Abstract
In order to analyse whether a C-terminal polybasic sequence represents a nuclear localization signal (NLS) we obtained several truncated and mutant forms of protein of regerating liver (PRL)-3 and evaluated their subcellular localization as compared to the wild-type form. Our results invalidate the hypothesis that this is an NLS. We also analysed the influence of the C- and N-terminal residues on the phosphatase activity of PRL-3. Our results provide in vitro evidence that the C-terminal CAAX motif, besides directing the protein farnesylation, plays an additional regulatory role by inhibiting the catalytic efficiency of PRL-3. Taking into account the results we obtained, as well as reported data, we propose a hypothetical molecular mechanism for the nucleocytoplasmic localization and transfer of PRL-3.
Keywords: PRL-3, subcellular localization, oligomerization, catalytic activity, steady-state parameters
Introduction
Human PRL-1, PRL-2 and PRL-3 (phosphatase of regenerating liver) represent a family of highly homologous protein tyrosine phosphatases encoded by genes located on chromosomes 6q12, 1p35 and 8q24.3, respectively [1]. Their expression in different human tissues is not well documented but it is known that in adult rodents [2] they are preferentially expressed in skeletal muscle; in addition, mRNA of PRL-1 was found in relatively significant amounts in brain while mRNA of PRL-3 was found at high levels in heart [2, 3].
The particular attention paid to PRL-3 is due to its involvement in tumour metastasis. Thus, it is consistently elevated in metastasis of colorectal cancer although in normal colorectal epithelium and in non-metastatic tumours PRL-3 is expressed at significantly lower levels [4]. Further studies reported the overexpression of PRL-3 in other cancer forms, like liver carcinoma [5], vasculature of invasive breast cancers [6], ovarian cancer [7] and in gastric carcinomas [8]. As an additional support for its involvement in cancer metastasis it was shown that Chinese hamster ovary cells overexpressing PRL-3 exhibited increased motility and invasiveness and promoted metastatic tumours formation in mice [9].
As concerning the intracellular localization, PRL-3 has been found at the cytoplasmic and nuclear membrane and as intracellular punctate structures scattered throughout the entire cytoplasm [10]. Also, PRL-3 was found at the metaphase plate in the progression of cells through mitosis [5]. Similarly, although PRL-1 was initially reported to be localized to the nucleus [3], most of later reports provide evidence for its localization at plasma membrane, early endosomes and endoplasmic reticulum [10–12]. Membrane association of all three forms of PRL family seems to be directly dependent on their C-terminal prenylation because in absence of prenylation they shifted into the nucleus [10]. Notably, it was reported that expression of PRL-1 became more consistently nuclear as development progressed [13]. Based on this finding it has been suggested that PRLs localized to the nucleus might play a role in terminal differentiation or its localization to the membranes may contribute to the cell growth and metastasis [1].
Despite the large number of reports related to the involvement of PRL-3 in cancer, the molecular determinants which control its subcellular localization and/or its catalytic activity are still poorly understood. Thus, it has been shown that the presence in PRL-1, -2 and -3 of the consensus C-terminal CAAX sequence (where C is cysteine, A is an aliphatic residue and X is any amino acid) promotes their farnesylation and consequently, as mentioned above, localization to membrane compartments [10]; however, there are no studies evaluating the importance of farnesylation on PRLs’ enzymatic activity although the pleiotropic effect of lipid modification on protein function is well documented [14]. Also, the presence of a potential (either mono- or bipartite) nuclear localization signals (NLS) close to the PRLs C-termini has been noted [10] but again, there are no reports regarding the significance of these putative localization signals. To add more experimental facts concerning the molecular determinants of PRL-3 we are investigating here a number of mutant forms of PRL-3 containing modifications with potential significance for the subcellular localization and the catalytic activity. Our results are in agreement with the fact that the putative NLS is insufficient for directing PRL-3 to the nucleus. On the other hand, the absence of the C-terminal CAAX prenylation motif leads to a significantly higher catalytic efficiency as compared to the wild-type (WT) form, suggesting the implication of this motif in regulation of the catalytic activity.
Materials and methods
Construction of PRL-3 truncated and mutant forms
Using as template the cDNA of PRL-3 WT various constructs were made for expression of 6xHis-PRL-3 fusion as following: inserts for PRL-3 ΔN10, PRL-3 ΔC4 and PRL-3 ΔN10C4 were obtained by PCR using the forward and reverse primers mentioned in Table 1. The PCR products thus obtained were digested with NdeI and XhoI and ligated into the NdeI/XhoI sites of pET-15b vector. For subcellular localization the following constructs were made: pEGFP-C2/PRL-3WT, pEGFP-C2/PRL-3ΔC4 and pEGFP-C2/PRL-3 ΔC4-NLS. The PCR fragments, corresponding to the PRL-3 WT, PRL-3 ΔC4 and PRL-3 ΔC4-NLS sequences, were amplified using as forward and reverse primers specified in Table 1. The PCR fragments were then digested with XhoI and BamHI and finally ligated into the XhoI/BamH1 sites of pEGFP-C2 vector. To obtain the pET15b/PRL-3 NLS mutant form the KRR amino acids from human PRL-3 (amino acid residues 136–138) were substituted for QLQ amino acids by site direct mutagenesis using as mutagenesis primer 5′-CATCCAGTTCATCCGCCAGCAGCTGCAGG GAGCCATCAACAGCAAGCAG-3′ and as selection primer 5′-GCGTCTTTATATCTGAA TTCGAATATTAAATCCTC-3′ according to the protocol provided by the producer, Clontech Laboratories, Inc., USA. Subsequent transfer of the mutated PRL-3 insert (amplified with primers and template mentioned in Table 1) into pEGFP-C2 vector led to pEGFP-C2/PRL-3 NLS.
Table 1.
Templates and primers used for amplification of inserts corresponding to various PRL-3 constructs
| Constructs | Template | Forward primer | Reverse primer |
|---|---|---|---|
| pET15/PRL-3 ΔN10 | pET15/PRL-3 WTO | CTGGGATCCCATATGG AGGTGAGCTACAAACACATGCGCTTC | TGGGAATTCCTCGAG TCACATAACGCAGCACCGGGTCTTG |
| pET15/PRL-3 ΔC4 | pET15/PRL-3 WTO | CATGCCATGGGCCACCATCATCA TCACCATATGGCTCGGATGAACCGCCC | CGAATTCTCGAGTCACCG GGTCTTGTGCGTGTGTGG |
| pET15/PRL-3 ΔN10C4 | pET15/PRL-3 WT | CTGGGATCCCATATGGAGGTGAG CTACAAACACATGCGCTTC | CGAATTCTCGAGTCACCGGG TCTTGTGCGTGTGTGG |
| pEGFP-C2/PRL-3 WT | pET15/PRL-3 WTO | GTTCCTCGAGCATGGCTCGGATGAACCGCCC | GCGGGATCCTCACATAACGCAGCACCGGGTCTTG |
| pEGFP-C2/PRL-3 ΔC4 | pET15/PRL-3 WTO | GTTCCTCGAGCATGGCTC GGATGAACCGCCC | GCGGGATCCTCACCG GGTCTTGTGCGTGTGTG |
| pEGFP-C2/PRL-3 NLS | pET15/PRL-3 NLS | GTTCCTCGAGCATGGCTCGGATGAACCGCCC | GCGGGATCCTCACATAACGCAGCACCGGGTCTTG |
| pEGFP-C2/PRL-3 ΔC4 -NLS | pEGFP-C2/PRL-3 NLS | GTTCCTCGAGCATGGCTCGGATGAACCGCCC | GCGGGATCCTCACCGGGTCTTGTGCGTGTGTG |
Protein expression and purification
WT, ΔN10, ΔC4 and ΔN10C4 forms of human PRL-3 were expressed as His-tagged fusion proteins using Escherichia coli BL21 (DE3) as host cells. The PRL-3WT and the deletion mutants (ΔN10-, ΔC4-,ΔN10C4-PRL-3) were expressed at 37°C after induction with 0.1 mM IPTG. The bacterial cells were lysed in buffer A (20 mM potassium phosphate buffer pH 7.4 containing 500 mM NaCl, 20 mM imidazole, 4 mM dithiotreitol (DTT) and 1 mM phenylmethylsulphonyl fluoride). The soluble proteins were purified on Ni-Sepharose High Performance resin (GE Healthcare, Uppsala, Sweden). The purified proteins were eluted with 250–300 mM imidazole in buffer A. The eluted fractions were dialyzed over night in 50 mM potassium phosphate buffer pH 6.8, 100 mM NaCl, 10 mM DTT. All purified proteins were electrophoretically homogeneous as checked by SDS-PAGE. Prenylation of the purified WT PRL-3 protein was performed as described [15]. Prenylated proteins were then cleaned by DyeEx spin columns. MALDI-TOF spectra were acquired with a Voyager DE-PRO workstation, and mass accuracies were obtained with external calibration.
Phosphatase activity assay
Enzymatic assays were carried out with 3-O-methylfluorescein phosphate (OMFP) and 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP) as substrates. In the case of OMFP absorbance at 450 nm was recorded for detection of the product. All assays were performed at 37°C in a reaction mixture containing 40 mM Tris, pH 6.2, 150 mM NaCl, 6 mM DTT and 2% dimethylsulfoxyde. The substrate concentration ranged from 0 to 200 μM. The enzyme concentrations were determined by Bradford method using bovine serum albumin as standard. The enzyme concentrations were 8.0: 20.8; and 10.4 μM for the WT, ΔN10- and ΔC4-mutants, respectively. Enzymatic reactions were initiated by the addition of the enzyme to the reaction mixture and monitored for 20 min. using an Ultrospec 3000 UV/VIS spectrophotometer (GE Healthcare). The product’s absorbance was converted in concentration using a molar extinction coefficient for 3-methylfluorescein (OMF) of 41,900/M/cm. The steady-state constants Km and V were obtained by using Hanes equation S/v =Km/V + S/V and as input data the initial rate values ‘v’ (calculated from the linear portion of the progress curves) corresponding to different substrate concentrations ‘S’. The catalytic constant kcat was obtained by dividing V by the total enzyme concentration. For enzymatic assays in the presence of DiFMUP as substrate, reaction mixture contained 25 mM imidazole pH 6.3, 150 mM NaCl, 5% glycerol and 6 mM DTT; substrate concentration varied between 0 and 1 mM. For all PRL-3 constructs 0.1 μM enzyme was used and all assay reactions were performed at 30°C. Excitation and emission wavelengths were 360 nm and 450 nm, respectively, and a JASCO FP-6500 spectrofluorometer was used for determinations. The initial rate values (calculated from the linear portion of the progress curves) were plotted against the corresponding substrate concentrations and the steady-state constants Km and V were obtained using GraphPad Prism 4 – Nonlinear regression applied directly to Michaelis-Menten type equation v = Vmax*S/(Km+S).
Cell culture and transfections
HeLa cells were grown on Lab-TekII chamber slide systems (Nalgene Nunc Int., Naperville, IL, USA) in RPMI1640 medium supplemented with 10% foetal bovine serum (Sigma Aldrich, St. Louis, MO, USA), 0.2 mM L-glutamine (Gibco) and antibiotics (50 U/ml penicillin/50 μg/ml streptomycin). EGFP-PRL-3 constructs were transfected with Lipofectamine2000 (Invitrogen, Eugene, OR, USA) according to the manufacturer protocol. The DNA-Lipofectamine2000 (0.8:2) mixture was prepared in RPMI 1640 incomplete medium. Twelve to sixteen hours after transfection, the mixture was exchanged with RPMI 1640 complete medium. Twelve to sixteen hours after transfection, the mixture was exchanged with RPMI 1640 complete medium. CHO and COS7 cells were grown on cover slips in six-well flasks in RPMI 1640 and DMEM medium, respectively. The media were supplemented with 10% foetal bovine serum and 0.2 mM L-glutamine. The cells were transfected with DNA: Lipofectamine2000 complexes (1:1.5) prepared in OptiMEM.
Transfected cells were observed 20–29 hrs after transfection. The cells were washed three times in phosphate buffered saline (PBS) and fixed with 4% paraformaldehyde for 15 min. at room temperature. After three washes in PBS, the cells were incubated with 4,6-diamino-2-phenylindole (DAPI) to visualize the nuclei. Finally, the slides were mounted in Vectashield (Vector Laboratories, Burtingame, CA, USA). Images were acquired with a Nikon Eclipse E600 microscope using a Plan Fluor 40× objective.
Results
One of the striking structural characteristic of PRLs as revealed by the crystal structure of PRL-1 [16] is their trimeric structure. Experimental facts support the idea that the oligomeric state is dependent on the farnesylation of PRL-1 and that trimerization may play a regulatory role on the phosphatase activity. In contrast, C-terminally truncated version of PRL-1 displayed only weak tendency for oligomeric association [16]. These findings raise the question whether the CAAX box, besides its role in C-terminal prenylation and in subcellular localization, could not play an additional role in regulating directly the tyrosine phosphatase activity. The N-terminus of PRLs may also be involved in the control of substrate specificity and enzymatic activity. Indeed, structural alignment of PRL-1 and PRL-3 and their closest homologue VHR indicates that the N-terminal helix α0 of VHR – with an important role in substrate recognition – is replaced in PRL-1 and PRL-3 by a disordered (but identical in both PRLs) segment of amino acids [16, 17]. To evaluate the significance of the N-terminal and C-terminal part for enzymatic activity of PRL-3 we obtained corresponding N- and C-terminal truncated forms (Fig. 1). A C-terminal truncation mutant was ectopically expressed in mammalian cells to study its subcellular localization. Additional mutant forms of PRL-3 were obtained in order to analyse the relevance of the putative, C-terminal NLS. Thus, the sequence KRRX12KYRPKQRLRFK (amino acid positions 136–161) suggests a bipartite NLS, being composed of an initial triplet of basic amino acids, a spacer of 10–12 residues and a final stretch of predominantly basic residues [18]. The N-terminal basic residues of a similar bipartite NLS are directly involved in binding importin α, the nuclear import receptor, forming salt bridges with negatively charged residues lining the binding groove [19, 20]. Substitution of these N-terminal basic residues of NLS for non-basic residues should therefore considerably modify the nucleocytoplasmic transport of the cargo protein. Following this line we substituted the KRR basic residues for the QLQ sequence of non-basic amino acidic residues (construct denoted PRL-3-NLS) and subsequently evaluated the subcellular localization of the corresponding mutant PRL-3 form (Figs 1 and 2). Glutamine residues, as relatively long residues containing polar but non-basic end groups, were chosen to replace the basic residues lysine and arginine of the putative signalling sequence.
Figure 1.

Schematic representation of wild-type and mutant forms of PRL-3. 6xHis – 6 histidine tag at N-terminal of the proteins expressed in prokaryotic system; CAAX box – C-terminal prenylation box; EGFP – enhanced green fluorescent protein at N-terminal of the proteins expressed in eukaryotic system; KRR/QLQ – substitution of KRR amino acids with QLQ amino acids in the putative nuclear localization sequence.
Figure 2.

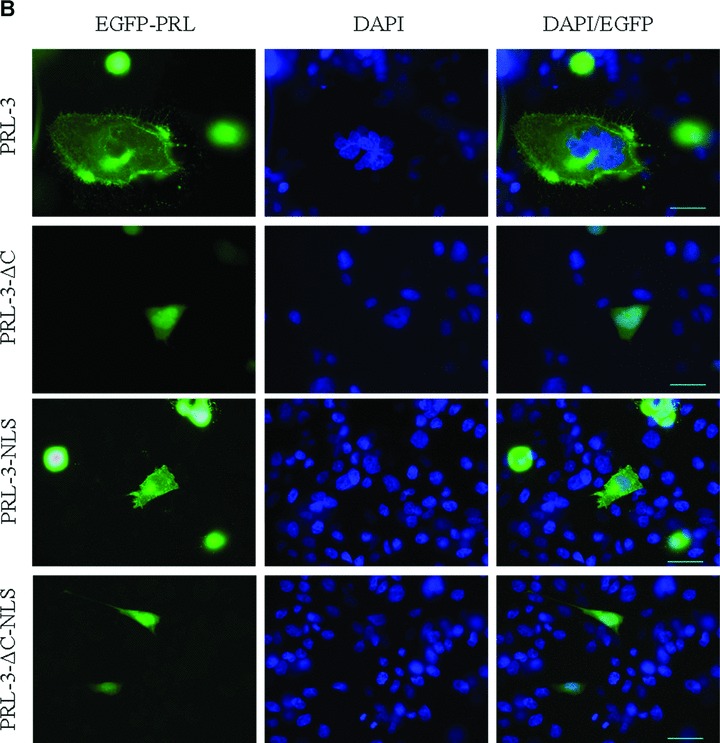

Cellular localization of wild-type and mutant forms of PRL-3, fused with EGFP. EGFP-tagged PRL-3 constructs were introduced by Lipofectamine transfection in HeLa (A), CHO (B) and COS (C) cell lines and visualized after approximately 24 hrs. DAPI stain was used for the visualization of the cell nucleus in fixed cells (second columns). Fluorescence of PRL-3 constructs is shown in the first columns while merge of DAPI staining in EGFP- constructs are in the third columns. Scalebar, 30 μM
HeLa (Fig. 2A), CHO (Fig. 2B), and COS7 (Fig. 2C) cells were used for transfection in order to analyse whether PRL-3 constructs might localize differently in different cell lines. As expected, EGFP-PRL-3 WT was found as punctate structures in the cytoplasm and also associated with plasma membrane (Fig. 2, PRL-3). Removal of the four C-terminal amino acids in EGFP-PRL-3 ΔC4 leads to redistribution of the protein throughout the cytosol and within the nucleus, confirming that in absence of farnesylation the protein has mainly nuclear localization (Fig. 2, PRL-3-ΔC). These findings are in agreement with previously published results of Zeng et al.[10] demonstrating that all three forms of PRL associate in a prenylation-dependent manner with plasma membrane and the early endosome. Surprisingly, the mutated form EGFP-PRL-3-NLS in which the stretch of the first three basic residues was replaced by a triplet of neutral amino acids, has a similar localization with WT form of PRL-3. On the other hand, additional removal of the C-terminal CCVM box within the EGFP-PRL-3 ΔC4-NLS construct seems to determine the localization of the protein to the nucleus. In cases of all three cell lines similar localization of the proteins were observed. These results support the fact that the KRRX12KYRPKQRLRFK sequence cannot be considered a NLS given that the presence of both basic domains in a bipartite NLS is essential for the nuclear targeting of a protein [18].
In principle, it can be imagined that the presence of the four C-terminal amino acids controls not only the prenylation of the protein but also its enzymatic activity. In order to evaluate this possibility as well as the possible involvement of the first 10 N-terminal amino acids we determined the steady-state kinetic parameters of the WT, the C-terminally truncated and of the N-terminally truncated PRL2-WT (Fig. 1). Corresponding proteins – including the PRL-3 – were expressed in prokaryotic expression system and the purified protein preparations displayed a high purity (Fig. 3A).
Figure 3.

Electrophoretic analysis of PRL-3 proteins. (A) 12% SDS-PAGE of the purified preparations of the PRL-3 constructs, under reducing conditions. (B) Evaluation of PRL-3 protein dimerization. For all PRL-3 constructs 5μg protein was applied on each lane of the 12% SDS – PAGE gel. Samples were processed under non-reducing condition. We refer to non-reducing conditions in terms of presence or absence of β-mercaptoethanol or DTT in the loading buffer. WT – wild type PRL-3, ΔN10 – PRL-3 without first 10 amino acids, ΔC4 –PRL-3 without CAAX box, ΔN10C4 – PRL-3 without first 10 and the last 4 amino acids.
The activity with para-nitrophenyl phosphate (pNPP) was extremely low so that determination of accurate values of kinetic parameters was not possible (data not shown). Subsequently alternative phosphatase substrates – OMFP and DiFMUP – were used because the values of the enzymatic activity in these cases were considerably higher than with pNPP.
The specificity constant for the N-terminally truncated PRL-3 ΔN10 construct was essentially similar to that of PRL2-WT, although both the turnover number kcat and the Michaelis constant Km slightly decreased as compared to those for the WT (Table 2). Thus, it can be stated that the 10 N-terminal amino acids sequence do not influence significantly the phosphatase activity of PRL-3. However, the C-terminally truncated form PRL-3 ΔC4 compared to the WT form, displayed different kcat values (slightly higher for OMFP and much lower for DiFMUP) and a significantly lower Km value. Altogether, the specificity constant was 2.3-fold higher for PRL-3 ΔC4 than for PRL-3 WT. These results indicate that the C-terminal CCVM box exerts an inhibitory effect on the catalytic activity of PRL-3, probably by decreasing the affinity of the substrate for the enzyme (assuming that Km approximates the dissociation constant of the enzyme-substrate complex). We further analysed the significance of N- and C-terminal sequences evaluating also the kinetic parameters for the doubly truncated construct ΔN10ΔC4 using DiFMUP as substrate. Interestingly, the Km decreased more than five times (Km= 816 μM) as compared the WT form but the kcat also decreased almost some four times (kcat= 116 ×10−4 s−1). Thus, the specificity constant of the ΔN10ΔC4 construct is situated between those for WT and ΔC4 constructs suggesting that the N-terminus might play an activating role on the catalytic efficiency when the C-terminal box is missing. To determine whether the inhibitory effect of the CCVM box could be a result of partial oligomerization due to disulfide bridge formation we tested the electrophoretic behaviour of PRL-3 constructs under non-reducing conditions. Figure 3B shows that in the absence of mercaptoethanol (as reducing agent) SDS-PAGE for PRL-3-WT, PRL-3-ΔN10 and PRL-3-ΔC4 display an additional band with a molecular weight corresponding to a PRL-3 dimer thus supporting the possibility of a partial oligomerization. Interestingly, the doubly truncated mutant ΔN10C4 does not have the band corresponding to the dimer thus suggesting that both the N-terminus and the C-terminal CAAX box contribute to the dimerization due to disulfide bridge formation. This fact is also supported by the trimeric crystal structure of PRL-3 [16] which shows that both the N-terminal and the C-terminal regions of the three protomers are sterically close to each other permitting their interaction towards formation of oligomers.
Table 2.
Steady-State Kinetic Constants for PRL-3 constructs with OMFP and DiFMUP as substrates. In parentheses the parameters values for DiFMUP are mentioned
| Construct | k cat (s−1) × 104 | K m (μM) | k cat/Km (s−1 M−1) |
|---|---|---|---|
| PRL-3 WT | 2.3 (374.6) | 21.7 (4.3 × 103) | 10.6 (8.7) |
| PRL-3 ΔC4 | 2.7 (100.7) | 11.0 (0.5 × 103) | 24.2 (20.5) |
| PRL-3 ΔN10 | 1.7 (190.5) | 17.7 (2.2 × 103) | 9.33 (8.8) |
Expectedly, the prenylated forms of PRL-3 may present a more pronounced oligomerization then the unprenylated WT form used in our experiments. Indeed, MALDI-TOF analyses of prenylated – both farnesylated form and geranyl-geranylated form – of PRL-3-WT (Fig. 4) clearly demonstrate the presence of dimers. In contrast, in the case of non-prenylated forms of PRL-3-WT the signals corresponding to dimers is much smaller (Fig. 4).
Figure 4.

Mass spectrometric analysis of prenylated and non-prenylated PRL-3 forms. MALDI-TOF analysis of wild-type (WT) (A), farnesylated (B) and geranyl-geranylated forms of PRL-3. Farnesylated and geranyl-geranylated forms were obtained by in vitro prenylation of the purified, WT form of PRL-3.
Discussion
The similar cellular location of WT and of EGFP-PRL-3-NLS mutant forms invalidates the hypothesis that KRRX12KYRPKQRLRFK is a NLS and at the same time raises the question what could be the role of this well-conserved – predominantly basic – C-terminal sequence. An interesting alternative formulated by Zeng et al.[10] is that the positively charged C-terminus could be involved in the membrane targeting of the protein through electrostatic interaction with negatively charged phospholipid heads located in the membrane. In a similar manner, the C-terminal polybasic region of Kras4B within the hypervariable domain, in combination with the prenylation CAAX motif, specifically direct the protein to the plasma membrane [21, 22]. Although this reasoning provides an explanation for the potential role of the C-terminal polybasic region of PRL-3, however the identity of the sequence which directs the protein to the nucleus remains still elusive.
Oligomerization may also play an important role in localization to the cellular membranes. Indeed, Jeong et al.[16] reported the crystal structure of PRL-1 evidencing that it forms trimers. In addition, they proved that oligomerization is a farnesylation-dependent process and it cooperates with membrane localization of the protein. The trimeric structure of PRL-1 (PDB ID: 1XM2) shows that the C-termini of the three monomers forming the trimer are relatively close to each other. Thus, it is reasonable to accept that the hydrophobic farnesyl tails attached to the cysteine of the CAAX box will interact through hydrophobic forces and in this way will contribute to the trimer formation. The high homology between PRL-1 and PRL-3 suggests that PRL-3 should display the same features. Thus, it is attractive to speculate that the inhibitory effect of the CCVM motif, as evidenced by our kinetic measurements (Table 2), is due to a partial oligomerization induced by the hydrophobic interactions induced by the two final hydrophobic residues (V and M). Clearly, in the presence of the long hydrophobic C-terminal prenyl tails, the hydrophobic interaction and consequently the oligomerization should be much more pronounced. In turn, the oligomerization expectedly will contribute to a limited access of the substrate to the enzyme active site and consequently will result in decreased enzyme activity. The N-terminal region – although seems to have a limited influence on the catalytic efficiency – due to its apparent involvement to the dimer formation (Fig. 3B) might indirectly contribute to the modulation of the PRL-3 phosphatase activity. Oligomerization induced inhibition of enzymatic activity due to restricted access of substrate is well documented for the family of protein tyrosine phosphatases: the crystal structure of RPTPα provided the first structural basis in this respect [23–25]. Oligomerization can be also involved in subcellular localization, p53 providing in this respect an interesting paradigm: it seems that tetramerization of p53 is able to regulate its nucleocytoplasmic transport by influencing the accessibility of NLS or of the nuclear export signal to bind to the corresponding receptors [26]. Assuming that a similar hypothetical mechanism could be valid for PRL-3 it would involve that PRL-3 oligomerization induces three distinct processes: (i) protein association to the membranes; (ii) masking of (a putative) NLS and (iii) inhibition of the phosphatase activity. Further experiments are necessary in order to test the validity of this hypothesis. Elucidation of the mechanism which governs PRL-3 subcellular localization and translocation could provide an important tool in understanding the physiological role played by PRL-3 in tumour metastasis and also could contribute to designing anti-metastatic drugs.
Acknowledgments
This work was financially supported by the VIASAN Program, grant number 353/2004, from the Ministry of Education and Research, Romania.
References
- 1.Stephens BJ, Han H, Gokhale V, et al. PRL phosphatases as potential molecular targets in cancer. Mol Cancer Ther. 2005;4:1653–61. doi: 10.1158/1535-7163.MCT-05-0248. [DOI] [PubMed] [Google Scholar]
- 2.Zeng Q, Hong W, Tan YH. Mouse PRL-2 and PRL-3, two potentially prenylated protein tyrosine phosphatases homologous to PRL-1. Biochem Biophys Res Commun. 1998;244:421–7. doi: 10.1006/bbrc.1998.8291. [DOI] [PubMed] [Google Scholar]
- 3.Diamond RH, Cressman DE, Laz TM, et al. PRL-1, a unique nuclear protein tyrosine phosphatase, affects cell growth. Mol Cell Biol. 1994;14:3752–62. doi: 10.1128/mcb.14.6.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saha S, Bardelli A, Buckhaults P, et al. A phosphatase associated with metastasis of colorectal cancer. Science. 2001;294:1343–6. doi: 10.1126/science.1065817. [DOI] [PubMed] [Google Scholar]
- 5.Wu X, Zeng H, Zhang X, et al. Phosphatase of regenerating liver-3 promotes motility and metastasis of mouse melanoma cells. Am J Pathol. 2004;164:2039–54. doi: 10.1016/S0002-9440(10)63763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker BS, Argani P, Cook BP, et al. Alterations in vascular gene expression in invasive breast carcinoma. Cancer Res. 2004;64:7857–66. doi: 10.1158/0008-5472.CAN-04-1976. [DOI] [PubMed] [Google Scholar]
- 7.Polato F, Codegoni A, Fruscio R, et al. PRL-3 phosphatase is implicated in ovarian cancer growth. Clin Cancer Res. 2005;11:6835–9. doi: 10.1158/1078-0432.CCR-04-2357. [DOI] [PubMed] [Google Scholar]
- 8.Miskad UA, Semba S, Kato H, et al. High PRL-3 expression in human gastric cancer is a marker of metastasis and grades of malignancies: an in situ hybridization study. Virchows Arch. 2007;450:303–10. doi: 10.1007/s00428-006-0361-8. [DOI] [PubMed] [Google Scholar]
- 9.Zeng Q, Dong JM, Guo K, et al. PRL-3 and PRL-1 promote cell migration, invasion, and metastasis. Cancer Res. 2003;63:2716–22. [PubMed] [Google Scholar]
- 10.Zeng Q, Si X, Horstmann H, et al. Prenylation-dependent association of protein-tyrosine phosphatases PRL-1, -2, and -3 with the plasma membrane and the early endosome. J Biol Chem. 2000;275:21444–52. doi: 10.1074/jbc.M000453200. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Kirby CE, Herbst R. The tyrosine phosphatase PRL-1 localizes to the endoplasmic reticulum and the mitotic spindle and is required for normal mitosis. J Biol Chem. 2002;277:46659–68. doi: 10.1074/jbc.M206407200. [DOI] [PubMed] [Google Scholar]
- 12.Gjorloff-Wingren A, Saxena M, Han S, et al. Subcellular localization of intracellular protein tyrosine phosphatases in T cells. Eur J Immunol. 2000;30:2412–21. doi: 10.1002/1521-4141(2000)30:8<2412::AID-IMMU2412>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 13.Kong W, Swain GP, Li S, et al. PRL-1 PTPase expression is developmentally regulated with tissue-specific patterns in epithelial tissues. Am J Physiol Gastrointest Liver Physiol. 2000;279:G613–21. doi: 10.1152/ajpgi.2000.279.3.G613. [DOI] [PubMed] [Google Scholar]
- 14.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–90. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 15.Dursina B, Reents R, Delon C, et al. Identification and specificity profiling of protein prenyltransferase inhibitors using new fluorescent phosphoisoprenoids. J Am Chem Soc. 2006;128:2822–35. doi: 10.1021/ja052196e. [DOI] [PubMed] [Google Scholar]
- 16.Jeong DG, Kim SJ, Kim JH, et al. Trimeric structure of PRL-1 phosphatase reveals an active enzyme conformation and regulation mechanisms. J Mol Biol. 2005;345:401–13. doi: 10.1016/j.jmb.2004.10.061. [DOI] [PubMed] [Google Scholar]
- 17.Kozlov G, Cheng J, Ziomek E, et al. Structural insights into molecular function of the metastasis-associated phosphatase PRL-3. J Biol Chem. 2004;279:11882–9. doi: 10.1074/jbc.M312905200. [DOI] [PubMed] [Google Scholar]
- 18.Robbins J, Dilworth SM, Laskey RA, et al. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–23. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 19.Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat Struct Biol. 1999;6:388–97. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- 20.Lange A, Mills RE, Lange CJ, et al. Classical nuclear localization signals: definition, function, and interaction with impor-tin alpha. J Biol Chem. 2007;282:5101–5. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hancock JF, Cadwallader K, Paterson H, et al. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 1991;10:4033–9. doi: 10.1002/j.1460-2075.1991.tb04979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–9. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 23.Bilwes AM, den Hertog J, Hunter T, et al. Structural basis for inhibition of receptor protein-tyrosine phosphatase-alpha by dimerization. Nature. 1996;382:555–9. doi: 10.1038/382555a0. [DOI] [PubMed] [Google Scholar]
- 24.Toledano-Katchalski H, Tiran Z, Sines T, et al. Dimerization in vivo and inhibition of the nonreceptor form of protein tyrosine phosphatase epsilon. Mol Cell Biol. 2003;23:5460–71. doi: 10.1128/MCB.23.15.5460-5471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabernero L, Aricescu AR, Jones EY, et al. Protein tyrosine phosphatases: structure-function relationships. FEBS J. 2008;275:867–82. doi: 10.1111/j.1742-4658.2008.06251.x. [DOI] [PubMed] [Google Scholar]
- 26.Liang SH, Clarke MF. Regulation of p53 localization. Eur J Biochem. 2001;268:2779–83. doi: 10.1046/j.1432-1327.2001.02227.x. [DOI] [PubMed] [Google Scholar]


