Abstract
Bacillus subtilis penicillin-binding protein PBP1 has been implicated in cell division. We show here that a PBP1 knockout strain is affected in the formation of the asymmetric sporulation septum and that green fluorescent protein-PBP1 localizes to the sporulation septum. Localization of PBP1 to the vegetative septum is dependent on various cell division proteins. This study proves that PBP1 forms part of the B. subtilis cell division machinery.
The cell wall is the principal shape-maintaining and stress-bearing element of the bacterial cell (for reviews, see references 9, 12, and 24). The Bacillus subtilis cell wall consists of peptidoglycan (PG), glycan strands with peptide side chains that are highly cross-linked, and covalently linked anionic polymers (9). The polymerization and cross-linking of PG is mediated by penicillin-binding proteins (PBPs). Various studies have indicated that PG synthesis is likely to be mediated by holoenzyme complexes, comprising PBPs and lytic transglycosylases that can perform the murein synthesis and hydrolysis reactions involved in PG synthesis and processing (e.g., 1, 2, 20, 21, 23). Since cell wall synthesis during elongation occurs parallel to the lateral wall, whereas cell wall synthesis during division occurs perpendicular to the lateral wall, at least two separate holoenzyme complexes have been proposed (12). The putative cell division specific complex of B. subtilis contains the class B transpeptidase PBP2b (6) and, together with known cell division proteins, forms the cell division machinery also known as the “divisome” (14). In B. subtilis, the assembly of the cell division machinery starts with the formation of the FtsZ ring and targeting of FtsA to this ring (reviewed in reference 7). Subsequently, all cell division proteins that contain a transmembrane span, localize to this ring in a concerted manner. DivIB, DivIC, FtsL, PBP2b, and probably FtsW (R. A. Daniel and L. J. Wu, unpublished data) are all completely interdependent for assembly at the division site, and mutation or depletion of any of these proteins prevents all of the others from assembling (reviewed in reference 7). This is in sharp contrast to Escherichia coli, in which division proteins assemble in a strictly ordered manner (reviewed in reference 3).
Another PBP implicated in cell division in B. subtilis is PBP1, which localizes to the cell division site (15, 19). PBP1 is a class A PBP with both transglycosylase and transpeptidase activities, encoded by the ponA gene (17). ponA knockout cells are not blocked in cell division, but grow more slowly, with an increase in mean cell length and a decrease in mean cell width (18), as well as having abnormal septal structures (15). These findings indicate that division is suboptimal in ponA knockout cells. Also, sporulation efficiency is severely reduced in a ponA knockout compared to single knockouts of genes encoding other class A PBPs, suggesting that PBP1 may play a role in correct asymmetric cell division at the start of spore development (18). We reasoned that if PBP1 is a true component of the cell division machinery, it should localize to the asymmetric division site that is formed during sporulation. Also, given the interdependency of cell division proteins for assembly at the cell division site in B. subtilis, we expected PBP1 septal localization to be dependent on other division proteins.
PBP1 is required for the efficient formation of the asymmetric sporulation septum.
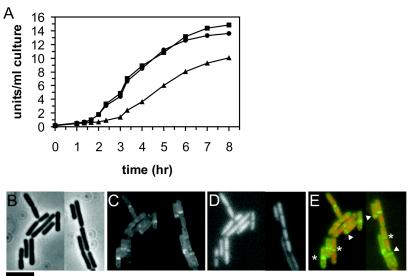
Although PBP1 is not an essential cell division protein, it may play an important role in the asymmetric cell division during sporulation. Sporulation efficiency has been reported to be markedly reduced in a ponA-null mutant (14% compared to wild type [18]). To test whether this sporulation defect is due to the inefficient formation of the asymmetric division septum, we studied the sporulation properties of a ponA-null mutant in more detail. Cells were grown in CH medium to an optical density at 600 nm of ∼0.8 before the induction of sporulation by resuspending the cells in sporulation medium essentially as described previously (22). Strain 2083 was grown in the presence of 0.75% xylose and washed with sporulation medium before resuspension to remove xylose during sporulation. Although formation of an “axial DNA filament,” a marker of sporulation stage I (16), was not affected in the ponA-null mutant, the formation of the asymmetric septum was greatly reduced (Table 1). A defect in asymmetric septation should be accompanied by reduced expression of σF- and σE-dependent genes since activation of these sigma factors is dependent on septation (reviewed in reference 16). To test this, the σE-dependent synthesis of alkaline phosphatase (APase) was measured as described previously (8, 10). As shown in Fig. 1A, the ponA-null mutant showed substantially delayed and reduced APase activity compared to both the wild-type strain and the strain expressing green fluorescent protein (GFP)-PBP1. Presumably, the defect in asymmetric septum formation is responsible for the reduced spore frequency observed (Table 1) (18).
TABLE 1.
Sporulation properties of a PB1 mutant
| Strain | Phenotype | % Cells (n) with an:
|
No. of heat-resistant spores (CFU/ml)c at:
|
||
|---|---|---|---|---|---|
| Axial filament at T1a | Asymmetric division septum at T2b | T10 | T25 | ||
| 168 | Wild type | 54 (190) | 45 (379) | 2.5 × 10−8 | 4.0 × 10−8 |
| 3511 | PBP1 null | 53 (183) | 18 (289) | 2.4 × 10−7 | 1.6 × 10−7 |
| 2083 | GFP-PBP1 | NDd | ND | 2.7 × 10−8 | ND |
Cells were examined 1 h after resuspension (T1), at which time the presence of axial filaments was determined by staining the DNA with Hoechst 33342 (1 μg/ml; Molecular Probes).
Asymmetric septa were visualized 2 h after resuspension (T2) by staining the membranes with Nile red (0.1 μg/ml; Molecular Probes).
Heat resistance was determined by incubating sporulating cells for 10 h (T10) or 25 h (T25) after resuspension for 20 min at 80°C.
ND, not done.
FIG. 1.
(A) Delayed and reduced APase activity in sporulating cells of a ponA-null mutant. APase activity was measured in strains 168 (wild type, ▪), 2083 (gfp-ponA, •), and 3511 (ΔponA, ▴), with T0 being the moment of resuspension. The results are representative of three independent experiments. (B) Localization of GFP-PBP1 to the asymmetric sporulation septum. Samples were obtained 90 min after the induction of sporulation. Images shown include phase contrast (B), GFP-PBP1 fluorescence (C), DNA staining with DAPI (4′,6′-diamidino-2-phenylindole) (D), and an overlay of the GFP-fluorescence in green and DNA staining in red (E). Arrowheads indicate GFP-PBP1 at asymmetric sporulation septa; asterisks denote GFP-PBP1 at vegetative division septa. Samples were prepared for fluorescence microscopy as described previously (19). Bar, 5 μm.
Recently, we constructed a fully functional GFP-PBP1 fusion protein in B. subtilis that localizes to the site of cell division (15, 19). This GFP-PBP1 strain sporulated with an efficiency similar to that of the wild-type strain (Table 1) and displayed a similar APase activity (Fig. 1A), showing that GFP-PBP1 is fully functional during sporulation as well. Xylose was not present in the sporulation medium, but GFP-PBP1 is presumably stable enough to function during sporulation, as previously described for PBP2b (6). Previously, lacZ fusions showed that transcription of ponA decreases gradually during sporulation to background levels (17), suggesting that PBP1 does not act later in sporulation.
GFP-PBP1 was visible at asymmetric sporulation septa during the early stages of sporulation (Fig. 1B to E). Fluorescence began to be detected at the asymmetric septum 60 min after the initiation of sporulation but disappeared after septum closure, following a localization pattern similar to that previously described for PBP2b (6). This makes it likely that, like PBP2b, PBP1 is targeted to the asymmetric division septum, where it forms part of the division machinery, and disappears after completion of the division event.
PBP1 localization depends on other cell division proteins.
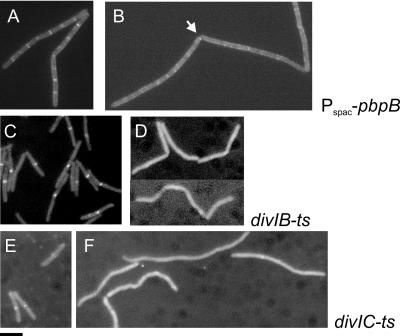
Previously, we have shown that GFP-PBP1 septal localization is dependent on FtsZ (19). Since FtsZ is essential for the localization for all other cell division proteins, we tested PBP1 localization in the absence of several membrane proteins that are components of the divisome, which are all interdependent for localization. Strains were constructed in which gfp-ponA as the only functional ponA copy was combined with (i) pbpB under the control of the Pspac promoter (strain 3542), (ii) a divIB temperature-sensitive (ts) mutation (strain 3526), and (iii) a divIC ts mutation (strain 3536) (Table 2). As shown in Fig. 2B, depletion of PBP2b led to the disappearance of GFP-PBP1 bands at septa. Interestingly, occasional spots of GFP-PBP1 could be observed at incomplete cell division sites (arrow in Fig. 2B), as previously described for PBP2b (6, 19). This would indicate that where there is a limiting amount of PBP2b, sufficient to start septation but not to support progression of septal ingrowth, PBP1 is also present as part of the division complex. In the divIB and divIC ts strains, GFP-PBP1 localization was normal at the permissive temperature, but upon a shift to the nonpermissive temperature midcell localization was abolished (Fig. 2D and F). A control experiment established that GFP-PBP1 in the wild-type background is stable at the nonpermissive temperature (results not shown). Therefore, we conclude that localization of PBP1 to the division septum depends on FtsZ, PBP2b, DivIB, and DivIC. FtsL was omitted from this analysis because the depletion strain for FtsL is dependent on xylose, as is GFP-PBP1 expression. However, since FtsL localizes to incomplete septa upon PBP2b depletion and is dependent on DivIB and DivIC (5), it seems likely that PBP1 localization will also be dependent on FtsL.
TABLE 2.
Strains used in this study
| Strain | Relevant characteristics | Source, reference, and/or constructiona |
|---|---|---|
| 168 | trpC2 | Laboratory collection |
| SU347 | divIC355 | 13 |
| MB76 | trpC2 divIBm12 | Laboratory stock, allele tms-12 (4) |
| 2083 | trpC2 ponA::pSG1492 (cat Pxyl-gfp-ponA1-394) | 19 |
| 3122 | trpC2 pbpB::pSG5061 (cat Pxyl-gfp-pbpB1-825) | 19 |
| 3295 | chr::(Pspac-pbpB neo) | 11 |
| 3511 | trpC2 ΔponA::spc | PS2062 (17) → 168 (Spcr)b |
| 3526 | trpC2 divIBm12 ponA::pSG1492 (cat Pxyl-gfp-ponA1-394) | 2083 → MB76 (Cmr) |
| 3536 | divIC355 ponA::pSG1492 (cat Pxyl-gfp-ponA1-394) | 2083 → SU347 (Cmr) |
| 3542 | trpC2 ponA::pSG1492 (cat Pxyl-gfp-ponA1-394) chr::(Pspac-pbpB neo) | 2083 → 3295 (Cmr) |
| 3907 | trpC2 ΔponA::spc chr::(Pspac-pbpB neo) | 3295 → 3511 (Kanr) |
| 3908 | trpC2 ΔponA::spc pbpB::pSG5061 (cat Pxyl-gfp-pbpB1-825) | 3122 → 3511 (Kanr) |
Spcr, spectinomycin resistance; Cmr, chloramphenicol resistance; Kanr, kanamycin resistance.
Strain 168 transformed to spectinomycin resistance with PS2062 chromosomal DNA.
FIG. 2.
Localization of PBP1 in conditional cell division mutants. (A and B) pbpB. Strain 3542 was grown in the presence of 0.5 mM IPTG and 0.75% xylose to exponential phase in S+ medium (19) at 30°C. The cells were harvested, washed twice, and resuspended in the same medium with (A) or without (B) IPTG and allowed to grow at 30°C for 90 min to deplete PBP2b. (C and D) divIB. Cells were grown to exponential phase at 30°C in Difco Antibiotic Medium 3 (17.5 g/liter; Penassay broth) with 0.75% xylose before the culture was split and grown for an additional 40 min at a permissive (30°C) (C) or nonpermissive (48°C) (D) temperature. (E and F) divIC. Cells were grown to exponential phase at 30°C in Penassay broth with 0.75% xylose before the culture was split and grown for an additional 40 min at a permissive (30°C) (E) or nonpermissive (48°C) (F) temperature. Samples were prepared for fluorescence microscopy as described previously (19). Bar, 5 μm.
Finally, PBP1 has both transglycosylase and transpeptidase activities. We tested whether PBP1 might at least partially substitute for the essential transpeptidase activity of PBP2b by testing the sensitivity of a ponA-null mutant to PBP2b depletion compared to a wild-type strain. ponA-null strains were constructed with either pbpB under control of Pspac (strain 3907) or gfp-pbpB under control of Pxyl (strain 3908). These strains were grown to exponential phase; the cells were then harvested, washed, and resuspended in fresh medium with various concentrations of inducer. Inducer became limiting at similar concentrations irrespective of the presence of PBP1, with growth abolished at 0.005 mM IPTG (isopropyl-β-d-thiogalactopyranoside) or 0.1% xylose. At slightly higher concentrations of inducer (0.01 mM IPTG or 0.2% xylose), pbpB expression was enough to allow wild-type growth rates for the parental strains 3295 and 3122. However, in the strains containing both inducible pbpB and the ponA-null mutation, this level of inducer did not restore the growth rate to the level of the ponA-null mutation on its own (a doubling time [Td] of 53 min for strain 3907 at 0.01 mM IPTG compared to a Td of 44 min for strain 3511 [mean of two experiments]). This shows that in the absence of PBP1 the effects of PBP2b depletion are slightly exacerbated.
We show here that PBP1 localization to the vegetative septum is dependent on various cell division proteins in B. subtilis and that PBP1 localizes to the asymmetric septum formed during sporulation. Furthermore, we show that PBP1 plays an important role in the assembly of the asymmetric division septum during sporulation. A ponA-null mutant is not inhibited in the entry of sporulation, as determined by the formation of the axial filament, but is strongly inhibited in the formation of asymmetric division septa compared to a wild-type strain. Although other cell division proteins localize in the absence of PBP1, we conclude that PBP1 forms a part of the B. subtilis cell division machinery, with a significant role in the synthesis of septal PG, located perpendicular to the lateral cell wall. This function is partially redundant during vegetative growth but becomes more critical during sporulation.
Acknowledgments
We thank members of our laboratory for discussions and advice.
D.-J.S. is supported by a Marie-Curie Postdoctoral Fellowship (HPMF-CT-2001-01421). This study was supported by a grant from the UK Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Alaedini, A., and R. A. Day. 1999. Identification of two penicillin-binding multienzyme complexes in Haemophilus influenzae. Biochem. Biophys. Res. Commun. 264:191-195. [DOI] [PubMed] [Google Scholar]
- 2.Bhardwaj, S., and R. A. Day. 1997. Detection of Intra-cellular protein-protein interactions: penicillin interactive proteins and morphogene proteins, p. 469-480. In D. Marshak (ed.), Techniques in protein chemistry, vol 8. Academic Press, Inc., New York, N.Y. [Google Scholar]
- 3.Buddelmeijer, N., and J. Beckwith. 2002. Assembly of cell division proteins at the E. coli cell center. Curr. Opin. Microbiol. 5:553-557. [DOI] [PubMed] [Google Scholar]
- 4.Callister, H., and R. G. Wake. 1981. Characterization and mapping of temperature-sensitive division initiation mutations of Bacillus subtilis. J. Bacteriol. 145:1042-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daniel, R. A., and J. Errington. 2000. Intrinsic instability of the essential cell division protein FtsL of Bacillus subtilis and a role for DivIB protein in FtsL turnover. Mol. Microbiol. 36:278-289. [DOI] [PubMed] [Google Scholar]
- 6.Daniel, R. A., E. J. Harry, and J. Errington. 2000. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol. Microbiol. 35:299-311. [DOI] [PubMed] [Google Scholar]
- 7.Errington, J., R. A. Daniel, and D.-J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Errington, J., and J. Mandelstam. 1983. Variety of sporulation phenotypes resulting from mutations in a single regulatory locus, spoIIA, in Bacillus subtilis. J. Gen. Microbiol. 129:2091-2101. [DOI] [PubMed] [Google Scholar]
- 9.Foster, S. J., and D. L. Popham. 2001. Structure and synthesis of cell wall, spore cortex, teichoic acids, S-layers, and capsules, p. 21-41. In L. Sonenshein, R. Losick, and J. A. Hoch (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 10.Glenn, A. R., and J. Mandelstam. 1971. Sporulation in Bacillus subtilis 168: comparison of alkaline phosphatase from sporulating and vegetative cells. Biochem. J. 123:129-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamoen, L. W., and J. Errington. 2003. Polar targeting of DivIVA in Bacillus subtilis is not directly dependent on FtsZ or PBP 2B. J. Bacteriol. 185:693-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Höltje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katis, V. L., E. J. Harry, and R. G. Wake. 1997. The Bacillus subtilis division protein DivIC is a highly abundant membrane-bound protein that localizes to the division site. Mol. Microbiol. 26:1047-1055. [DOI] [PubMed] [Google Scholar]
- 14.Nanninga, N. 1998. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:110-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pedersen, L. B., E. R. Angert, and P. Setlow. 1999. Septal localization of penicillin-binding protein 1 in Bacillus subtilis. J. Bacteriol. 181:3201-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piggot, P. J., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-517. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 17.Popham, D. L., and P. Setlow. 1995. Cloning, nucleotide sequence, and mutagenesis of the Bacillus subtilis ponA operon, which codes for penicillin-binding protein (PBP) 1 and a PBP-related factor. J. Bacteriol. 177:326-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popham, D. L., and P. Setlow. 1996. Phenotypes of Bacillus subtilis mutants lacking multiple class a high-molecular-weight penicillin-binding proteins. J. Bacteriol. 178:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheffers, D.-J., L. J. F. Jones, and J. Errington. 2004. Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis. Mol. Microbiol. 51:749-764. [DOI] [PubMed] [Google Scholar]
- 20.Schiffer, G., and J. V. Holtje. 1999. Cloning and characterization of PBP 1C, a third member of the multimodular class A penicillin-binding proteins of Escherichia coli. J. Biol. Chem. 274:32031-32039. [DOI] [PubMed] [Google Scholar]
- 21.Simon, M. J., and R. A. Day. 2000. Improved resolution of hydrophobic penicillin-binding proteins and their covalently linked complexes on a modified C18 reversed-phase column. Anal. Lett. 33:861-867. [Google Scholar]
- 22.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to the development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vollmer, W., M. von Rechenberg, and J. V. Holtje. 1999. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 274:6726-6734. [DOI] [PubMed] [Google Scholar]
- 24.Young, K. D. 2003. Bacterial shape. Mol. Microbiol. 49:571-580. [DOI] [PubMed] [Google Scholar]