Abstract
Current UK national standards recommend routine bacteriology surveillance in severe antibody-deficient patients, but less guidance exists on virology screening and viral infections in these patients. In this retrospective audit, we assessed the proportion of positive virology or bacteriology respiratory and stool samples from patients with severe, partial or no immune deficiency during a 2-year period. Medical notes were reviewed to identify symptomatic viral infections and to describe the course of persistent viral infections. During the 2-year period, 31 of 78 (39·7%) severe immune-deficient patients tested had a positive virology result and 89 of 160 (55.6%) had a positive bacteriology result. The most commonly detected pathogens were rhinovirus (12 patients), norovirus (6), Haemophilus influenzae (24), Pseudomonas spp. (22) and Staphylococcus aureus (21). Ninety-seven per cent of positive viral detection samples were from patients who were symptomatic. Low serum immunoglobulin IgA levels were more prevalent in patients with a positive virology sample compared to the total cohort (P = 0·0078). Three patients had persistent norovirus infection with sequential positive isolates for 9, 30 and 16 months. Virology screening of symptomatic antibody-deficient patients may be useful as a guide to anti-microbial treatment. A proportion of these patients may experience persistent viral infections with significant morbidity.
Keywords: antibody deficiency, IgA, infection, norovirus, viral
Introduction
The hallmark of primary immune deficiencies (PID) is increased susceptibility to infection. Patients with severe antibody deficiency (defined as requiring immunoglobulin replacement), such as those with common variable immune deficiency (CVID), are known to experience recurrent infections especially of the respiratory tract and to a lesser extent the gastrointestinal (GI) tract 1. Treatment with immunoglobulin replacement has been effective at reducing the frequency of serious infections 2,3. Adjusting the immunoglobulin dose on an individual patient basis to limit breakthrough infections [rather than targeting a specific trough immunoglobulin IgG level] has further improved the control of infections 4. However, current practice has been unable to prevent all breakthrough infections, which can result in direct infection-related morbidity and mortality as well as complications such as bronchiectasis 5,6. Inflammatory complications such as autoimmunity and lymphoproliferation are known to occur in a subset of CVID patients 7–9. Persistent infections result in chronic antigen stimulation which, in the context of a dysregulated immune system, could exacerbate inflammatory or lymphoproliferative disorders 10. An association between cytomegalovirus (CMV) infection, an aberrant T cell compartment and inflammatory disorders in CVID patients has been hypothesized previously 11,12.
Patients with antibody deficiency are most susceptible to infections with encapsulated bacteria (e.g. Haemophilus influenzae, Streptococcus pneumoniae) 13, whereas combined immune deficiency patients are also susceptible to viral infections 14,15. Previous studies have identified the presence of encapsulated bacteria 16,17 and common viruses such as rhinovirus 18 in respiratory secretions of X-linked agammaglobulinaemia (XLA) and CVID patients. Some severe viral infections in antibody-deficient patients have also been noted, such as astrovirus encephalitis in XLA patients 19,20 and disseminated astrovirus infection in severe combined immune deficiency (SCID) patients 21. However, given the common view of antibody deficiency patients being most susceptible to bacterial infections, the importance of viral infections in these patients may be underappreciated. As well as direct virus-related pathology and increased susceptibility to secondary bacterial infections, viral infections could drive local and/or systemic inflammatory responses resulting in further pathology in a subset of antibody-deficient patients.
Here we report an audit of bacteriology and virology requests for patients attending our adult immunology clinic during a period of 2 years. This included a comparison of patients with severe immune deficiency, partial immune deficiency or no immune deficiency. The frequency and type of viral and bacterial infections are reported, as well as accompanying immunological features. Case reports of three CVID patients with persistent viral infection demonstrate potential consequences of viral infections and implications for changes to current practice are discussed.
Patients and methods
A retrospective audit of all virology and bacteriology samples from patients attending the immunodeficiency clinic at Barts Health NHS Trust (London, UK) during a 2-year period (1 July 2011–30 June 2013) was carried out. A 2-year period was used to account for seasonal and annual variations. The aim of the audit was to ensure that microbiological testing conformed to international and national recommendations for infection surveillance, and where recommendations were not available that testing provided diagnostic benefit, defined as ‘reasonable’ likelihood of a positive finding leading to change in patient management 22,23. Patients were categorized as having a severe immune deficiency (patients requiring immunoglobulin replacement), a partial immune deficiency (mannan-binding ligand deficiency or specific antibody deficiency not requiring immunoglobulin replacement) or no immune deficiency (hereditary angioedema or new patients under investigation found subsequently to have no immune deficiency). The majority of patients in the severe immune deficiency group have a diagnosis of antibody deficiency (CVID, XLA or secondary hypogammaglobulinaemia), with only one patient having a combined deficiency and one with Good’s syndrome.
The local policy at the time of the study was for bacteriology and virology surveillance of sputum or cough swab samples from asymptomatic known severe immunodeficiency patients at each routine visit, with additional sampling if symptomatic. Sampling from other sites (e.g. stool) was carried out only if patients were symptomatic.
Data for the clinical summaries were collected with written informed consent and in accordance with approval by the City and East London Research Ethics Committee. Ethical approval for the audit section was not required, under the guidance of the Research Ethics Committee.
Bacteriology and virology analysis
Samples included in the study for microbiological analysis were respiratory (cystic sputum, cystic cough swab, bronchial washings) and stool samples. Local policy is to request extended culture (‘cystic’) for respiratory samples, as low pathogenicity organisms are relevant in severe immunodeficiency. For virology analysis respiratory (nose/throat swabs, throat swabs, nasopharyngeal aspirate, bronchoalveolar lavage) and stool samples were included.
A respiratory panel and a gastroenteritis panel of multiplex real-time polymerase chain reaction (PCR) were used for virological testing. The respiratory multiplex panel was modified from a previously published method 24, and included respiratory syncytial virus (RSV), influenza A and B viruses, parainfluenza viruses 1–3, adenoviruses, human metapneumovirus, rhinovirus and enterovirus. The gastroenteritis multiplex panel was based on the method developed by the Health Protection Agency for the 2012 Olympic preparedness 25, and included norovirus genotype I and II, rotavirus, adenoviruses type 40/41, astrovirus and sapovirus. For bacteriology testing, the organisms were cultured on selective media and identified by matrix assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (Bruker, Coventry, UK). Antibiotic susceptibility testing was undertaken using the British Society for Anti-microbial Chemotherapy (BSAC) method.
Statistical analysis
Categorical data counts were analysed by the χ2 test and P-values < 0·05 were considered significant. Data were analysed using GraphPad Prism software (La Jolla, CA, USA).
Results
A 2-year retrospective audit of bacteriology and virology samples from patients with a severe immune deficiency, partial immune deficiency or no immune deficiency was carried out. The number and proportion of patients in each group with a positive result during the 2-year period was compared. The number of patients with a positive result rather than the number of positive samples was used to exclude the effect of few patients with multiple samples. There was a significant difference in the proportion of patients with a positive virology or bacteriology result between the severe, partial and no immune deficiency groups, with 39·7% (31 of 78) of the severe immune deficiency group having a positive virology result and 55·6% (89 of 160) having a positive bacteriology result (Table 1).
Table 1.
The number of patients with a positive virology or bacteriology result during a 2-year period
| Virology | Bacteriology | |
|---|---|---|
| Total number of patients tested | 153 | 401 |
| Total number of samples (samples per patient) | 1111 (7·26) | 1917 (4·78) |
| Number of patients with | ||
| Severe antibody deficiency | 78 | 160 |
| Number of patients with a positive result | 31 (39·7%) | 89 (55·6%) |
| Partial immune deficiency | 15 | 49 |
| Number of patients with a positive result | 4 (26·7%) | 14 (28·6%) |
| No immune deficiency | 60 | 192 |
| Number of patients with a positive result | 11 (18·3%) | 50 (26·0%) |
| χ2 value, P-value | 7·485 *P = 0·0237 | 34·54 ***P < 0·0001 |
The χ2 test was used to compare the number of severe, partial and no deficiency patients with a positive result, for virology and bacteriology tests.
The respiratory virus screen was equivalent to nine samples and the gastrointestinal (GI) virus screen was equivalent to seven samples.
Respiratory samples were the most common type, accounting for 76% (2321 of 3037) of virology results reported and 45% (1033 of 2294) of bacteriology results, followed by stool samples accounting for 17% (520 of 3037) of virology results and 10% (234 of 2294) of bacteriology results reported. A greater proportion of the severe immune deficiency group had a positive virology or bacteriology respiratory sample compared to the partial and no immune deficiency patients (Table 2). For stool samples, the majority of patients with a positive bacteriology sample were from the severe deficiency group (four of 27), with two of 14 patients with no immune deficiency having a positive bacteriology result. Positive virology stool samples were found only in the severe antibody deficiency group.
Table 2.
Patients in each immune deficiency group with a positive virology or bacteriology result from respiratory or stool samples
| Severe | Partial | None | χ2 test | |
|---|---|---|---|---|
| Respiratory | ||||
| Total virology patients | 60 | 12 | 38 | 4·856 P = 0·0882 |
| Positive virology patients (%) | 25 (41·7%) | 3 (25·0%) | 8 (21·1%) | |
| Total bacteriology patients | 140 | 34 | 53 | 16·05 ***P = 0·0003 |
| Positive bacteriology patients (%) | 78 (55·7%) | 11 (32·4%) | 14 (26·4%) | |
| Stool | ||||
| Total virology patients | 21 | 1 | 2 | 1·412 P = 0·4937 |
| Positive virology patients (%) | 7 (33·3%) | 0 (0·0%) | 0 (0·0%) | |
| Total bacteriology patients | 27 | 6 | 14 | 1·009 P = 0·6038 |
| Positive bacteriology patients (%) | 4 (14·8%) | 0 (0·0%) | 2 (14·3%) |
We identified the most frequently detected organisms and the sampling site for the severe immune deficiency patients. The most frequently identified viruses were rhinovirus, human metapneumovirus and parainfluenza virus 3 in respiratory samples and norovirus genotype II from stool samples (Table 3). The most frequently cultured bacteria were H. influenzae, Pseudomonas spp. and Staphylococcus aureus, but surprisingly only one severe immune deficiency patient had a S. pneumoniae-positive culture during the 2-year period (Table 4).
Table 3.
The number of severe immune deficiency patients with each virus and their sample site
| Virus | Respiratory (severe deficiency) | Stool (severe deficiency) | Number of all patients |
|---|---|---|---|
| Rhinovirus | 12 | – | 17 |
| Metapneumovirus | 6 | – | 6 |
| Norovirus genotype II | – | 6 | 6 |
| Parainfluenza 3 | 3 | – | 5 |
| Adenovirus | 2 | 2 | 4 |
| Enterovirus | 2 | 1 | 4 |
| Influenza A | 2 | – | 2 |
| Influenza B | 2 | – | 2 |
| HSV1 and HSV2 | 1 | – | 10 |
| Astrovirus | – | 2 | 2 |
| Norovirus genotype I | – | 1 | 1 |
| Parainfluenza 1 | 0 | – | 1 |
| RSV | 0 | – | 1 |
Table 4.
The number of severe immune deficiency patients with each bacterium or fungus and their sample site
| Organism | Respiratory (severe deficiency) | Stool (severe deficiency) | Number of all patients |
|---|---|---|---|
| Haemophilus influenzae | 24 | 0 | 27 |
| Pseudomonas spp. | 22 | 0 | 30 |
| Staphylococcus aureus | 21 | 0 | 46 |
| Aspergillus spp. | 10 | 0 | 12 |
| Candida spp. | 8 | 0 | 12 |
| Serratia spp. | 7 | 0 | 9 |
| Stenotrophomonas maltophilia | 7 | 0 | 9 |
| Moraxella spp. | 6 | 0 | 6 |
| Methicillin-resistant S. aureus | 5 | 0 | 8 |
| Enterobacter spp. | 3 | 0 | 5 |
| Proteus spp. | 2 | 0 | 7 |
| Streptococcus pneumoniae | 1 | 0 | 4 |
| Klebsiella spp. | 1 | 0 | 3 |
| Citrobacter spp. | 1 | 0 | 2 |
| Achromobacter xylosoxidans | 1 | 0 | 1 |
| Enterococcus | 0 | 0 | 4 |
| Giardia lamblia | 0 | 2 | 2 |
Local policy has been to screen known immunodeficient patients for surveillance even if asymptomatic, as well as to sample during symptomatic episodes. However, of the positive virology isolates, 97% (98 of 101) of samples were from patients who were symptomatic at the time. There were three samples from asymptomatic patients, two of whom had no immune deficiency. Additionally, most of the positive virology samples (95 of 101; 94·1%) were from patients with a negative bacteriology result, suggesting that most positive virology episodes were not associated with secondary bacterial infections. C-reactive protein (CRP) levels were raised in 55·1% (27 out of 49 values available) of episodes associated with a positive virology result (Fig. 1a).
Figure 1.
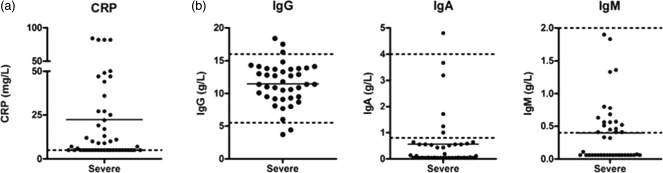
C-reactive protein (CRP) and serum immunoglobulin levels at the time of positive virology result. CRP levels at the time of positive virology result are shown in (a). Serum immunoglobulin IgG, IgA and IgM (b) at the time of each positive virology result are shown. Dotted lines indicate reference ranges.
The severe immune deficiency patients were all receiving immunoglobulin replacement at the time. During the episodes with a positive virology result, the majority of patients (96·1%) had adequate IgG replacement, with a median serum IgG level of 11·4 g/l (normal range 5·5–16·5 g/l). Immunoglobulin replacement products contain minimal amounts of other immunoglobulin isotypes, and as such 88·2% of patients had low serum IgA levels (median 0·11 g/l, normal range 0·8–4·0 g/l) and 62·7% had low serum IgM levels (median 0·11g/l, normal range 0·4-–2·0 g/l) (Fig. 1b). Immunoglobulin levels from severe immune deficiency patients with a positive virology result were compared to the total cohort of severe immune deficiency patients described previously 26. Low levels of IgA, but not IgG or IgM, were significantly more prevalent in the patients with a positive virology result (Supporting information, Fig. S1), suggesting a role for IgA in protection from viral infection. Moreover, positive virology results were more frequent in the severe immune deficiency patients with low IgA (Supporting information, Table S1).
We identified severe immune deficiency patients with ‘persistent’ viral detection, defined as two or more positive detections of the same virus >8 weeks apart. Three CVID patients during the 2-year period were found to have a persistent norovirus infection, with one of these patients additionally having a persistent rhinovirus infection (Table 5). All patients were symptomatic. Patients 1 and 3 experienced severe weight loss and both required large increases in immunoglobulin dose to maintain trough IgG levels. Patient 1 cleared norovirus after 9 months, regaining weight, and has reduced immunoglobulin requirement (Fig. 2a). The other two patients continue to excrete norovirus after 30 and 16 months, respectively; both remain intermittently symptomatic and one has required long-term parenteral nutrition (Fig. 2b,c). Further details of these three cases can be found in the Supporting information.
Table 5.
Characteristics of three severe antibody deficiency patients with a persistent viral infection
| Antibody deficiency | Patient 1‡CVID | Patient 2†CVID | Patient 3†CVID | |
|---|---|---|---|---|
| Virus | Norovirus genotype 2 | Norovirus genotype 2 | Norovirus genotype 2 | Rhinovirus |
| Months of infection | 9 | 30 | 16 | 11 |
| Virus clearance | Yes | No | No | Yes |
| Number of positive results | 13 | 12 | 29 | 10 |
| Number of negative results since clearance | 1 | – | – | 13 |
| At the time of first positive result | ||||
| IgG (5·5–16·5) | 10.0 | 13·9 | 8·7 | 9·1 |
| IgA (0·8–4·0) | 0·06 | 0·14 | <0·04 | <0·04 |
| IgM (0·4–2·0) | <0·06 | 1·36 | 0·45 | 0·45 |
| Lymphocyte count × 109/l | 1·2 | 1·4 | 8·1 | 9·5 |
| CD3+ count × 106/l (918–2023) | 971 | 720 | 9974 | 8927 |
| CD4+ count × 106/l (455–1820) | 402 | 466 | 2668 | 2750 |
| CD8+ count × 106/l (140–906) | 577 | 261 | 7109 | 5892 |
| CD56+ count × 106/l (90–600) | 94 | 122 | 345 | 278 |
Reference ranges are shown in parentheses. CVID = common variable immune deficiency; Ig = immunoglobulin.
Lymphocyte count reference ranges: †(1·2–3·5) × 109/l ‡(1·5–4·0) × 109/l.
Figure 2.
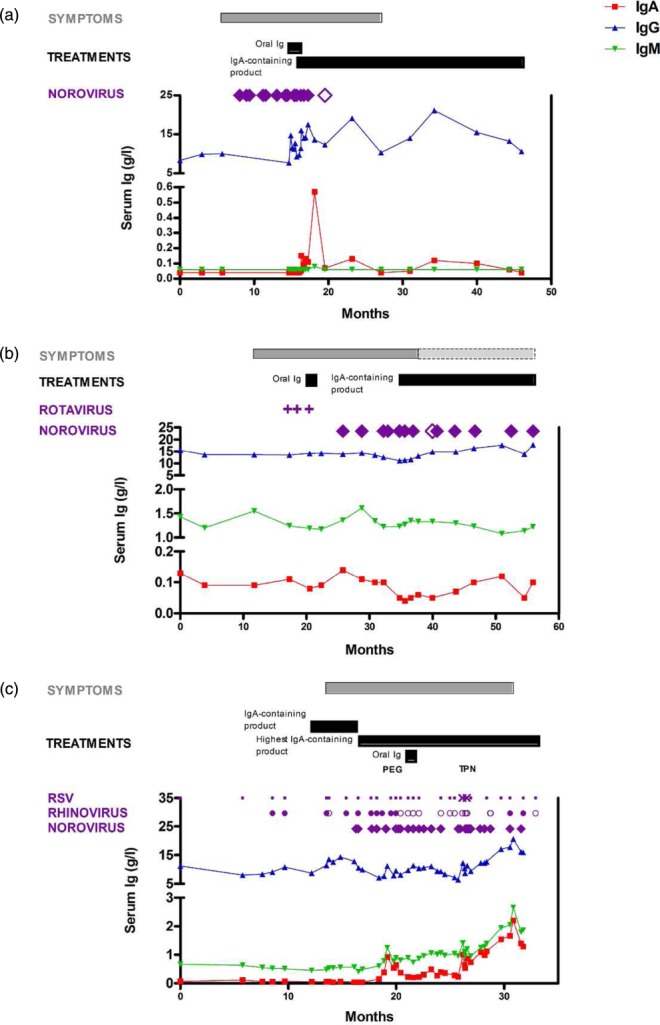
Three common variable immune deficiency (CVID) patients with persistent viral infections. Serum immunoglobulin IgG, IgA, IgM and the time–course of diarrhoeal symptoms and treatments are shown for patient 1 (a), patient 2 (b) and patient 3 (c). Positive norovirus samples (filled diamonds), negative norovirus samples (open diamonds), positive rhinovirus samples (filled circles), negative rhinovirus samples (open circles), positive rotavirus antigen (plus signs), positive respiratory syncytial virus (RSV) samples (crosses) and negative RSV samples (dots) are shown. Light grey bars indicate intermittent symptoms.
Discussion
Current national standards recommend routine bacteriology screening of CVID patients, but recommend virology screening only for SCID patients 22,23. We observed a greater prevalence of both bacterial and viral infections in severe immune deficient patients. A high prevalence of viral infections in antibody deficiency patients has been identified previously 27,28, and our audit suggests that screening for viruses in symptomatic patients and for infection control purposes is useful.
In agreement with several other studies, the most frequently cultured bacteria in severe immunodeficient patients was H. influenzae 16,18,29. However, we observed a lower frequency of other common upper respiratory tract pathogens such as S. pneumoniae than previous studies 17,30, which may reflect our policy of increasing immunoglobulin dose to prevent breakthrough infection 2,4. Opportunistic infections such as Pseudomonas spp. and S. aureus were relatively more common. A relatively high proportion of our cohort has bronchiectasis 26, which may partially explain the more common occurrence of these pathogens. Although some isolates may have represented upper airway sample contamination, the majority occurred in symptomatic patients and may require more aggressive management. For those with viral infection, there was only a low prevalence of co-existing or secondary bacterial infections in this study compared to others 27, which could reflect our practice of prescribing patient-held antibiotics to be used as soon as patients are symptomatic, in accordance with national and international consensus 23,31.
The most common detected viruses, rhinovirus and norovirus, probably reflect the high prevalence of these viruses in the general population, as infections were community-acquired. Rhinovirus was identified as the most common viral pathogen in sinus lavage samples from asymptomatic antibody-deficient patients 18 and in sputum samples from symptomatic antibody-deficient patients 27. Norovirus was also the most common faecal pathogen identified in antibody-deficient children, although almost half were asymptomatic 28, which is in contrast to our study where all positive patients were symptomatic, as stool sampling was carried out only on symptomatic patients. There was a surprisingly low occurrence of respiratory syncytial virus (RSV) in the patients with severe antibody deficiency. Palivizumab is a monoclonal antibody administered intramuscularly to prevent RSV infection in high-risk children, suggesting that systemic immunoglobulins can protect against RSV. The high level of replacement immunoglobulin treatment used in our patients should contain some level of anti-RSV antibodies, as the majority of the adult population are seropositive 32, which may similarly offer protection against certain pathogens such as RSV. However, for other pathogens, serum IgG replacement may not offer protection at the mucosal surface and mucosal IgA, which is not replaced with treatment, may be more important. Although antibody deficiency is not usually thought to result in an increased risk of common viral infections, CVID is a heterogeneous group of diseases with varying molecular mechanisms. Several studies have noted defects in T cell number and function in some CVID patients 8,33. Additionally, patients with an inflammatory/lymphoproliferative CVID phenotype may be on immunosuppressive medication that could further suppress cell-mediated immunity. As such, a subset of CVID patients may be more susceptible to viral infections than considered previously.
The three case reports demonstrate that in a small proportion of antibody-deficient patients viral infections may become persistent. Persistent norovirus infection has been described in hypogammaglobulinaemic patients with chronic lymphocytic leukaemia 34 and following allogeneic haematopoetic stem cell transplantation, and has been associated with significant morbidity and mortality 35. A study of paediatric PID patients also noted that norovirus shedding persisted for a median of 9·5 months, additionally posing an infection control risk 28. Similarly, rhinovirus was also persistently isolated for >2 months from half of symptomatic antibody-deficient patients 27. In hypogammaglobulinaemic patients rhinovirus shedding was found to last for a mean of 40 days, compared to 10 days in healthy controls 36. In the absence of molecular typing it is difficult to conclude whether the sequential positive viral isolates reflect genuine persistence of a single strain or reinfection with multiple strains, as shown for rhinovirus 36.
A limitation of this study is that it was retrospective, and so may be open to sampling bias. Additionally, the virology-positive results rely on detection of viral RNA by PCR, whereas bacteriology-positive results are from cultured growth, which may overestimate the prevalence of viral infection caused by the pathogens tested, while failing to detect other viruses not specifically screened for. However, the majority of virology positive results in this study were from symptomatic episodes in the absence of positive bacteriology culture. A further limitation of this study is that the sensitivity of multiplex detection assays may be lower than that of monoplex assays, as has been shown for a rhinovirus assay 37, leading to an underestimation of positive viral detections.
Treatment of norovirus with oral immunoglobulin has been shown to lead to a faster resolution of symptoms after 7 days of treatment, probably through local mucosal blocking of virus 38. The three case reports here suggest that intravenous replacement with immunoglobulin products with a higher IgA content may also be helpful, although it was not effective in all patients, so further study is needed. Treatment of persistent rhinovirus in antibody-deficient patients with IFN-α2a and ribavirin has also been shown to clear rhinovirus rapidly from sputum samples 39.
As a result of this audit, we have made changes to our practice. Bacteriology and virology sampling of symptomatic immunodeficient patients has proved useful, but asymptomatic patients will not be sampled routinely for virology, except for follow-up of previously detected virus. Active treatment of viral infection with appropriate anti-virals for high-risk patients, or where virus is persistent, would be considered. For patients with persistent infection we consider the options of oral immunoglobulin or IgA-containing immunoglobulin replacement on a case-by-case basis. For infection control measures during clinic visits, patients known to be infected with, e.g. norovirus, are isolated and there is continued emphasis on hand-washing and surface decontamination for all patients. There is continued recommendation for influenza vaccine for at-risk patients and household contacts. Our audit suggests that viral infection may be important for patients with severe antibody deficiency. Further work is required to assess the benefit of a more proactive approach to diagnosis and treatment of viral infection in this patient group.
Acknowledgments
We thank Chris Scott for retrieving the general laboratory data, Melvyn Eydmann for retrieving the bacteriology data and Spiro Pereira for retrieving the virology data. S. D.’s salary was funded by CSL Behring. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author contributions
S. D. collected, analysed and interpreted the data, performed statistical analyses and wrote the manuscript; A. M. collected data, wrote the clinical summaries and critically read the manuscript; M. B. and S. G. cared for the involved patients and critically read the manuscript; C. Y. W. T. interpreted the data and critically read the manuscript; H. J. L. cared for the involved patients and provided clinical data, designed and organized the study, interpreted data and wrote the manuscript.
Disclosure
S. G. has received support for meeting attendances from BPL and CSL Behring and has been the Chief Investigator/Lead Investigator/Co-investigator for several immunoglobulin clinical trials with the following companies: Baxter, CSL Behring and Octapharma. H. J. L. has received educational funding, consultation fees, participated in research with or received staff support funding from the following immunoglobulin manufacturers: Baxter, Biotest, CSL Behring, Grifols and Octapharma. S. D.’s salary was funded by CSL Behring.
Supporting Information
Additional Supporting information may be found in the online version of this article at the publisher’s web-site:
Fig. S1. The number of patients with low immunoglobulin (Ig)G, IgA or IgM from the total severe immune deficiency Barts cohort is compared to the severe immune deficiency patients with a positive virology result. For the total Barts cohort, IgG values are the average during the year up to June 2013; for the IgA and IgM levels values, where available, were prior to treatment.
Table S1. Number of virus detections in severe immune deficiency patients with different serum IgA levels.
Supporting Information
REFERENCES
- Wood P. Stanworth S. Burton J. Recognition, clinical diagnosis and management of patients with primary antibody deficiencies: a systematic review. Clin Exp Immunol. 2007;149:410–23. doi: 10.1111/j.1365-2249.2007.03432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orange JS. Grossman WJ. Navickis RJ. Wilkes MM, et al. Impact of trough IgG on pneumonia incidence in primary immunodeficiency: a meta-analysis of clinical studies. Clin Immunol. 2010;137:21–30. doi: 10.1016/j.clim.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Quinti I. Soresina A. Spadaro G. Long-term follow-up and outcome of a large cohort of patients with common variable immunodeficiency. J Clin Immunol. 2007;27:308–16. doi: 10.1007/s10875-007-9075-1. [DOI] [PubMed] [Google Scholar]
- Lucas M. Lee M. Lortan J. Lopez-Granados E. Misbah S. Chapel H, et al. Infection outcomes in patients with common variable immunodeficiency disorders: relationship to immunoglobulin therapy over 22 years. J Allergy Clin Immunol. 2010;125:1354–60.e4. doi: 10.1016/j.jaci.2010.02.040. [DOI] [PubMed] [Google Scholar]
- Baris S. Ercan H. Cagan HH. Efficacy of intravenous immunoglobulin treatment in children with common variable immunodeficiency. J Investig Allergol Clin Immunol. 2011;21:514–21. [PubMed] [Google Scholar]
- Gathmann B. Mahlaoui N. Gerard L, et al. Clinical picture and treatment of 2212 patients with common variable immunodeficiency. J Allergy Clin Immunol. 2014;134:116–260. doi: 10.1016/j.jaci.2013.12.1077. [DOI] [PubMed] [Google Scholar]
- Chapel H. Lucas M. Lee M, et al. Common variable immunodeficiency disorders: division into distinct clinical phenotypes. Blood. 2008;112:277–86. doi: 10.1182/blood-2007-11-124545. [DOI] [PubMed] [Google Scholar]
- Cunningham-Rundles C. Bodian C, et al. Common variable immunodeficiency: clinical and immunological features of 248 patients. Clin Immunol. 1999;92:34–48. doi: 10.1006/clim.1999.4725. [DOI] [PubMed] [Google Scholar]
- Edgar JD. Buckland M. Guzman D. The United Kingdom Primary Immune Deficiency (UKPID) Registry: report of the first 4 years’ activity 2008-2012. Clin Exp Immunol. 2014;175:68–78. doi: 10.1111/cei.12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva SP. Resnick E. Lucas M, et al. Lymphoid proliferations of indeterminate malignant potential arising in adults with common variable immunodeficiency disorders: unusual case studies and immunohistological review in the light of possible causative events. J Clin Immunol. 2011;31:784–91. doi: 10.1007/s10875-011-9565-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marashi SM. Raeiszadeh M. Enright V, et al. Influence of cytomegalovirus infection on immune cell phenotypes in patients with common variable immunodeficiency. J Allergy Clin Immunol. 2012;129:1349–56.e3. doi: 10.1016/j.jaci.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Marashi SM. Raeiszadeh M. Workman S, et al. Inflammation in common variable immunodeficiency is associated with a distinct CD8(+) response to cytomegalovirus. J Allergy Clin Immunol. 2011;127:1385–93.e4. doi: 10.1016/j.jaci.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried AJ. Bonilla FA, et al. Pathogenesis, diagnosis, and management of primary antibody deficiencies and infections. Clin Microbiol Rev. 2009;22:396–414. doi: 10.1128/CMR.00001-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries E. Patient-centred screening for primary immunodeficiency: a multi-stage diagnostic protocol designed for non-immunologists. Clin Exp Immunol. 2006;145:204–14. doi: 10.1111/j.1365-2249.2006.03138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A. Notrangelo LD. Ochs HD. Winkelstein JA. In: Combined immunodeficiencies in Immunologic Disorders in Infants and Children. 5th Edn. Stiehm ER, editor; Philadelphia, PA: Elsevier Saunder; pp. 447–479. [Google Scholar]
- Pettit SJ. Bourne H. Spickett GP. Survey of infection in patients receiving antibody replacement treatment for immune deficiency. J Clin Pathol. 2002;55:577–80. doi: 10.1136/jcp.55.8.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelstein JA. Marino MC. Lederman HM. X-linked agammaglobulinemia: report on a United States registry of 201 patients. Medicine (Balt) 2006;85:193–202. doi: 10.1097/01.md.0000229482.27398.ad. [DOI] [PubMed] [Google Scholar]
- Kainulainen L. Suonpaa J. Nikoskelainen J, et al. Bacteria and viruses in maxillary sinuses of patients with primary hypogammaglobulinemia. Arch Otolaryngol Head Neck Surg. 2007;133:597–602. doi: 10.1001/archotol.133.6.597. [DOI] [PubMed] [Google Scholar]
- Quan PL. Wagner TA. Briese T, et al. Astrovirus encephalitis in a boy with X-linked agammaglobulinemia. Emerg Infect Dis. 2010;16:918–25. doi: 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster AD, et al. Virus infections in primary immunodeficiency. J Clin Pathol. 1994;47:965–7. doi: 10.1136/jcp.47.11.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderli W. Meerbach A. Gungor T. Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PLoS One. 2011;6:e27483. doi: 10.1371/journal.pone.0027483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UK Primary Immunodeficiency Network (UKPIN) 2009. Severe combined immunodeficiency (SCID): initial diagnosis and management. Standards of Care,. Available at: http://ukpin.org.uk/home/standards/standards-SCID.pdf. Accessed 7 July 2014.
- UK Primary Immunodeficiency Network (UKPIN) 2009. CVID diagnosis and management. Standards of Care,. Available at: http://ukpin.org.uk/home/standards/standards-CVID.pdf. Accessed 7 July 2014.
- Bibby DF. McElarney I. Breuer J. Clark DA, et al. Comparative evaluation of the Seegene Seeplex RV15 and real-time PCR for respiratory virus detection. J Med Virol. 2011;83:1469–75. doi: 10.1002/jmv.22125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Gilad J. Chand M. Brown C. Microbiological aspects of public health planning and preparedness for the 2012 Olympic Games. Epidemiol Infect. 2012;140:2142–51. doi: 10.1017/S0950268812001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingham SS. Buckland M. Dempster J. Lorenzo L. Grigoriadou S. Longhurst HJ, et al. Primary vs. secondary antibody deficiency: clinical features and infection outcomes of immunoglobulin replacement. PLOS ONE. 2014;9:e100324. doi: 10.1371/journal.pone.0100324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainulainen L. Vuorinen T. Rantakokko-Jalava K. Osterback R. Ruuskanen O. Recurrent and persistent respiratory tract viral infections in patients with primary hypogammaglobulinemia. J Allergy Clin Immunol. 2010;126:120–6. doi: 10.1016/j.jaci.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frange P. Touzot F. Debre M. Prevalence and clinical impact of norovirus fecal shedding in children with inherited immune deficiencies. J Infect Dis. 2012;206:1269–74. doi: 10.1093/infdis/jis498. [DOI] [PubMed] [Google Scholar]
- Kainulainen L. Nikoskelainen J. Vuorinen T. Tevola K. Liippo K. Ruuskanen O, et al. Viruses and bacteria in bronchial samples from patients with primary hypogammaglobulinemia. Am J Respir Crit Care Med. 1999;159:1199–204. doi: 10.1164/ajrccm.159.4.9807067. [DOI] [PubMed] [Google Scholar]
- Resnick ES. Moshier EL. Godbold JH. Cunningham-Rundles C. Morbidity and mortality in common variable immune deficiency over 4 decades. Blood. 2012;119:1650–7. doi: 10.1182/blood-2011-09-377945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla FA. Bernstein IL. Khan DA. Practice parameter for the diagnosis and management of primary immunodeficiency. Ann Allergy Asthma Immunol. 2005;94:S1–63. doi: 10.1016/s1081-1206(10)61142-8. [DOI] [PubMed] [Google Scholar]
- Sastre P. Ruiz T. Schildgen O. Schildgen V. Vela C. Rueda P, et al. Seroprevalence of human respiratory syncytial virus and human metapneumovirus in healthy population analyzed by recombinant fusion protein-based enzyme linked immunosorbent assay. Virol J. 2012;9:130. doi: 10.1186/1743-422X-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin-Proulx D. Santos BA. Carvalho KI. IVIg immune reconstitution treatment alleviates the state of persistent immune activation and suppressed CD4 T cell counts in CVID. PLOS ONE. 2013;8:e75199. doi: 10.1371/journal.pone.0075199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capizzi T. Makari-Judson G. Steingart R. Mertens WC, et al. Chronic diarrhea associated with persistent norovirus excretion in patients with chronic lymphocytic leukemia: report of two cases. BMC Infect Dis. 2011;11:131. doi: 10.1186/1471-2334-11-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roddie C. Paul JP. Benjamin R. Allogeneic hematopoietic stem cell transplantation and norovirus gastroenteritis: a previously unrecognized cause of morbidity. Clin Infect Dis. 2009;49:1061–8. doi: 10.1086/605557. [DOI] [PubMed] [Google Scholar]
- Peltola V. Waris M. Kainulainen L. Kero J. Ruuskanen O, et al. Virus shedding after human rhinovirus infection in children, adults and patients with hypogammaglobulinaemia. Clin Microbiol Infect. 2013;19:E322–7. doi: 10.1111/1469-0691.12193. [DOI] [PubMed] [Google Scholar]
- Karhu J. Ala-Kokko TI. Vuorinen T. Ohtonen P. Syrjala H. Lower respiratory tract virus findings in mechanically ventilated patients with severe community-acquired pneumonia. Clin Infect Dis. 2014;59:62–70. doi: 10.1093/cid/ciu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florescu DF. Hermsen ED. Kwon JY. Is there a role for oral human immunoglobulin in the treatment for norovirus enteritis in immunocompromised patients? Pediatr Transplant. 2011;15:718–21. doi: 10.1111/j.1399-3046.2011.01556.x. [DOI] [PubMed] [Google Scholar]
- Ruuskanen O. Waris M. Kainulainen L, et al. Treatment of persistent rhinovirus infection with pegylated interferon alpha2a and ribavirin in patients with hypogammaglobulinemia. Clin Infect Dis. 2014;58:1784–6. doi: 10.1093/cid/ciu169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The number of patients with low immunoglobulin (Ig)G, IgA or IgM from the total severe immune deficiency Barts cohort is compared to the severe immune deficiency patients with a positive virology result. For the total Barts cohort, IgG values are the average during the year up to June 2013; for the IgA and IgM levels values, where available, were prior to treatment.
Table S1. Number of virus detections in severe immune deficiency patients with different serum IgA levels.
Supporting Information


