Abstract
Overexpression of HMGA2 is common in uterine leiomyomas (ULM). The expression of HMGA2 in its malignant counterpart – uterine leiomyosarcomas (ULMS) remains undetermined. Recently it has been shown that repression of HMGA2 by microRNA let-7s is a critical molecular regulatory mechanism associated with tumour growth in many tumours and cell types, including leiomyomas. To test whether HMGA2 and let-7s play a role in ULMS, we examined the levels of endogenous HMGA2 and let-7 expression and found a significant correlation between these two molecules in a case-matched cohort of human ULMS. We found that overexpression of HMGA2 and let-7-mediated HMGA2 repression is a relevant molecular alteration in ULMS. Disrupting the control of HMGA2 and let-7 pairs promotes ULMS cell growth in vitro.
Introduction
Uterine leiomyosarcomas (ULMS) are rare neoplasms, representing 1 in 200 to 800 smooth muscle tumours or ∼1% of all malignancies of the uterus [1]. Although the reported 5-year ULMS survival rates are variable, these tumours are clinically aggressive and have a high risk of recurrence and overall poor prognosis [2]. The cause or the molecular basis of human ULMS is largely unknown. While it was believed that most ULMS could arise de novo, some reports suggest that ULMS was progressive from pre-existing uterine leiomyomas (ULM) [3, 4]. However, the conclusive evidence documenting the transformation of ULM to ULMS is lacking in human beings and it is thought that the incidence of transformation is <0.1%[5].
HMGA2, a high-mobility-group AT-hook (HMGA) protein, is considered to function as an oncogene strongly associated with many malignant epithelial and mesenchymal neoplasms [6]. It has three AT-hook DNA binding domains, through which HMGA2 binds to AT-rich sequences in the minor groove of the DNA helix. HMGA2 is expressed in embryonic tissue but not in most adult tissues [7, 8]. HMGA2 is overexpressed in ULM due to chromosomal 12q15 changes [9–12]. Overexpression of HMGA2 in ULMS has been suggested, but not fully investigated [4, 13]. HMGA2 is an important regulator of cell growth, differentiation, apoptosis and transformation [14]. New evidence suggest that dysregulation of microRNAs (miRNAs) may play a central role in tumour development [15]. Members of the let-7 miRNA family function as tumour suppressors through specific repression of its target gene, particularly of HMGA2 expression in some tumour cells both in vivo and in vitro[4, 16, 17]. The biological importance of molecular pairing of HMGA2::let-7 was further illustrated by the demonstration that repression of HMGA2 by let-7s impairs tumour cell proliferation in many different tumour types, including ULM [4, 16–19]. It would be of great interest to characterize whether overexpression of HMGA2 and disruption of the HMGA2::let-7 pairs contributes to the aggressive growth behaviour of ULMS.
In this study, we examined the levels of endogenous HMGA2 and let-7 expression and analysed a potential correlation between these two molecules in a case-matched cohort of human ULMS. We demonstrate that overexpression of HMGA2 and let-7-mediated HMGA2 repression is important for molecular changes in ULMS. Disrupting the HMGA2::let-7 pairs promote ULMS cell growth in vitro.
Materials and methods
Patient and tissue samples
A total of 35 hysterectomies for ULMS were collected in this study. The mean ages at hysterectomies were 59.9 years old (ranging from 38 to 83 years old). Tumour sizes were ranged from 2 to 22 cm with mean tumour size of 11.6 cm. Of all 35 cases, 23 were solitary and 12 coexisted with leiomyomas (Table 1).
Table 1.
HMGA2 and let-7s expression in 35 uterine leiomyosarcomas
| Case no. | Age (years) | Size (cm) | HMGA2 (IHC) | HMGA2 (RT) | Let-7c (ISH) | Let-7 (c, d, f-2) | Ki-67 index (%) |
|---|---|---|---|---|---|---|---|
| ULMS1 | 43 | 5 | 0 | 0 | 30 | ||
| ULMS2 | 57 | 4 | 0 | −2 | 15 | ||
| ULMS3 | 55 | 4 | 0 | 1 | 50 | ||
| ULMS4 | 53 | 10.5 | 0 | 0 | 60 | ||
| ULMS5 | 54 | 11# | 0 | −1 | 50 | ||
| ULMS6 | 79 | 30 | 0 | −1 | 25 | ||
| ULMS7 | 83 | 13 | 2+ | −1 | 10 | ||
| ULMS8 | 38 | 17 | 0 | 0 | 25 | ||
| ULMS9 | 82 | 9 | 0 | 0 | 35 | ||
| ULMS10 | 69 | 8.5 | 3+ | 1 | 25 | ||
| ULMS11 | 85 | ?*** | 3+ | −1 | 25 | ||
| ULMS12 | 59 | 15 | 0 | 0 | 30 | ||
| ULMS13 | 74 | 5 | 0 | 0 | 5 | ||
| ULMS14 | 50 | 6.8 | 0 | 0 | 50 | ||
| ULMS15 | 52 | 22# | 3+ | −2 | 40 | ||
| ULMS16 | 60 | 12 | 2+ | −1 | 15 | ||
| ULMS17 | 55 | 9 | 0 | −1 | 30 | ||
| ULMS18 | 73 | 11 | 2+ | −2 | 60 | ||
| ULMS19 | 46 | ? | 0 | 1 | 20 | ||
| ULMS20 | 64 | ? | 3+ | −1 | 35 | ||
| ULMS21 | 64 | 8 | 0 | 0 | 30 | ||
| ULMS22 | 48 | 7 | 0 | 0 | 5 | ||
| ULMS23 | 63 | ? | 0 | 1 | |||
| ULMS24 | 59 | 4.5 | 0 | 1 | 60 | ||
| ULMS25 | 55 | ? | 0 | 0 | 60 | ||
| ULMS26 | 43 | 20 | 3+ | 0 | 20 | ||
| ULMS27 | 57 | 12 | 4+ | 0 | 30 | ||
| ULMS28 | 49 | 14 | 3+ | −1 | 50 | ||
| ULMS29 | 74 | ? | 3+ | −1 | 40 | ||
| ULMS30 | 60 | 2 | 0 | ||||
| ULMS31 | 55 | 16 | 3 | 1, 0, 1 | |||
| ULMS32 | 63 | 17 | 2* | 2 | 2, 0, 1 | ||
| ULMS33 | 51 | 20 | 3 | 3, 1, 3 | |||
| ULMS34 | 53 | 21** | 2* | 2 | 1, 0, 1 | ||
| ULMS35 | 73 | 12 | 4 | 2, 1, 1 | |||
| Mean | 59.94 | 11.62 | 1.13 | 2.50 | −0.35 | − | 33.21 |
| Sem | 2.00 | 1.07 | 0.24 | 0.18 | 0.15 | − | 3.06 |
Western blot results.
Karyotype: t(12; 14) and tri(12) (see Fig. 2); IHC: immunohistochemistry; ISH: in situ hybridization.
: Tumour size is not documented in pathological report.
: Tumours with extrauterine extension.
High-density tissue microarray (TMA) was prepared from formalin fixed and paraffin-embedded (FFPE) tissue cores (0.6 mm) in ULMS (n= 30) and matched myometrium (n= 30) in triplicate. Fresh frozen tissues from ULMS and matched myometrium were collected in five cases for RT-PCR and Western blot analysis (Table 1).
Three uterine leiomyosarcoma cell lines were used for the study including: SK-LMS-1, SK-UT-1 and SK-UT-1b.
The study was approved by the NYU Medical Center institutional review board.
Let-7 mimic and inhibitor
Mature double stranded miRNAs of let-7 and let-7 inhibitors were purchased from Dharmacon, Inc. (Lafayette, CO, USA). All experiments were controlled using a non-functional double-stranded random 22 nt RNA (Block-iT, Invitrogen, Carlsbad, CA, USA).
Primers and antibodies
Primers from HMGA2 and its alternative spliced transcripts were reported previously [19]. Primers for mature let-7 family (a–i) were purchased from Ambion, Inc. (Austin, TX, USA). Antibodies includes HMGA2 (provided by Dr. Masashi Norita and Biocheck, CA, USA), Ki-67, ERα and PR-A (Ventana Medical System, Inc., Tucson, AZ, USA), and P53 (Neomarkers, CA, USA).
Let-7c miRNAs in situ hybridization
The hybridization system and probes, miRCURY LNA, Let-7c, and U6, were purchased from Exiqon (Vedbaek, Denmark). The detailed procedure for in situ hybridization was followed as per manufacturer’s protocol [20]. In brief, 4-μm TMA slides were prepared. Following deparaffinization and deproteinization, the slides were pre-hybridized with 1× hybridization buffer without probe. The hybridization was carried out overnight in a 1× hybridization buffer (30–70 μl) with pre-denatured miRCURY LNA, let-7c or U6 probes. After washing, the slides were blocked and incubated with AP conjugated anti-DIG Fab fragments (1:1500, Roche, Indianapolis, IN, USA) and visualized for colour detection.
qRT-PCR
For the detection of mature miRNAs, mirVana qRT-PCR primers and the mirVana qRT-PCR Detection Kits (Ambion, Inc.) were used and optimized according to the abundance of miRNAs in the samples. In brief, 50 to 100 ng of total RNA were reverse transcribed with specific miRNA primers. A total of 15–30 cycles were performed for quantitation. Primers for the common domain of HMGA2, the dominant transcript (HMGA2a), and the cryptic HMGA2 transcripts were described [19]. The abundances of cDNA products were detected by qRT-PCR and were normalized by the internal control products of U6 and α-Actin.
Immunohistochemistry
The TMA blocks from FFPE tissues were sectioned at 4 microns. After deparaffinization and antigen retrieval, all immunohistochemical staining was performed on a Ventana Nexus automated system.
Western blot analysis
Fresh frozen tissue or culture cell samples were homogenized at 4°C in a protein lysis buffer (0.5 g tissue in 1–2 ml). Identical amounts of total proteins from each sample were separated through a 12% SDS-PAGE gel and then transferred to a PVDF membrane (Perkin Elmer Life Scientific Inc.). Development of the immunoblot with antisera against HMGA2 and negative control HMGA2 blocking peptide (provided by Dr. Masashi Norita and Santa Cruz Biotechnology, Inc., CA, USA) was tested and a single specific HMGA2 band at 25 kD was detected, as previously described.
Cell culture and miRNAs transfection
LMS cell lines were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) containing 10% foetal bovine serum (Gemini, Calabasas, CA, USA) in 37°C incubators with 5% CO2 until the cells reached 30–40% confluence. Prior to transfection, cells were placed in standard media without antibiotics for 24 hrs. As per manufacturers’ protocol, transfection was performed with the Lipofectamine system with miRNAs concentrations of 20–60 pmol/well in either 6- or 24-well plates. To estimate transfection efficiency, cotransfection with the block-iT fluorescent double-stranded random 22mer RNA from Invitrogen was performed. The FITC fluorescence was visualized by lex= 494 nm and lem= 519 nm to assess the percentage of cells that were successfully transfected. Cells receiving only the tagged random sequence double-strand 22mer were used as non-specific references at all data points. Following transfection, cells were harvested and analysed at the indicated times.
Cellular proliferation assay
PC3 and LNCaP cell lines of control (with pBabe) and of tests (stable overexpression of HMGA2a with and without let-7 LCS) were passed in 24-well plates in triplicate at densities of 5 × 103 cells/well for LNCaP and 1 × 104 for PC3 cells. Cells were subsequently transfected with control RNA (non-function, Invitrogen), let-7c, and/or let-7 inhibitors (Dharmacon, Inc.) at a dose of 40 pmol/well. Cellular proliferation was counted at 24, 48, 72 and 96 hrs using the colorimetric WST-1 assay (Cell proliferation Reagent, Roche). Briefly, the cells were incubated with 10% WST-1 reagent in normal medium for 2 hrs. Aliquots (100 μl) were then transferred to 96-well plates and the samples were read in a spectrophotometric plate reader at 450 nm (FLUOstar OPTIMA, BMG Lab Technologies, Durham, NC, USA).
Statistical analysis
Mean and standard errors were calculated for the quantitative values. Statistical significance was analysed by a paired t-test and P-value <0.05 was considered significant.
Results
HMGA2 and let-7 expression in ULMS
To evaluate whether elevated HMGA2 expression is common in ULMS, we examined HMGA2 expression in 30 ULMS by immunohistochemistry analysis. With internal negative controls of matched myometrium and staining done with two different sources of HMGA2 antibodies (see section ‘Materials and methods’), we found up to 38% of ULMS (11/29) immunoreactive for HMGA2 (Fig. 1). We then examined HMGA2 expression at transcription and translational levels by RT-PCR and Western blot, respectively, in five randomly selected ULMS collected from fresh frozen tissues. With a negative (matched myometrium) and a positive (HMGA2 positive ULM) controls, a moderate to high level of HMGA2 mRNA expression (32 cycles) was detected in all five cases (Fig. 2A). Of same five tumours, two of them were positive for HMGA2 by Western blot analysis (Table 1, Fig. 2A).
Figure 1.
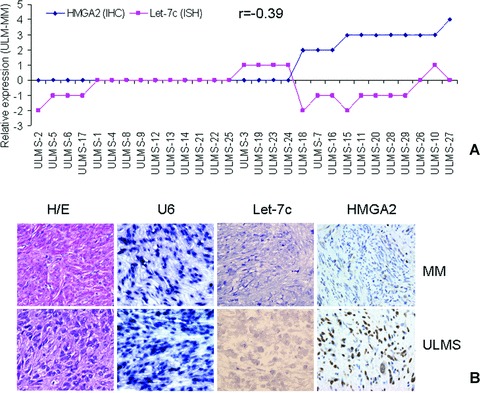
HMGA2 and let-7c expression in uterine leiomyosarcoma (ULMS) in a tissue microarray (TMA)-based analysis. A. Case-matched comparison of HMGA2 and let-7c expression in 30 ULMS. Net changes of let-7c (pink dots, detected by TMA-based tissue in situ hybridization with LNA let-7c probe (see section ‘Materials and methods’) and HMGA2 (blue dots, detected by immunohistochemistry) were scored in ULMS against matched myometrium. Among those ULMS with immunoreactivity for HMGA2, loss of let-7c was more frequent (r=−0.39). B. Photomicrographs illustrating examples of tissue cores from ULMS (bottom) and matched myometrium (MM, top) in haematoxylin and eosin staining, miRNA in situ hybridization for U6 and let-7c, and immunohistochemistry for HMGA2.
Figure 2.
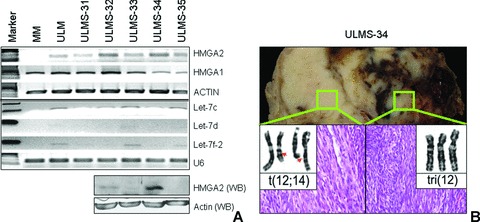
Semi-quantitative RT-PCR (upper panel) and Western blot (lower panel) analyses of HMGA2 and some let-7 family members in fresh frozen tissue samples of five ULMS. (A) HMGA2 mRNA was detectable in all 5 ULMS. One matched myometrium (MM) and one leiomyoma (ULM) were used as HMGA2 negative and positive control, respectively. Actin was used as loading control. Among let-7 members, let-7c was relatively higher than others in all ULMS. U6 was used as small RNA loading control. Western blot analysis of HMGA2 was examined in 4 ULMS (bottom). (B) Photographs illustrate gross appearance of a ULMS cross-section in case ULMS-34 and the corresponding karyotype (inserts with chromosome alterations) obtained in two different tumour regions.
The karyotype analysis was carried out in the latter five cases (ULMS31–35). One (ULMS-34) of five ULMS had non-random chromosomal changes involving chromosome 12 (Fig. 2B). These non-random chromosome 12 changes were commonly seen in usual type ULM. Further analysis of this tumour revealed a high level of HMGA2 mRNA and protein (Fig. 2A). This tumour had all the features of malignant uterine smooth muscle tumour, including tumour necrosis, cytological atypia, invasion and high mitotic counts (Fig. 2B). We speculated that overexpression of HMGA2 in ULMS-34 was contributed by chromosome 12 changes same as those seen in usual ULM.
The levels of Let-7 expression in ULMS were low based on semi-quantitative RT-PCR analysis in 5 ULMS (Fig. 2A). Among five let-7 family members (let-7a, b, c, e, f-2) examined, let-7c transcript was detectable in all ULMS. We therefore selected let-7c to analyse its expression in 30 ULMS by TMA based in situ hybridization (see section ‘Materials and methods’). We used U6 as tissue quality control (Fig.1B). The relative levels of let-7c were first normalized by U6 (let-7c/U6 × 100%), and the net changes of let-7c in ULMS against matched myometrium (ULMS myometrium) were further calculated. Overall, there were slightly net losses of let-7c in ULMS in comparison to matched myometrium (−0.35 ± 0.15). A case-matched analysis of HMGA2 and let-7c expression was performed. As illustrated in Fig. 1A, in those ULMS with positive immunoreactivity for HMGA2, net loss of let-7c expression was quite more common than in tumours negative for HMGA2. Correlation analysis showed an inverse association between HMGA2 and let-7c expression (r=−0.39) (Table 1, Fig. 1A). In those five cases with RT-PCR data, higher levels of HMGA2 mRNA in ULMS-32 and -34 were accompanied by lower levels of let-7s (Fig. 2A).
Correlation analyses were performed between HMGA2 expression and tumour size, tumour markers of Ki-67, P53, ERα and PR-A. We found a weak positive correlation of HMGA2 expression and tumour sizes (r= 0.35). There was not significant correlation of HMGA2 with other immunomarkers.
Translational regulation of HMGA2 by let-7s in ULMS cell lines
To test whether endogenous let-7s play a major role in repression of HMGA2 in ULMS, we selected three ULMS cell lines for the study. We first examined HMGA2 and its cryptic transcript (isoforms) [12, 21] expression in these cell lines by RT-PCR analysis. Since HMGA2a and its cryptic transcripts can be induced in primary cultured leiomyoma cells [12], we used cultured leiomyoma cells as a positive control. We found all ULMS cell lines had a high abundance of HMGA2a mRNA (Fig. 3A). In addition, HMGA2 cryptic transcripts c, d and f showed moderate levels in these cell lines (Fig. 3A). In contrast, expression of let-7s was quite low in comparison to normal myometrium and ULM based on a semi-quantitative analysis (Fig. 3B). Let-7c and f were the most abundant transcripts of let-7 family in these cell lines.
Figure 3.
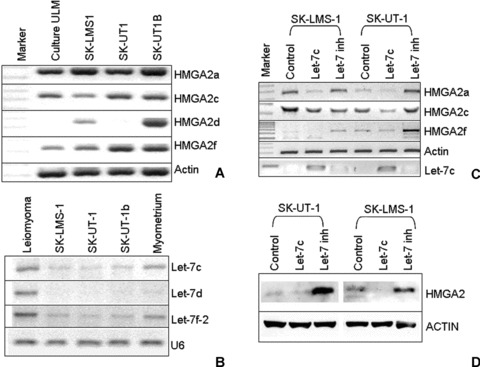
Molecular analysis of HMGA2 and let-7 expression in three ULMS cell lines. (A) Semi-quantitative analysis of HMGA2a and its cryptic transcripts in ULMS cell lines of SK-LMS-1, SK-UT-1 and SK-UT-1b. Uterine leiomyoma (ULM) primary culture cells were used as a positive control. Actin was used as RNA loading control. (B) photonegative image showing relative expression of let-7s by RT-PCR in ULMS cell lines. One ULM and one myometrium were used as positive controls. U6 was used as loading control for small RNAs. (C) Transient transfection of exogenous let-7c and let-7c inhibitor and HMGA2 expression in LMS cell lines. Two LMS cell lines (SK-LMS1, SK-UT1) were treated with control miRNA (block-iT), let-7c and let-7 inhibitor. Let-7 levels in the treated cells were assessed by RT-PCR (bottom). Repression of HMGA2 mRNA by let-7c was examined by semi-quantitative RT-PCR. Actin was used as control. (D) Western blot analysis of HMGA2 gene product in LMS cell lines with different let-7 levels, induced by transient transfection of control, let-7c or let-7c inhibitors.
To test the role of let-7s in repression of HMGA2 after transcription and translation, we transfected the exogenous let-7c mimic and let-7 inhibitor in all cell lines. In untreated cell lines (controls), low levels of endogenous let-7s seemed not enough to destabilize HMGA2a and other cryptic mRNA (Fig. 3A and B), but were sufficient to repress HMGA2 translation (Fig. 3D, control lanes). When applying 40 pmol of exogenous let-7c, a significant reduction of HMGA2a, c, and f mRNAs (Fig. 3C) was observed, and a complete repression of HMGA2 protein was evident by a Western blot analysis (Fig. 3D, let-7c channel). When the cells treated with exogenous let-7 inhibitor (which can completely block endogenous let-7s), up to fivefold increases of HMGA2 protein were observed in comparison to the untreated cells (Fig. 3D). The findings further supported that HMGA2 repression by let-7s is specific regulatory mechanism for HMGA2 expression in uterine leiomyosarcoma cells.
Repression of HMGA2 inhibited ULMS cell growth
To test whether HMGA2 in ULMS cell lines plays a role in regulation of tumour growth rate, we examined the tumour cell proliferation by introducing exogenous let-7 and let-7 inhibitor, respectively. The cells were treated in triplicate with Block-iT (control), let-7c and let-7 inhibitor. Repression of HMGA2 by let-7c and induction of HMGA2 overexpression by let-7 inhibitor were validated as shown in Fig. 3C and D, we counted for the tumour growth rates at days 1, 2 and 3 in all three ULMS cell lines (Fig. 4). Starting from day 2 (48 hrs), cells treated with let-7c had significantly lower rates of tumour cell growth than control (P < 0.05). The differential growth rates were even wider at day 3 (72 hrs). Particularly, the growth rates in tumour cells treated with let-7 inhibitor were significantly higher than those in let-7c treated cells (P < 0.05), but at least two cell lines lower than those in control groups (Fig. 4). The later findings suggested that let-7 mediated tumour growth in ULMS is not solely through repression of HMGA2, and other let-7 target genes may be involved [22] (Fig. 4).
Figure 4.
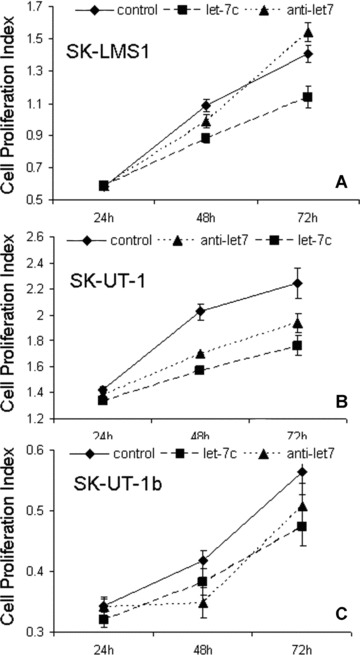
Anti-proliferative effects of let-7 in ULMS cell lines SK-LMS-1 (A), SK-UT-1 (B) and SK-UT-1b (C). The cell lines were treated in three different conditions: (1) non-functional 20 nt RNA (block-iT, labelled as control, solid lines); (2) 40 pmol of exogenous let-7c (Dharmacon, Inc.) (long dash lines) and (3) exogenous let-7 inhibitor (anti-let-7, short dash lines). Cell proliferation rates were calculated (y-axis) by WST-1 staining at 24, 48 and 72 hrs after transfection. Small t-bars were standard errors.
Discussion
ULMS usually have very complicated cytogenetic alterations involving many different chromosomal regions. Cytogenetic changes involving chromosomal 12q13–15 region in leiomyosarcomas had been occasionally reported through traditional karyotype [23] and comparative genomic hybridization (CGH) analysis [4], in which a gain of chromosome 12q15 region including HMGA2 was identified. Examination of HMGA2 expression in anatomic sites other than the uterus revealed overexpression of HMGA2 mRNA in 57% of leiomyosarcomas by RT-PCR [13]. Examination of HMGA2 expression in ULMS has not been reported in the literature. This study is the first attempt to examine HMGA2 expression in ULMS. In this study, we observed that over one third of ULMS were immunoreactive for HMGA2. Furthermore, overexpression of HMGA2 mRNA was observed in all native ULMS and ULMS cell lines in a small scale analysis. Given that only one of five ULMS showed chromosomal 12 changes, overexpression of HMGA2 in ULMS seemed to be through an alternative mechanism.
The specific role of HMGA2 in tumourigenesis remains to be determined. Recent characterization of let-7 regulation of HMGA2 indicates that this molecular pair plays a critical role in the control of mitogenic activity in many different cell types and tumour cells [17, 18, 24, 25]. Particularly, it has been found that dysregulation of let-7s is associated with HMGA2 expression in ULM and related to cell proliferation [17]. The later findings provided the evidence that this molecular pair is involved in benign uterine smooth muscle tumours. Further characterization of this molecular pair in its malignant counterpart ULMS may provide a new inside for our understanding of ULMS.
In this study, a seemly inverse association of HMGA2 protein and let-7c expression was observed. Although the global expression pattern of miRNA in ULMS remains to be determined, a loss of let-7 expression was identified in a proportion of ULMS. Particularly, we illustrate that exogenous let-7 can significantly reduce the tumour cell proliferation in leiomyosarcoma cell lines, largely through repression of HMGA2 expression (Figs 3 and 4). Therefore, it is speculated that disrupting of HMGA2 and let-7 pairs in ULMS could be an important molecular change responsible for ULMS growth. In our future study, it will be important to compare whether overexpression of HMGA2 and loss of let-7 are associated with unfavourable clinical outcome and specific histological grade in a large cohort of ULMS cases.
Another interesting finding is that the karyotype characterized by specific chromosome 12 changes, commonly seen in benign ULM, was also identified in one ULMS in our study. Further analysis revealed an overexpression of HMGA2 mRNA and protein in this tumour. The findings raise two questions:
Can overexpression of HMGA2 in ULMS be induced through specific chromosome changes involving chromosome 12, at least in a small proportion of tumours? A recent study of CGH in seven ULMS showed that one of the highest levels of genomic gains was in chromosome region 12q13–15 [4], where HMGA2 is located. This study suggested that overexpression of HMGA2 is commonly associated with increase genomic material in this region. Findings of two types of chromosome 12 changes involving different tumour regions in one leiomyosarcoma in this study apparently are not coincidence.
Is there a molecular connection between benign and malignant uterine smooth muscle tumours to a non-random chromosome 12 changes? It is generally recognized that ULMS may develop independently from ULM, as it is (i) commonly a solitary mass of uterus, (ii) commonly in the peri- and post-menopausal women and (iii) composed of different genetic alterations as compared to leiomyomas. However, it is not uncommon that ULMS contain usual or atypical leiomyoma-like areas (LLA) [26]. The nature of LLA within ULMS has not yet been established. In the case with chromosome 12 changes, the reason we selected two different regions for cytogenetic analysis was that this tumour consists of both histological malignant and LLA regions. It remains a challenge to characterize whether the chromosome 12q changes in ULMS arises from the possible LLA area or frankly from well-differentiated ULMS.
Acknowledgments
This study was supported by New York University and Northwestern University institutional funds (JJW). This work is partially present in National conference of United States and Canadian Academy of Pathology in March 2008.
References
- 1.Leibsohn S, d’Ablaing G, Mishell DR, Jr, et al. Leiomyosarcoma in a series of hysterectomies performed for presumed uterine leiomyomas. Am J Obstet Gynecol. 1990;162:968–74. doi: 10.1016/0002-9378(90)91298-q. [DOI] [PubMed] [Google Scholar]
- 2.Giuntoli RL, Metzinger DS, DiMarco CS, et al. Retrospective review of 208 patients with leiomyosarcoma of the uterus: prognostic indicators, surgical management, and adjuvant therapy. Gynecol Oncol. 2003;89:460–9. doi: 10.1016/s0090-8258(03)00137-9. [DOI] [PubMed] [Google Scholar]
- 3.Rijhwani K, Wei JJ, Zhu H, et al. Histological and Immunohistochemical phenotypes of atypical leiomyoma like areas within uterine leiomyosarcoma. Mod Pathol. 2006;19:194A. [Google Scholar]
- 4.Lee WY, Tzeng CC, Chou CY. Uterine leiomyosarcomas coexistent with cellular and atypical leiomyomata in a young woman during the treatment with luteinizing hormone-releasing hormone agonist. Gynecol Oncol. 1994;52:74–9. doi: 10.1006/gyno.1994.1014. [DOI] [PubMed] [Google Scholar]
- 5.Morton CC. Genetic approaches to the study of uterine leiomyomata. Environ Health Perspect. 2000;5:775–8. doi: 10.1289/ehp.00108s5775. [DOI] [PubMed] [Google Scholar]
- 6.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 7.Rogalla P, Drechsler K, Frey G, et al. HMGI-C expression patterns in human tissues. Implications for the genesis of frequent mesenchymal tumors. Am J Pathol. 1996;149:775–9. [PMC free article] [PubMed] [Google Scholar]
- 8.Gattas GJ, Quade BJ, Nowak RA, et al. HMGIC expression in human adult and fetal tissues and in uterine leiomyomata. Genes Chromosomes Cancer. 1999;25:316–22. [PubMed] [Google Scholar]
- 9.Flake GP, Andersen J, Dixon D. Etiology and pathogenesis of uterine leiomyomas: a review. Environ Health Perspect. 2003;111:1037–54. doi: 10.1289/ehp.5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ligon AH, Morton CC. Leiomyomata: heritability and cytogenetic studies. Hum Reprod Update. 2001;7:8–14. doi: 10.1093/humupd/7.1.8. [DOI] [PubMed] [Google Scholar]
- 11.Tallini G, Vanni R, Manfioletti G, et al. HMGI-C and HMGI(Y) immunoreactivity correlates with cytogenetic abnormalities in lipomas, pulmonary chondroid hamartomas, endometrial polyps, and uterine leiomyomas and is compatible with rearrangement of the HMGI-C and HMGI(Y) genes. Lab Invest. 2000;80:359–69. doi: 10.1038/labinvest.3780040. [DOI] [PubMed] [Google Scholar]
- 12.Gross KL, Neskey DM, Manchanda N, et al. HMGA2 expression in uterine leiomyomata and myometrium: quantitative analysis and tissue culture studies. Genes Chromosomes Cancer. 2003;38:68–79. doi: 10.1002/gcc.10240. [DOI] [PubMed] [Google Scholar]
- 13.Hisaoka M, Sheng WQ, Tanaka A, et al. HMGIC alterations in smooth muscle tumors of soft tissues and other sites. Cancer Genet Cytogenet. 2002;138:50–5. doi: 10.1016/s0165-4608(02)00568-x. [DOI] [PubMed] [Google Scholar]
- 14.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 15.Fedele M, Pentimalli F, Baldassarre G, et al. Transgenic mice overexpressing the wild-type form of the HMGA1 gene develop mixed growth hormone/prolactin cell pituitary adenomas and natural killer cell lymphomas. Oncogene. 2005;24:3427–35. doi: 10.1038/sj.onc.1208501. [DOI] [PubMed] [Google Scholar]
- 16.Hebert C, Norris K, Scheper MA, et al. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:1–11. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T, Zhang X, Obijuru L, et al. A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes Chromosomes Cancer. 2007;46:336–47. doi: 10.1002/gcc.20415. [DOI] [PubMed] [Google Scholar]
- 18.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–9. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng Y, Laser J, Shi G, et al. Antiproliferative effects by let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol Cancer Res. 2008;6:663–73. doi: 10.1158/1541-7786.MCR-07-0370. [DOI] [PubMed] [Google Scholar]
- 20.Kloosterman WP, Wienholds E, de Bruijn E, et al. in situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–9. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 21.Hauke S, Leopold S, Schlueter C, et al. Extensive expression studies revealed a complex alternative splicing pattern of the HMGA2 gene. Biochim Biophys Acta. 2005;1729:24–31. doi: 10.1016/j.bbaexp.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 22.Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 23.Nilbert M, Mandahl N, Heim S, et al. Complex karyotypic changes, including rearrangements of 12q13 and 14q24, in two leiomyosarcomas. Cancer Genet Cytogenet. 1990;48:217–23. doi: 10.1016/0165-4608(90)90123-r. [DOI] [PubMed] [Google Scholar]
- 24.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shell S, Park SM, Radjabi AR, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. 2007;104:11400–5. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y, Chen Y, Ito H, et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]


