Abstract
Recent evidence suggests that interference competition between bacteria shapes the distribution of the opportunistic pathogen Staphylococcus aureus in the lower nasal airway of humans, either by preventing colonization or by driving displacement. This competition within the nasal microbial community would add to known host factors that affect colonization. We tested the role of toxin-mediated interference competition in both structured and unstructured environments, by culturing S. aureus with toxin-producing or nonproducing Staphylococcus epidermidis nasal isolates. Toxin-producing S. epidermidis invaded S. aureus populations more successfully than nonproducers, and invasion was promoted by spatial structure. Complete displacement of S. aureus was prevented by the evolution of toxin resistance. Conversely, toxin-producing S. epidermidis restricted S. aureus invasion. Invasion of toxin-producing S. epidermidis populations by S. aureus resulted from the evolution of toxin resistance, which was favoured by high initial frequency and low spatial structure. Enhanced toxin production also evolved in some invading populations of S. epidermidis. Toxin production therefore promoted invasion by, and constrained invasion into, populations of producers. Spatial structure enhanced both of these invasion effects. Our findings suggest that manipulation of the nasal microbial community could be used to limit colonization by S. aureus, which might limit transmission and infection rates.
Keywords: community ecology, experimental evolution, interference competition, invasion, spatial structure, staphylococci, toxin production
Introduction
Staphylococcus aureus colonizes the lower portion of the nasal airway (anterior nares) persistently in around 20% of the human population (Van Belkum et al. 2009). Although persistent nasal colonization by S. aureus (carriage) is typically asymptomatic, it is a risk factor for infection in specific patient groups (Von Eiff et al. 2001). These infections can be recurrent and respond poorly to treatment (Kreisel et al. 2006), while the risk of infection is significantly higher for immunocompromised carriers, with increased severity and mortality rates (Yu et al. 1986; Hoen et al. 1995; Senthilkumar et al. 2001).
Studies have revealed many diverse host, bacterial and environmental factors that influence S. aureus carriage. Host factors include genetic variation of the immune response (Van den Akker et al. 2006; Ruimy et al. 2010) and being part of certain patient groups give higher rates of carriage (Atela et al. 1997; Lederer et al. 2007). S. aureus determinants that affect carriage include secreted components associated with immune system interaction (De Haas et al. 2004; Genestier et al. 2005; Rooijakkers et al. 2005) or components of the bacterial cell surface (Kreikemeyer et al. 2002; Clarke et al. 2004; Heilmann et al. 2004).
The nasal microbial community is mainly comprised of Corynebacterium,Propionibacterium and Staphylococcus, with the latter genus constituting between 15% and 60% of the nasal microbial community and mainly comprising the species S. aureus and Staphylococcus epidermidis (Wos-Oxley et al. 2010). There is increasing evidence that the nasal microbial community may contribute to determining S. aureus carriage (Peacock et al. 2001; Frank et al. 2010; Wos-Oxley et al. 2010; Yan et al. 2013; Libberton et al. 2014). One well-described staphylococcal mechanism is via competition arising from allelic variation within agr-dependent signal transduction (Regassa et al. 1992; Yarwood et al. 2002; Weinrick et al. 2004; Schlievert et al. 2007; Horswilll and Nauseef 2008; Peterson et al. 2008). Several studies report negatively associated distributions of S. epidermidis and S. aureus across nasal communities, suggesting that these species engage in one-way or mutual exclusion (Lina et al. 2003; Frank et al. 2010; Wos-Oxley et al. 2010; Libberton et al. 2014). Several potential biochemical mechanisms for these observed patterns have been suggested. Iwase et al. (2010) identified that S. epidermidis can displace S. aureus from the nasal niche by serine protease-mediated biofilm disruption; Lina et al. (2003) showed that quorum sensing interference could contribute to competition whereby different agr types of S. aureus and S. epidermidis could not inhabit the same community. In addition, S. aureus and S. epidermidis both secrete a variety of toxins, which can kill interspecific competitors (Nascimento et al. 2012; Sandiford and Upton 2012; Peschel and Otto 2013)
Here, we constructed simple in vitro communities of S. epidermidis and S. aureus to explore the hypothesis that toxin-mediated killing of competitor species (interference competition) could contribute to the observed negatively associated distributions of these species in nasal communities. Theory predicts that interference competition can both promote and prevent invasion of resident communities. Invasion is promoted when invading populations produce toxin(s) that can kill the resident. However, the cost of producing toxins must be lower than the benefits gained from producing them, and the benefits must not be shared between invader and resident populations. If these criteria are not met, then the interference competition will reduce the chance of invasion (Chao and Levin 1981). Resident populations that produce toxins have been shown to restrict invasion by toxin-sensitive populations (Adams et al. 1979; Chao and Levin 1981; Durrett and Levin 1994; Frank 1994; Duyck et al. 2006; Allstadt et al. 2012). We explored two scenarios in which toxin production by S. epidermidis could drive exclusion of S. aureus: first, where resident toxin-producing S. epidermidis prevent invasion by susceptible S. aureus, and second, where invading toxin-producing S. epidermidis displace a resident susceptible S. aureus population. In addition, we manipulated two ecological parameters that influence the success of toxin-mediated interference competition, specifically, the spatial structure of the environment and the starting frequency of invaders.
In bacteria, interference competition is typically mediated by environmentally secreted toxins, and therefore, it is likely to be affected by environmental spatial structure. Experiments with Escherichia coli have demonstrated that in spatially structured environments (agar plates), bacteriocin producers invaded from very low starting frequency (0.001) into bacteriocin-sensitive populations. By contrast, in the absence of spatial structure (shaken liquid broth), much higher initial frequencies of producers (0.1) were required for successful invasion (Chao and Levin 1981). Spatially structured environments were proposed to promote invasion of toxin producers because clustering of producers enables toxins to reach higher local concentrations (Majeed et al. 2011). As such, the benefits of costly toxin production can accrue to small founding populations. By contrast, in spatially unstructured environments, rapid diffusion of the bacteriocin and quorum sensing molecules away from producing cells of E. coli required bacteriocin producers to exceed a higher threshold frequency before the benefits of bacteriocin production could be realized (Chao and Levin 1981; Tait and Sutherland 2002; Greig and Travisano 2004). Similar frequency-dependent invasion effects of toxin producers were demonstrated in spatially structured populations of the yeast Saccharomyces cerevisiae (Greig and Travisano 2004). We predicted therefore that toxin-producing S. epidermidis strains would be better able to invade-from-rare than nonproducing strains and would do so from lower starting frequencies in more highly spatially structured populations.
Ecological theory proposes that interference competition by a resident species should prevent invasion by a susceptible species irrespective of spatial structure (Adams and Traniello 1981; Doyle et al. 2003). When a toxin kills susceptible immigrants, invaders are unable to sustain a viable population; in population ecology, such hostile environmental patches are often termed black hole sinks (Holt and Gaines 1992). Evolutionary theory also proposes that there is potential for a susceptible invading population to evolve resistance to a toxin and that the probability of this will depend upon the frequency of invaders and the spatial structure of the environment (Chao and Levin 1981; Holt et al. 2003). Several theoretical models predict that the likelihood of adaptation to a black hole sink environment increases with the frequency of immigrants from the source population (Gomulkiewicz et al. 1999; Holt et al. 2003). Higher immigration rates will increase the probability that immigrants carry beneficial mutations that are pre-adapted to survive the conditions of the black hole sink (Holt and Gaines 1992; Perron et al. 2008). Therefore, invading S. aureus populations are more likely to contain mutants resistant to S. epidermidis toxins when invading from higher starting frequencies. However, the spread of these beneficial resistance mutations is likely to be impeded in more highly spatially structured environments. This is because competition of the beneficial mutant can only occur at the edge of a colony, and as the colony grows, a smaller proportion of the mutant population will be competing with the ancestral genotype (Habets et al. 2007). Taken together, we predict therefore that nonproducing residents will be more easily invaded, that resistance of the invader to inhibitory toxins is more likely to evolve when invaders are at a high starting frequency and that resistant mutants that evolve will be more likely to invade in unstructured environments.
To test these predictions, we performed competition experiments whereby toxin-producing and nonproducing nasal isolates of S. epidermidis were invaded from three starting frequencies (0.1, 0.01 and 0.001) into resident populations of toxin-sensitive S. aureus. Conversely, to test whether S. aureus invasion could be restricted by S. epidermidis toxin production, we performed the reciprocal invasion of S. aureus from three starting frequencies (0.1, 0.01 and 0.001) into resident populations of toxin-producing and nonproducing S. epidermidis. All competitions were propagated for 7 days on solid agar with daily transfer of communities to fresh medium; in half of the replicates, population structure was maintained at each transfer, whereas in the other half of the replicates, the population structure was homogenized at each transfer.
Materials and methods
Culture conditions
All bacterial strains used in this study were cultured at 37°C in 10 mL BHI broth shaken at 200 rpm and on agar-solidified BHI medium (brain–heart infusion solids (porcine), 17.5 g/L; tryptose, 10.0 g/L; glucose, 2.0 g/L; sodium chloride, 5.0 g/L; disodium hydrogen phosphate, 2.5 g/L) (Lab M, Heywood, UK). Chemicals were obtained from Sigma-Aldrich Co., UK.
Selection of nasal isolates
Four independent S. epidermidis isolates were selected from a previous study that sampled the anterior nares of 60 healthy volunteers (Libberton et al. 2014): two isolates were toxin producers as revealed in a deferred inhibition assay by their killing of S. aureus [zone of clearing when a lawn of S. aureus strain SH1000 was sprayed over them (Nascimento et al. 2012)]; two isolates were toxin nonproducers based on not reducing viability of strain SH1000. SH1000 displayed no growth inhibition activity against any of the selected S. epidermidis strains in the deferred inhibition assay (Nascimento et al. 2012). Of the two toxin-producing S. epidermidis strains, B180 produced an inhibition area that was around ten times greater than that of B155. We first established that the S. epidermidis strains had comparable growth rates to SH1000. An overnight culture of each strain (Table1) was inoculated (1% inoculum) into 200 μL of BHI broth in a 96-well plate. The 96-well plates were incubated at 37°C for 8 h, and OD600 readings were taken at 20-min intervals. The doubling time (min) was then calculated (Table2) using the following formula where Td is the doubling time; t1 and t2 are two consecutive time points throughout the bacterial growth; and d1 and d2 are the corresponding OD600 readings at t1 and t2.
Table 1.
Strains used in this study
| Species | Strain identification | Reference |
|---|---|---|
| S. aureus | SH1000 | Horsburgh et al. (2002) |
| S. epidermidis | B155 (inhibitor producing) | Libberton et al. (2014) |
| S. epidermidis | B180 (inhibitor producing) | Libberton et al. (2014) |
| S. epidermidis | B035 (noninhibitor producing) | Libberton et al. (2014) |
| S. epidermidis | B115 (noninhibitor producing) | Libberton et al. (2014) |
Table 2.
Doubling times of strains used in this study. The doubling times in minutes were compared to SH1000 (S. aureus) as a control using a post hoc Dunnett’s test. There is no significant difference between any of the S. epidermidis strains tested and the S. aureus strain SH1000 used in this study
| Doubling time (min) | T-value | P-value | |
|---|---|---|---|
| SH1000 | 116.45 | NA | NA |
| B180 | 116.06 | −0.074 | 1.0000 |
| B155 | 110.11 | −1.203 | 0.5689 |
| B115 | 120.49 | 0.767 | 0.8579 |
| B035 | 123.34 | 1.307 | 0.4970 |
Competition experiments
All strains were cultured on BHI agar plates prior to competition experiments. Bacteria were cultured for 18 h on 50-mm-diameter BHI agar plates, and the lawns of S. aureus (SH1000) and S. epidermidis strains (resident and invader – Table1) were then scraped off the agar plates and suspended in 10 mL of PBS by vortexing thoroughly. The cfu/mL in each tube was equalized by diluting the cell suspensions in PBS and comparing the OD600 of each suspension (approximately 5 × 108 cfu/mL for S. aureus and S. epidermidis, determined by viable count). Both species were then mixed together in a final volume of 10 mL PBS, with the invader at different frequencies (ratios) to the resident (0.1:1, 0.01:1, 0.001:1). For brevity, these ratios are referred to in this manuscript as frequencies, and only the first number in the ratio pair is used to define each frequency. The mixtures were vortexed thoroughly before 50 μL (containing approximately 2.5 × 106 cells) was plated onto 25 mL BHI agar and incubated at 37°C. Six replicate communities (structured and unstructured, in triplicate) were established at each starting frequency. The communities were transferred to a new agar plate every day for 7 days. Half of the replicates underwent a regime whereby the transfers were made by replica plating with velvet (Lederberg and Lederberg 1952) to maintain spatial structure. While the other half of replicates underwent a mixed regime whereby the spatial structure was destroyed every 24 h transfer by scraping the entire bacterial lawn off the plate and transferring to 10 mL of sterile PBS, before thoroughly vortexing and pipetting 50 μL onto a new plate to complete the transfer. Each set was performed in triplicate. Viable counts for each isolate were calculated every second day. On the structured plates, this was achieved after replica plating from viable counts of the remaining lawn; colonies were differentiated by colony morphology and pigmentation. S. aureus SH1000 possesses a distinct yellow carotenoid pigment which was stable over the course of these experiments. Raw data for the experiments are presented in appendices (Figs 7 and 8).
Deferred inhibition spray assay
A deferred inhibition spray assay was performed to determine whether S. aureus clones had developed resistance to the toxin-producing S. epidermidis strains. The assay was performed on 10 clones from each experiment. A 25-μL spot (approximately 108 cells) of an overnight bacterial culture was pipetted onto the centre of an agar plate containing 15 mL of BHI agar (Lab M). The plates were incubated for 18 h at 37°C before 250 μL of a 10-fold diluted overnight culture of a different strain (106 cfu) was sprayed over the plate. The plates were incubated for a further 18 h after when the size of the inhibition zones produced by the central spot on the overlaid strain was assessed. The clarity of the inhibition zone was scored based on a simple scoring system of 1–4, 4 being completely clear and 1 being no detectable zone. The areas of any detectable zones were also recorded by measuring the diameter of the inhibition zone and the central colony.
Data analysis
To quantify the success of the invasion, we calculated the selection rate constant for each invader using relative bacterial frequencies from day 0 and day 7 with the following equation.
where Ni (0) and Nr (0) represent the initial densities of the competing populations i (invader) and r (resident), and Ni (1) and Nr (1) represent their densities after 1 day (Travisano & Lenski, 1996).
Negative values indicated that invasion was not possible, whereas positive values indicated invasion was possible. The invasion time-course data were visualized using plots of the natural log of the invader to resident ratio over time; selection rate constants were analysed in a three-way anova.
Results
Spatial structure promotes invasion by inhibitor-producing S. epidermidis
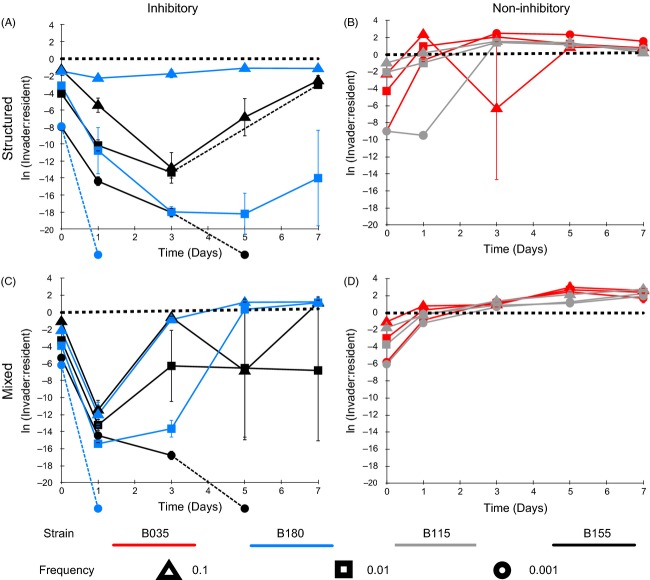
Environmental structure promoted S. epidermidis invasion (structure, F1,64 = 322.77, P < 0.001) (Fig.1 and Table3), and this effect was stronger for S. epidermidis toxin producers than for nonproducers (structure × inhibition, F1,64 = 14.29, P < 0.001) (Table3). S. epidermidis was never able to successfully invade under mixed conditions (Fig.1 and Table3). However, S. epidermidis was more likely to persist at low frequencies and avoid extinction in mixed environments when initiated at a higher starting frequency (frequency × structure, F1,64 = 13.55, P < 0.001).
Figure 1.
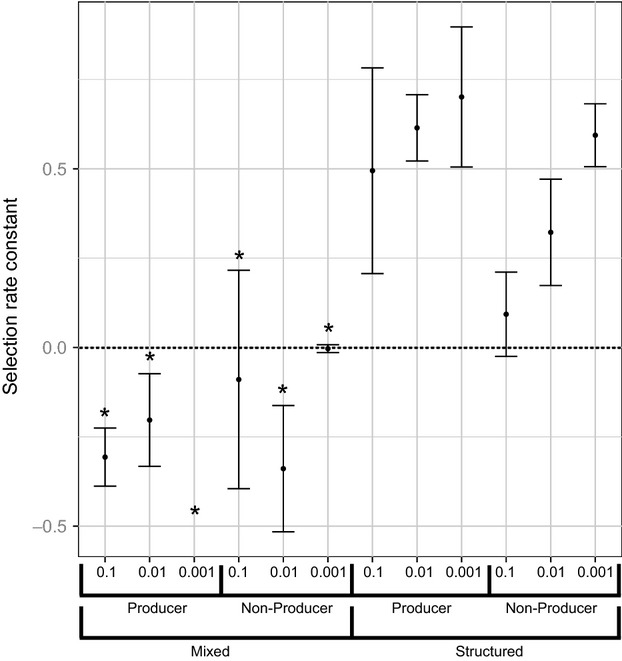
Selection rate coefficients for Staphylococcus epidermidis invading populations of S. aureus (SH1000). Toxin-producing S. epidermidis isolates (155 and 180) and nonproducing isolates (035 and 115) were invaded into populations of S. aureus (SH1000) at relative frequencies of 10, 100 and 1000. Each of the invasions was also carried out under a spatially structured treatment and a mixed treatment. Asterisks mark negative selection rate coefficients where invasion did not occur. Error bars represent the standard error of the mean.
Table 3.
Analysis of variance testing the main effects of successful invasion of S. epidermidis into populations of S. aureus. The table shows the results of a multifactorial anova. Both main effects and interactions are shown
| df | Sum sq | Mean sq | F value | P value | |
|---|---|---|---|---|---|
| Frequency | 1 | 0.1382 | 0.1382 | 3.4920 | 0.0662444 |
| Structure | 1 | 12.7710 | 12.7710 | 322.7662 | <2.2e-16*** |
| Inhibition | 1 | 0.1452 | 0.1452 | 3.6690 | 0.0599020 |
| Frequency × Structure | 1 | 0.5359 | 0.5359 | 13.5452 | 0.0004798*** |
| Frequency × Inhibition | 1 | 0.0243 | 0.0243 | 0.6141 | 0.4361327 |
| Structure × Inhibition | 1 | 0.5652 | 0.5652 | 14.2852 | 0.0003474*** |
| Frequency × Structure × Inhibition | 1 | 0.0746 | 0.0746 | 1.8858 | 0.1744717 |
df, Degrees of freedom; Sum sq, sum of squares; Mean sq, Mean of squares; F value, F statistic for terms in the row; P value, significance.
Asterisks indicate the significance levels at different thresholds. ***P < 0.001.
Invasion was impeded by evolution of resistance
Under structured conditions, the two invading, toxin-producing strains of S. epidermidis show different dynamics over time (Fig.2A). All starting frequencies of strain B155 increase after day 1 and approach a 1:1 invader to resident ratio, whereas strain B180 (starting frequencies 0.1 and 0.01) increases until day 3, after which they decrease. Spray assays were performed to test whether the decline in frequency of strain B180 populations (of starting frequency 0.1 and 0.01) was caused by resistance evolution in the resident S. aureus population. Ancestral and evolved resident S. aureus clones were sprayed over ancestral and evolved S. epidermidis strain B180 (Fig.3). These assays show that after 7 days, the resident S. aureus had evolved resistance to the invading S. epidermidis under structured conditions at starting frequencies of 0.1 and 0.01 (Fig.3) (Fisher’s exact test, P = 0.0022). Resistance was not seen in the S. aureus resident population when invaded with strain B180 at a starting frequency of 0.001 (Fig.3) (Fisher’s exact test, P = 1). Of note, evolved S. epidermidis strains (Fig.3B) produced larger inhibition zones against susceptible S. aureus than the ancestral S. epidermidis strains (Fig.3B) (paired t-test: T = 2.69, P = 0.03).
Figure 2.
Toxin-producing (blue and black) and nonproducing (red and grey) isolates of Staphylococcus epidermidis invading populations of S. aureus (SH1000) at frequencies of 0.1 (triangle), 0.01 (square) and 0.001 (circle). Toxin-producing S. epidermidis isolates (155 and 180) and nonproducing S. epidermidis isolates (035 and 115) were introduced into a population of S. aureus (SH1000) at three different frequencies. This was carried under a spatially structured regime (A and B) and under a mixed regimen (C and D). The x-axis is the time in days, and the y-axis is the natural log of the invader to resident ratio. A dotted line in the time course shows when the population dipped below the experiment detection threshold (for clarity, these lines also cross the x-axis if the population went to extinction). There is a heavy dotted line at 0 on the y-axis to indicate an equal invader to resident ratio. The line crossing the x-axis symbolizes that the population went to extinction. Error bars represent the standard error of the mean (n = 3).
Figure 3.
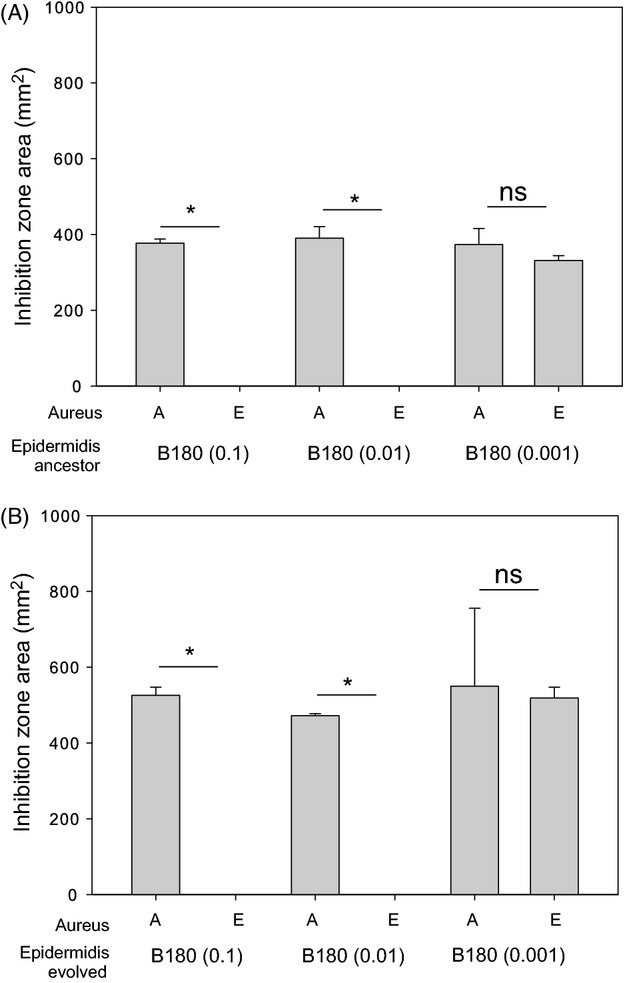
Resistance of evolved SH1000 resident after Staphylococcus epidermidis (B180) invasion. Panel A shows inhibition zone produced by the ancestral S. epidermidis strains, and panel B shows the inhibition zones produced by the evolved S. epidermidis strains. Both panels A and B show the inhibition zone area (mm2) produced by the toxin-producing S. epidermidis strains against the ancestral SH1000 (A) and the evolved SH1000 (E). Asterisks represent a significant difference between the inhibition zone areas of ancestral (A) and evolved (E) S. aureus strains as determined by a Fisher’s exact test. Each significance star represents a P value of 0.0022 which is significant when Bonferroni corrected for multiple comparisons with an alpha value of 0.1. Error bars represent the standard error of the mean.
Toxin-producing S. epidermidis strains resist invasion, especially in structured environments
Toxin-producing S. epidermidis strains were more resistant to invasion than nonproducing strains (inhibition, F1,64 = 124.95, P < 0.0001, Table4) and restricted invasion more effectively under structured environmental conditions (Figs4 and 5A) (structure × inhibition, F1,64 = 6.14, P < 0.05, Table4). Invasion of S. aureus into a toxin-producing S. epidermidis resident was positively frequency-dependent with highest initial frequencies invading the fastest and lower initial frequencies going to extinction (Fig.5A,C) (frequency × inhibition, F1,64 = 46.5, P < 0.001).
Table 4.
Analysis of variance testing the main effects of successful invasion of S. aureus into populations of S. epidermidis. The table shows the results of a multifactorial anova. Both main effects and interactions are shown
| df | Sum sq | Mean sq | F value | P value | |
|---|---|---|---|---|---|
| Frequency | 1 | 2.721 | 2.721 | 8.1457 | 0.005810** |
| Structure | 1 | 2.949 | 2.949 | 8.8266 | 0.004177** |
| Inhibition | 1 | 41.744 | 41.744 | 124.9525 | <2.2e-16*** |
| Frequency × Structure | 1 | 0.007 | 0.007 | 0.0198 | 0.88449 |
| Frequency × Inhibition | 1 | 15.554 | 15.554 | 46.5589 | 3.794e-09*** |
| Structure × Inhibition | 1 | 2.051 | 2.051 | 6.1382 | 0.015880* |
| Frequency × Structure × inhibition | 1 | 0.398 | 0.398 | 1.1900 | 0.279418 |
df, Degrees of freedom; Sum sq, sum of squares; Mean sq, Mean of squares; F value, F statistic for terms in the row; P value, significance.
Asterisks indicate the significance levels at different thresholds. *P < 0.05; **P < 0.01, ***P < 0.001.
Figure 4.
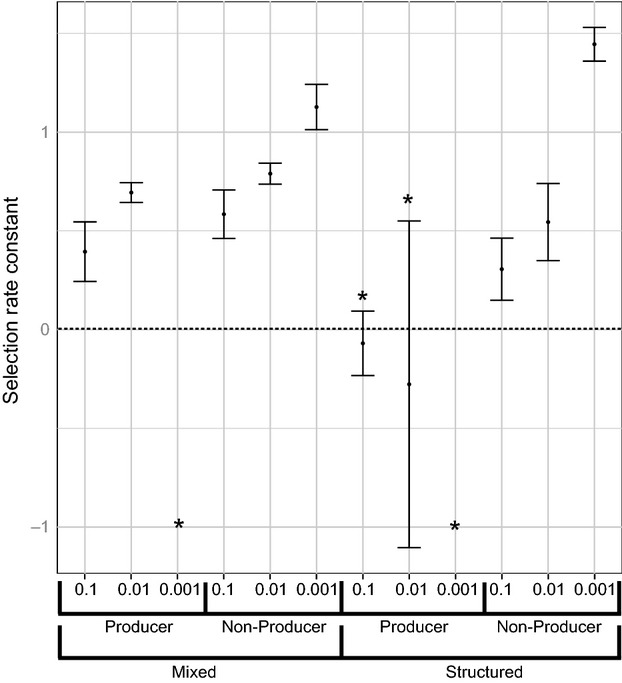
Selection rate coefficients for Staphylococcus aureus (SH1000) invading populations of S. epidermidis. S. aureus was introduced into populations of toxin-producing S. epidermidis isolates (155 and 180) and nonproducing isolates (035 and 115) at relative frequencies of 10, 100 and 1000. Each of the invasions was also carried out under a spatially structured treatment and a mixed treatment. Asterisks mark negative selection rate coefficients where invasion did not occur. Error bars represent the standard error of the mean.
Figure 5.
Staphylococcus aureus invading populations of toxin-producing (blue and black) and nonproducing (red and grey) S. epidermidis at frequencies of 0.1 (triangle), 0.01 (square) and 0.001 (circle). S. aureus strain (SH1000) was introduced into two different toxin-producing S. epidermidis populations (155 and 180), and two different nonproducing populations (035 and 115) at three different frequencies. This was carried under a spatially structured regime (A and B) and under a mixed regime (C and D). The x-axis is the time in days, and the y-axis is the natural log of the invader to resident ratio. A dotted line in the time course shows when the population dipped below the experiment detection threshold (for clarity, these lines also cross the x-axis if the population went to extinction). There is a heavy dotted line at 0 on the y-axis to indicate an equal invader to resident ratio. The line crossing the x-axis symbolizes that the population went to extinction. Error bars represent the standard error of the mean (n = 3).
Evolved resistance promotes S. aureus invasion
Staphylococcus aureus was only able to invade toxin-producing S. epidermidis under mixed conditions (Fig.5C). To test whether the evolution of inhibitory toxin resistance by S. aureus was responsible for the invasion in a mixed environment (Figs4 and 5C), ancestral and evolved S. aureus strains were sprayed over ancestral and evolved S. epidermidis toxin-producing residents. In all cases, evolved S. aureus were resistant to the S. epidermidis toxin (Fig.6) (Fisher’s exact test, P = 0.0022).
Figure 6.
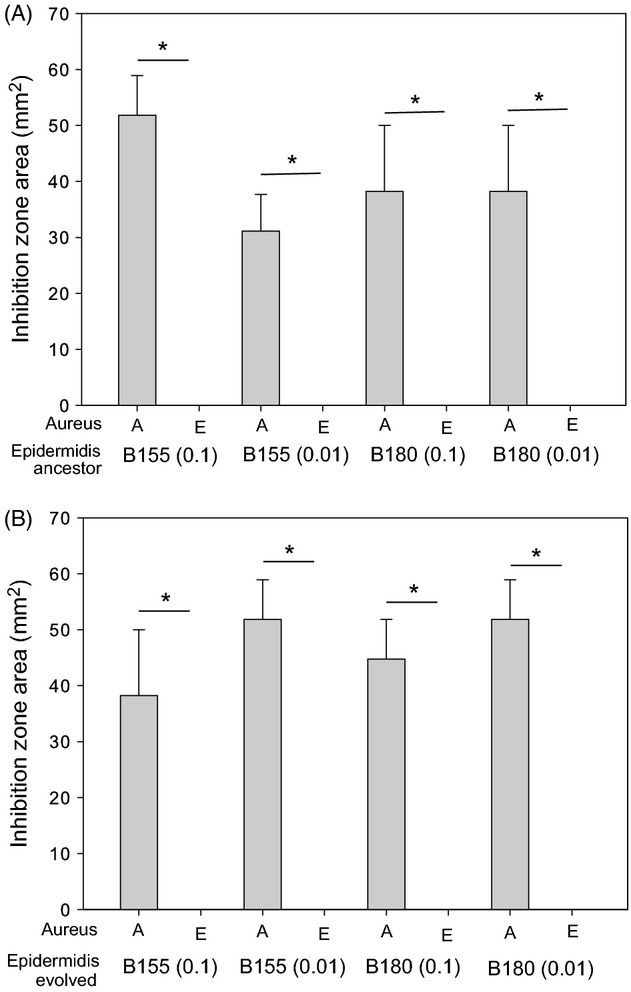
Resistance of evolved SH1000 after successful invasion. Panel A shows inhibition zone produced by the ancestral Staphylococcus epidermidis strains, and panel B shows the inhibition zones produced by the evolved S. epidermidis strains. Both panels A and B show the inhibition zone area (mm2) produced by the inhibitory S. epidermidis strains against the ancestral SH1000 (A) and the evolved SH1000 (E). The * represents a significant difference between the inhibition zone areas of ancestral (A) and evolved (E) S. aureus strains as determined by a Fisher’s exact test. Each significance star represents a P value of 0.0022, which is significant when Bonferroni corrected for multiple comparisons with an alpha value of 0.1. Error bars represent the standard error of the mean.
Discussion
We show that antimicrobial toxin production by S. epidermidis nasal isolates can have important effects on competition with S. aureus: interference competition acts both to promote invasion by, and to prevent invasion into, S. epidermidis populations. This supports the growing body of evidence that species interactions can play an important role determining species distributions in nasal microbial communities (Lina et al. 2003; Frank et al. 2010; Wos-Oxley et al. 2010; Yan et al. 2013), and more specifically that trait variation, in this case in the toxins mediating interference competition, could act to prevent S. aureus nasal carriage (Libberton et al. 2014).
Our findings highlight the critical role for spatial structure in determining the outcome of interference competition. If spatial structure is maintained, then inhibitor-producing bacteria can better prevent the invasion-from-rare of S. aureus, whereas unstructured environments generally do not favour the production of inhibitory toxins. Spatial structure is likely to be an important component of life in the anterior nares. While nutrient agar is clearly not equivalent to this environment and transfers using velvet may select those bacteria growing nearest to the colony surface, our manipulation of spatial structure is arguably more relevant to the nasal environment than comparing agar plates to liquid culture (Chao and Levin 1981). The macrotopography of the nares is irregular, with ridges and recesses providing spatially discrete surfaces. The base layer of the nares is comprised of a squamous epithelium that microbes colonize and form spatially discrete groups (Uraih and Maronpot 1990; Yuki et al. 2000; Dongari-Bagtzoglou et al. 2009). Spatial population structure is therefore expected to be present, but is likely to be disrupted by changes to the squamous epithelium, the flow of air (Churchill et al. 2004) and mucus (Proctor et al. 1973) through the nasal passages and mechanical disruptions (e.g. nose picking). Factors that reduce the spatial structuring in nasal communities could weaken the ability of inhibitory resident species to prevent invasion by S. aureus.
Further, our data demonstrate that rapid evolution of resistance to antimicrobial toxins can determine the outcome of interference competition. Positive frequency-dependent fitness was observed for S. aureus invading inhibitory residents. This may have occurred due to a protective effect from a lager inoculum neutralizing the effect of the toxin. This is unlikely, however, as protection caused by large bacterial densities is typically a result of quenching and lowering the local concentration of available toxin. In this experimental set-up, toxin would be produced during growth phases every time the population is transferred, which would overcome any possible quenching effect. The more likely explanation for resistance evolution is that the higher inoculation frequencies increased the chance of these invading populations containing beneficial resistance mutations. This outcome is similar to theory predicting conditions for adaptation to black hole sink environments where an increased immigration rate (i.e., increased frequency of the invading population) increases the probability of adaptation (Holt and Gaines 1992; Perron et al. 2008). Moreover, this scenario suggests that an inhibitor-producing community could resist invasion from rare by S. aureus, because of a low probability of resistance evolution. When toxin-producing S. epidermidis were invading resident S. aureus, evolution of resistance in S. aureus resident populations was most likely when the toxin-producing invaders were relatively common (Fig.3). Higher frequencies of invading toxin producers would have produced more of the toxin, generating stronger selection for resistance; additionally, a larger fraction of the S. aureus populations would have been exposed to these toxins. This relationship suggests that resistance evolution by residents may frequently impede invasion by toxin-producing strains, because resident populations are unlikely to be mutation-limited and selection for resistance progressively strengthens as an invasion proceeds. Intriguingly, in spatially structured environments, the evolved invading S. epidermidis (B180) showed greater inhibitory activity on ancestral SH1000 than the ancestral B180 genotype (Fig.3). This suggests that the toxin producer coevolved to meet the survival challenges posed by increasingly resistant S. aureus populations. The evolved S. epidermidis may have upregulated production of the inhibitory toxin, or alternatively initiated production of alternative toxins. However, in the absence of knowledge of the mechanism of inhibition, this remains unclear.
One strength of this study is the use of toxin-producing strains isolated from the nares of healthy volunteers and not isogenic toxin-producing and nonproducing laboratory strains. Although nasal isolates are more difficult to compare with well-characterized laboratory strains, they have greater relevance to future development of therapeutic strategies and provide added realism to laboratory models of colonization. There are also other limitations, for example the zones of inhibition produced from B180 and B155 had different areas, which implies differential expression of the same toxin or discrete toxins. Resource competition and adaptation to the growth medium could affect the outcomes described and contribute to the interactions between the pairs of bacteria, but these aspects would require further study to describe.
It is stated that preventing S. aureus carriage significantly reduces the risk of infection (Von Eiff et al. 2001). Our findings support the possibility that manipulation of the microbial community in the human nose to increase the frequency of inhibitor-producing residents could reduce S. aureus colonization. The human gut has been a model system for therapeutic manipulation of the microbial flora for many years (Borody et al. 1989; Landy et al. 2011). If the models of colonization and their outcomes can be replicated in the nasal environment, it would represent a novel way to limit S. aureus carriage that is correlated with associated life-threatening infections.
Acknowledgments
This study was supported by a BBSRC studentship (BB/D526529/1) to B.L. awarded to M.A.B. and M.J.H.
A Appendix
Figure 7.
Toxin-producing ( and
and  ) and nonproducing (
) and nonproducing ( and
and  ) isolates of Staphylococcus epidermidis invading populations of S. aureus (SH1000) at frequencies of 0.1 (
) isolates of Staphylococcus epidermidis invading populations of S. aureus (SH1000) at frequencies of 0.1 ( ), 0.01 (
), 0.01 ( ) and 0.001 (
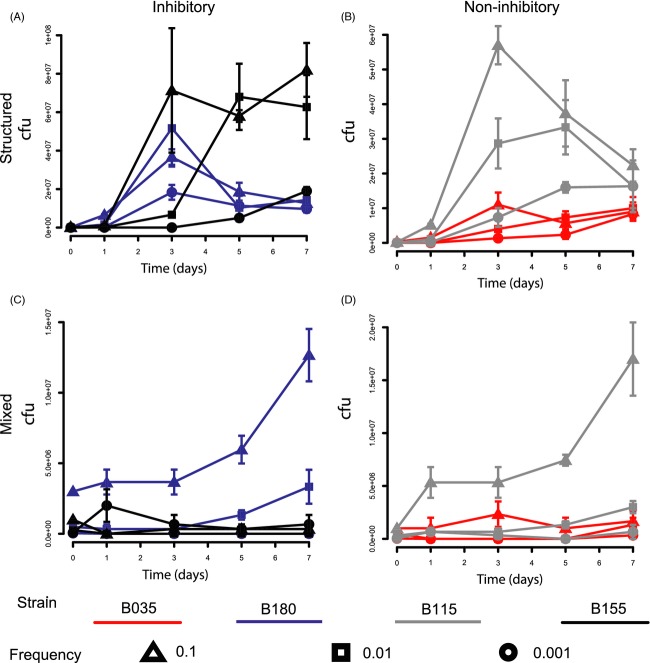
) and 0.001 ( ). Toxin-producing S. epidermidis isolates (155 and 180) and nonproducing S. epidermidis isolates (035 and 115) were introduced into a population of S. aureus (SH1000) at three different frequencies. This was carried under a spatially structured regime (A and B) and under a mixed regimen (C and D). The x-axis is the time in days, and the y-axis is the colony-forming units (cfu) per plate. Error bars represent the standard error of the mean (n = 3).
). Toxin-producing S. epidermidis isolates (155 and 180) and nonproducing S. epidermidis isolates (035 and 115) were introduced into a population of S. aureus (SH1000) at three different frequencies. This was carried under a spatially structured regime (A and B) and under a mixed regimen (C and D). The x-axis is the time in days, and the y-axis is the colony-forming units (cfu) per plate. Error bars represent the standard error of the mean (n = 3).
B Appendix
Figure 8.
Staphylococcus aureus invading populations of toxin-producing ( and
and  ) and nonproducing (
) and nonproducing ( and
and  ) S. epidermidis at frequencies of 0.1 (
) S. epidermidis at frequencies of 0.1 ( ), 0.01 (
), 0.01 ( ) and 0.001 (
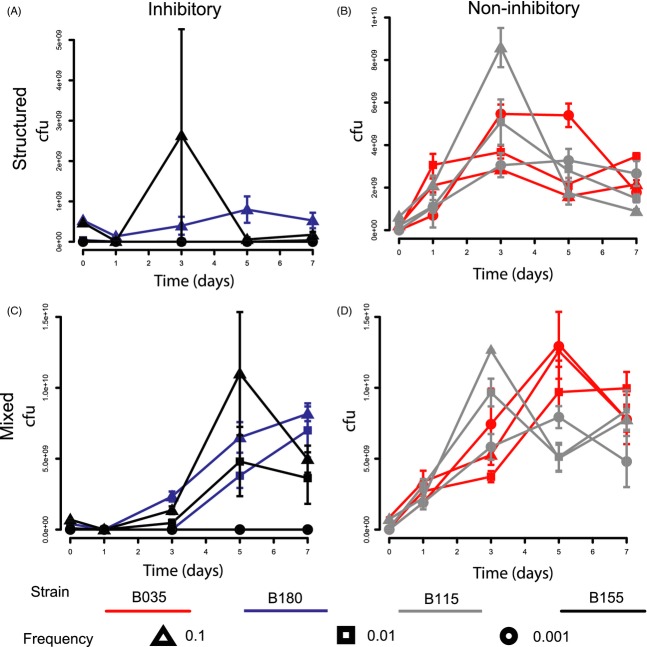
) and 0.001 ( ). S. aureus strain (SH1000) was introduced into two different toxin-producing S. epidermidis populations (155 and 180), and two different nonproducing populations (035 and 115) at three different frequencies. This was carried under a spatially structured regime (A and B) and under a mixed regime (C and D). The x-axis is the time in days, and the y-axis is the colony-forming units (cfu) per plate. Error bars represent the standard error of the mean (n = 3).
). S. aureus strain (SH1000) was introduced into two different toxin-producing S. epidermidis populations (155 and 180), and two different nonproducing populations (035 and 115) at three different frequencies. This was carried under a spatially structured regime (A and B) and under a mixed regime (C and D). The x-axis is the time in days, and the y-axis is the colony-forming units (cfu) per plate. Error bars represent the standard error of the mean (n = 3).
Data archiving statement
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.nb535.
Conflict of interest
The authors declare no competing commercial interests relating to the data presented in this manuscript.
Literature cited
- Adams ES. Traniello JFA. Chemical interference competition by Monomorium minimum (Hymenoptera: Formicidae) Oecologia. 1981;51:265–270. doi: 10.1007/BF00540612. [DOI] [PubMed] [Google Scholar]
- Adams J, Kinney T, Thompson S, Rubin L. Helling RB. Frequency dependent selection for plasmid containing cells of Escherichia coli. Genetics. 1979;91:627–637. doi: 10.1093/genetics/91.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allstadt A, Caraco T, Molnár F. Korniss G. Interference competition and invasion: spatial structure, novel weapons and resistance zones. Journal of Theoretical Biology. 2012;306:46–60. doi: 10.1016/j.jtbi.2012.04.017. [DOI] [PubMed] [Google Scholar]
- Atela I, Coll P, Rello J, Quintana E, Barrio J, March F, Sanchez F, et al. Serial surveillance cultures of skin and catheter hub specimens from critically ill patients with central venous catheters: molecular epidemiology of infection and implications for clinical management and research. Journal of Clinical Microbiology. 1997;35:1784–1790. doi: 10.1128/jcm.35.7.1784-1790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borody TJ, George L, Andrews P, Brandl S, Noonan S, Cole P, Hyland L, et al. Bowel-flora alteration: a potential cure for inflammatory bowel disease and irritable bowel syndrome. Medical Journal of Australia. 1989;150:604. doi: 10.5694/j.1326-5377.1989.tb136704.x. [DOI] [PubMed] [Google Scholar]
- Chao L. Levin BR. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:6324–6328. doi: 10.1073/pnas.78.10.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill SE, Shackelford LL, Georgi JN. Black MT. Morphological variation and airflow dynamics in the human nose. American Journal of Human Biology. 2004;16:625–638. doi: 10.1002/ajhb.20074. [DOI] [PubMed] [Google Scholar]
- Clarke SR, Wiltshire MD. Foster SJ. IsdA of Staphylococcus aureus is a broad spectrum, iron-regulated adhesin. Molecular Microbiology. 2004;51:1509–1519. doi: 10.1111/j.1365-2958.2003.03938.x. [DOI] [PubMed] [Google Scholar]
- De Haas CJC, Veldkamp KE, Peschel A, Weerkamp F, Van Wamel WJB, Heezius ECJM, Poppelier MJ, et al. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. Journal of Experimental Medicine. 2004;199:687–695. doi: 10.1084/jem.20031636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dongari-Bagtzoglou A, Kashleva H, Dwivedi P, Diaz P. Vasilakos J. Characterization of mucosal Candida albicans biofilms. PLoS One. 2009;4:e7967. doi: 10.1371/journal.pone.0007967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle RD, Francis MD. Smart RM. Interference competition between Ludwigia repens and Hygrophila polysperma: two morphologically similar aquatic plant species. Aquatic Botany. 2003;77:223–234. [Google Scholar]
- Durrett R. Levin S. The importance of being discrete (and spatial) Theoretical Population Biology. 1994;46:363–394. [Google Scholar]
- Duyck PF, David P, Junod G, Brunel C, Dupont R. Quilici S. Importance of competition mechanisms in successive invasions by polyphagous tephritids in La Reunion. Ecology. 2006;87:1770–1780. doi: 10.1890/0012-9658(2006)87[1770:iocmis]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Frank SA. Spatial polymorphism of bacteriocins and other allelopathic traits. Evolutionary Ecology. 1994;8:369–386. [Google Scholar]
- Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN. Pace NR. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One. 2010;5:e10598. doi: 10.1371/journal.pone.0010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genestier AL, Michallet MC, Prevost G, Bellot G, Chalabreysse L, Peyrol S, Thivolet F, et al. Staphylococcus aureus Panton-Valentine leukocidin directly targets mitochondria and induces Bax-independent apoptosis of human neutrophils. Journal of Clinical Investigation. 2005;115:3117–3127. doi: 10.1172/JCI22684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomulkiewicz R, Holt RD. Barfield M. The effects of density dependence and immigration on local adaptation and niche evolution in a black-hole sink environment. Theoretical Population Biology. 1999;55:283–296. doi: 10.1006/tpbi.1998.1405. [DOI] [PubMed] [Google Scholar]
- Greig D. Travisano M. The Prisoner’s Dilemma and polymorphism in yeast SUC genes. Proceedings of the Royal Society of London. Series B: Biological Sciences. 2004;271:S25–S26. doi: 10.1098/rsbl.2003.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habets MGJL, Czaran T, Hoekstra RF. de Visser J. Spatial structure inhibits the rate of invasion of beneficial mutations in asexual populations. Proceedings of the Royal Society of London. Series B: Biological Sciences. 2007;274:2139–2143. doi: 10.1098/rspb.2007.0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C, Niemann S, Sinha B, Herrmann M, Kehrel BE. Peters G. Staphylococcus aureus fibronectin-binding protein (FnBP)-mediated adherence to platelets, and aggregation of platelets induced by FnBPA but not by FnBPB. Journal of Infectious Diseases. 2004;190:321–329. doi: 10.1086/421914. [DOI] [PubMed] [Google Scholar]
- Hoen B, Kessler M, Hestin D. Mayeux D. Risk-factors for bacterial-infection in chronic heamodialysis adult patients – a multicenter prospective survey. Nephrology Dialysis Transplant. 1995;10:377–381. [PubMed] [Google Scholar]
- Holt RD. Gaines MS. Analysis of adaptation in heterogeneous landscapes – implications for the evolution of fundamental niches. Evolutionary Ecology. 1992;6:433–447. [Google Scholar]
- Holt RD, Gomulkiewicz R. Barfield M. The phenomenology of niche evolution via quantitative traits in a “black-hole” sink. Proceedings of the Royal Society of London. Series B: Biological Sciences. 2003;270:215–224. doi: 10.1098/rspb.2002.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK. Foster SJ. SigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325–4. Journal of Bacteriology. 2002;184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horswilll AR. Nauseef WM. Host interception of bacterial communication signals. Cell Host & Microbe. 2008;4:507–509. doi: 10.1016/j.chom.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- Kreikemeyer B, McDevitt D. Podbielski A. The role of the Map protein in Staphylococcus aureus matrix protein and eukaryotic cell adherence. International Journal of Medical Microbiology. 2002;292:283–295. doi: 10.1078/1438-4221-00212. [DOI] [PubMed] [Google Scholar]
- Kreisel K, Boyd K, Langenberg P. Roghmann MC. Risk factors for recurrence in patients with Staphylococcus aureus infections complicated by bacteremia. Diagnostic Microbiology Infectious Disease. 2006;55:179–184. doi: 10.1016/j.diagmicrobio.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Landy J, Al-Hassi HO, McLaughlin SD, Walker AW, Ciclitira PJ, Nicholls RJ, Clark SK, et al. Review article: faecal transplantation therapy for gastrointestinal disease. Alimentary Pharmacology and Therapeutics. 2011;34:409–415. doi: 10.1111/j.1365-2036.2011.04737.x. [DOI] [PubMed] [Google Scholar]
- Lederberg J. Lederberg EM. Replica plating and indirect selection of bacterial mutants. Journal of Bacteriology. 1952;63:399–406. doi: 10.1128/jb.63.3.399-406.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer SR, Riedelsdorf G. Schiffl H. Nasal carriage of methicillin resistant Staphylococcus aureus: the prevalence, patients at risk and the effect of elimination on outcomes among outclinic haemodialysis patients. European Journal of Medical Research. 2007;12:284–288. [PubMed] [Google Scholar]
- Libberton B, Coates RE, Brockhurst MA. Horsburgh MJ. Evidence that intraspecific trait variation among nasal bacteria can shape the distribution of Staphylococcus aureus. Infection and Immunity. 2014;82:3811–3815. doi: 10.1128/IAI.02025-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina G, Boutite F, Tristan A, Bes M, Etienne J. Vandenesch F. Bacterial competition for human nasal cavity colonization: role of Staphylococcal agr alleles. Applied and Environmental Microbiology. 2003;69:18–23. doi: 10.1128/AEM.69.1.18-23.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed H, Gillor O, Kerr B. Riley MA. Competitive interactions in Escherichia coli populations: the role of bacteriocins. The ISME Journal. 2011;5:71–81. doi: 10.1038/ismej.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento JD, Coelho MLV, Ceotto H, Potter A, Fleming LR, Salehian Z, Nes IF, et al. Genes involved in immunity to and secretion of aureocin A53, an atypical class II bacteriocin produced by Staphylococcus aureus A53. Journal of Bacteriology. 2012;194:875–883. doi: 10.1128/JB.06203-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock SJ, de Silva I. Lowy FD. What determines nasal carriage of Staphylococcus aureus. Trends in Microbiology. 2001;9:605–610. doi: 10.1016/s0966-842x(01)02254-5. [DOI] [PubMed] [Google Scholar]
- Perron GG, Gonzalez A. Buckling A. The rate of environmental change drives adaptation to an antibiotic sink. Journal of Evolutionary Biology. 2008;21:1724–1731. doi: 10.1111/j.1420-9101.2008.01596.x. [DOI] [PubMed] [Google Scholar]
- Peschel A. Otto M. Phenol-soluble modulins and staphylococcal infection. Nature Reviews Microbiology. 2013;11:667–673. doi: 10.1038/nrmicro3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson MM, Mack JL, Hall PR, Alsup AA, Alexander SM, Sully EK, Sawires YS, et al. Apolipoprotein B is an innate barrier against invasive Staphylococcus aureus infection. Cell Host & Microbe. 2008;4:555–566. doi: 10.1016/j.chom.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor D, Andersen I. Lundqvist G. Clearance of inhaled particles from the human nose. Archives of Internal Medicine. 1973;131:132–139. [PubMed] [Google Scholar]
- Regassa LB, Novick RP. Betley MJ. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infection and Immunity. 1992;60:3381–3388. doi: 10.1128/iai.60.8.3381-3388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooijakkers SHM, Ruyken M, Roos A, Daha MR, Presanis JS, Sim RB, van Wamel WJ, et al. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nature Immunology. 2005;6:920–927. doi: 10.1038/ni1235. [DOI] [PubMed] [Google Scholar]
- Ruimy R, Angebault C, Djossou F, Dupont C, Epelboin L, Jarraud S, Lefevre LA, et al. Are host genetics the predominant determinant of persistent nasal Staphylococcus aureus carriage in humans? Journal of Infectious Diseases. 2010;202:924–934. doi: 10.1086/655901. [DOI] [PubMed] [Google Scholar]
- Sandiford S. Upton M. Identification, characterization, and recombinant expression of epidermicin NI01, a Novel unmodified bacteriocin produced by Staphylococcus epidermidis that displays potent activity against Staphylococci. Antimicrobial Agents and Chemotherapy. 2012;56:1539–1547. doi: 10.1128/AAC.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlievert PM, Case LC, Nemeth KA, Davis CC, Sun YP, Qin W, Wang F, et al. Alpha and beta chains of hemoglobin inhibit production of Staphylococcus aureus exotoxins. Biochemistry. 2007;46:14349–14358. doi: 10.1021/bi701202w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthilkumar A, Kumar S. Sheagren JN. Increased incidence of Staphylococcus aureus bacteremia in hospitalized patients with acquired immunodeficiency syndrome. Clinical Infectious Diseases. 2001;33:1412–1416. doi: 10.1086/322656. [DOI] [PubMed] [Google Scholar]
- Tait K. Sutherland IW. Antagonistic interactions amongst bacteriocin-producing enteric bacteria in dual species biofilms. Journal of Applied Microbiology. 2002;93:345–352. doi: 10.1046/j.1365-2672.2002.01692.x. [DOI] [PubMed] [Google Scholar]
- Travisano M. Lenski RE. Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics. 1996;143:15–26. doi: 10.1093/genetics/143.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraih LC. Maronpot RR. Normal histology of the nasal cavity and application of special techniques. Environmental Health Perspectives. 1990;85:187–208. doi: 10.1289/ehp.85-1568325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belkum A, Verkaik NJ, de Vogel CP, Boelens HA, Verveer J, Nouwen JL, Nouwen JL, et al. Reclassification of Staphylococcus aureus nasal carriage types. Journal of Infectious Diseases. 2009;199:1820–1826. doi: 10.1086/599119. [DOI] [PubMed] [Google Scholar]
- Van den Akker ELT, Nouwen JL, Melles DC, van Rossum EFC, Koper JW, Uitterlinden AG, Hofman A, et al. Staphylococcus aureus nasal carriage is associated with glucocorticoid receptor gene polymorphisms. Journal of Infectious Diseases. 2006;194:814–818. doi: 10.1086/506367. [DOI] [PubMed] [Google Scholar]
- Von Eiff C, Becker K, Machka K, Stammer H. Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. New England Journal of Medicine. 2001;344:11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- Weinrick B, Dunman PM, McAleese F, Murphy E, Projan SJ, Fang Y. Novick RP. Effect of mild acid on gene expression in Staphylococcus aureus. Journal of Bacteriology. 2004;186:8407–8423. doi: 10.1128/JB.186.24.8407-8423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wos-Oxley ML, Plumeier I, von Eiff C, Taudien S, Platzer M, Vilchez-Vargas R, Becker K, et al. A poke into the diversity and associations within human anterior nare microbial communities. The ISME Journal. 2010;4:839–851. doi: 10.1038/ismej.2010.15. [DOI] [PubMed] [Google Scholar]
- Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S. Relman DA. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host & Microbe. 2013;14:631–640. doi: 10.1016/j.chom.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood JM, McCormick JK, Paustian ML, Kapur V. Schlievert PM. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. Journal of Bacteriology. 2002;184:1095–1101. doi: 10.1128/jb.184.4.1095-1101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu VL, Goetz A, Wagener M, Smith PB, Rihs JD, Hanchett J. Zuravleff JJ. Staphylococcus aureus nasal carriage and infection in patients on hemodialysis – efficacy of antibiotic prophylaxis. New England Journal of Medicine. 1986;315:91–96. doi: 10.1056/NEJM198607103150204. [DOI] [PubMed] [Google Scholar]
- Yuki N, Shimazaki T, Kushiro A, Watanabe K, Uchida K, Yuyama T. Morotomi M. Colonization of the stratified squamous epithelium of the nonsecreting area of horse stomach by lactobacilli. Applied and Environmental Microbiology. 2000;66:5030–5034. doi: 10.1128/aem.66.11.5030-5034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]