2 that modulate aggregation of endosomal compartments. Cell. Microbiol. 4:813-824. [DOI] [PubMed] [Google Scholar]
- 27.Lee, H. S., and J. E. Galán. 2004. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol. Microbiol. 51:483-495. [DOI] [PubMed] [Google Scholar]
- 28.Lee, S. H., and J. E. Galán. 2003. InvB is a type III secretion-associated chaperone for the Salmonella enterica effector protein SopE. J. Bacteriol. 185:7279-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikolaus, T., J. Deiwick, C. Rappl, J. A. Freeman, W. Schroder, S. I. Miller, and M. Hensel. 2001. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J. Bacteriol. 183:6036-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Page, A. L., and C. Parsot. 2002. Chaperones of the type III secretion pathway: jacks of all trades. Mol. Microbiol. 46:1-11. [DOI] [PubMed] [Google Scholar]
- 34.Parales, R. E., and C. S. Harwood. 1993. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for gram-bacteria. Gene 133:23-30. [DOI] [PubMed] [Google Scholar]
- 35.Rang, C., M. Bes, W. J. Moar, and R. Frutos. 1997. Simultaneous production of the 34-kDa and 40-kDa proteins from Bacillus thuringiensis subsp. thompsoni is required for the formation of inclusion bodies. FEBS Lett. 412:587-591. [DOI] [PubMed] [Google Scholar]
- 36.Ruiz-Albert, J., R. Mundy, X. J. Yu, C. R. Beuzon, and D. W. Holden. 2003. SseA is a chaperone for the SseB and SseD translocon components of the Salmonella pathogenicity-island-2-encoded type III secretion system. Microbiology 149:1103-1111. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz-Albert, J., X. J. Yu, C. R. Beuzon, A. N. Blakey, E. E. Galyov, and D. W. Holden. 2002. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol. 44:645-661. [DOI] [PubMed] [Google Scholar]
- 38.Salcedo, S. P., and D. W. Holden. 2003. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 22:5003-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sampson, J. S., S. P. O'Connor, A. R. Stinson, J. A. Tharpe, and H. Russell. 1994. Cloning and nucleotide sequence analysis of psaA, the Streptococcus pneumoniae gene encoding a 37-kilodalton protein homologous to previously reported Streptococcus sp. adhesins. Infect. Immun. 62:319-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slonim, L. N., J. S. Pinkner, C. I. Branden, and S. J. Hultgren. 1992. Interactive surface in the PapD chaperone cleft is conserved in pilus chaperone superfamily and essential in subunit recognition and assembly. EMBO J. 11:4747-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, B. P., L. Da Roden, M. C. Thurmond, G. W. Dilling, H. Konrad, J. A. Pelton, and J. P. Picanso. 1994. Prevalence of salmonellae in cattle and in the environment on California dairies. J. Am. Vet. Med. Assoc. 205:467-471. [PubMed] [Google Scholar]
- 43.Stebbins, C. E., and J. E. Galán. 2001. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414:77-81. [DOI] [PubMed] [Google Scholar]
- 44.Steele-Mortimer, O., J. H. Brumell, L. A. Knodler, S. Meresse, A. Lopez, and B. B. Finlay. 2002. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell. Microbiol. 4:43-54. [DOI] [PubMed] [Google Scholar]
- 45.Steele-Mortimer, O., S. Meresse, J. P. Gorvel, B. H. Toh, and B. B. Finlay. 1999. Biogenesis of Salmonella typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cell. Microbiol. 1:33-49. [DOI] [PubMed] [Google Scholar]
- 46.Tucker, S. C., and J. E. Galán. 2000. Complex function for SicA, a Salmonella enterica serovar Typhimurium type III secretion-associated chaperone. J. Bacteriol. 182:2262-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uchiya, K., M. A. Barbieri, K. Funato, A. H. Shah, P. D. Stahl, and E. A. Groisman. 1999. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 18:3924-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 49.Wattiau, P., S. Woestyn, and G. R. Cornelis. 1996. Customized secretion chaperones in pathogenic bacteria. Mol. Microbiol. 20:255-262. [DOI] [PubMed] [Google Scholar]
- 50.Yu, X. J., J. Ruiz-Albert, K. E. Unsworth, S. Garvis, M. Liu, and D. W. Holden. 2002. SpiC is required for secretion of Salmonella pathogenicity island 2 type III secretion system proteins. Cell. Microbiol. 4:531-540. [DOI] [PubMed] [Google Scholar]
- 51.Zhou, D., and J. E. Galán. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 3:1293-1298. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, D., M. S. Mooseker, and J. E. Galán. 1999. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science 283:2092-2095. [DOI] [PubMed] [Google Scholar]
- 53.Zurawski, D. V., and M. A. Stein. 2003. SseA acts as the chaperone for the SseB component of the Salmonella pathogenicity island 2 translocon. Mol. Microbiol. 47:1341-1351. [DOI] [PubMed] [Google Scholar]