Abstract
Vibrio cholerae causes the life-threatening diarrheal disease cholera. This organism persists in aquatic environments in areas of endemicity, and it is believed that the ability of the bacteria to form biofilms in the environment contributes to their persistence. Expression of an exopolysaccharide (EPS), encoded by two vps gene clusters, is essential for biofilm formation and causes a rugose colonial phenotype. We previously reported that the lack of a flagellum induces V. cholerae EPS expression. To uncover the signaling pathway that links the lack of a flagellum to EPS expression, we introduced into a rugose flaA strain second-site mutations that would cause reversion back to the smooth phenotype. Interestingly, mutation of the genes encoding the sodium-driven motor (mot) in a nonflagellated strain reduces EPS expression, biofilm formation, and vps gene transcription, as does the addition of phenamil, which specifically inhibits the sodium-driven motor. Mutation of vpsR, which encodes a response regulator, also reduces EPS expression, biofilm formation, and vps gene transcription in nonflagellated cells. Complementation of a vpsR strain with a constitutive vpsR allele likely to mimic the phosphorylated state (D59E) restores EPS expression and biofilm formation, while complementation with an allele predicted to remain unphosphorylated (D59A) does not. Our results demonstrate the involvement of the sodium-driven motor and suggest the involvement of phospho-VpsR in the signaling cascade that induces EPS expression. A nonflagellated strain expressing EPS is defective for intestinal colonization in the suckling mouse model of cholera and expresses reduced amounts of cholera toxin and toxin-coregulated pili in vitro. Wild-type levels of virulence factor expression and colonization could be restored by a second mutation within the vps gene cluster that eliminated EPS biosynthesis. These results demonstrate a complex relationship between the flagellum-dependent EPS signaling cascade and virulence.
Vibrio cholerae causes the diarrheal disease cholera. This organism is introduced into human populations through the ingestion of contaminated food or water. Within the human, it colonizes the small intestine through the action of a type IV pilus (TCP) and expresses cholera toxin (CT), which causes the electrolyte imbalance and profuse watery diarrhea that is characteristic of this disease. The expression of TCP and CT is coordinated through a complex regulatory cascade that is referred to frequently as the ToxR regulon (for reviews, see references 24 and 31).
V. cholerae is a natural inhabitant of the aquatic environment. Epidemic strains can be found in both fresh- and saltwater locations in areas of endemicity and are the cause for the initiation of new cholera epidemics. V. cholerae can form biofilms in the laboratory, and it is believed that this is a likely persistent form of the bacteria within the environment, since biofilms are more resistant to environmental stresses, e.g., chlorine and antibiotics (41, 44). A great deal of interest and research has recently been focused on V. cholerae biofilm formation, which can be considered a primitive developmental process.
V. cholerae biofilm development is dependent upon the expression of an exopolysaccharide (EPS). Expression of EPS is believed to occur after the bacteria have attached to an abiotic surface and formed microcolonies, and the EPS allows the bacteria to build three-dimensional structures characteristic of mature biofilms (41, 44). However, natural phase variation can also occur upon passage in the laboratory, which leads to an altered wrinkled colonial variant referred to as a rugose variant, and the rugose phenotype is due to EPS expression (44). The rugose phenotype also occurs in some strains with the inactivation of hapR (a luxR homologue) (15) and in some strains with the inactivation of flagellar genes (42) (see below). Two large operons encode the vps genes necessary for EPS expression (44). A response regulator, VpsR, which has homology with σ54-dependent activators (21), has been identified as a positive regulator of vps gene transcription (43), while a CytR homologue has been identified as a repressor of vps gene transcription (11). The exact manner in which these factors stimulate EPS expression has not yet been elucidated, but some details of the HapR-dependent signaling cascade have recently been reported.
V. cholerae has multiple signaling cascades that respond to quorum-dependent molecules and ultimately converge on regulating the phosphorylation state of the response regulator LuxO (28, 46), which has homology with σ54-dependent activators (21). LuxO (presumably in the phosphorylated state, as mimicked by constitutive mutant forms [8, 36]) exerts an effect on virulence factor expression by repressing the expression of HapR, which in turn acts as a repressor of the TcpP/ToxR virulence cascade. Thus, CT and TCP expression is reduced in a luxO strain and elevated in hapR and luxO hapR strains (28, 46). The LuxO/HapR quorum-dependent signaling cascade also regulates the expression of EPS and biofilm formation (8, 36, 45). HapR represses vps gene transcription and biofilm formation, and thus hapR strains express elevated amounts of EPS (and exhibit the rugose phenotype, as mentioned above) and form thicker biofilms. The authors of these previous studies have suggested that quorum sensing regulates biofilm development in an unusual manner, in that high cell density (represented by unphosphorylated LuxO and/or high levels of HapR) would appear to promote the dissolution of biofilms; high cell density would also appear to promote the cessation of virulence factor expression by the same rationale. We have found (see below) that the HapR-dependent pathway appears to control EPS expression in a subset of strains, while a flagellum-dependent pathway controls EPS expression in another distinct subset of strains.
Mutations that disrupt flagellar synthesis in the O139 strain MO10 cause elevated EPS expression (and a rugose phenotype) (42). Interestingly, the expression of EPS also causes a decrease in O139 intestinal colonization, and this decrease is specifically due to EPS expression rather than to a lack of motility. Zhu and Mekalanos (45) also found a defect in intestinal colonization of a biofilm-associated hapR strain, suggesting that EPS expression interferes in some manner with V. cholerae virulence. We have further investigated the flagellum-dependent signaling cascade that controls EPS expression and have found that the sodium-driven motor and active VpsR (presumed to be phospho-VpsR) play important roles in signal transduction that leads to EPS expression. Moreover, we have found that there is a complex relationship between EPS expression, motility, and intestinal colonization.
MATERIALS AND METHODS
Plasmid construction.
The ΔflaA::Cm, ΔflrA::Cm, Δ(flrBC)::Cm, pGP704 ′motY′, pGP704 ′vpsF′, pGP704 ′rpoN′, pGP704 ′vpsR′, and pGP704 ′motB′ plasmids have been described previously (7, 19-21, 42). The ΔhapR::Kn, ΔmotA::Kn, ΔmotX::Kn, ΔvpsR, ΔfliF, ΔfliA, and ΔflhF plasmids were constructed by using the same general strategy. A ∼500-bp fragment 5′ of the deletion was PCR amplified with the corresponding primers (primers 1 and 2 [Table 1 ]) and then cleaved with EcoRI and BamHI or HindIII and ligated into the corresponding sites in pWSK30 (40). A ∼500-bp fragment 3′ of the deletion was PCR amplified with the corresponding primers (primers 3 and 4 [Table 1]); digested with BamHI or EcoRI and XbaI, HindIII, or SalI; and ligated into the plasmids that already contained the 5′ fragment of the corresponding gene. The ΔvpsR, ΔfliF, ΔfliA, and ΔflhF mutations were designed to be in-frame deletions. The plasmids containing ΔmotX, ΔmotA, and ΔhapR were then digested with BamHI or EcoRI, and the BamHI or EcoRI Kanr fragment from pUC4K (Pharmacia) was ligated into this site. Finally, the constructs were ligated into pKEK229 (4), a pir-dependent derivative of pCVD442 (5), which resulted in the plasmids listed in Table 2.
TABLE 1.
Oligonucleotide primer sequences
| Primer | Sequence (5′→3′) |
|---|---|
| HapR1 | GCGGATCCGCGACCTCTTGCTCAGAAATC |
| HapR2 | GCGAATTCGCGTTTTTCGATTGATGCGTC |
| HapR3 | GCGAATTCCAAGTCTCCGTTGCAACAGTG |
| HapR4 | GCGCGTCGACGCTGGCCATGTTATCGACATC |
| MotX1 | GCGAATTCGCAAAAACGCTGGCTGAACTG |
| MotX2 | GCGGATCCGGAAGCGGCTACCGTTCGTAG |
| MotX3 | GCGGATCCCGAGCCAAACGCCGAGAAACG |
| MotX4 | GCACTAGTTACAGCTACATTCCTGACAAAG |
| MotA1 | GCGAATTCTCGCCACTTTCTAGCTGTTCG |
| MotA2 | GCGGATCCAACCAGTGTTGCTAAATCCAC |
| MotA3 | GCGGATCCGACGGTGTTTTAGCGATTCAAG |
| MotA4 | GCACTAGTGAATTCCAAGGTTTGTTGGGTG |
| FliD1 | GCGGATCCGATATTATCCGTGGAATCAATGGT |
| FliD2 | GCAAGCTTCGATAAGGTCGCCACCCCATCCAG |
| FliA1 | GCGGATCCAGCAAAGAACATCAAGTTCAAC |
| FliA2 | GCGAATTCGATACGCTTAACCAATACAGAG |
| FliA3 | GCGAATTCGAAATTGGTGAGGTACTTGGAG |
| FliA4 | GCGAAGCTTAGCGGCTTCGATGATTTGCTCAC |
| FlhF1 | GCGGATCCATGGAAATAAAACGATTTTTTGCCAAG |
| FlhF2 | ATTGCTCATGCTGCAGTGCATCGGCATCTTCTTGCAG |
| FlhF3 | TGCCGATGCACTGCAGCATCAGCAATTGTCGATTTATG |
| FlhF4 | GCGCGTCGACCTAGAATCTCTCTGAATCACTG |
| FliF1 | GCGAAGCTTGAATGGAATTTTTGAGTCAAAC |
| FliF2 | GCGAATTCGCCTGCCATCAAGGCATGCTC |
| FliF3 | GCGAATTCCAGGTTCTGATCGGCACCGTAG |
| FliF4 | GCTCTAGATTAGCCATTTTGCATCCAGTTC |
| VpsR1 | GCGGATCCAGAAATAATCGTGCCAAGTCG |
| VpsR2 | GCGAATTCGGTATCTGAACTGAGCTGCGC |
| VpsR3 | GCGAATTCCTGATCGATGGTGATTTTAAC |
| VpsR4 | GCGAAGCTTCAAAACTTAGAAGTTTTCATC |
| VC0916p1 | GCGAATTCGCAGAGCTCAACCATGAGCTG |
| VC0916p2 | GCGGATCCCTCGAGTACTGATAAACCTTTAACCTTC |
| VC0934p1 | GCGAATTCATATTGTTCTGTTTTTCCTTTC |
| VC0934p2 | GCGGATCCCTCGAGTATTCTGCTTTTTTCCTTCATC |
| VpsRregionD | GCGAAGCTTTGAACGATGCTGAAGACCAAG |
| VpsRregionU | GCGGATCCCCGCAGCCTAATAAGAGGTTAC |
| D59A Up | CATGGCTTAAAGCCACAATACC |
| D59A Down | GGTATTGTGGCTTTAAGCCATG |
| D59E Up | CATGGCTTAACTCCACAATACC |
| D59E Down | GGTATTGTGGAGTTAAGCCATG |
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| V. cholerae strains | ||
| P27459 | Wild-type O1 E1 Tor | 30 |
| KKV1101 | P27459 ΔflaA::Cm ΔlacZ | This study |
| KKV1507 | P27459 ΔhapR::Kn ΔlacZ | This study |
| C6706 | Wild-type O1 E1 Tor | 35 |
| KKV1599 | C6706 ΔflaA::Cm | This study |
| KKV1612 | C6706 ΔhapR::Kn | This study |
| C6709 | Wild-type O1 E1 Tor | 37 |
| KKV89 | C6709 ΔflaA::Cm | This study |
| KKV1562 | C6709 ΔhapR::Kn | This study |
| 2740-80 | Wild-type O1 E1 Tor | 30 |
| KKV1608 | 2740-80 ΔflaA::Cm | This study |
| KKV1610 | 2740-80ΔhapR::Kn | This study |
| N16961 | Wild-type O1 E1 Tor | 13 |
| KKV832 | N16961 ΔflaA::Cm | This study |
| KKV1198 | N16961 hapR::pGP704 (Ampr) | This study |
| E7946 | Wild-type O1 E1 Tor | 26 |
| KKV1601 | E7946 ΔflaA::Cm | This study |
| KKV1611 | E7946 ΔhapR::Kn | This study |
| A1552 | Wild-type O1 E1 Tor | F. Yildiz |
| KKV1600 | A1552 ΔflaA::Cm | This study |
| KKV1613 | A1552 ΔhapR::Kn | This study |
| MO10 | Wild-type O139 | 39 |
| KKV927 | MO10 motY::pGP704 (Ampr) | 42 |
| KKV955 | MO10 ΔflaA::Cm ΔlacZ | 42 |
| KKV1004 | MO10 ΔflrA::Cm ΔlacZ | This study |
| KKV1026 | MO10 Δ(flrBC)::Cm ΔlacZ | This study |
| KKV1028 | MO10 vpsF::pGP704 (Ampr) | 42 |
| KKV1029 | MO10 ΔflaA::Cm vpsF::pGP704 (Ampr) | 42 |
| KKV1029 | MO10 ΔflaA::Cm vpsR::pGP704 (Ampr) | This study |
| KKV1046 | MO10 ΔfliD::Cm ΔlacZ | This study |
| KKV1082 | MO10 rpoN::pGP704 (Ampr) | This study |
| KKV1114 | MO10 ΔfliA ΔlacZ | This study |
| KKV1184 | MO10 motB::pGP704 (Ampr) | This study |
| KKV1273 | MO10 ΔmotA::Kn ΔlacZ | This study |
| KKV1495 | MO10 ΔmotX::Kn ΔlacZ | This study |
| KKV1502 | MO10 ΔflaA::Cm ΔmotX::Kn ΔlacZ | This study |
| KKV1504 | MO10 ΔflaA::Cm motY::pGP704 (Ampr) ΔlacZ | This study |
| KKV1505 | MO10 ΔhapR::Kn ΔlacZ | This study |
| KKV1520 | MO10 ΔflaA::Cm ΔmotA::Kn ΔlacZ | This study |
| KKV1536 | MO10 ΔfliF ΔlacZ | This study |
| KKV1559 | MO10 ΔflaA::Cm motB::pGP704 (Ampr) ΔlacZ | This study |
| KKV1561 | MO10 ΔflhF | This study |
| KKV1578 | MO10 ΔvpsR ΔlacZ | This study |
| KKV1579 | MO10 ΔflaA::Cm ΔvpsR ΔlacZ | This study |
| KKV1842 | MO10 ΔmotX::Kn ΔvpsR ΔlacZ | This study |
| KKV1843 | MO10 ΔflaA::Cm ΔmotX::Kn ΔvpsR ΔlacZ | This study |
| KKV1862 | MO10 vpsR::pGP704 (Ampr) | This study |
| Plasmids | ||
| pKEK229 | R6K ori sacB mob Ampr | 4 |
| pKEK428 | ΔmotA::Kn in pKEK229 | This study |
| pKEK436 | ΔmotX::Kn in pKEK229 | This study |
| pKEK479 | ΔhapR::Kn in pKEK229 | This study |
| pKEK311 | fliD::Cm in pKEK229 | This study |
| pKEK374 | ΔfliA in pKEK229 | This study |
| pKEK518 | ΔvpsR in pKEK229 | This study |
| pKEK516 | ΔflhF in pKEK229 | This study |
| pKEK424 | ΔfliF in pKEK229 | This study |
| pGP704 | R6K ori mob Ampr | 29 |
| pCG1050 | ′motB′ in pGP704 | 7 |
| PKEK129 | ′vpsR′ in pGP704 | 21 |
| pKEK328 | ′motY′ in pGP704 | 42 |
| pKEK349 | ′vpsF′ in pGP704 | 42 |
| pKEK370 | ′hapR′ in pGP704 | This study |
| pRS551 | Transcriptional lacZ fusion vector, Ampr Kanr | 32 |
| pKEK343 | VC0916 promoter in pRS551 | This study |
| pKEK396 | VC0934 promoter in pRS551 | This study |
| pWSK30 | pSC101 ori Ampr | 40 |
| pKEK725 | vpsR WT allele in pWSK30 | This study |
| pKEK662 | vpsR D59A allele in pWSK30 | This study |
| pKEK663 | vpsR D59E allele in pWSK30 | This study |
The fliD::Cm construct was made by PCR amplifying a ′fliD′ fragment with primers FliD1 and FliD2, digesting with BamHI and HindIII, ligating into the corresponding sites in pWSK30 (40), then digesting with EcoRI, which cleaves at a site within the fliD sequence, and ligating to an MfeI fragment containing Cmr (19); the fliD::Cmr fragment was subsequently ligated into pKEK229 as described above. Plasmid pKEK370 was constructed by digesting the 3′ fragment used to generate ΔhapR (see above; amplified with HapR3 and HapR4) with EcoRI and SalI and ligating into pGP704 (29) digested similarly; this creates a suicide plasmid with an internal ′hapR′ fragment used to insertionally inactivate the gene. The vps promoter transcriptional fusion plasmids were made by PCR amplification with the primer pairs VC0916p1 and -2 and VC0934p1 and -2, digestion with EcoRI and BamHI, and ligation into the corresponding sites in pRS551 (32).
The D59A and D59E alleles of vpsR were constructed by a two-step PCR technique in which overlapping PCR fragments containing the mutation of interest were generated and then used as a template in a second PCR. In the first step, two separate fragments were generated by PCR amplification with primers VpsRregionD and VpsRD59A Up or VpsRD59E Up and with primers VpsRregionU and VpsRD59A Down or VpsRD59E Down. In the second step, the two fragments corresponding to the mutation to be generated were used as a template in a second PCR amplification with primers VpsRregionD and VpsRregionU. The corresponding wild-type vpsR allele was generated by a single PCR amplification with VpsRregionD, VpsRregionU, and MO10 chromosomal DNA. These amplicons were digested with BamHI and HindIII and then ligated into pWSK30 digested similarly to form plasmids which express the various vpsR alleles from the native promoter in a low-copy-number vector.
All PCRs were performed with either KOD HiFi DNA polymerase or XL DNA polymerase (Novagen), with MO10 chromosomal DNA as a template. All primer sequences were designed based on the complete V. cholerae genome sequence (13).
Bacterial strains and media.
The V. cholerae strains used in this study are listed in Table 2. Strain construction with pGP704 and pCVD442 derivatives has been described previously (5, 19, 29); the correct construction of all strains was verified by PCR, sequencing, and/or Southern blot analysis. Bacteriophage CP-T1ts-mediated transduction was used to construct some strains, and the protocol of Hava and Camilli (12) was followed. Escherichia coli strain DH5α (9) was used for plasmid construction, and SM10λpir (29) was used for propagation of pir-dependent plasmids and conjugation into V. cholerae.
V. cholerae were grown in Luria-Bertani (LB) broth, supplemented with the following concentrations of antibiotics, when appropriate: 2 μg of chloramphenicol per ml for smooth strains, 20 μg of chloramphenicol per ml for rugose strains, 25 μg of kanamycin per ml, 100 μg of streptomycin per ml, and 50 μg of ampicillin per ml. For counterselection with sacB-containing plasmids, LB broth without NaCl and with 10% sucrose was used. For virulence-factor-inducing conditions, strains were grown in modified AKI medium (1.5% tryptone, 0.4% yeast extract, 0.5% NaCl) overnight at 37°C and then normalized to identical densities based on the optical density at 600 nm (OD600), and 100 μl was inoculated into a 10-ml tube filled completely to the top with modified AKI medium. These tubes were incubated statically at 37°C for 4 h, and then 5 ml was removed and added to a 25-ml culture tube, which was then incubated on a roller drum at 37°C for 18 to 20 h.
Biofilm assay.
The basic biofilm protocol used previously (42) was followed, with some modifications. Strains were grown overnight in LB broth and then normalized to identical densities based on OD600, and 5 μl was inoculated into 500 μl of LB broth in 10-ml borosilicate glass tubes. The tubes were then incubated statically at 30°C for 22 h. The tubes were rinsed with distilled water, incubated with 600 μl of 0.1% crystal violet for 30 min, and rinsed again with distilled water. One milliliter of dimethyl sulfoxide was then added, the tube was vortexed and allowed to stand for 10 min, and the OD570 measured.
β-Galactosidase assays.
ΔlacZ V. cholerae strains were transformed with plasmids pKEK343 and pKEK396 (pBR322 derivatives) (33) and then grown in LB broth, harvested at OD600 of ∼0.2 to 0.4, permeabilized with chloroform and sodium dodecyl sulfate, and assayed for β-galactosidase activity according to the method of Miller (27).
In vitro and in vivo virulence assays.
CT was measured by GM1-ganglioside enzyme-linked immunosorbent assay, as described previously (34). For TCP detection, whole-cell lysates were matched by OD600, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and probed with TcpA polyclonal antiserum by utilizing ECL detection reagent (Amersham Pharmacia). Mouse intestinal competition assays to measure colonization have been described previously (7). The inocula consisted of ∼105 wild-type and ∼105 mutant organisms.
RESULTS
EPS expression is regulated differently in V. cholerae O1 El Tor strains.
We previously reported (42) that flagellar mutations which produce nonflagellated cells caused EPS expression and a rugose colony phenotype in the V. cholerae O139 strain MO10. The O139 serogroup is believed to have arisen from an O1 El Tor strain following acquisition of the O139 biosynthetic gene cluster (2, 38). We therefore reasoned that a lack of flagellar synthesis would likely lead to EPS expression and a rugose phenotype in O1 El Tor strains. A mutation in flaA, which encodes the “core” flagellin of the flagellum, results in a nonflagellated cell (19) and causes a rugose phenotype in MO10 (Fig. 1, top panel), as previously reported (42). We introduced a ΔflaA::Cm mutation into a panel of O1 El Tor strains via CP-T1ts-mediated transduction (12) and found that a flaA mutation in the O1 El Tor strains P27459, C6706, and E7946 also caused a rugose colonial phenotype (Fig. 1, middle panel, and data not shown). However, flaA mutants of the O1 El Tor strains N16961, A1552, 2740-80, and C6709 maintained a smooth colony phenotype, even though these strains were nonflagellated (not shown). The MO10, P27459, and C6706 flaA strains were clearly rugose after 24 to 48 h of growth on LB agar, while the E7946 flaA strain was not obviously rugose until approximately 72 h of growth.
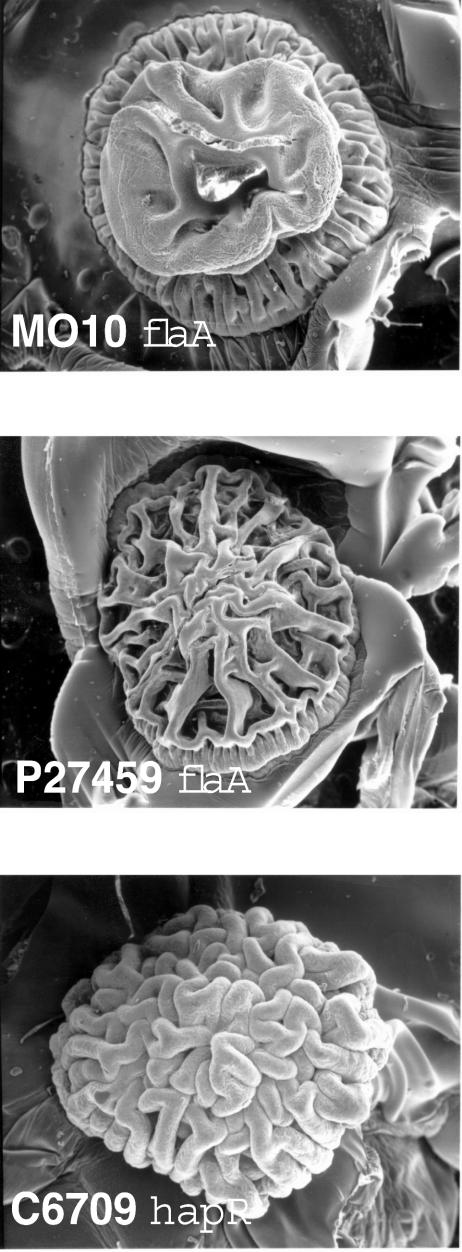
FIG. 1.
Rugose colonial phenotypes. Colonies of the MO10 flaA (KKV955) (top), P27459 flaA (KKV1101) (middle), and C6709 hapR (KKV1562) (bottom) strains were visualized by scanning electron microscopy. The rugose colonial phenotype is well-preserved by this technique, but the underlying agar medium becomes somewhat corrugated upon dehydration.
Jobling and Holmes (15) reported that a mutation in hapR, which encodes the transcriptional regulator of the HA protease gene, results in a rugose colonial phenotype in the O1 El Tor strain 3083. We introduced a ΔhapR::Kn mutation via CP-T1ts-mediated transduction into strains MO10, P27459, C6709, C6706, E7946, A1552, 2740-80, and N16961. The C6709, A1552, C6706, and 2740-80 hapR strains displayed a rugose phenotype (Fig. 1, bottom panel, and data not shown), while the other hapR mutant strains remained smooth (not shown). The C6709, A1552, and 2740-80 hapR strains were clearly rugose after 24 h of growth on LB agar, while the C6706 hapR strain was not obviously rugose until approximately 48 h of growth. The rugose phenotype of the C6706 hapR strain is consistent with the enhanced biofilm development reported for this strain (8, 45).
The rugose phenotype in the P27459 flaA and C6709 hapR strains is caused by expression of the vps genes encoding the EPS, because the introduction of a polar mutation into one of the EPS biosynthetic gene clusters (in vpsF) in these strains restored a smooth phenotype (not shown), as we had shown previously for the MO10 flaA strain (42). While neither the flaA nor the hapR N16961 strain was rugose, it has been demonstrated previously that rugose variants of this strain can be isolated upon nutrient starvation (43). Our results demonstrate that there are apparently at least three distinct genetic pathways for rugose EPS expression in V. cholerae O1 El Tor strains: one initiated by a lack of flagellar synthesis (seen in the O139 strain MO10 and O1 El Tor strains C6706, P27459, and E7946), one initiated by a lack of HapR (seen in the O1 El Tor strains C6706, C6709, A1552, and 2740-80), and one independent of both flagellum- and HapR-dependent pathways (seen in spontaneous rugose variants of O1 El Tor strain N16961; this strain carries a frameshift mutation in hapR, so no HapR-dependent pathway was anticipated). Interestingly, both HapR- and flagellum-dependent pathways seem to be operational in strain C6706.
The HapR-dependent pathway to EPS expression in strain C6706 has been the subject of recent investigations in several laboratories (8, 45). We have undertaken a more in-depth analysis of the flagellum-dependent pathway to EPS expression, utilizing MO10 as our model strain.
Mutations in the sodium-driven motor or VpsR reduce EPS expression and biofilm formation in nonflagellated V. cholerae.
We had previously demonstrated that a mutation in motY, which encodes one of the components of the sodium-driven flagellar motor, results in flagellated but nonmotile MO10 cells which have a smooth colonial phenotype and do not express EPS (42). We have subsequently constructed MO10 strains with mutations in the other three motor genes, motA, motB, and motX, and all three of these strains are flagellated but nonmotile and maintain a smooth phenotype (not shown). These results are consistent with our previous hypothesis that the lack of a complete flagellum, rather than a lack of motility, stimulates EPS expression. However, we had not considered an alternative possibility, namely, that perhaps the sodium-driven motor has a dual function: as an integral motility component and also as a component of the signal transduction cascade that leads to EPS expression.
Interestingly, the introduction of any of the mot mutations (motA, motB, motX, or motY) into a flaA MO10 strain results in reversion to a smooth colonial phenotype, suggestive of reduced EPS expression (Fig. 2), similar to the phenotype of a flaA strain that has been disrupted in one of the vps gene clusters (flaA vpsF). Considering that these strains already lack a complete flagellum, this indicates that the sodium-driven motor may participate in transduction of the EPS inducing signal, in addition to its role in flagellar rotation.
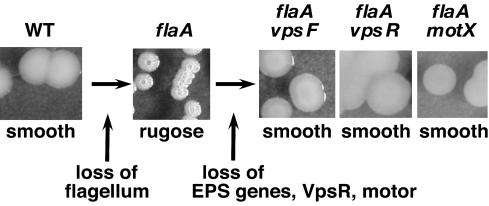
FIG. 2.
Mutations in EPS, vpsR, and mot genes suppress the rugose phenotype of a flaA strain. Shown are photographs of the colonial phenotypes of MO10 (wild type [WT]), KKV955 (flaA), KKV1029 (flaA vpsF), KKV1579 (flaA vpsR), and KKV1502 (flaA motX).
The MO10 flaA strain can form a biofilm (Fig. 3A), as demonstrated previously (42). Presumably, high-level EPS expression in this strain overcomes any need for motility and microcolony formation, because the strain forms aggregates in solution that likely settle onto the surface and serve as microcolonies to initiate mature biofilm development. However, the flaA motA and flaA motX strains are defective for biofilm development. The motX and motA mutant strains are also defective for biofilm development, as we showed previously for a motY mutant strain (42). These results are consistent with an important role for the sodium-driven motor in EPS expression and biofilm formation by both flagellated and nonflagellated MO10 cells.
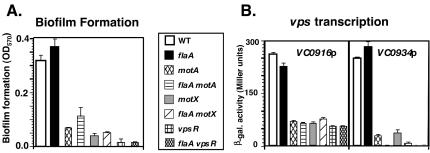
FIG. 3.
The sodium-driven motor and VpsR are necessary for enhanced biofilm formation and high-level vps transcription. Biofilm formation (A) and VC0916p and VC0934p transcription (B) in the MO10 (wild type [WT]), KKV955 (flaA), KKV1273 (motA), KKV1520 (flaA motA), KKV1495 (motX), KKV1502 (flaA motX), KKV1578 (vpsR), and KKV1579 (flaA vpsR) V. cholerae strains were measured as described in Materials and Methods. β-gal., β-galactosidase. Error bars indicate standard deviations.
Yildiz et al. have identified a response-regulatory protein, VpsR, that is important for biofilm formation in the O1 El Tor strain A1552 (43). To determine whether VpsR is involved in biofilm formation in nonflagellated MO10 cells, we constructed a flaA vpsR MO10 strain. This strain demonstrated a smooth colonial phenotype (Fig. 2) and had greatly reduced biofilm formation (Fig. 3A). A vpsR mutation in a wild-type (i.e., flagellated) background also led to a reduction in biofilm formation. These results suggest an important role for VpsR in EPS expression and biofilm formation of both flagellated and nonflagellated MO10 cells.
VpsR and the sodium-driven motor are essential for high-level vps transcription.
Yildiz and Schoolnik (44) identified a number of polysaccharide biosynthetic genes (vps) that are necessary for EPS expression and biofilm formation. The vps genes are organized in two large operons corresponding to VC0916-VC0928 and VC0934-VC0939. It was shown (43) that VpsR was necessary for high-level transcription of the two vps operons in A1552 O1 El Tor cells. To determine whether transcription of the two vps operons correlated with the colonial phenotype and biofilm-forming abilities of mot and vpsR MO10 strains, we measured vps transcription from the two vps operon promoters (corresponding to VC0916p and VC0934p) (Fig. 3B).
Our results showed a strong correlation between the ability of the strains to form biofilms (Fig. 3A) and high levels of vps gene transcription (Fig. 3B). Interestingly, the wild-type (smooth) MO10 strain had high levels of transcription of both vps gene clusters, which were not altered by the introduction of a flaA mutation (causing the rugose phenotype). This suggests that the smooth-to-rugose transition is not caused by an increase in vps transcription, unlike in a spontaneous rugose O1 El Tor A1552 strain (43) or the O1 El Tor C6706 hapR strain (45). However, transcription of both vps gene clusters in motX, motA, and vpsR mutant strains was reduced, either in a flagellated (wild-type) or nonflagellated (flaA) background. Our results demonstrate that high-level vps transcription may be necessary, but not sufficient, for EPS expression and that VpsR and the sodium-driven motor are necessary for high-level vps gene transcription.
Phenamil inhibits vps transcription and biofilm formation.
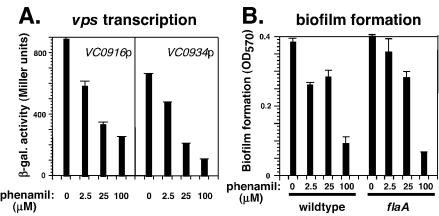
Phenamil specifically poisons the sodium-driven motor and inhibits motility in Vibrio spp., including V. cholerae (16, 23). We have confirmed that phenamil reduces the motility of the wild-type (flagellated) MO10 strain in a dose-dependent manner, as determined in a motility assay (not shown). Transcription of the two vps operons in wild-type and flaA MO10 strains is inhibited by phenamil in a dose-dependent manner (Fig. 4A). Likewise, biofilm formation by wild-type and flaA MO10 strains is inhibited by phenamil in a dose-dependent manner (Fig. 4B). These observations are consistent with the function of the sodium-driven motor being critical for vps transcription and biofilm formation in both flagellated and nonflagellated MO10 cells.
FIG. 4.
Phenamil inhibits vps transcription and biofilm formation. (A) Transcription of VC0916p and VC0934p in the wild-type MO10 strain with increasing concentrations of phenamil was measured as described in Materials and Methods. β-gal., β-galactosidase. (B) Biofilm formation by the MO10 (wild-type) and KKV955 (flaA) strains was measured as described Materials and Methods. Error bars indicate standard deviations.
Mutant forms of VpsR suggest that phosphorylation is necessary for EPS expression and biofilm formation.
As shown above, the response-regulatory protein VpsR is necessary for high-level vps transcription, EPS expression, and biofilm formation in the MO10 O139 strain, as was shown previously for spontaneous rugose colonies of the O1 El Tor strain A1552 (43). VpsR has a response-regulatory domain in its amino terminus with the conserved aspartate residue (D59) that is predicted to be the site of phosphorylation. Because VpsR is an “orphan” response regulator (i.e., no gene encoding a histidine kinase is located nearby) and its cognate histidine kinase has not yet been identified, we have no biochemical proof that D59 is the site of phosphorylation in VpsR. However, it has been shown for numerous response regulators that an alteration of this conserved aspartate residue to an alanine (D59A) prevents phosphorylation and results in a protein that represents the unphosphorylated state. Interestingly, the substitution of a glutamate residue (D59E) can mimic aspartyl-phosphate even though it prevents phosphorylation (i.e., it can act as a “constitutive” mutation), at least in some response regulators that share homology with σ54-dependent activators (e.g., VpsR, NtrC, and LuxO) (4, 6, 22); this is the “locked-on” constitutively active mutation that has been used extensively to analyze LuxO function (6, 8). Thus, a D59A allele would be predicted to behave like unphosphorylated VpsR, while a D59E allele would be predicted to behave like phospho-VpsR.
To determine the effect of substitutions at the putative phosphorylation site of VpsR on the flagellum-dependent EPS signaling pathway, we constructed vpsR alleles containing alanine and glutamate substitutions (D59A and D59E, respectively) and complemented ΔvpsR, flaA ΔvpsR, ΔmotX ΔvpsR, and ΔflaA ΔmotX ΔvpsR strains with these alleles expressed from the native vpsR promoter in a low-copy-number vector. We complemented these same strains with the wild-type vpsR allele in the same manner.
The ability of these strains to form biofilms was measured (Fig. 5). Provision of the wild-type VpsR protein expressed from the plasmid stimulated biofilm formation in both a wild-type and a flaA mutant background, and to approximately the same level as when the wild-type VpsR protein is expressed from the chromosome, as expected. Strains complemented with the D59A mutant VpsR behaved similarly to a ΔvpsR strain in both wild-type and flaA backgrounds; i.e., this allele fails to stimulate biofilm development. Interestingly, complementation with the D59E mutant VpsR allowed biofilm formation at a level two- to threefold greater than that for complementation with the wild-type VpsR in both wild-type and flaA mutant backgrounds. Our results suggest that phosphorylation of VpsR at aspartate 59 is necessary for the flagellum-dependent EPS signaling cascade that leads to biofilm formation (due to inhibition of biofilm formation by the D59A allele) and that the D59E mutant protein is active, i.e., mimics phospho-VpsR.
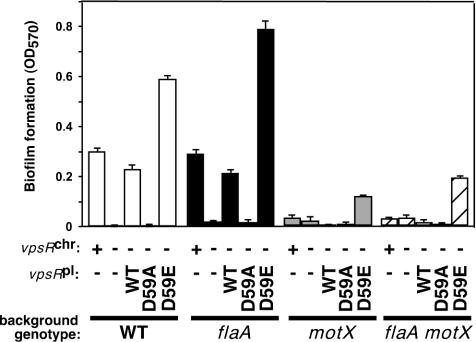
FIG. 5.
Mutant forms of VpsR predicted to affect phosphorylation alter biofilm formation. Biofilm formation in the following sets of strains was measured as described Materials and Methods. (i) MO10 (wild type [WT]), KKV955 (flaA), KKV1495 (motX), and KKV1502 (flaA motX) contain the native vpsR allele on the chromosome and are designated vpsRchr +. (ii) KKV1578 (WT), KKV1579 (flaA), KKV1842 (motX), and KKV1843 (flaA motX) additionally carry an in-frame deletion within the chromosomal vpsR allele and are designated vpsRchr −. (iii) Strains KKV1578, KKV1579, KKV1842, and KKV1843 were transformed with low-copy-number plasmids carrying either the wild-type vpsR allele (pKEK725, WT vpsRpl), the D59A vpsR allele (pKEK662, D59A vpsRpl), or the D59E vpsR allele (pKEK663, D59E vpsRpl). Error bars indicate standard deviations.
Provision of the wild-type or D59A VpsR protein failed to stimulate significant biofilm formation in the motX or flaA motX background, as expected, since we have shown above that the motor is an important component of the flagellum-dependent EPS signaling cascade. Interestingly, the D59E VpsR protein stimulated biofilm development in both motX and flaA motX backgrounds, at a level 12- to 20-fold greater than that for the wild-type VpsR protein. This suggests that VpsR is unphosphorylated in the absence of the sodium-driven motor. Thus, the requirement for the sodium-driven motor for biofilm development is in part likely due to its stimulation of the phosphorylation of VpsR, which can be bypassed by a constitutive (D59E) vpsR mutation.
The colonial phenotypes of the nonmotile strains, indicative of the level of EPS expression, correlated with their abilities to form biofilms. The ΔflaA ΔvpsR strain complemented with either the wild-type or D59E vpsR allele was rugose, while complementation of this strain with the D59A vpsR allele resulted in a smooth phenotype (not shown). Complementation of the ΔmotX, and ΔflaA ΔmotX strains with the D59E vpsR allele also led to a modest rugose phenotype (rough center and smooth edges). All strains complemented with the D59A vpsR allele remained smooth. These results suggest that phosphorylation of VpsR is necessary for the rugose phenotype and hence for EPS expression.
Effects of VpsR and the sodium-driven motor on in vivo colonization and in vitro virulence factor expression.
We have previously shown that a flaA strain (rugose, nonmotile) is defective for intestinal colonization in the infant mouse competition assay (42) and that this defect is specifically due to the expression of EPS, since a flaA vpsF mutant strain (smooth, nonmotile) colonizes similarly to the wild-type strain (Fig. 6A). Mutations in the sodium-driven motor and vpsR also diminish EPS expression in a nonflagellated (flaA) strain, as shown above. To determine whether mot and vpsR mutations can restore intestinal colonization by the flaA strain, similar to a vpsF mutation, we measured the ability of motX, flaA motX, vpsR, and flaA vpsR strains to colonize the infant mouse intestine in a competition assay (Fig. 6A).
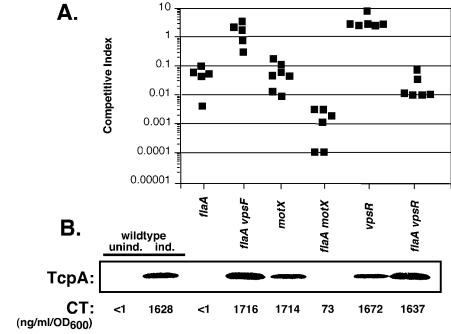
FIG. 6.
Effects of the flagellum, sodium-driven motor, VpsR, and EPS on intestinal colonization and virulence factor expression. (A) Infant mouse intestinal colonization competition assay. Strains KKV955 (flaA), KKV1029 (flaA vpsF), KKV1495 (motX), KKV1502 (flaA motX), KKV1862 (vpsR), and KKV1029 (flaA vpsR) were coinoculated with MO10 perorally into infant mice at ratio of ∼1:1; intestinal homogenates were recovered at 24 h postinoculation, and the CFU of wild-type and mutant strains were determined. The competitive index is given as the output ratio of mutant to wild type divided by the input ratio of mutant to wild-type; each value shown is from an individual mouse. Strains KKV955, KKV1495, KKV1502, and KKV1029 colonized the intestine significantly less than the wild-type strain (P < 0.01 as determined by Student's two-tailed t test). (B) In vitro expression of CT and TcpA. The same strains as in panel A were grown under AKI-inducing conditions, and values correspond to the strain designations in panel A. TcpA was detected by Western immunoblotting with anti-TcpA antiserum, and CT in the supernatant was measured by GM1-ganglioside ELISA. Also shown is CT and TcpA detection for the wild-type MO10 strain grown under noninducing (unind.) and AKI-inducing (ind.) conditions.
The motX and flaA motX mutants were defective for intestinal colonization; the motX strain colonized similarly to a flaA strain, while the flaA motX mutant colonized worse than either single mutant strain. These strains are smooth and nonmotile and thus would be predicted to colonize the intestine similarly to the smooth, nonmotile flaA vpsF strain. However, the motX mutation likely alters sodium flux across the membrane, which Hase and Mekalanos have shown can alter virulence factor expression (10); this may explain the colonization deficiencies of these strains. The vpsR mutant was competent for colonization, even slightly outcompeting the wild-type strain, indicating that VpsR is not essential for intestinal colonization. However, the flaA vpsR mutant showed a defect for colonization that was similar to that of the flaA strain, demonstrating that the vpsR mutation, while able to disrupt EPS expression in the flaA strain, was unable to restore wild-type levels of colonization to the flaA strain.
These strains were grown under in vitro conditions that promote virulence factor expression (AKI growth conditions) (14) (Fig. 6B); the inability of mutant strains to colonize the infant mouse intestine is frequently correlated with a lack of in vitro expression of CT and TcpA, the major component of TCP. Growth of the wild-type MO10 strain under identical AKI-inducing conditions results in detectable CT and TcpA expression, and there is no detectable expression of either CT or TcpA when MO10 is grown under noninducing conditions. The in vitro CT and TcpA expression of the flaA and flaA vpsF strains could be correlated with their colonization patterns; i.e., the flaA strain failed to express CT or TcpA in vitro, but the introduction of the vpsF mutation into this strain, which abolishes EPS expression, restored wild-type levels of both CT and TcpA expression. There was also a good correlation between the very low levels of CT and TcpA expression in vitro by the flaA motX strain and poor colonization in vivo and between detectable levels of CT and TcpA expression in vitro by the vpsR strain and wild-type levels of colonization in vivo.
Interestingly, the motX and flaA vpsR strains expressed wild-type levels of CT and detectable TcpA in vitro yet colonized poorly in vivo. This discrepancy between virulence factor expression in vitro and virulence in vivo emphasizes the difficulty in replicating the intestinal environment in a test tube, as suggested by Lee et al. (25). Perhaps although the flaA vpsR strain is nonrugose and thus should be competent for colonization, VpsR is required to regulate some other factor that facilitates intestinal colonization by nonflagellated (but not flagellated) cells. Our results suggest a complex relationship between flagellar synthesis, motor function, EPS expression, and intestinal colonization.
DISCUSSION
The ability of V. cholerae to form biofilms has been postulated to contribute to cholera epidemics by enhancing environmental persistence of the organisms in aquatic reservoirs. Expression of the EPS encoded by the vps genes is necessary to form the mature biofilms seen when V. cholerae is grown under the laboratory conditions utilized in this study (42, 44). A recent report (17) has shown that this particular EPS may be utilized only by the O139 strain MO10 found in freshwater biofilms, while a vps-independent MO10 biofilm, dependent on the O139 antigen, appears to form in saltwater environments (18). Considering that cholera infections are frequently derived from freshwater sources, especially in areas of endemicity, understanding the regulation of vps-dependent EPS expression is likely to be directly relevant to understanding the environmental persistence of epidemic strains.
Some of the details of the induction of EPS in V. cholerae are beginning to be understood. Two recent reports (8, 45) have demonstrated that a quorum-sensing signaling cascade controls EPS expression in the V. cholerae O1 El Tor C6706 strain. This cascade converges on controlling the expression of HapR, a LuxR homologue that represses both virulence factor expression and vps gene transcription (28, 46). Thus, a hapR mutant of this strain is derepressed for both virulence factor and EPS expression, explaining the rugose phenotype associated with a hapR mutant (15). However, natural frameshift mutations in hapR have been found in several (smooth) clinical isolates of V. cholerae which can still induce EPS and form biofilms in the laboratory (13, 43), suggesting that there are HapR-independent pathways for this process.
We previously identified a second pathway that leads to EPS expression and biofilm formation in the O139 MO10 strain (42). We have shown here that a flagellum-dependent pathway also regulates EPS expression in several O1 El Tor strains, and thus this signaling pathway is not unique to O139 V. cholerae. The absence of the flagellum is the inducing signal for EPS expression, which leads to the question of how an intracellular signaling cascade can recognize the lack of an extracellular organelle. In the studies presented here, we have identified the sodium-driven motor as an essential component of this signal cascade, since mutations in the sodium-driven motor abolish vps gene transcription, EPS expression, and biofilm formation. Phenamil, a specific poison of the sodium-driven motor, has the same effect as a mutation in one of the motor components, suggesting that there is a functional, rather than structural, role for the motor in this EPS signal cascade. While our studies identify the sodium-driven motor as a component in the EPS signaling cascade, it still remains unclear how the loss of the flagellum stimulates EPS expression, since the two vps operons are transcribed at high levels even in the smooth wild-type strain. We hypothesize that vps transcription is necessary but not sufficient for EPS expression and that transcription of an additional necessary gene(s) is stimulated by the lack of a flagellum.
A sodium gradient exists across the Vibrio membrane, and the sodium-driven motor allows an influx of sodium ions, which is coupled to flagellar rotation (1). Flux of sodium ions through the sodium-driven motor of Vibrio parahaemolyticus, which is predicted to change upon a decrease in the flagellar rotation rate, has been shown to be coupled to the induction of lateral flagellum transcription (16); the authors of that study characterized the motor as a mechanosensor. Our results suggest that the V. cholerae motor also acts as a mechanosensor to induce EPS expression. We hypothesize that the function of the sodium motor as a mechanosensor of flagellar rotation has been conserved among Vibrio spp. and has been adapted to induce appropriate behavior on solid surfaces, e.g., swarming behavior in V. parahaemolyticus and biofilm formation in V. cholerae.
One of the downstream events involved in this EPS signaling cascade appears to be the phosphorylation of the regulatory protein VpsR. Because VpsR with an alteration predicted to prevent phosphorylation (D59A) failed to stimulate biofilm formation, our results suggest that VpsR must be phosphorylated to induce biofilm formation. Also, VpsR with an alteration predicted to mimic phosphorylation (D59E) stimulated biofilm formation even in the absence of the motor. These results suggest that the motor is involved in stimulating the formation of phospho-VpsR, which in turn stimulates vps gene transcription and biofilm formation (Fig. 7). Because no cognate histidine kinase for VpsR has been identified, it is unclear what phosphorylates VpsR and whether this responds directly to sodium influx through the motor. We have also been unable to demonstrate a direct effect of VpsR at the two vps gene cluster (VC0916 and VC0934) promoters; chromosomal transcriptional reporter fusions of these promoters in a heterologous system failed to be transcribed in the presence of the constitutive D59E VpsR allele (not shown). Therefore, we hypothesize that phospho-VpsR may directly activate the transcription of some other factor that, in turn, activates vps gene transcription; this unknown factor may be the recently identified VpsT (3).
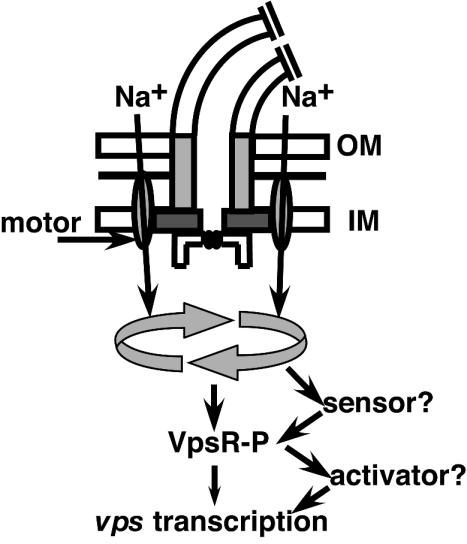
FIG. 7.
Proposed flagellum-dependent EPS signaling cascade. The sodium-driven motor couples the flux of Na+ across the membrane to flagellar rotation (depicted as circular arrows). Our results suggest that the motor may act as a mechanosensor, possibly by altering Na+ flux, which stimulates the phosphorylation of VpsR, which in turn stimulates vps transcription and EPS production. The sensor responsible for phosphorylation of VpsR, as well as the direct activator of vps transcription, has not yet been identified (see text for details). OM, outer membrane; IM, inner membrane.
While the flagellum-dependent EPS signaling cascade is operational in some O1 El Tor and O139 strains, other O1 El Tor strains regulate EPS expression via the HapR-dependent pathway. The presence of two distinct EPS signaling pathways in these closely related isolates seems odd, and we suspect that the pathways are linked in some manner. One manner in which the two pathways may converge would be for both signaling pathways to regulate the phosphorylation of VpsR; this hypothesis is currently being tested. Our evidence already suggests that the sodium-driven motor does not control the HapR-dependent signaling pathway, because we have inactivated a mot gene in a rugose hapR strain (C6709), which had no effect on the rugose phenotype.
Evidence also suggests that the MO10 strain has a functional HapR. Sequence analysis revealed that the MO10 HapR contains a single R12L substitution (compared to the functional HapR from strain 3083) (15). Expression of MO10 hapR from its native promoter in a low-copy-number plasmid in the (rugose) C6709 hapR strain or in strain N16961 (which has a natural frameshift mutation in hapR) complements these strains for increased HA protease expression and causes a reversion of C6709 hapR to the smooth phenotype (not shown). Moreover, the MO10 hapR strain shows decreased protease expression that can be complemented back to wild-type levels by providing hapR from MO10 or C6709 on a plasmid, indicating that the MO10 hapRR12L allele is functional. Interestingly, Hammer and Bassler (8) identified an R12Q hapR mutation as a suppressor of a luxO mutant C6706 strain that demonstrated reduced protease activity and increased EPS expression, suggesting that mutation of R12 to Q results in decreased activity; perhaps changing this residue to L is less deleterious to HapR function.
The flagellum-dependent EPS signaling cascade affects the virulence of V. cholerae in some unexpected ways. We had previously shown that the nonflagellated rugose flaA MO10 strain is defective for intestinal colonization and that this defect was specifically due to EPS expression, since a nonrugose but still nonflagellated flaA vpsF strain could colonize to wild-type levels (42) (Fig. 6A). One reason for this may be that the rugose strain forms an aggregate that is unable to effectively contact the intestinal epithelia to result in productive colonization. However, our in vitro results (Fig. 6B) suggest that induction of virulence factor expression is defective in the rugose strain and that this defect can be alleviated by disruption of the EPS (via mutation of vpsF). Likewise, disruption of EPS in the flaA strain via mutation of vpsR also allows for CT and TCP expression in vitro. Thus, the EPS itself likely disrupts the ToxR/TcpP signaling cascade that induces TCP and CT, perhaps by altering the microenvironment surrounding the cell.
Mutations in the sodium-driven motor also disrupt EPS expression in a nonflagellated strain, so one might expect that a flaA motX strain, like a flaA vpsF strain, would colonize similarly to a wild-type strain. However, the motX mutation in both flagellated and nonflagellated cells leads to decreases in intestinal colonization. This defect is clearly not due to a lack of motility, since the nonmotile flaA vpsF strain can colonize at wild-type levels, but rather might be linked to the altered sodium signaling induced by the lack of the motor. Hase and Mekalanos (10) found that disruptions in the sodium motive force across the membrane alter transcription of the virulence-regulatory gene toxT, suggesting that virulence factor expression would also be altered. However, this effect of the motor on virulence factor expression may occur only in vivo, since the motX strain expressed detectable CT and TCP under in vitro inducing conditions.
A strain with a mutation in vpsR exhibited normal intestinal colonization and in vitro virulence factor expression, yet the introduction of the vpsR mutation into the flaA strain did not restore normal intestinal colonization, even though this mutation disrupts EPS expression in this strain (similar to the vpsF mutation) and allows for normal induction of CT and TCP in vitro. Since VpsR is a regulatory factor, its absence may have pleiotropic effects that may not be evident except in certain genetic backgrounds (e.g., nonflagellate). Thus, perhaps the lack of EPS allows the flaA vpsR strain to induce CT and TCP under inducing in vitro conditions, like the flaA vpsF strain, but the lack of some other VpsR-dependent factor causes a reduction in colonization of the nonflagellated strain; this VpsR-dependent factor is not necessary for colonization by flagellated cells. While this scenario is speculative, it suggests a role for VpsR in V. cholerae virulence, at least under certain circumstances. Given the homology of VpsR and LuxO with σ54-dependent activators (21), our results and the recent results of others (28, 46) suggest the involvement of σ54 in multiple aspects of V. cholerae virulence that are distinct from flagellar regulation, as we had previously hypothesized (20).
Acknowledgments
This work was supported by NIH grant AI43486 to K.E.K.
REFERENCES
- 1.Atsumi, T., L. McCarter, and Y. Imae. 1992. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355:182-184. [DOI] [PubMed] [Google Scholar]
- 2.Bik, E. M., A. E. Bunschoten, R. D. Guow, and F. R. Mooi. 1995. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 14:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186:1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Correa, N. E., C. M. Lauriano, R. McGee, and K. E. Klose. 2000. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol. Microbiol. 35:743-755. [DOI] [PubMed] [Google Scholar]
- 5.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31:665-677. [DOI] [PubMed] [Google Scholar]
- 7.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:577-580. [DOI] [PubMed] [Google Scholar]
- 10.Hase, C. C., and J. J. Mekalanos. 1999. Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 96:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haugo, A. J., and P. I. Watnick. 2002. Vibrio cholerae CytR is a repressor of biofilm development. Mol. Microbiol. 45:471-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hava, D. L., and A. Camilli. 2001. Isolation and characterization of a temperature-sensitive generalized transducing bacteriophage for Vibrio cholerae. J. Microbiol. Methods 46:217-225. [DOI] [PubMed] [Google Scholar]
- 13.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA Sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwanaga, M., K. Yamamoto, N. Higa, Y. Ichinose, N. Nakasone, and M. Tanabe. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol. Immunol. 30:1075-1083. [DOI] [PubMed] [Google Scholar]
- 15.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023-1034. [DOI] [PubMed] [Google Scholar]
- 16.Kawagishi, I., M. Imagawa, Y. Imae, L. McCarter, and M. Homma. 1996. The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol. Microbiol. 20:693-699. [DOI] [PubMed] [Google Scholar]
- 17.Kierek, K., and P. I. Watnick. 2003. Environmental determinants of Vibrio cholerae biofilm development. Appl. Environ. Microbiol. 69:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kierek, K., and P. I. Watnick. 2003. The Vibrio cholerae O139 O-antigen polysaccharide is essential for Ca2+-dependent biofilm development in sea water. Proc. Natl. Acad. Sci. USA 100:14357-14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klose, K. E., and J. J. Mekalanos. 1998. Differential regulation of multiple flagellins in V. cholerae. J. Bacteriol. 180:303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28:501-520. [DOI] [PubMed] [Google Scholar]
- 21.Klose, K. E., V. Novick, and J. J. Mekalanos. 1998. Identification of multiple σ54-dependent transcriptional activators in Vibrio cholerae. J. Bacteriol. 180:5256-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klose, K. E., D. S. Weiss, and S. Kustu. 1993. Glutamate at the site of phosphorylation of nitrogen-regulatory protein NTRC mimics aspartyl-phosphate and activates the protein. J. Mol. Biol. 232:67-78. [DOI] [PubMed] [Google Scholar]
- 23.Kojima, S., K. Yamamoto, I. Kawagishi, and M. Homma. 1999. The polar flagellar motor of Vibrio cholerae is driven by an Na+ motive force. J. Bacteriol. 181:1927-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krukonis, E. S., and V. J. DiRita. 2003. From motility to virulence: sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 6:186-190. [DOI] [PubMed] [Google Scholar]
- 25.Lee, S. H., D. L. Hava, M. K. Waldor, and A. Camilli. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625-634. [DOI] [PubMed] [Google Scholar]
- 26.Mekalanos, J. J., D. J. Swartz, G. D. Pearson, N. Harford, F. Groyne, and M. de Wilde. 1983. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306:551-557. [DOI] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1992. A short course in bacterial genetics, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 28.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 29.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson, G. D., A. Woods, S. L. Chiang, and J. J. Mekalanos. 1993. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc. Natl. Acad. Sci. USA 90:3750-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reidl, J., and K. E. Klose. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol. Rev. 26:125-139. [DOI] [PubMed] [Google Scholar]
- 32.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 33.Sutcliffe, J. G. 1979. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harbor Symp. Quant. Biol. 43:77-90. [DOI] [PubMed] [Google Scholar]
- 34.Svennerholm, A. M., and J. Holmgren. 1978. Identification of the Escherichia coli heat-labile enterotoxin by means of a ganglioside immunosorbent assay (GM1-ELISA) procedure. Curr. Microbiol. 1:19-23. [Google Scholar]
- 35.Thelin, K. H., and R. K. Taylor. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 64:2853-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vance, R. E., J. Zhu, and J. J. Mekalanos. 2003. A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect. Immun. 71:2571-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wachsmuth, I. K., G. M. Evins, P. I. Fields, O. Olsvik, T. Popvic, C. A. Bopp, J. G. Wells, C. Carrillo, and P. A. Blake. 1993. The molecular epidemiology of cholera in Latin America. J. Infect. Dis. 167:621-626. [DOI] [PubMed] [Google Scholar]
- 38.Waldor, M. K., R. Colwell, and J. J. Mekalanos. 1994. The Vibrio cholerae O139 serogroup antigen includes an O-antigen capsule and lipopolysaccharide virulence determinants. Proc. Natl. Acad. Sci. USA 91:11388-11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waldor, M. K., and J. J. Mekalanos. 1994. ToxR regulates virulence gene expression in non-O1 strains of Vibrio cholerae that cause epidemic cholera. Infect. Immun. 62:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, R. F., and S. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 41.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development, and virulence in Vibrio cholerae O139. Mol. Microbiol. 39:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647-656. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]